Significance
This work demonstrates that it is possible to alter the prey target range of the antibacterial activity of the type 6 secretion system (T6SS). Being able to change the T6SS target specificity is the first step toward using the T6SS as an antipathogen or commensal therapeutic, prophylactic, or probiotic. This work also uncovers a cryptic secretion mechanism(s) for delivering protein substrates from the bacterial cytosol to the periplasm. This mechanism is exploited by some T6SS effectors as an alternative pathway for reaching periplasmic targets when they are by chance delivered into the cytosol of target cells.
Keywords: type 6 secretion system, VgrG, interbacterial competition, effector engineering, effector trafficking
Abstract
The type 6 secretion system (T6SS) is used by many Gram-negative bacterial species to deliver toxic effector proteins into nearby bacteria prey cells to kill or inhibit their growth. VgrG proteins are core conserved secretion substrates of the T6SS and one subset of T6SS effectors consists of VgrG proteins with C-terminal extension domains carrying various enzymatic activities. In Vibrio cholerae, VgrG3 has a hydrolase extension domain and degrades peptidoglycan in the periplasm of target bacteria. In this study, we replaced this domain with a nuclease domain from Salmonella enterica subsp. arizonae. This modified V. cholerae strain was able to kill its parent using its T6SS. This result also demonstrated that V. cholerae T6SS is capable of delivering effectors that could attack substrates found either in the periplasm or cytosol of target bacteria. Additionally, we found that effectors VgrG3 and TseL, despite lacking a classical Sec or TAT secretion signal, were able to reach the periplasm when expressed in the bacterial cytosol. This effector trafficking likely represents an evolutionary strategy for T6SS effectors to reach their intended substrates regardless of which subcellular compartment in the target cell they happen to be delivered to by the T6SS apparatus.
The type 6 secretion system (T6SS) (1–3) is a bacterial organelle present in a wide range of Gram-negative bacteria (4). This nanomachine delivers toxic effector proteins into target eukaryotic and bacterial prey cells. Mechanistically, the T6SS consists of a transmembrane-baseplate complex (5) and a central Hcp tube sharpened by a VgrG trimer and capped by a PAAR domain-containing protein (the T6SS spike) (6, 7). A sheath composed of VipA and VipB forms around the Hcp tube (8, 9). After assembly, a conformation change occurs in the sheath, which results in the propulsion of the Hcp tube capped by the VgrG/PAAR spike out of the cell and into target cells (10). The association of toxic effectors with components of this VgrG/PAAR/Hcp tube complex is thought to be critical for their cotranslocation into target cells.
Different effectors can be classified based on the mechanism by which they are delivered along with the tube/spike complex. These include effectors delivered within the Hcp tube (e.g., Tse2 from Pseudomonas aeruginosa) (2, 11), fused to the VgrG spike protein (e.g., VgrG-1 from Vibrio cholerae) (12), fused to the PAAR domain protein [e.g., RhsA from Dickeya dadantii (13) and Tde2 from Agrobacterium tumefaciens (14)] and effectors that bind directly or indirectly to the VgrG complex (15–17). In each of these T6SS delivery mechanisms a single secretion event (defined by contraction of the T6SS sheath) is thought to result in the delivery of a single toxic payload of effectors into a target cell (2). However, it remains unclear exactly where in bacterial target cells this payload is delivered.
Understanding where effectors are delivered into target cells is an important aspect to T6SS function because different toxic effectors attack components of the target cell that reside in very different subcellular locations. For example, peptidoglycan hydrolases, like VgrG3 from V. cholerae, have a periplasmic substrate (18), whereas nucleases like RhsA from D. dadantii (13) have a cytosolic one. For these effectors to kill target cells, the translocated effectors need to end up in the correct subcellular compartment of the prey cell. It is unclear whether the T6SS delivers its effectors directly into all prey cell compartments, or whether effectors are delivered to the large cytosolic compartment and then subsequently move to other subcellular locations by using prey cell machinery or their own intrinsic trafficking determinants. For V. cholerae at least, a recent report has shown that V. cholerae can use the secreted T6SS components delivered by the T6SS of adjacent V. cholerae cells (19). This recycling of T6SS substrates suggests that protein substrates of the T6SS may be able to traffic back into organelle assembly pathways after they have been delivered by an exogenous T6SS. Furthermore, it has been previously hypothesized that effectors attached to the C terminus of VgrG proteins can act as modular domains that can be exchanged horizontally between organisms (12, 20), and in at least one case, swapping the C-terminal domain of different VgrG proteins results in different VgrG-associated effectors being secreted (17). In order for effectors with different subcellular target locations to be horizontally exchangeable and functional between organisms, VgrG proteins and their T6SS would need to be capable of delivering effectors into different subcellular locations.
In this study, we followed up on our previous report that when the full-length VgrG3 protein is expressed in the Escherichia coli cytosol in the absence of its cognate immunity protein, the cells round up and die, suggesting that the lysozyme effector domain of VgrG3 can traffic from cytosol to periplasm (21). We provide direct evidence that the V. cholerae T6SS is capable of delivering effector domains into the cytosol by functionally replacing this lysozyme domain with a nuclease domain from a VgrG effector in a heterologous species, Salmonella enterica subsp. arizonae. We identified a portion of the VgrG3 protein located in the linker region between the core VgrG domain and the lysozyme effector domain that is responsible for delivery of an active, toxic effector to the periplasm. Removal of this portion of the protein eliminates cellular toxicity but does not affect the enzymatic function of the effector in that adding an exogenous Sec-secretion signal to the truncated effector restores its toxicity. This cryptic internal periplasmic localization/activity signal is a potential mechanism by which effector molecules randomly delivered into the cytosol compartment can be redirected to the periplasm. Additionally, a second T6SS effector in V. cholerae was found to also contain a cryptic periplasmic localization signal. Collectively, our data suggest that the T6SS of V. cholerae is capable of delivering both homologous and heterologous effector domains directly to the target cell cytosol and that additional signals within these effectors can lead to their trafficking to other subcellular locations. This result also shows that the T6SS can be engineered to deliver heterologous effectors for a variety of applications in disease control.
Results
Based on the geometry of the T6SS, previous studies have observed that VipA/B sheaths of V. cholerae extend the entire width of the cell and contract to approximately half their length during each secretion event (10). This observation suggests that the “reach” of the T6SS is about half the width of the bacterial cell, a sufficient distance to possibly penetrate through the entire cell envelop of a target cell that is in close proximity. In fact, given that the thickness of the periplasmic space is roughly 10–25 nm (22), while the width of the whole cell is roughly 500 nm, the periplasmic space only occupies ∼5% of the potential target space of any given T6SS attack, suggesting that the cytosol is the most likely repository of T6SS-delivered effectors. However, the only effector found in V. cholerae with a clearly identified bacterial target is the peptidoglycan-degrading VgrG3 effector, which has a periplasmic substrate. As such it was unclear whether VgrG effectors of the V. cholerae T6SS could actually reach the target cytosol or if they could only breach the outer membrane before being stopped by the peptidoglycan layer in the periplasm.
To address this question, we replaced the enzymatic C-terminal domain of VgrG3 with a heterologous effector domain with a different subcellular target. Although there has been precedent for delivering heterologous domains by attaching extra domains to the end of different VgrG proteins (23), this heterologous domain was not an effector domain and was not being delivered into bacterial target cells. As such, we wanted to take a more conservative approach and turned to a gene (SARI_02603) that apparently encodes a VgrG in S. enterica subsp. arizonae carrying a C-terminal extension domain predicted to be an HNH-family nuclease (24, 25). Although this heterologous VgrG (here after designated VgrG-NucSe1) has not been confirmed to be a functional VgrG effector in its original Salmonella species, we were not able to clone the protein in the absence of a cognate immunity protein without spontaneous mutations emerging, suggesting the domain was indeed toxic to cells. Because this predicted immunity protein (encoded by SARI_02603) lacked a recognizable signal sequence, we further postulated that this effector likely targets nucleic acid substrates in the bacterial cytosol. The C-terminal extension of VgrG-NucSe1 bears strong homology to the C-terminal cytoxicity domain of S2/SD2 pyocins found in P. aeruginosa (26, 27). S2/SD2 pyocins require interaction with the ferripyoverdine receptor FpvAI to enter cells (27, 28). However, the receptor-binding domain and about half of the translocation domain of the pyocin involved in this interaction is not conserved in VgrG-NucSe1. Thus, we speculated that the effector domain of this Salmonella VgrG was not likely trafficking in target cells by a mechanism analogous to pyocins and other protein antibiotics.
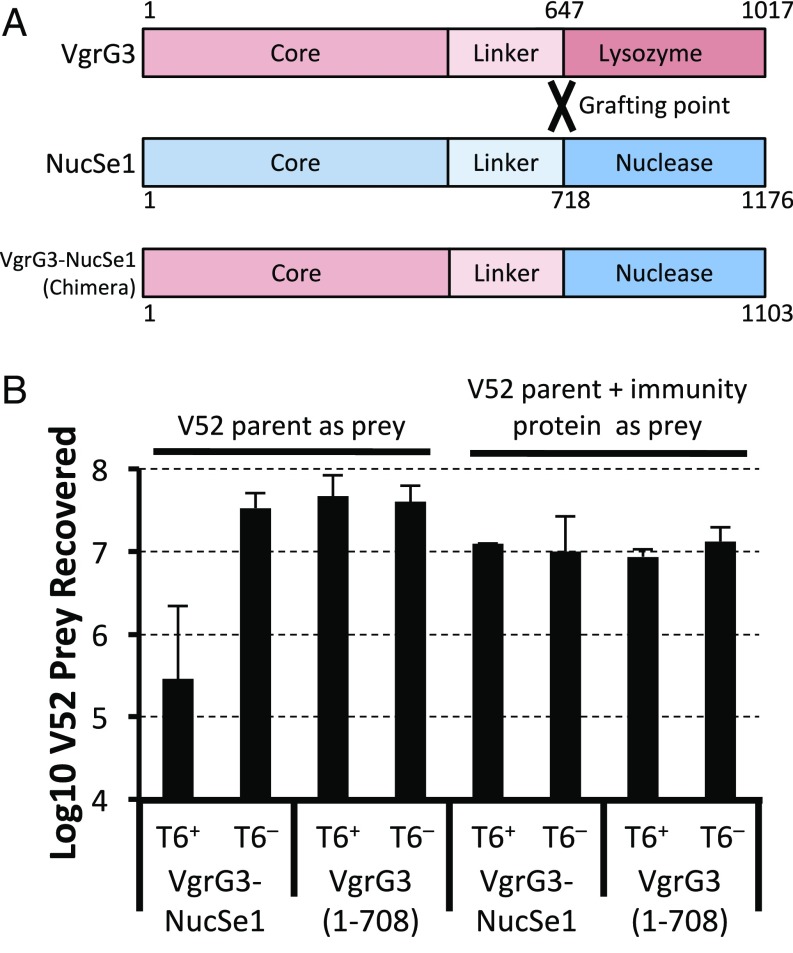
To test whether this nuclease effector could be delivered to the cytosol of target cells using a heterologous T6SS, we constructed a chimeric vgrG operon encoding the VgrG core domain from V. cholerae VgrG3 fused to the predicted nuclease domain of the S. arizonae VgrG-NucSe1 (Fig. 1A), followed by the sequence predicted to encode the putative cognate immunity protein for this nuclease effector found immediately downstream of the VgrG-NucSe1 coding sequence. When we expressed this chimeric VgrG in V. cholerae V52, and grew it in competition with V52 lacking the cognate immunity protein, the original parent strain was readily eliminated (Fig. 1B). As a control, expression of only the VgrG3 core domain, VgrG3 (1–708), truncated after the linker domain conferred no additional killing activity. To confirm that the toxic activity of the VgrG3-NucSe1 was due to the newly added toxic domain, we also expressed the cognate immunity protein in the cytosol of the prey strain and found that this eliminated any observed killing activity (Fig. 1B). These data show that (i) this putative Salmonella nuclease VgrG effector is a functional bacterial toxin that can be delivered by the heterologous V. cholerae T6SS; (ii) a V. cholerae T6SS VgrG effector can serve as a scaffold for delivering heterologous effector domains from other bacterial species; and (iii) a V. cholerae T6SS VgrG is capable of delivering toxic effectors with both cytosolic and periplasmic targets into prey bacteria to kill them.
Fig. 1.
Replacing the lysozyme effector domain of VgrG3 with the nuclease domain of a heterologous VgrG. (A) The C-terminal enzymatic domain of a VgrG effector from S. arizonae (NucSe1) was grafted onto the core domain of VgrG3 from V. cholerae at the flexible linker region of each VgrG (1–647 of VgrG3 with 718-END of NucSe1). (B) Killing assay results after mixing killer and prey strains at a 10:1 ratio. Killer strains were V. cholerae V52 expressing chimeric VgrG and its cognate immunity protein (VgrG3-NucSe1) or just the core domain of VgrG3 (residues 1–708). T6SS-negative (T6−) ∆vipA mutants were also included. Prey strains were either the wild-type V52 or V52 expressing the cognate immunity protein for the nuclease domain. Data shown represent the average of at least three experiments. Error bars represent 1 SD.
In previous work, we made the observation that VgrG3 was toxic when expressed in the cytosol of E. coli (21). This result was somewhat surprising, given that the toxic domain of VgrG3 is a known peptidoglycan-degrading lysozyme and would be expected to be innocuous in the cytosol. In light of our observation that the V. cholerae T6SS could deliver effectors into the cytosol of target cells, we wondered whether VgrG3’s unexpected toxicity could reflect the existence of a trafficking signal that redirects cytosolic VgrG3 or its effector domain to the periplasm. Because our previous work showed that this toxicity was dependent on the enzymatic activity of the VgrG3 lysozyme domain (21), we also wanted to eliminate the formal possibility that the enzymatic domain might have an unexpected cytosolic target (such as a peptidoglycan precursor) whose degradation might lead to cell death.
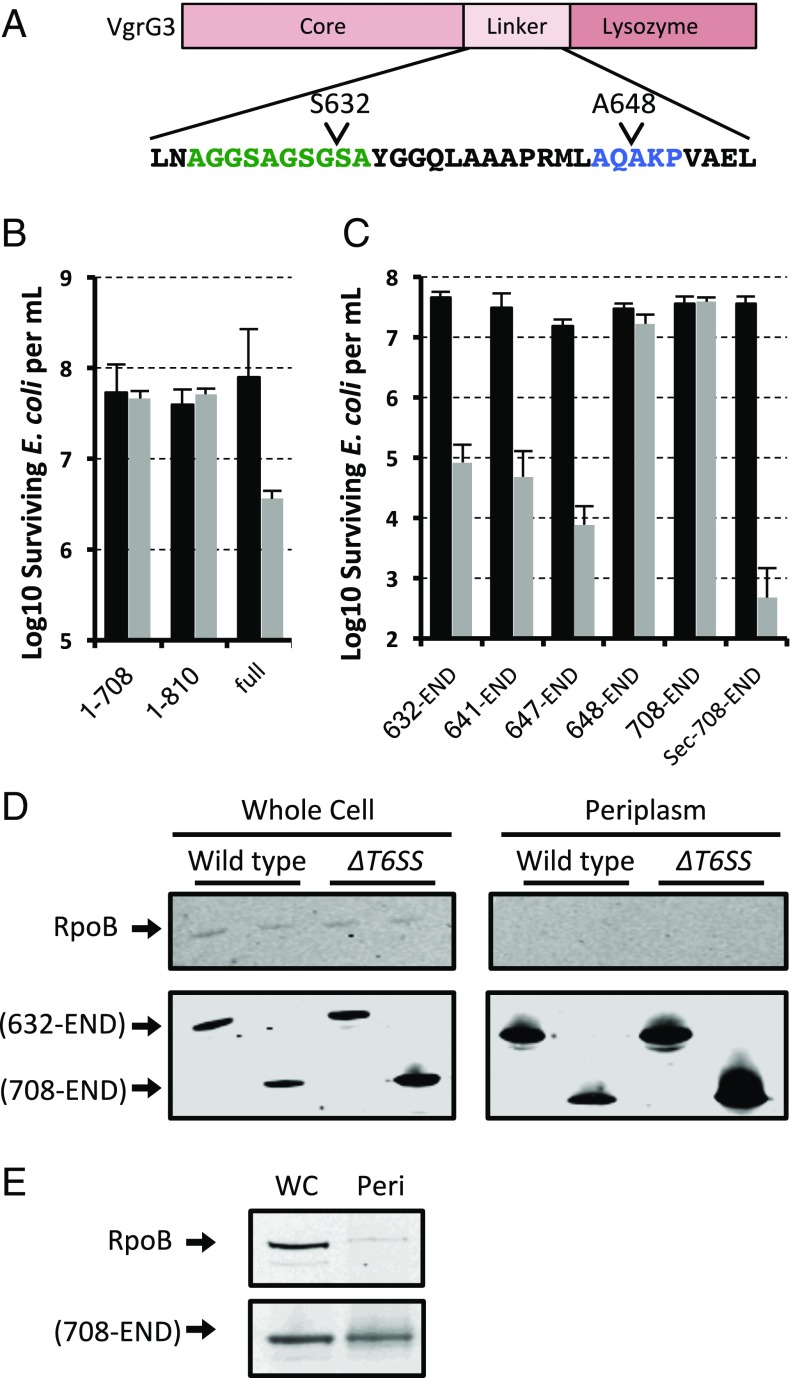
To address these questions, we made a series of truncation deletions removing the core VgrG domain and parts of the linker region connecting it to the enzymatic domain (Fig. 2A). We also made two additional mutations consisting of the VgrG core domain truncated after the linker region (1–708) and at the lysozyme domain (1–810) to confirm that the VgrG core conferred no toxicity. Each of these constructs was cloned into a pBAD33 vector where expression is tightly repressed in the presence of glucose but induced in the presence of arabinose. To avoid the emergence of resistance mutations against these highly toxic constructs, each experiment was performed immediately after a fresh transformation of the vector into E. coli DH10B to minimize passaging. Colonies from this transformation were resuspended in LB, used to inoculate cultures containing either glucose or arabinose, grown for 2 h, and then serially diluted and plated to count cfus (Fig. 2B). Using this assay, we were able to identify a minimal domain that still retained toxicity when expressed in the E. coli cytosol. To confirm that enzymatic activity was not lost, we cloned a Sec-secretion signal from E. coli dsbA to the N terminal of our smallest truncation mutant and observed a restoration of toxic activity, indicating that the truncation mutants are still enzymatically active if properly localized to the periplasm (Fig. 2C). Because VgrG3 and its various truncation mutants that carry the 648–708 region are still toxic, we hypothesized that the VgrG3 effector domain when expressed in the cytosol can traffic to the periplasm. We wanted to confirm that we could biochemically detect VgrG3 in the periplasm. To avoid complications, such as cell lysis and strong selective pressure for mutation that arise from the enzymatic activity of VgrG3 and incomplete protection by exogenous immunity proteins that would need to also be expressed along with the effector, we looked for periplasmic localization of VgrG3 within V. cholerae. As expected, we were able to observe the periplasmic localization of the toxic VgrG3 truncation mutant (Fig. 2D). Further, this localization was not dependent on the presence of the T6SS as a T6SS-negative ∆vipA mutant exhibited similar VgrG3 localization (Fig. 2D). We next wondered whether the truncation mutant of VgrG3 that was not toxic (residues 708-END) might have lost its toxicity due to being trapped in the cytosol. Surprisingly, this nontoxic truncation mutant could also be found in the periplasm when expressed in both V. cholerae (Fig. 2D) and E. coli (Fig. 2E). We further confirmed that the 708-END construct was not toxic when expressed in a V. cholerae mutant lacking the cognate immunity protein of VgrG3 (data not shown), suggesting that the periplasmic localization in the two species is likely following a similar mechanism. Together, these data suggest that residues between 648 and 708 confer some activating effect on the protein allowing it to retain its enzymatic activity when it is transported into the periplasm. The requirement for these amino acid residues is circumvented when the activity domain is directly transported into the periplasm via Sec (Fig. 2C), suggesting that Sec is likely not responsible for the natural delivery of VgrG3 into the periplasm.
Fig. 2.
Toxicity of VgrG3 truncation mutants in E. coli. (A) Domain layout of VgrG3. The residues highlighted in green are the flexible linker region, and the residues highlighted in blue are a canonical signal peptidase cleavage motif. Different truncation mutants were expressed from a plasmid under the control of an arabinose-inducible promoter, where expression is tightly repressed in the presence of glucose (black bars) and strongly expressed in the presence of arabinose (gray bars). (B) Survival of E. coli expressing full-length VgrG3 (full) compared with the same strain expressing different VgrG3 truncation mutants lacking the effector domain and containing only the conserved core domain (1–708 and 1–810). (C) Survival of E. coli expressing different truncation mutants of VgrG3. The smallest domain of VgrG3 was also fused to an N-terminal Sec-secretion signal to confirm that these truncations fully retain their enzymatic activity. Data shown represent the average of at least three experiments. Error bars represent 1 SD. (D) Western blot of VgrG3 truncations from cell fractionation of V. cholerae V52 and T6SS-negative (T6−) ∆vipA mutant. (E) Western blot of VgrG3 truncations from the whole cell (WC) or periplasm (Peri) of E. coli expressing the 708-END truncation mutant of VgrG3. Truncation mutants in D and E were tagged at the C terminal with an HA-tag epitope. RNA polymerase beta subunit RpoB is a cytosolic protein used as a lysis control. Blots are representative of at least two replicates.
Interestingly, the truncated mutants removing the core VgrG domain were greater than 100-fold more toxic to bacteria than the full-length VgrG3 protein. Furthermore, the toxicity of the VgrG3 lysozyme domain is 10,000-fold greater in toxicity when delivered to the periplasm by a Sec-secretion signal. Together these data suggest that rate limiting cytosolic proteolysis might be needed to release a highly toxic fragment of VgrG that contains both the periplasmic localization/activity signal and the intact lysozyme effector domain. Some degree of proteolysis of VgrG is apparent in E. coli when it is expressed in the cytosol; however, we have not been able to document the formation of fragments of VgrG that would correspond to the products postulated above (data not shown).
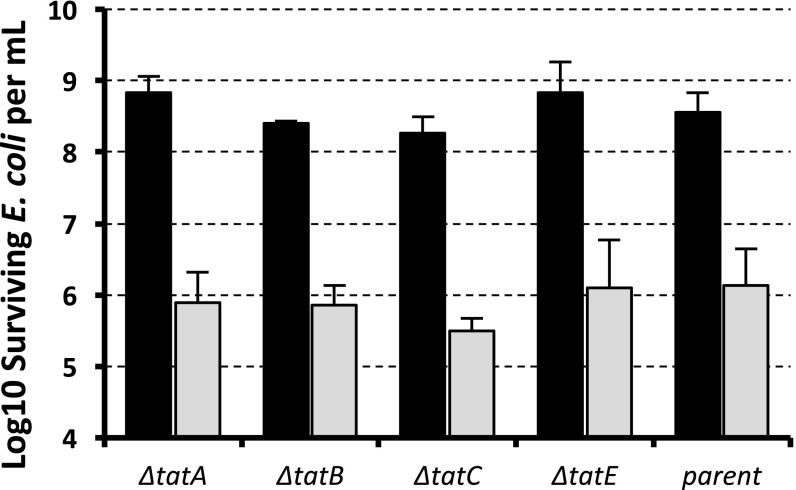
We next attempted to look more carefully at the amino acid sequence of this linker region (Fig. 2A). Using signal sequence neural-network prediction software TatP (29), this region is actually predicted to be a Tat signal peptide despite lacking the conical twin arginine residues. Interestingly, this region also contains the canonical A-X-A motif associated with signal peptidase substrates. However, when our VgrG3 constructs were expressed in E. coli strains carrying individual deletions in the tatA, tatB, tatC, or tatE genes, there was no observable reduction in the toxicity, indicating that, despite bioinformatic prediction that the Tat secretion pathway is involved, some other secretion mechanism is likely responsible for its periplasmic delivery (Fig. 3). An additional deletion of the whole operon of tatABC in multiple E. coli strain backgrounds similarly had no effect on the observable toxicity of VgrG3 (data not shown).
Fig. 3.
Deletion of the TAT secretion system does not eliminate VgrG3 toxicity in E. coli. VgrG3 truncation including the cryptic periplasmic localization signal was expressed in knockouts of each component of the TAT system. VgrG3 (632-END) was expressed from an arabinose-inducible promoter, where expression is tightly repressed in the presence of glucose (black bars) and strongly expressed in the presence of arabinose (gray bars). Data shown represent the average of at least three experiments. Error bars represent 1 SD.
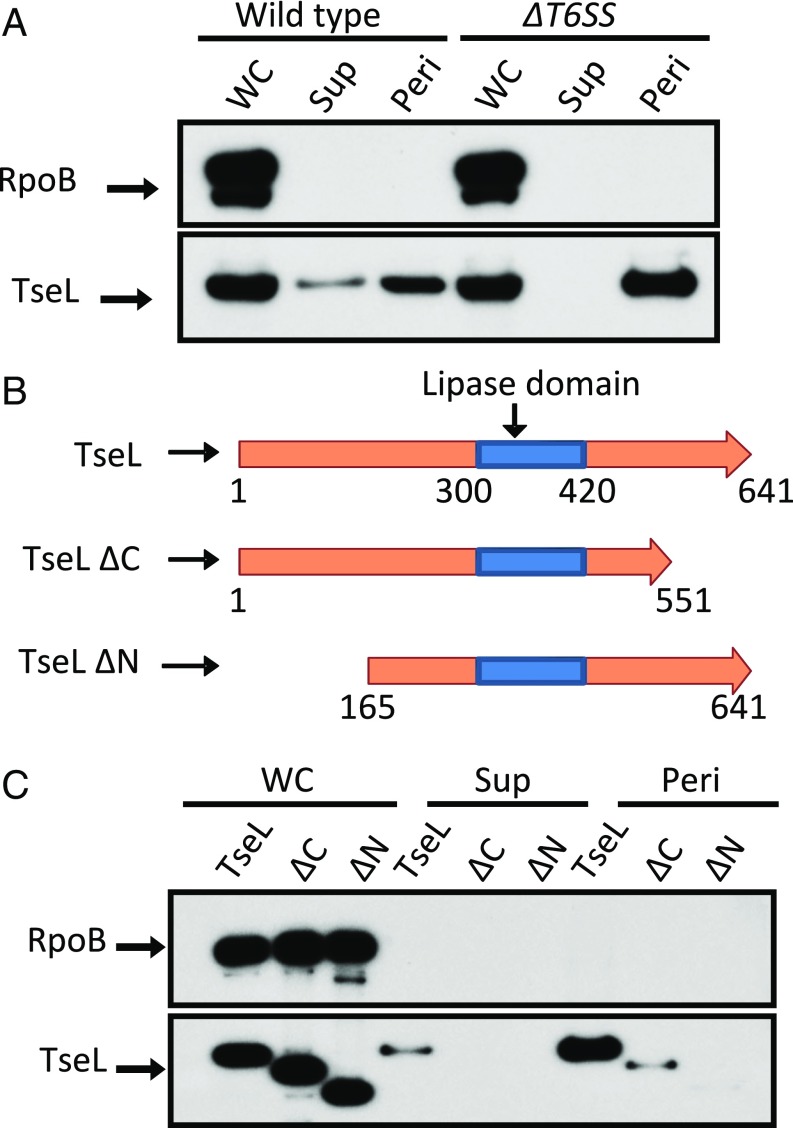
Based on the toxicity of VgrG3 and our various truncation mutants, we conclude that the VgrG3 effector domain when expressed in the cytosol can traffic to the periplasm of a Gram-negative cell. Given this result, we wondered if periplasmic relocalization might be a more general strategy used by other T6SS effectors. One of the T6SS effectors of V. cholerae is the lipase TseL (21). TseL has no predicted secretion signals, and although the precise subcellular target for either is not known, using PRED-TAT (30) signal prediction software, its cognate immunity protein has a single predicted transmembrane domain near its N terminus and a predicted Sec-secretion signal. If the innate protection against the lipase is at least partly localized to the periplasm, it would be reasonable to expect that the lipase effector itself might also find its way into the periplasm. To test this, we performed subcellular fractionation of V. cholerae cells isolating whole cells, periplasmic contents, and the secreted contents from the supernatant and assayed for the presence of TseL (Fig. 4B). As expected, TseL was only secreted into the supernatant when the T6SS system was functional. However, independent of the T6SS functionality, TseL could be found in the periplasm. We made truncation mutants of the N and C termini before and after the lipase domain to see if we could identify a cryptic periplasmic localization signal (Fig. 4B). We found that deletion of the first 165 residues from the N terminus prevented TseL from localizing to the periplasm. Although deletion of the C terminus significantly lowered the amount of TseL present in the periplasm, it could still be detected there, suggesting that an intact N terminus domain is critical for TseL to reach the periplasm (Fig. 4C). The C terminal may be playing a role in the overall stability and function of the protein, as deletion of either the N or the C terminus blocked secretion by T6SS (Fig. 4C). As such, the construct lacking the C terminus might simply be less stable in the periplasm.
Fig. 4.
TseL traffic to the periplasm. (A) Western blot of whole cell (WC), supernatant (Sup), and periplasmic (Peri) fractions of V. cholerae V52 in wild type and a T6SS− mutant (∆vasK). Blot against RNA polymerase beta subunit (RpoB) is presented as a lysis control. (B) Domain layout of different truncation mutants of TseL. (C) Whole cell, supernatant, and periplasmic fractions of the different truncation mutants. Blots are representative of at least three replicates.
Discussion
In this study we show that the V. cholerae T6SS is capable of delivering toxic molecules targeting different essential cellular components that reside in different subcellular compartments of Gram-negative prey cells. Specifically, our data show that the V. cholerae T6SS effector VgrG3 is a highly refined bacterial weapon capable of finding its enzymatic target even if it is delivered into the wrong subcellular compartment. As such, V. cholerae VgrG3 can serve as an efficient scaffold onto which novel heterologous bacterial effectors can be linked. Because the precise location of delivery is not particularly important in this case, the V. cholerae T6SS seems particularly primed to be able to accept heterologous exchange of T6SS effectors. Furthermore, these exchanges can confer changes in target range and thus drive antagonistic behavior toward both homologous as well as heterologous species. The fact that immunity proteins are so tightly linked with effector domains is further evidence that exchange may be occurring naturally. Although bioinformatics analysis observing tight linkage between effectors and their immunity genes suggests that these domains could be exchanged between heterologous organisms as a single unit (12, 20), this study demonstrates that a properly positioned recombination event could fuse a heterologous effector domain to an endogenous VgrG effector while bringing along the gene for the cognate immunity of the heterologous effector. This may be particularly relevant, given the recent report of Borgeaud et al. that the expression of V. cholerae T6SS is coordinated in some strains with the expression of the natural competence system governing DNA uptake (31). Thus, T6SS-mediated target cell lysis might be coordinated with a DNA uptake event that allows Vibrio species to scavenge its prey for possible effector–immunity gene pairs.
Although there have been reports of T6SS effectors inserting into target membranes, such as VgrG5 from Burkholderia pseudomallei (32) and Tse6 from P. aeruginosa (33), the data presented here suggest that T6SS effectors can be trafficked within a prey cell from one subcellular compartment to another with surprising ease. Although we were not able to determine the precise mechanism by which either TseL or VgrG3 translocate to the periplasm, we did uncover a protein sequence determinant in VgrG (in the vicinity of residues 632–708) that contributes to periplasm activity of the effector when it trafficks there through this undefined or “cryptic” secretion pathway. A mutant protein that lacks this sequence determinant but carries the predicted peptidoglycan hydrolytic domain (i.e., residues 708-END) was not toxic to E. coli unless a Sec-secretion signal was added to the beginning of the protein. Despite lacking a Sec-signal sequence, the 708-END construct retained its ability to traffic to the periplasm of both E. coli and V. cholerae by a T6SS-independent mechanism. Because this same construct lacks toxicity, we conclude that the 632–708 determinant contributes to maintaining the activity of the enzyme specifically when it gets delivered to the periplasm by the cryptic secretion pathway. Given that Sec-mediated transport unfolds substrates during transport, it is tempting to speculate that the cryptic pathway may act analogously with the 632–708 determinant being needed to correctly refold the hydrolase domain once it is transported to the periplasm.
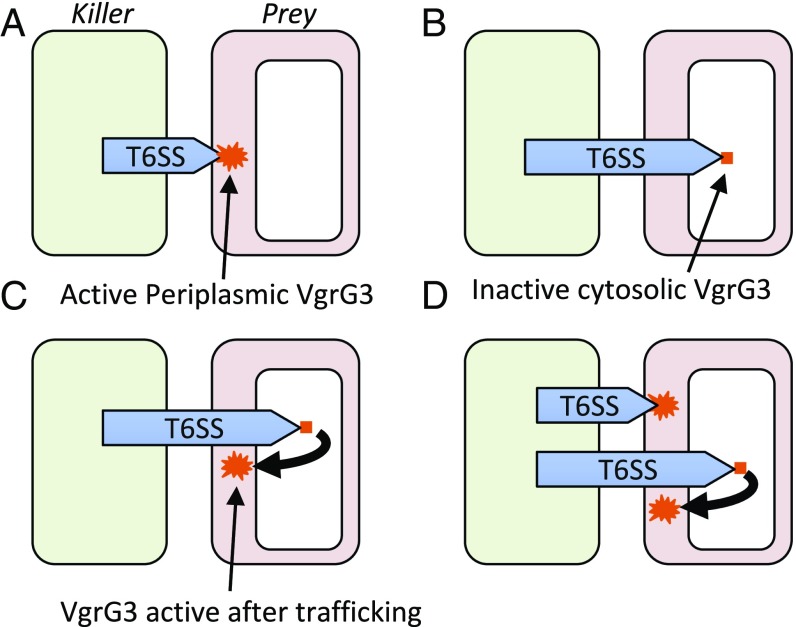
Regardless of how these effectors reach the periplasm, the fact that at least two V. cholerae T6SS effectors have intrinsic periplasmic localization properties suggests that the T6SS of V. cholerae has sufficient penetrating power to deliver many of its effector payloads to the cytosol. We envision the following scenarios are all possible subsequent to V. cholerae T6SS attack: If the VgrG/PAAR spike complex on the tip of the Hcp tube is delivered into periplasm, then VgrG3 can have its effect there (Fig. 5A); if the spike complex penetrates all of the way into the cytosol, then the VgrG3 effector will initially have no function (Fig. 5B); however, if the target cell then traffics the VgrG3 effector to the periplasm, then the effector will be able to degrade its peptidoglycan substrate localized there (Fig. 5C); clearly both modes of attack can also occur if the bacterial target cell remains in close proximity long enough as might be the case in a stable structure such as a biofilm (Fig. 5D).
Fig. 5.
VgrG3 delivery into target bacteria. Shown are all possible outcomes subsequent to V. cholerae T6SS attack. (A) If the VgrG/PAAR spike complex on the tip of the Hcp tube is delivered into periplasm, then VgrG3 can have an effect there. (B) If the spike complex penetrates all the way into the cytosol then the VgrG effector will initially have no function. (C) If the target cell then traffics the VgrG3 effector to the periplasm, then the effector will be able to attack its peptidoglycan substrate there. (D) Both cytosolic and periplasmic delivery of effectors can occur if bacterial target cells remain in close proximity long enough as might be the case in a stable structure such as a biofilm.
It is known that certain protein antibiotics (colicins and pyocins) use short peptide sequences called the TolB or TonB boxes to engage corresponding transporters that drive their translocation through the inner membrane using proton motive force (34, 35). However, the S2 pyocin-like T6SS effector domain we used in this study lacks these domains, suggesting that this effector is seldom naturally delivered to the periplasm and that the cytoplasm is the default location of most T6SS-mediated delivery events. However, T6SS-mediated delivery of an effector that has a cytoplasmic target may occasionally occur into the periplasm and thus, like colicins, there could be other mechanisms for translocation of these effectors into the cytosol (36). Thus, trafficking of T6SS effectors both from cytosol to periplasm or from periplasm to cytosol, could be intrinsically mediated by determinants in a given T6SS effector, depending on the subcellular location of the effector’s target.
Ultimately, it should not be too surprising that bacterial effectors have evolved to exploit their prey’s cellular machinery to better kill prey cells, given how well adapted microbial toxins are in exploiting the endogenous cellular protein trafficking systems of eukaryotic target cells (37). Unfortunately, we do not yet know the mechanism through which either intact VgrG3 or a proteolytically released C-terminal fragment actually traffics to the periplasm of prey cells. Although bioinformatics predictions about the linker region of the VgrG protein suggest that there might be an internal twin-arginine translocation signal, deletion of components required for the TAT system had no effect on VgrG toxicity. One possibility worth considering is that the sequences within VgrG3 and the N-terminal region of TseL have intrinsic membrane-penetrating activity that allows certain protein domains to be translocated from cytosol to periplasm. This would allow some fraction of effector proteins that were delivered to the cytosol to reach the periplasm without needing to be recognized by the cellular machinery in the prey cell. Additional experiments will need to be done to identify what the nature of this cryptic translocation pathway/mechanism is and whether other T6SS effectors carry such signals for subcellular trafficking in target prey cells.
From a more practical point of view, our studies provide an interesting approach for the repurposing of T6SS in applications such as therapeutics, vaccines, and probiotics. Here we have shown that by introducing a new effector function into a VgrG, one can construct a strain that can kill its isogenic parental strain using the T6SS apparatus (Fig. 1B). Thus, an innocuous organism (for example, a live, attenuated vaccine or commensal organism) can in theory be designed to kill either its pathogenic kin or a heterologous species by engineering these apathogenic organisms to express a toxic T6SS effector domain that kills unwanted target pathogens lacking immunity to the corresponding effector. We plan to test this concept by engineering live, attenuated V. cholerae vaccines that can kill their wild-type parental strains while colonizing vaccinated animals. These engineered prophylactic (and potentially therapeutic) vaccines might provide a window of short-term protection after immunization, during which, adaptive immune responses would develop and then provide long-term protection against the target pathogen. We hope that this sort of synthetic biology applied to repurposing the T6SS will usher in a new generation of prophylactic organisms that will find applications in not only human health but also in agriculture, aquaculture, and other ecological settings where microbial antagonism can be exploited to our societal benefit.
Materials and Methods
Extended materials and methods can be found in SI Materials and Methods.
Bacteria Strains and Growth Conditions.
V. cholerae V52 and its derivatives were used in this study. T6SS mutants were clean deletions of vipA (38) or vasK (1) as indicated. E. coli K12 strain DH10B was used for toxicity assays. Tat secretion system mutants were from the Keio collection (39).
Chimeric VgrG Construction.
DNA encoding residues 1–647 of VgrG3 were inserted into pBAD33. The nuclease domain (residues 718–1176) of SARI_02603 and the entire trailing gene, SARI_02602 of S. enterica subsp. arizonae serovar 62:z4,z23:- str. RSK2980 (ATCC BAA-731) was inserted in-frame into the plasmid carrying the VgrG3 (1–647). The immunity protein alone was also cloned into pBAD33 for use in the V. cholerae competition assays.
Toxicity Assay.
Strains were freshly transformed with plasmid before each experiment. Colonies from the transformation were resuspended in 1 mL of LB, which was then added into 2 mL of LB with either 0.1% (wt/vol) arabinose or 0.1% (wt/vol) glucose. After 2–4 h, the cultures were serially diluted and plated onto agar plates to count surviving colony forming units (cfus).
Bacterial Killing Assay.
Killing assays were performed as described previously (38). Briefly, log-phase V. cholerae strains growing at an OD600 of 0.8–1.0 were concentrated to an OD600 of 10.0. Killer and prey strains were mixed at a 10:1 ratio, spotted onto LB agar plates, and incubated at 37 °C for 2 h. Cells were then collected, resuspended in LB, serially diluted, and plated to count surviving cfus of the prey.
Periplasmic Localization Assays.
TseL and VgrG3 constructs were tagged at their C terminus with V5 and HA epitope tags, respectively. V. cholerae V52 cells carrying these constructs were grown in LB to an OD600 of 0.5 and then induced by 0.1% (wt/vol) arabinose for 30 min. A total of 1 mL of culture was pelleted, and the supernatant was collected, filtered to remove cells, and precipitated by TCA (20% wt/vol). Periplasmic contents of V. cholerae were prepared as previously described (40). Samples of each fraction were run on SDS/PAGE and Western blotted against HA (Cell Signaling), V5 (Sigma), or RNA polymerase beta subunit RpoB (Abcam).
SI Materials and Methods
Bacteria Strains and Growth Conditions.
V. cholerae O37 serogroup strain V52 and its derivatives were used in this study. The T6SS− mutants were a clean deletion of the vipA gene (38) or the vasK gene (1) as indicated. Escherichia coli K12 strain DH10B was used for VgrG3 toxicity assays. Tat secretion system mutants used were from the Keio collection (39). Cultures were grown in LB at 37 °C with streptomycin (100 µg/mL). All protein constructs were made in pBAD33 with chloramphenicol (5 μg/mL for V. cholerae and 20 μg/mL for E. coli). Strains were grown in 0.1% arabinose to induce and 0.1% glucose to suppress expression. Mutants of V. cholerae were constructed as described previously (1, 38). All constructs were verified by sequencing.
Chimeric VgrG Construction.
DNA encoding residues 1–647 of VgrG3 were amplified by PCR from genomic DNA of V. cholerae V52 and inserted into the multiple cloning sites of pBAD33. The nuclease domain (residues 718–1176) of SARI_02603 and the entire trailing immunity protein, SARI_02602, were amplified together by PCR from the genomic DNA of Salmonella enterica subsp. arizonae serovar 62:z4,z23:- str. RSK2980 (ATCC BAA-731) and inserted in-frame into the plasmid carrying the VgrG3 (1–647). Restriction sites were designed such that an NheI and a SpeI site would generate the Ala–Ser corresponding to residues 646 and 647 of VgrG3. The immunity protein alone was also cloned into pBAD33 for use in the V. cholerae competition assays.
Toxicity Assay.
To avoid spontaneous resistance mutations, strains were freshly transformed with plasmid before each experiment. Overnight colonies from the transformation were resuspended in 1 mL of LB. A total of 0.5 mL of this cell suspension was then added to 2 mL of LB containing chloramphenicol to select for the plasmid and either 0.1% arabinose or 0.1% glucose. After 2–4 h, the cultures were then serially diluted and plated onto agar plates carrying chloramphenicol and glucose to count surviving colony forming units (cfus).
Bacterial Killing Assay.
Prey V. cholerae strains were transformed with pBAD24 to confer carbenicillin resistance. Overnight cultures of V. cholerae strains were diluted 100:1 and grown for 2.5 h at 37 °C to an OD600 of around 0.8–1.0. Cultures were then concentrated to an OD600 of 10.0. Killer and prey strains were mixed at a 10:1 ratio, spotted onto semidry LB agar plates, and incubated at 37 °C for 2 h. Cells were then collected and resuspended in LB, serially diluted and plated on 100 µg/mL of carbenicillin to count surviving cfus of the prey.
Bioinformatics.
The online tool for TatP (www.cbs.dtu.dk/services/TatP/) (29) and PRED-TAT (www.compgen.org/tools/PRED-TAT) (30) were used to predict Tat secretion signals and transmembrane domains, respectively.
Periplasmic Localization Assays.
TseL constructs were tagged at their C terminus with V5 epitope tag and cloned into pBAD33. VgrG3 truncation domains were cloned with an HA-epitope tag at the C terminus and cloned into pBAD33. V. cholerae V52 cells carrying these constructs were grown in LB to an OD600 of 0.5 and then induced by 0.1% (wt/vol) arabinose for 30 min. One milliliter of culture was then pelleted at 10,000 × g for 2 min. The supernatant was collected and filtered through a 0.2-μm filter to remove all cells and precipitated by TCA (20% wt/vol). Periplasmic contents of V. cholerae were prepared as previously described (40). Briefly, 1 mL culture was pelleted and resuspended in LB containing polymyxin B (1 mg/mL). After a 20-min incubation at room temperature, the cells were pelleted and the supernatant was TCA precipitated (20% wt/vol). Samples of each fraction were run on SDS/PAGE and Western blotted against HA (Cell Signaling), V5 (Sigma), or RNA polymerase beta subunit RpoB (Abcam). Periplasmic contents of E. coli were prepared following the same protocol except using only 200 µg/mL polymyxin B and only incubating for 5 min.
Acknowledgments
We thank the members of the J.J.M. laboratory for helpful discussions regarding this project. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-01845 (to J.J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711219114/-/DCSupplemental.
References
- 1.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durand E, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 6.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoued A, et al. Priming and polymerization of a bacterial contractile tail structure. Nature. 2016;531:59–63. doi: 10.1038/nature17182. [DOI] [PubMed] [Google Scholar]
- 10.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverman JM, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koskiniemi S, et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci USA. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unterweger D, et al. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015;34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, et al. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci USA. 2015;112:9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bondage DD, Lin JS, Ma LS, Kuo CH, Lai EM. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc Natl Acad Sci USA. 2016;113:E3931–E3940. doi: 10.1073/pnas.1600428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vettiger A, Basler M. Type VI secretion system substrates are transferred and reused among sister cells. Cell. 2016;167:99–110 e112. doi: 10.1016/j.cell.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 20.Unterweger D, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Wielink JE, Duine JA. How big is the periplasmic space? Trends Biochem Sci. 1990;15:136–137. doi: 10.1016/0968-0004(90)90208-s. [DOI] [PubMed] [Google Scholar]
- 23.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleanthous C, et al. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 25.Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okawa I, Kageyama M, Egami F. Purification and properties of pyocin S2. J Biochem. 1973;73:281–289. [PubMed] [Google Scholar]
- 27.McCaughey LC, et al. Discovery, characterization and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa. Biochem J. 2016;473:2345–2358. doi: 10.1042/BCJ20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denayer S, Matthijs S, Cornelis P. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol. 2007;189:7663–7668. doi: 10.1128/JB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 31.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 32.Toesca IJ, French CT, Miller JF. The T6SS-5 VgrG spike protein mediates membrane fusion during intercellular spread by pseudomallei-group Burkholderia species. Infect Immun. 2014;82:1436–1444. doi: 10.1128/IAI.01367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney JC, et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015;163:607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Housden NG, Kleanthous C. Colicin translocation across the Escherichia coli outer membrane. Biochem Soc Trans. 2012;40:1475–1479. doi: 10.1042/BST20120255. [DOI] [PubMed] [Google Scholar]
- 35.Loftus SR, et al. Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc Natl Acad Sci USA. 2006;103:12353–12358. doi: 10.1073/pnas.0603433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker D, Mosbahi K, Vankemmelbeke M, James R, Kleanthous C. The role of electrostatics in colicin nuclease domain translocation into bacterial cells. J Biol Chem. 2007;282:31389–31397. doi: 10.1074/jbc.M705883200. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y, et al. Retrograde transport of cholera toxin from the plasma membrane to the endoplasmic reticulum requires the trans-Golgi network but not the Golgi apparatus in Exo2-treated cells. EMBO Rep. 2004;5:596–601. doi: 10.1038/sj.embor.7400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]