In a paper published in PNAS, Li et al. (1) solve the long-awaited crystal structure of kindlin-2, which plays a central role in integrin activation, clustering, and signaling (2, 3). Integrins are a large family of heterodimeric adhesion molecules composed of α- and β-subunits and expressed on almost all cells. They mediate cell matrix adhesion by binding extracellular matrix proteins, such as collagens, fibronectin, and laminins, and cell–cell adhesion by interacting with counterreceptors such as VCAM, ICAM, and cadherins (Fig. 1A). Upon ligand binding, integrins cluster and assemble an enormous number of proteins at their short cytoplasmic domains, which are collectively called the adhesome (4). The adhesome transmits biochemical signals into the cell and connects integrins to the actin cytoskeleton, which, in turn, enables the transduction of myosin-II–generated traction forces to ligated integrins.
Fig. 1.
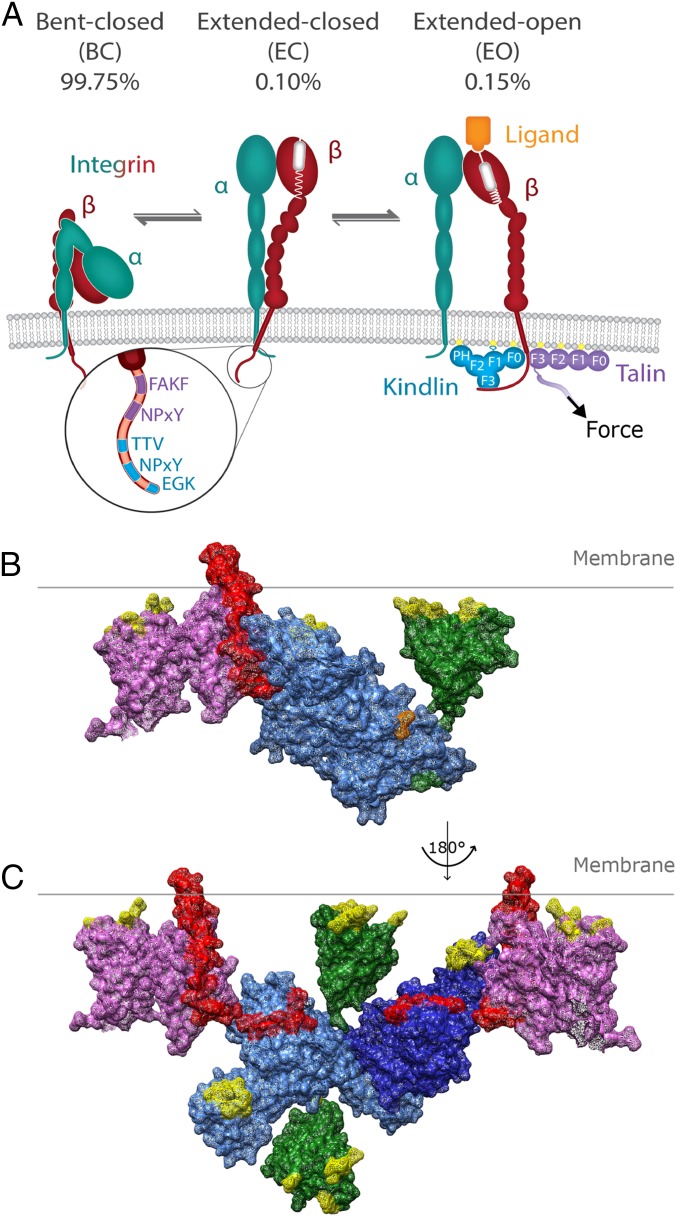
Kindlin and talin promote integrin activation. (A) Integrins are heterodimers (α, turquoise; β, red) that can adopt three conformations: BC, EC, or EO. The EO conformation, corresponding to the active conformation, is induced and/or stabilized by talin (violet) and kindlin (cyan), which bind to specific motifs in the β-integrin tail (magnified) and to charged lipids in the plasma membrane (yellow). The distribution of BC, EC, and EO conformations in equilibrium on K652 cells is depicted for α5β1 integrins. A model of a kindlin-2 monomer [F0–F3 modules (Protein Data Bank [PDB] ID code 5XPY, cyan)] and PH domain (PDB ID code 4F7H, green) (B) and a model of dimeric kindlin-2 (PDB ID code 5XQ0, cyan) (C) interacting with talin (PDB ID code 3G9W, violet) and the β1-integrin tail (red) are shown. Regions known to interact with charged lipids are highlighted in yellow. The PH domain was modeled into the structure to demonstrate its size and potential localization, whereas the flexible loop inserted into the F1 domain is absent. In B, positions where the PH domain and the F1 loop would attach to kindlin-2 are highlighted in green and orange, respectively.
The integrin α/β-subunits have large ectodomains that associate at their head domains to form the ligand-binding site, single-span transmembrane domains, and short cytoplasmic tails. A hallmark of integrins is that the affinity of their ligand-binding sites is tightly regulated by complex conformational changes affecting the entire integrin molecule: They reversibly shift from a low-affinity (or inactive) state, in which they adopt a bent-closed (BC) or extended-closed (EC) conformation, to the high-affinity (or active) state characterized by an extended-open (EO) conformation (2, 5) (Fig. 1A). The BC and EC conformations are characterized by a “closed” ligand-binding site, a loose connection between the transmembrane helices and cytoplasmic tails of α- and β-subunits, and either an association of the ectodomain headpiece with the legs (BC) or an extension of the α/β-subunits (EC). The conversion of integrins into the EO conformation is characterized by the separation of the legs, transmembrane, and tail domains; the swing-away of the hybrid domain from the α-subunit; and the opening of the ligand-binding site, which results in a massive increase in affinity for ligand. A recent study measured the intrinsic affinity of α5β1 integrin on K562 cells for fibronectin and the free energy for each activation state of α5β1 (6). The measurements revealed that the affinity is 4,000-fold higher for the EO conformation compared with the EC or BC conformation. Furthermore, the free energy required to extend α5β1 is about 4 kcal/mol, indicating that extension is the major obstacle that needs to be overcome to activate α5β1 integrin (6).
A vast body of evidence indicates that talin and kindlin induce the high-affinity state of integrins (7). Talins consist of two isoforms (talin-1 and talin-2) that are composed of an N-terminal FERM (protein 4.1, ezrin, radixin, moesin) and a C-terminal rod domain. The talin FERM domain is composed of classical F1, F2, and F3 modules and an extra, atypical F0 module. The F3 module contains a phosphotyrosine-binding motif that binds to the first (or membrane-proximal) of two conserved NxxY motifs in β-integrin tails, as well as to two phenylalanine residues located in the juxtamembrane region (8, 9). The rod domain harbors multiple Vinculin and two F-actin–binding sites that link integrin to the actomyosin system. Interestingly, the crystal structure of the talin F0–F3 modules revealed an extended conformation rather than the compact cloverleaf-shaped structure that is typical for FERM domains (10, 11). The kindlin family consists of three isoforms (kindlin-1–kindlin-3), which are expressed in a tissue-specific manner (3, 7, 12). Kindlins consist of a FERM domain and a pleckstrin homology (PH) domain that is inserted into the F2 module. Biochemical interaction studies revealed that the F3 modules of kindlins bind to the membrane-distal NxxY motif; the adjacent threonine/serine residue; and, at least in integrin β1, the C-terminal carboxylate moiety (13).
The prevailing view is that agonist-induced signaling (e.g., chemokine signaling) activates talin and kindlin, which then bind the integrin β-tails (also called inside-out signaling). Subsequently, talin activates integrins (EO state) by inducing the separation of the transmembrane domains of the α- and β-subunits, whereas kindlin induces integrin clustering (3). However, integrins can also be activated upon binding to extracellular ligands, which cannot be explained with the current model. An alternative activation model for α5β1 binding to fibronectin, which would enable ligand-induced integrin activation, was put forward in PNAS by Li and Springer (14). Based on the affinity and free-energy measurements mentioned above, they concluded that the different conformational states distribute on the cell surface according to thermodynamic equilibrium (99.75% for BC, 0.10% for EC, 0.15% for EO) and that the stabilization of the rare active state (EO) is achieved with relatively small pulling forces (in the range of 0–4 pN) applied by the actomyosin system via talin and/or kindlin to ligand-bound integrins (14).
Irrespective of the “integrin activation” model that operates in vivo, talin and kindlin must play an essential role for each model, as lack of either protein results in complete deadhesion of cells (15). Due to the early discovery of talin, much more is known of its structure and functions than of kindlin. After the talin F0–F3 structure was solved, it was assumed, based on high-sequence similarities, that kindlin also shares the atypical extended conformation and, together with talin, forms a subclass of FERM domain-containing proteins. Contrary to expectations, however, Li et al. (1) solve the crystal structure of a “tailored” kindlin-2 lacking the flexible loop in the F1 module and the PH domain in monomeric and dimeric forms and in complexes with and without β1- and β3-integrin tails. The structure revealed that kindlin-2 adopts a classical cloverleaf-like shape and also disclosed how kindlin-2 achieves specificity for certain motifs and residues in integrin β-tails. Like the F3 module of talin, the kindlin-2 F3 module also contains a narrow groove on one side of the globular protein surface that accommodates the integrin tail-binding sites. The structure shows that Y795 of the membrane-distal NxxY motif is coordinated by K613, while the three C-terminal amino acids (EGK) form backbone–backbone interactions with the F3 domain. Interestingly, the conserved residues N616 and W619 within the groove interact with three amino acids (TTV in β1 or STF in β3) located directly upstream of the membrane-distal NxxY motif, hence strongly increasing binding affinity. The absence of the conserved residues N616 and W619 in talin F3 provides a reasonable explanation for the exclusive binding of kindlin to the membrane-distal NxxY motif. Despite the use of three contact sites, the affinity between F3 and β1-tail peptide is rather weak, with a Kd of 17 μM. The affinity of talin for β1-tails is even lower (Kd of 150–200 μM) but can undergo a 20-fold increase when talin simultaneously interacts with negatively charged phospholipids in the plasma membrane (16). Kindlin also binds phosphatidylinositides with its PH domain and the charged loop in F1 (17). Whether this interaction also strengthens the binding to integrin tails has still to be tested.
An astonishing finding of the work by Li et al. (1) is the observation that kindlin-2 forms homodimers via an interaction between two specific sequences. One sequence motif locates to the N-terminal half of the F2 module (before the PH domain), and the second locates to the C-terminal half of the F2 module (after the PH domain). Interestingly, the sequences are conserved between all three kindlin isoforms, suggesting that all isoforms are able to form homodimers and maybe even heterodimers. Furthermore, Li et al. (1) demonstrate that the absence of key residues in the interaction interface abrogates dimerization and decreases activation of integrins in CHO cells by ∼20–30%, suggesting that the dimer induces and/or stabilizes the EO form of integrins. The dimerization of kindlin has already been presumed for UNC-112, the single kindlin ortholog of Caenorhabditis elegans (18). Also in UNC-112, the dimerization interface is distributed into both halves of the F2 module. These findings indicate that the dimer formation is most likely relevant in vivo, despite the amazingly slow dynamics of the dimerization reaction observed for purified kindlin-2 proteins that Li et al. (1) found to be in the range of several days. Also, talin contains a dimerization domain at the C-terminal end of the rod domain, whose function is still unclear, however. It has been suggested to promote integrin clustering, a role also conceivable for kindlin dimers (19).
Would a kindlin dimer fit with talin onto the integrin tails? With the help of the structural information, we modeled kindlin-2 and talin onto the β1-integrin. Our model shows that the kindlin-2 monomer can interact simultaneously with a talin-bound β1-tail and charged lipids in the plasma membrane (Fig. 1B). This is also possible for one molecule of the kindlin-2 dimer, while the F0 module and PH domain of the second kindlin-2 molecules extend into the cytoplasm (Fig. 1C). Although this orientation prevents contacts with plasma membrane lipids, it would allow recruitment of binding partners such as paxillin, integrin-linked kinase, or migfilin. In addition, the kindlin-2 dimer might link two β1-tails and promote integrin clustering (Fig. 1C).
The dimerization of kindlin-2 may also increase the local kindlin-2 concentration, and thereby enhance the probability of integrin tail rebinding. The relevance for an increase in local adaptor protein concentrations is evident from the simulations by Li and Springer (14). They predict that the stabilization of the active conformation of integrins at a given actomyosin-mediated pulling force is very sensitive toward changes in talin/kindlin concentration close to the Kd of these proteins for the integrin tail.
The crystal structure is a first step toward a better understanding of how kindlin binds and activates integrins. The information allows researchers to perform well-defined mutagenesis studies in cell and animal models. It will be important to further investigate and define the relevance of the dimer formation, whether monomers play a role in integrin biology, and how kindlins cooperate with talin. It is still not known whether the two adaptors bind simultaneously or sequentially to integrin tails, and whether inside-out signaling activates the two adaptor proteins to then activate integrins or stabilize active integrins. Clearly, the players are known, but how they play the game is still poorly understood.
Acknowledgments
We thank Monika Krause for her excellent artwork. The R.F. laboratory is supported by European Research Council (Grant 322652), Deutsche Forschungsgemeinschaft (Grant SFB-914), the Munich Heart Alliance, and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
See companion article on page 9349.
References
- 1.Li H, et al. Structural basis of kindlin-mediated integrin recognition and activation. Proc Natl Acad Sci USA. 2017;114:9349–9354. doi: 10.1073/pnas.1703064114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moser M, Legate KR, Zent R, Fässler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 3.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: The end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller HB, Friedel CC, Boulegue C, Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–1666. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, et al. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 2017;36:629–645. doi: 10.15252/embj.201695803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: Partners in integrin-mediated adhesion. Nat Rev Mol Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Alvarez B, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 9.Wegener KL, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 10.Goult BT, et al. Structure of a double ubiquitin-like domain in the talin head: A role in integrin activation. EMBO J. 2010;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott PR, et al. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moser M, et al. Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat Med. 2009;15:300–305. doi: 10.1038/nm.1921. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick P, Shattil SJ, Ablooglu AJ. C-terminal COOH of integrin β1 is necessary for β1 association with the kindlin-2 adapter protein. J Biol Chem. 2014;289:11183–11193. doi: 10.1074/jbc.M113.535369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Springer TA. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc Natl Acad Sci USA. 2017;114:4685–4690. doi: 10.1073/pnas.1704171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theodosiou M, et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. doi: 10.7554/eLife.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z, et al. Implications of the differing roles of the β1 and β3 transmembrane and cytoplasmic domains for integrin function. eLife. 2016;5:5. doi: 10.7554/eLife.18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu H, et al. Kindlin-2 regulates podocyte adhesion and fibronectin matrix deposition through interactions with phosphoinositides and integrins. J Cell Sci. 2011;124:879–891. doi: 10.1242/jcs.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qadota H, Moerman DG, Benian GM. A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (Kindlin) to integrin adhesion sites. J Biol Chem. 2012;287:28537–28551. doi: 10.1074/jbc.M112.354852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingras AR, et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]