Abstract
From microbial biofilms to human migrations, spatial competition is central to the evolutionary history of many species. The boundary between expanding populations is the focal point of competition for space and resources and is of particular interest in ecology. For all Escherichia coli strains studied here, these boundaries move in a counterclockwise direction even when the competing strains have the same fitness. We find that chiral growth of bacterial colonies is strongly suppressed by the expression of extracellular features such as adhesive structures and pili. Experiments with other microbial species show that chiral growth is found in other bacteria and exclude cell wall biosynthesis and anisotropic shape as the primary causes of chirality. Instead, intimate contact with the substratum is necessary for chirality. Our results demonstrate that through a handful of surface molecules cells can fundamentally reorganize their migration patterns, which might affect intra- and interspecific competitions through colony morphology or other mechanisms.
Introduction
From the structure of amino acids to the shape of our galaxy, chirality is ubiquitous in nature and has fascinated scientists for centuries. The accepted mechanism for the emergence of left–right asymmetry is a fluctuation producing a chiral state followed by positive feedback favoring homochirality (Frank, 1953; Gayathri and Rao, 2005; Saito et al., 2007). More recent work further demonstrates that intrinsic noise in autocatalytic reactions is also sufficient to produce and stabilize left–right asymmetry (Jafarpour et al., 2015). While these physical mechanisms might explain chirality in the inanimate world, chirality in living systems could be a product of natural selection.
Although the ecological and evolutionary role of chirality is largely unexplored, there are several examples that point to the possibility that chirality could be advantageous. Extensive work with Paenibacillus showed that this microbe switches from chiral to non-chiral phenotypes in response to changing environmental conditions (Ben-Jacob et al., 2000). More recently, Wan et al. (2011) found that cancer cells are chiral and have the opposite chirality of the normal tissue from the same patient. In both cases, molecular changes propagate to population-level scales and alter the ecology and evolution of the species. The details of these processes are poorly understood. These striking examples motivated us to develop a molecular understanding of chirality in a simple model system of an Escherichia coli colony.
In the lab, E. coli typically grows as a compact circular colony when inoculated on an agar plate; yet internal dynamics of those colonies are often chiral. These dynamics can be revealed by fluorescently labeling subpopulations of growing cells. Strong genetic drift at the growing edge of the colony promotes local loss of genetic diversity and results in spatial de-mixing of the subpopulations producing a characteristic pattern of flaring sectors on long timescales (>30 generations) (Kreft, 2004; Hallatschek et al., 2007; Ali and Grosskinsky, 2010; Hallatschek and Nelson, 2010; Korolev et al., 2010). Without chirality, the boundaries between the sectors extend radially, but, in chiral species, boundaries appear as spirals emerging from the center of the colony (Korolev et al., 2011).
While the existence of chirality in bacterial colony growth has been established, there are still open and interesting questions regarding the origin and evolutionary importance of sector boundary chirality: (i) why and when is chirality beneficial, (ii) how does microscopic chirality translate into a macroscopic chirality and (iii) what are the molecular origins of the chirality? The first question is of great interest and, while our work does not address this question directly, it provides important foundation for future studies. The second question has been largely resolved for bacteria via thorough mathematical modeling and experimentation (Ben-Jacob et al., 2000; Xue et al., 2011). Here, we primarily focus on the last question and uncover the key molecular players shaping chirality of bacterial growth.
Already in 1848, Louis Pasteur related macroscopic chirality of a crystal to a microscopic symmetry breaking of constitutive molecules (Flack, 2009). Similar mechanisms operate in biological systems; for example, Lymnaea stagnalis snail shell chirality could be reversed by altering asymmetry in the blastomere (Kuroda et al., 2009). Although the general principle that macroscopic chirality originates at cellular or subcellular level is well established, a detailed molecular and biophysical understanding of these processes is still lacking for all but a handful of systems. Perhaps the best-studied example of chiral behavior in bacteria is that of E. coli, which turns clockwise while swimming near surfaces due to the chiral motion of its flagella (DiLuzio et al., 2005).
To uncover the molecular mechanisms of chiral growth, we studied several microbial species and E. coli mutants lacking certain surface structures under varying environmental conditions. Our results ruled out shape and the direction of cell wall biosynthesis as the primary drivers of chiral growth. Instead, the data suggest that chirality relies on intimate contact between the substratum and the E. coli cell surface. This contact can be masked by surface structures; thus, the deletion of surface factors progressively increases E. coli chirality compared with wild type. Not only the cell surface, but also the substratum itself is important for the interaction mediating chirality. We found maximal chiral behavior at intermediate agar concentration (1–1.5%) and a significant dependence on substrate composition. Collectively, our results reveal how molecular mechanisms control spatial patterning during colony growth and suggest new ecological roles for several proteins expressed on cell surfaces.
Materials and methods
Cell culture
The strains used in this study are listed in Table 1. E. coli strains were grown at 37 °C in Luria-Bertani (LB) broth (1% tryptone, 0.5% NaCl, 0.5% yeast extract) or on LB agar (1.5% agar) supplemented with the appropriate antibiotics, unless otherwise stated. Bacillus subtilis and Schizosaccharomyces pombe were grown at 30 °C in LB and YPD broth supplemented with uracil, respectively. For competition experiments, differently labeled E. coli K-12 strains were obtained by transformation with plasmids pVS132 and pVS130 expressing yellow fluorescent protein or cyan fluorescent protein (Hallatschek et al., 2007), respectively. Antibiotics were used at the following concentrations: kanamycin (25 μg ml−1), ampicillin (E. coli: 100 μg ml−1, B. subtilis: 10 μg ml−1). For the MS528+pHHA13 strain, ampicillin (100 μg ml−1) and chloramphenicol (10 μg ml−1) were added to the agar and medium. For cells grown on trypsin plates we added 0.025% trypsin w/v (Sigma Aldrich, Darmstadt, Germany) to the LB agar together with Isopropyl β-D-1-thiogalactopyranoside (IPTG) and ampicillin.
Table 1. Description of the strains and plasmids used in the study.
| Name, relevant characteristics | Reference | |
|---|---|---|
| E. coli strains | ||
| DH5α | LuxS supE44 ΔlacU169, (φ80 lacZΔM15) hsdR17, recA1, endA1, gyrA96, thi-1, relA1 | Invitrogen |
| DH5α-H | LuxS supE44 ΔlacU169, (φ80 lacZΔM15) hsdR17, recA1, endA1, gyrA96, thi-1, relA1 | Hallatschek et al. (2007) |
| MS613 | MG1655 K-12 reference (F-lambda- ilvG- rfb-50 rph-1) strain | Bachmann (1996) |
| MS427 | MG1655 Δflu | Reisner et al. (2003) |
| MS428 | MG1655 Δfim | Kjærgaard et al. (2000) |
| MS528 | MG1655 Δflu,Δfim | Kjærgaard et al. (2000) |
| RMV340 | MG1655cps::tet | Beloin et al. (2006) |
| RMV359 | MG1655fliC ::kan | This study |
| RMV612 | MG1655csgAB::kan | This study |
| S. pombe strains | ||
| PG3732 | h+ ura4D::mCherry | G Thon |
| PG3733 | h+ ura4D::YFP | G Thon |
| B. subtilis strains | ||
| 168phymKATE | eps−: epsG | van Gestel et al. (2014) |
| 168phymGFP | eps−: epsG | van Gestel et al. (2014) |
| Plasmids | ||
| pVS130 | eCFP | V Sourjik |
| pVS132 | YFP | V Sourjik |
| pKD4 | ApKm, kan cassette template | Datsenko and Wanner (2000) |
| pKD46 | Ap, λ Red recombinase | Datsenko and Wanner (2000) |
| pCP20 | ApCm, FLP recombinase expression | Datsenko and Wanner (2000) |
| pACYC184 | Cloning vector, CamR | Chang and Cohen (1978) |
| pHHA13 | Wild-type fim gene cluster in pACYC184 | H Hasman |
Abbreviations: CFP, cyan fluorescent protein; YFP, yellow fluorescent protein.
Competition experiments
The strains were grown as separate cell cultures. Overnight cultures were mixed 1:1 and 1 μl droplets were inoculated on plates with ampicillin (100 μg ml−1) and IPTG (0.5 mm for induction of fluorophores). The seeding of a droplet on an agar plate is illustrated in Figure 1a. B. subtilis was grown on plates with ampicillin (10 μg ml−1) with or without IPTG (0.5 mm for exo-polysaccharide induction (van Gestel et al., 2014)). We also did line inoculations by dipping the side of a cover slip in the cell culture and gently placing it on the agar plate afterwards. Plates with partly absorbed inoculation droplets were incubated at 37 °C for up to 11 days or until the circular colony had obtained a diameter of at least 1.5–2 cm. The cell density used for the inoculation influenced the patchiness of the homeland (Hallatschek et al., 2007) and the number of resulting sectors (van Gestel et al., 2014), but had no apparent effect on the sectoring pattern as also reported (Hallatschek et al., 2007).
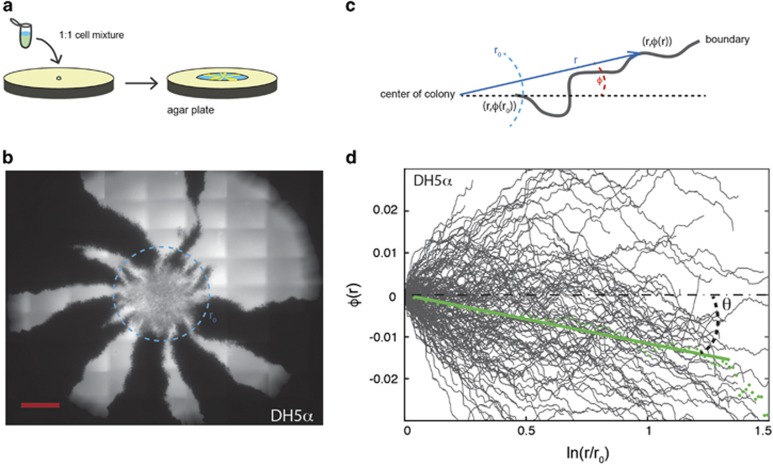
Figure 1.
Competition experiments between neutral E. coli strains expressing different colors, either yellow fluorescent protein (YFP) (yellow) or cyan fluorescent protein (CFP) (cyan). (a) Sketch of the experiment where the two species are mixed 1:1, diluted 1/10, and inoculated on an agar plate where the borders between competing expanding strains are visible. (b) An example of an expanding E. coli DH5α colony is shown from the agar side; the cells transformed with CFP appear white and those with YFP black in this image. The radius of the light-blue dashed circle equals r0. The scale bar corresponds to 1 mm. (c) Illustration of the parameterization, (r, φ(r)), of a point on the sector boundary where r is the length of the radial vector (blue) from the center of the colony and φ(r) is the angle (denoted with orange) between the direction of the sector boundary (dashed orange line) and the radial vector. (d) Parametrization, φ(r), of individual E. coli DH5α sector boundaries (N=146) plotted on semilog scales. The green dots show the average boundary, 〈φ(r)〉, and the full green line shows the best linear fit to the average boundary. As clear from Equation (1), the slope of the fit is the dimensionless measure of chirality: v⊥/v∥=tan(θ). Note that, although the behavior of individual boundaries appear erratic due to random wandering of the boundaries, the average over 146 colonies shows a clear negative slope due to chirality. The values of v⊥/v∥ and θ for all strains investigated are given in Table 2.
Recombinant DNA techniques
Specific mutants (MG1655fliC and MG1655csgAB) of E. coli MG1655 were constructed using the λ-red recombinase-based gene inactivation system as described (Datsenko and Wanner, 2000; Yu et al., 2000). Following amplification of the kanamycin cassette from pKD4, the PCR products were transformed into MG1655 carrying the helper plasmid pKD46 and the transformations were plated on selective plates (50 μg ml−1 kanamycin; primer sequences are given in supplementary Table S1). Deletion mutants were verified by PCR using the test primers given in supplementary Table S1. The kanamycin-resistance cassette was subsequently removed by use of the Flippase-encoding plasmid, pCP20 (Datsenko and Wanner, 2000). For complementation experiments, the pHHA13 plasmid was transformed into the MS528 strain.
Trypsin assay
To test whether trypsin modulates the cell surface or the substratum, autoclaved agar and water were mixed into freshly filter-sterilized LB broth with or without trypsin (0.025%) and left overnight at room temperature. On the following day, the media were autoclaved and IPTG and ampicillin were added. As trypsin under some conditions is heat-resistant (Edie, 1914), trypsin-inhibitor AEBSF (0.02%) was added to half of the medium before pouring plates (1.2% agar). A third medium was prepared from fresh powders and no trypsin.
Substratum substitutions
To identify which of the ingredients of LB agar are responsible for chiral growth, we substituted each ingredient one by one (while always maintaining IPTG and ampicillin). Yeast extract was replaced by 1 μg ml−1 vitamin B1, 1 mm MgCl2 and 10 mm sodium phosphate. Tryptone and NaCl were replaced by 1% decolorized casamino acids (which also contained NaCl). Agar (1.2%) was replaced by 1.2% agarose. Notably, these experiments were done with a viscosity of 1.2% agar/agarose.
Fluorescence microscopy
The colonies were imaged with an inverted Nikon Eclipse Ti fluorescent microscope (Nikon, Tokyo, Japan) using a × 4 air objective and an Andor Neo camera (Andor, Belfast, UK). Emission from yellow fluorescent protein/cyan fluorescent protein was collected at 535/485 nm (30 nm bandwidth) upon excitation at 500/405 nm (20 nm bandwidth). The agar plates were placed upside down directly on the microscope and images were obtained in an array of 12 × 12 sub-images. Individual sub-images were automatically stitched after acquisition with a 15% overlap to image the entire colony, which typically had a diameter of 2–4 cm.
Transmission electron microscopy
Bacteria were harvested from semisolid agar plates and analyzed with respect to their cell morphology and the possible presence of flagella using transmission electron microscopy. The cells were adsorbed onto carbon-coated grids and negatively stained with 2% phosphotungsten acid. The samples were then visualized with a Philips CM 100 TEM (Philips, Eindhoven, The Netherlands) equipped with a OSIS Veleta digital slow scan 2 k × 2 k CCD camera. Digital images were recorded using the ITEM software package (Irvine, CA, USA).
Tracking of sector boundaries
In fluorescent microscopy images of bacterial colonies, the sectors were visible as shown in Figure 1b. The images were converted to black and white by an automated Matlab thresholding procedure (see, for example, left part of Figure 2). Next, we identified the center of the colony and the radius, r0, at which clear sector boundaries first appear, by fitting a circle to the colony rim using the Taubin method in Matlab. A circle with radius r0 is marked on the image in Figure 1b, the strains are mixed inside this region, but sectors can be clearly identified outside r0, this radius being limited by nutrients (Mitri et al., 2015). The mixed region was typically larger than the original inoculated droplet and r0 should be subscript. Then, each colony boundary was identified by edge detection and each pixel on a boundary was assigned a radius (distance to center), r, and an angle, φ. The latter denotes the angle between the boundary and the radial vector, see definitions of r and φ in Figure 1c. Thus, the individual boundaries were parametrized by custom-made Matlab procedures and the average boundary for each data set, 〈φ(r)〉, was found by binning and averaging over all boundaries. The sector boundaries were found to twist as an equiangular, that is, logarithmic, spiral (Huntley, 1970), and each average boundary, 〈φ(r)〉, was therefore least-squared fitted to Equation (1) (Korolev et al., 2011), which describes the evolution of an equiangular spiral:
Figure 2.
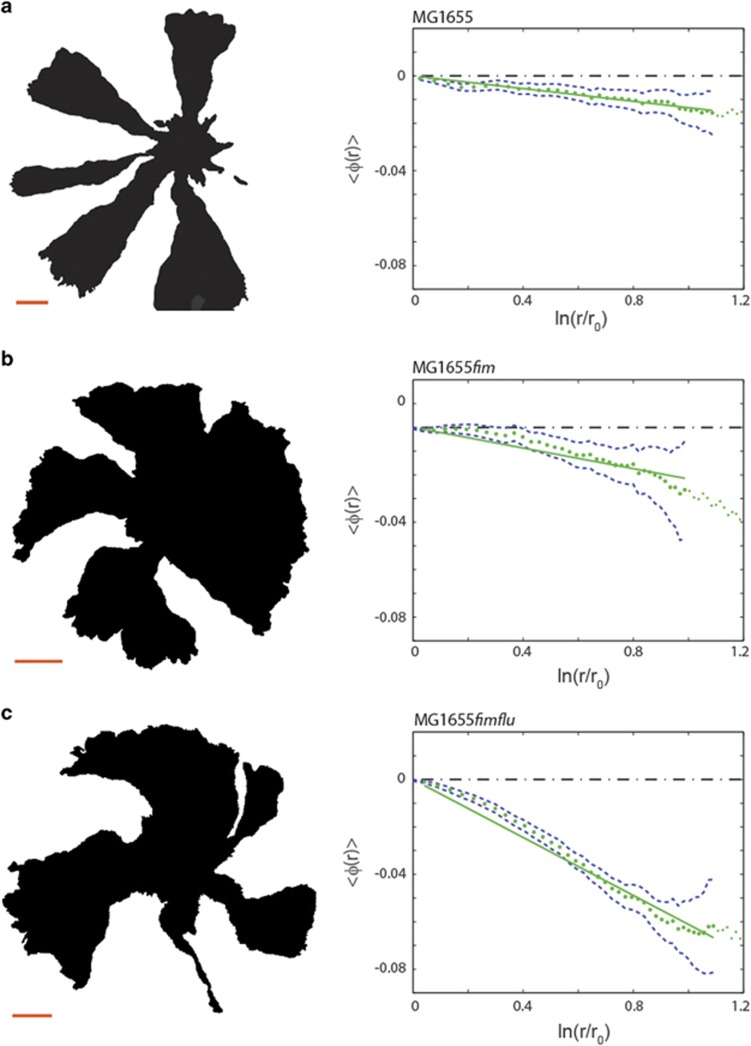
The sector boundaries show different degrees of chirality depending on expression of surface structures. The left panels show examples of intensity thresholded images (displayed from the agar side) of different E. coli MG1655 mutants in competition experiments; the scale bars are 1 mm. The right panels show the average boundary angle, 〈φ(r)〉 (green dotted line), a linear fit (full green line) and one standard error of the mean (blue punctuated line). (a) Results from competition experiments with the mother strain, MG1655; this colony expansion is only slightly chiral (N=369). (b) Competition experiments with the MG1655fim strain lacking pili (N=831). This strain exhibits more chiral borders than MG1655. (c) Competition experiments with the double deleted MG1655fimflu strain lacking both Antigen 43 and pili (N=636). This double deletion causes even more pronounced chirality compared with the single deletion mutants.
Here, v∥ is the velocity in the radial direction, and v⊥ the velocity in the direction perpendicular to the expanding frontier. In the following, we only consider the average change in boundary angle φ(r)−φ(r0) as a function of the radius r, but denote it as 〈φ(r)〉 for simplicity. This can be interpreted as simply choosing the reference direction along 〈φ(r0)〉. For the DH5α strain, the spatial pattern of rotating sector boundaries is shown in Figure 1b and its analysis in terms of Equation (1) is shown in Figure 1d where individual boundaries are plotted as full gray lines and the average boundary is given by green dots. The data are indeed consistent with a linear dependence of a semilog plot as predicted by Equation (1). The slope of the line equals v⊥/v∥, and thus quantifies chirality. In the following, we use a more convenient measure of chirality θ defined as v⊥/v∥=tan(θ) (see definition of θ in Figure 1d). θ denotes the angel between the outward expansion of the colony and the direction of boundary motion. We only observed left-handed (sinistral) motion of sector boundaries when seen from the bottom of the colony (the agar side), so all chiral angles reported are negative. The larger the absolute value of θ, the more chiral the colonies.
Results
Observation of chiral boundaries between competing strains
In this study, we used pairs of isogenic E. coli K-12 strains expressing two different fluorescent proteins (yellow fluorescent protein/cyan fluorescent protein) and the strains with the two labels have similar (neutral) fitness (Hallatschek and Nelson, 2010; Korolev et al., 2011). For competition experiments, a mixture of the two neutral strains was inoculated on an agar surface as illustrated in Figure 1a and colony expansion was quantified by fluorescence microscopy. The growing colonies exhibit a striking segregation of the two populations (cyan fluorescent protein and yellow fluorescent protein) over time in accordance with previous observations. The boundaries, resulting from population segregation, were identified and the chirality angle, θ, determined. The example shown in Figures 1b and d is from an E. coli DH5α colony. The dynamics of segregation was restricted to the edge of the colony, whereas, except for a gradual thickening, the interior distribution of cells was frozen as previously reported (Hallatschek and Nelson, 2010; Korolev et al., 2011). Our analysis shows that DH5α colonies have chiral sector boundaries with θ=−6.1° (s.e.m.=0.05°, N=146), where N is the number of boundaries over all investigated colonies. This compares well to the interval (from −5° to −7°) reported for the DH5α-H strain in Hallatschek et al. (2007). The same strain was later shown to have a chirality angle, θ, within the interval from −13° to −17° (Korolev and Nelson, 2011), which is reasonably similar to the value obtained in the current study for the DH5α-H strain (all θ-values are given in Table 2). This demonstrates that even between similar strains, or strains with the same original ancestry, differences in pedigree or growth conditions might affect the chirality angle.
Table 2. Chirality of the different strains.
| Strain | Name | N | θ±s.e.m. |
|---|---|---|---|
| DH5α | — | 146 | −6.1°±0.05° |
| DH5α-H | — | 183 | −10.2°±0.05° |
| MS613 | MG1655 | 369 | −0.8°±0.003° |
| MS427 | MG1655flu | 192 | −0.4°±0.005° |
| MS428 | MG1655fim | 831 | −1.4°±0.01° |
| MS528 | MG1655fimflu | 636 | −3.5°±0.008° |
| RMV340 | MG1655cps | 989 | −1.3°±0.008° |
| RMV612 | MG1655csgAB | 646 | −1.3°±0.009° |
| RMV359 | MG1655fliC | 542 | −0.4°±0.005° |
| MS528Complemented | MG1655fimflu + pHHA13 | 50 | −1.8°±0.05° |
| MS528trypsin | MG1655fimflu + trypsin | 1952 | −1.0°±0.005° |
| B. subtilis 168 | B. subtilis | 286 | −1.1°±0.03° |
Using different strains, we did both line inoculations (see Supplementary Figure S1) and circular inoculations (as shown in Figure 1) and observed that the sectoring occurs in both types of assays with a similar chirality of the boundaries. Hence, chirality is not linked to radial expansions of the colony front per se but is a more general feature of the species.
Systematic removal of extracellular structures
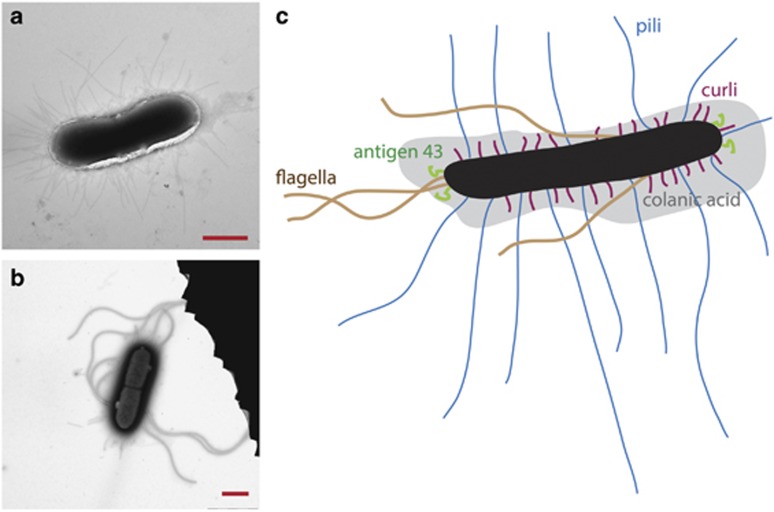
It is a possibility that the sector boundaries observed in bacterial colonies are related to the composition of the cell wall or the extracellular matrix. Obvious determinants could be bacterial cell adhesive surface structures such as exo-polysaccharides, fimbriae, flagella and extracellular DNA, all of which have previously been shown to play a role in cell–surface and cell–cell interactions (see O’Toole et al., 2000 for a review on biofilm formation). Based on this hypothesis, we examined the chirality of expanding colonies of a collection of E. coli strains with specific deletions in genes encoding various bacterial surface structures. Information on all strains and relevant plasmids used is listed in Table 1 and Figure 3 shows transmission electron microscopy images of a MG1655 cell as well as a sketch illustrating the relevant surface structures, that is, flagella, pili, colanic acid, curli fimbriae and antigen 43.
Figure 3.
Electron microscopy images as well as a sketch of the E. coli and the surface structures investigated. (a, b) Transmission electron microscopy images of MG1655. The scale bar is 1 μm. (c) A sketch of E. coli with its extracellular constituents: Antigen 43, pili, curli fimbriae, colanic acid and flagella are explicitly drawn (not to scale). The body of the bacteria is shown in black.
To examine the influence of such structures, a systematic examination of these cell surface components was undertaken, using the parental strain MG1655 (Bachmann, 1996). The sectoring during expansion of MG1655 is shown in Figure 2a. As visible both in the (left) image and in the (right) plot of the average boundary, MG1655 is only slightly chiral with θ=−0.8° (s.e.m.=0.003°, N=369) (the values of θ for all strains are listed in Table 2). Thus, E.coli MG1655 (Figure 3a) displays significantly less chirality than DH5α (Figure 1b). As DH5α and MG1655 both are K-12 strains of E. coli, we find that even among closely related strains the chirality can vary substantially.
We then examined the effect of deleting different adhesion proteins: We first tested MG1655flu, which carries a deletion in the gene encoding the auto-transporter protein antigen 43, a handshake protein which is important for cell–cell aggregation and cellular chain formation (Diderichsen, 1980; Vejborg and Klemm, 2009). Antigen 43 appears in the outer membrane of various bacterial species, including E. coli, and come in copy numbers up to 50 000 per cell (Owen et al., 1987). For MG1655flu θ=−0.4° (s.e.m.=0.05°, N=192), hence, deletion of the antigen 43 gene, flu, lowered chirality slightly, but not significantly, with respect to the parental strain MG1655. We then tested MG1655fim, a strain that does not express pili (also known as type I fimbriae) which are important for cell–cell and cell–surface adhesion. Figure 2b shows that MG1655fim θ=−1.4° (s.e.m.=0.01°, N=831), which is significantly more chiral than MG1655flu and MG1655. Chirality was even more pronounced in the double mutant, MG1655flufim; the expansion of this mutant is shown in Figure 2c and it had θ=−3.5° (s.e.m.= 0.008°, N=636). Hence, the chirality of the double mutant was significantly larger than a simple superposition of the chirality angles of the two individual deletion mutants. Complementing the double deletion mutant with a fim-encoding plasmid (MG1655flufim + pHHA13) reduced the chirality as expected, although not to parental level, thus suggesting that the phenotype is multifactorial. Given the inverse regulation of pili and antigen 43 (Hasman et al., 1999, 2000), the slight loss of chirality for MG1655flu could be a secondary effect, for example, the flu mutant could be overexpressing fim.
As may be apparent from the results with MG1655flu, MG1655fim and MG1655fimflu, the measure of chirality used here, θ, is not additive. Given that θ is small, one might expect that Taylor’s approximation holds and that the contributions from independent mutations should simply add up. However, the deviation from additivity observed here suggests that flu and fim mutations have epistatic interactions, possibly due to induced changes in gene expression, which is in line with the fact that many of the bacterial surface structures display complex regulation (Saldaña et al., 2009).
Colanic acid is an exo-polysaccharide important for biofilm formation while curli fimbriae are important for cell–cell and cell–surface adhesion (Vidal et al., 1998; Prigent-Combaret et al., 1999, 2000; Danese et al., 2000; Oh et al., 2016). Both of these are linked to biofilm formation, although predominantly at ambient temperature (Prigent-Combaret et al., 2000; Gualdi et al., 2008). To examine the effect of the biofilm matrix structures, mutants carrying deletions in genes encoding colanic acid, MG1655cps, and curli fimbriae, MG1655cgsAB, were tested. MG1655cps and MG1655cgsAB both exhibited an increased chirality in comparison with MG1655.
We also examined the effect of flagella expression. For swimming motility in E. coli, a motor rotates the peritrichous flagella controlling the formation of a corkscrew-shaped bundle. By rotating the flagella counterclockwise or clockwise, and by bundling or de-bundling the flagella E. coli performs its characteristic run-tumble-run (Wolfe and Berg, 1989) and is able to move rapidly in response to chemical gradients. This is also the case for (chemotactic) E. coli cells on semisolid agar (0.20–0.35%) (Wolfe and Berg, 1989). The exact function of the flagella on solid agar, as used for this study (1.5%), is still unknown although recent studies suggest that they may have a structural role in biofilm formation. The sector boundaries in colony formation of the flagella mutant, MG1655fliC, had a slightly, but not significantly, lower degree of chirality than the parental strain, θ=−0.4° (s.e.m.=0.005°, N=542). Hence, flagella do not appear to play an important role for colony chirality. This is consistent with the results obtained for DH5α, which is poorly motile, but shows significant chirality. In soft agar swimming assays, we also did not find any correlation between motility and chirality (data not shown). Supporting this observation is the fact that the highly chiral mutant, MG1655fimflu, showed lower motility than the parental strain MG1655 (data not shown).
In summary, the colony chirality significantly increased upon deletion of pili, colanic acid and curli fimbriae. In addition, it was enhanced upon double deletion of both pili and antigen 43. Hence, it appears that the more adhesive structures that are stripped off the cell, the more chiral the colony formation. Deletion of antigen 43 alone or of flagella lowered the chirality slightly, but not significantly, during colony expansion compared with the expansion of the mother strain. Supporting the small effect of flagella removal, chirality also is independent of swimming motility. We found a lower chirality of the strain MG1655flufim when grown at 30 °C (data not shown), and this may be linked to an increased expression of colanic acid and curli fimbriae at 30 °C (Prigent-Combaret et al., 2000; Gualdi et al., 2008).
Chirality and cell wall biosynthesis
During growth of E. coli an MreB guided insertion of material into the peptidoglycan has been suggested to cause a chiral ordering of the bacterial cell wall and a left-handed twisting motion of the elongating E. coli cell (Wang et al., 2012). This twisting motion of individual cells could affect the overall chirality of a colony. To investigate a possible relation between the twisting of bacteria during growth and chirality of the corresponding colony formation, we performed competition experiments with B. subtilis colonies. The cell wall structure of B. subtilis cells twist with an opposite handedness in comparison with E. coli; hence, we wanted to investigate whether this would cause the sector boundaries to twist in a direction opposite to that in E. coli colonies. Therefore, we performed competition experiments with the B. subtilis strains described in van Gestel et al. (2014). The sector boundaries between competing B. subtilis strains are slightly chiral returning a chirality angle θ=−1.1° (s.e.m.=0.03°, N=286), an image from a B. subtilis competition experiment is shown in Supplementary Figure S2b. Hence, we did not see a correlation between the twisting motion of the underlying structure of the cell wall and the overall chirality during colony expansion; this could, however, be because the underlying cell wall structures were masked by extracellular constituents in B. subtilis. In addition, it should be noted that the B. subtilis grow with a morphology quite different from E. coli (see example in Supplementary Figure S2) which complicates a direct comparison. Interestingly, we did find that sector boundaries became coarser and less chiral when exo-polysaccharides were expressed.
Effect of varying agar concentration
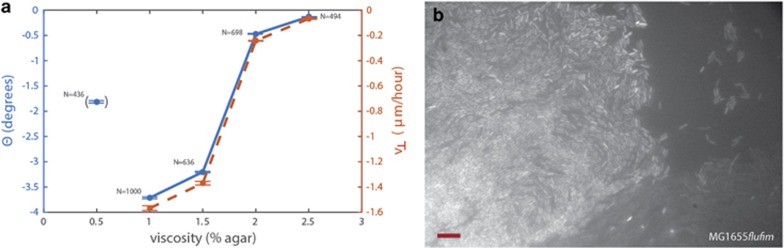
We also investigated the dependence of colony chirality on agar concentration. To this end MG1655flufim was plated on agar plates with agar concentrations in the interval 0.5–2.5%. The observed relation between θ and viscosity is shown in Figure 4a. For an agar concentration of 0.5% the sector boundaries became less clear and the colonies tended to spread into the agar (as opposed to spreading on top of the agar); hence, the nature of colony growth differed significantly from growth at other concentrations. With this point excluded, we found that increasing the agar concentration decreased chirality and θ approached zero at the highest agar concentration tested. These results suggest that the interaction with the surface may become too strong to allow the emergence of chiral behavior. In addition, agar concentration may affect expression of surface structures and thereby influence colony chirality.
Figure 4.
Effect of agar on chirality of sector boundaries and a microscopy image of the sector–sector and colony–agar boundaries. (a) Chirality measured θ (blue points and full line) and the border velocity, v⊥ (red points and punctuated line) are plotted for different agar concentrations; here MG1655fimflu E. coli strain with high chirality was used. The number of boundaries analyzed, N, are noted for each experiment and the error bars signify one s.e.m. At 0.5 % agar, the colony growth was severely compromised compared with other experiments. To indicate this, we put the corresponding θ value in parenthesis. (b) Fluorescent microscopy image (the YFP channel) of sector–sector and colony–agar boundaries in the MG1655fimflu strain. The scale bar is 10 μm.
Colony chirality, θ, is a composite measure of chiral velocity tangential to the expansion front, v⊥ and the radial outward velocity of colony growth, v∥ (v⊥/v∥=tan (θ)). Since both velocities could depend on the agar concentration, we directly measured v∥ and then computed v⊥ from the measured values of θ. The radial velocity component, v∥, increased by 25% (24 –30 μm h−1), as the agar concentration increased from 1 to 2.5%. Simultaneously, the vertical velocity component, v⊥, decreased by 96% from (−1.6 to −0.07 μm h−1). Hence, the change of v⊥ was substantially larger than the change of v∥ and it is clear from Figure 4a that v⊥ is proportional to θ. The radial colony growth is only slightly perturbed by changing the viscosity and diffusion of nutrients in accordance with mechanical predictions (Farrell et al., 2013). However, the chirality is highly affected.
Microscopic alignment during colony expansion
Individual rod-shaped E. coli cells align themselves parallel to the boundaries between the two competing strains; see Figure 4b for an image for the MG1655fimflu. This parallel alignment of mother and daughter cells can be explained by a mechanism where a large number of cells competing for space causes cells to push against each other, and this mechanical instability leads to buckling and folding of the parallel cells lines (Rudge et al., 2012). Away from the boundaries, we often observed cell alignment at the rim of the colony towards the empty agar plate (Figure 4b) as reported previously (Shapiro and Dworkin, 1988; Shapiro and Hsut, 1989; Shapiro, 1995). In contrast, colonies of the spherical S. cerevisiae (budding yeast) show neither appreciable alignment nor chirality (Hallatschek et al., 2007; Korolev et al., 2012; Müller et al., 2014).
To investigate whether macroscopic chirality is determined by the geometry of the microorganism, we performed chirality assays with S. pombe (fission yeast). S. pombe cells are rod shaped albeit larger than E. coli and with a smaller aspect ratio. The boundaries separating competing S. pombe strains are not chiral (see Supplementary Figure S2a); hence, the rod-like shape is not sufficient to cause chiral growth of microorganism colonies. An elongated shape could, however, still be necessary for asymmetric growth as shown, for example, for morphogenesis in Drosophila (Taniguchi et al., 2011).
Trypsin-sensitivity assays
Having observed that systematic removal of features from the E. coli cell surface gave rise to increasingly chiral colony growth, we searched for the mediator of the chirality. To this end, we grew the double deletion strain, MG1655fimflu, on agar plates with trypsin (0.025% w/v). Trypsin is a pancreatic serine protease that cleaves peptide chains on the carboxyl side of lysine and arginine and digests proteins on the cell surface. The overall patterning and the radial growth rate of the colony were unaffected by the presence of trypsin; however, it had a large effect on the chirality of the population boundaries: The chirality angle of the MG1655fimflu strain was reduced to θ=−1.0° (s.e.m.=0.005°, N=1952) (Supplementary Figure S3) similar to the parental level (MG1655). Hence, a non-essential protein component of the bacterial cell surface or the substratum mediates the chiral growth. To test whether trypsin enzymatically modulated the substratum, we let trypsin (0.025%) incubate with LB agar (1.2%) before pouring plates with or without the trypsin-inhibitor AEBSF (0.02%). We found that even in the presence of AEBSF chirality of the MG1655fimflu strain decreased to a level below parental level (data not shown). This observation suggests trypsin enzymatically modulated the substratum, and maybe the cell surface, and indirectly modulates chirality.
Substratum substitution experiments
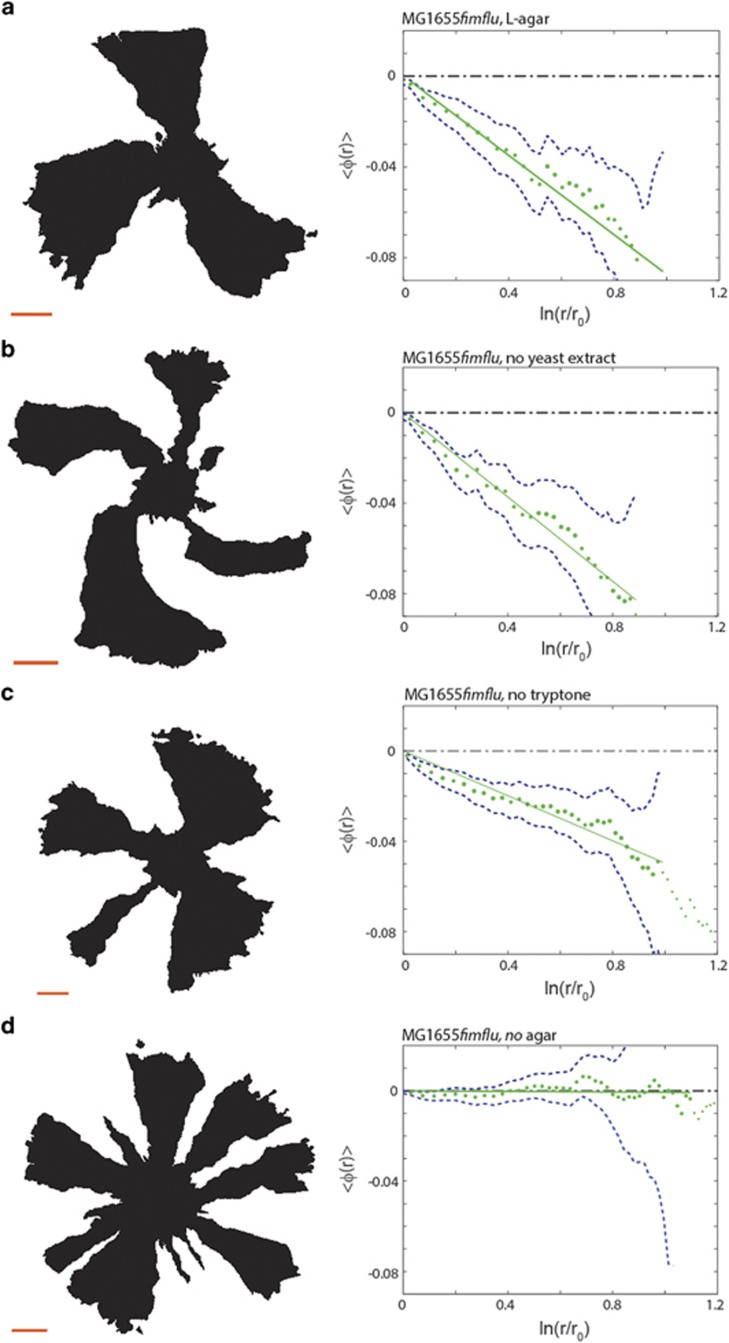
To identify which component of the substratum mediates chiral growth we performed a stepwise substitution of each LB agar constituent. For each substitution, we performed competition experiments using the MG1655fimflu strain and the results are shown in Figure 5. The following experiments were performed: A control experiment, Figure 5a, on normal LB agar (1.2%) which gave θ=−4.0° (s.e.m.=0.05°, N=222). Replacement of yeast extract by vitamin B1, magnesium chloride and sodium phosphate (Figure 5b) gave θ=−6.7° (s.e.m.=0.07°, N=223). Replacement of tryptone by decolorized casamino acids (Figure 5c) gave θ=−4.0° (s.e.m.= 0.05°, N=337), a chirality identical to the control. However, replacing 1.2% agar with 1.2% agarose (Figure 5d) reduced chirality dramatically to θ=−0.1° (s.e.m.=0.01°, N=410), independent of viscosity of the plates (data not shown). Taken together, these results show that the substratum composition modulates chirality either directly, for example, via an intrinsic chiral structure, or indirectly, for example, by altering gene expression or by mediating adhesion of bacteria with accessible chiral surface structures. We cannot exclude the possibility that the change in composition also affects the metabolism/phentoype of the cells. It is well known that small changes in growth medium and solidifying agents can alter gene expression dramatically. This is also true for the expression of cell surface structures. Hence, the change in medium composition may also affect the expression of certain cell surface structures in the cells.
Figure 5.
Effect of substratum substitution. The left panels show examples of intensity thresholded images (displayed from the agar side) of the E. coli MG1655fimflu mutant in competition experiments on LB medium with systematic replacements of constituents; the scale bars are 1 mm. The right panels show the average boundary angle, 〈φ(r)〉 (green dotted line), a linear fit (full green line) and one s.e.m. (blue punctuated line). (a) Results from control competition experiments on normal LB medium (1.2% agar) (N=222). (b) Replacement of yeast extract by 1 μg ml−1 B1, 1 mm MgCl2, and 10 mm sodium phosphate (N=223). (c) Replacement of tryptone and NaCl by 1% decolorized casamino acids (N=337). (d) Replacement of agar by 1.2% agarose (N=410).
Discussion
Some E. coli strains display chiral borders between competing subpopulations during colony expansion. We found that a systematic removal of extracellular constituents involved in biofilm formation, cell–cell and cell–surface adhesion increased chirality. Deletion of pili, curli fimbriae and colanic acid in E.coli MG1655 produced sector boundaries that were more chiral. Loss of two extracellular constituents (antigen 43 and pili) increased chirality further. Interestingly, E.coli MG1655 colony formation displayed relatively low chirality compared with the other K-12 strain, DH5α. This could be due to inadvertent selection in the construction of E. coli DH5α, which is known to be a poor biofilm former and likely lacks certain extracellular structures present in MG1655. These results support a view that intimate contact with the substratum is responsible for the chirality in E. coli, and the adhesive surface structures are masking it by physical shielding or by reducing cell movement during colony growth.
Some of the largest surface structures on E. coli are the pili, which are thin, rigid and adhesive organelles. They are found on many E. coli strains and other members of the Enterobacteriaceae (Klemm and Krogfelt, 1994) and are important for adherence to mammalian host tissues (Krogfelt et al., 1990; Connell et al., 1996) and for biofilm formation (Pratt and Kolter, 1998; Schembri and Klemm, 2001). On a piliated cell, these peritrichous surface structures can radiate to a distance of up to 2 μm from the cell surface (Figure 3a), making them prime candidates for a physical masking of an underlying cell envelope structure. Accordingly, we found that the deletion of piliating genes in MG1655 (MG1655fim) significantly increased colony chirality. Interestingly, pili expression, which is subject of phase variation, that is, individual cells can switch between a piliated and non-piliated state, is normally considered low on solid media. Nevertheless, the presence of pili clearly decreases chirality.
Antigen 43, the product of the flu gene, is a self-recognizing auto-transporter protein. It confers auto-aggregation giving rise to a characteristic frizzy colony morphology on solid media as well as flocculation of cells in static liquid cultures (Hasman et al., 1999). Antigen 43 is known to be important for bacterial biofilm formation (Danese et al., 2000; Kjærgaard et al., 2000; O’Toole et al., 2000) and for cellular chain formation (Vejborg and Klemm, 2009). Interestingly, antigen 43 mediated auto-aggregation is inhibited by the presence of pili on the cells (Hasman et al., 1999). Consistent with this finding, we observed little change of chirality in the MG1655flu mutant, but a significant enhancement in θ of the double flu, fim mutant (MG1655flufim). Overexpression of pili in the MG1655flufim also could not fully reverse the phenotype, clearly suggesting that antigen 43 also plays an important role, and both surface structures mask the underlying chirality of the cell.
The deletion of curli fimbriae (MG1655csgAB) or colanic acid (MG1655cps) also made sector boundaries significantly more chiral. This increase in chirality was somewhat surprising, given that curli and colanic acid are not typically expressed in K-12 strains at 37 °C (Olsén et al., 1989). Given that the expression of many surface structures is intricately connected, we cannot exclude the possibility that the deletion of these genes could affect the expression of other surface structures. It should also be noted that MG1655cps was independently derived and could have acquired additional genetic and phenotypic variations, which might obscure direct comparison with our version of E.coli MG1655.
Flagella-dependent motility is known to play a major role in chiral spreading in other bacteria. For example, Proteus mirabilis produces chiral swarming patterns via this mechanism (Xue et al., 2011). E. coli also use flagella for active swimming, which can produce chiral motion due to hydrodynamic interactions (DiLuzio et al., 2005). In addition, flagella are involved in biofilm formation (Pratt and Kolter, 1998), and they may play a structural role in colony biofilms (Serra et al., 2013). Not surprisingly, flagella have been put forward as the primary suspect for chiral behavior in bacteria. For instance, for Paenibacillus dendritiformis, it is proposed in Ben-Jacob et al. (1995) that chirality is caused by flagella and is mediated by strong cell–cell interaction. We found that chirality decreases slightly, and not statistically significantly, upon flagella removal. However, there was no link between the magnitude of chirality and flagella-mediated motility (as determined in swimming motility assays); in particular, the most chiral strains (DH5α and MG1655flufim) were poorly motile. Taken together, our data suggest that flagella, for these types of assays, contributes little to chirality in E. coli colonies under the studied growth conditions.
The macroscopic handedness of the chiral borders between competing E. coli strains is opposite of the microscopic handedness of the growth of the peptidoglycan layer (Wang et al., 2012). Hence, although chirality is increased upon the removal of surface structures that possibly mask underlying cell wall structures, there is no direct link between the spiral growth direction of the peptidoglycan and the chirality of colony borders. This was confirmed by the same chirality direction of E. coli and B. subtilis despite the opposite directions of cell wall biosynthesis in these two species. However, spiral growth has in Myxococcus xanthus been linked to another kind of motility (gliding) than employed by E. coli.
Similar to chirality in regular matter (de Gennes and Prost, 1995), chirality in bacterial colonies may require parallel geometric ordering of cells during cell division and colony expansion (Shapiro and Hsut, 1989; Su et al., 2010), which is clearly visible at the border between competing strains (Figure 4b). The composition of the substratum has been shown to control cell morphogenesis (Su et al., 2010). Hence, the alignment of mother–daughter cells is probably affected by the cells’ ability to attach to the substratum and possibly to each other. Supporting the importance of the substratum for chiral growth, we also found that interchanging agar with agarose in the substratum reduced chirality. This suggests that the agar polymer network itself could be chiral or that it includes chiral entities. However, it could also be that cell–surface adhesion were modulated or that gene expression was changed.
Active chiral processes play an important role in pattern formation as well as during embryonic development (Coutelis et al., 2014). While the ecological role of chirality in the microbial world is still poorly understood, chiral phenotypes could be functional or even advantageous to microbial communities as they are during embryogenisis. Indeed, one can view microbial colonies as a stepping-stone towards multi-cellularity (Shapiro and Dworkin, 1988), and, therefore, related pattern forming mechanisms could be required. Consistent with these ideas, a switch of chirality has been reported in bacteria as a response to changes in the environmental conditions (Ben-Jacob et al., 2000) and during tumorigenesis (Wan et al., 2011). We found that E. coli employs a mechanisms to generate chirality distinct from higher organisms, which often rely on the asymmetric orientation of the mitotic spindle during cell division (Grande and Patel, 2009) or, in the case of Caenorhabditis elegans, on chiral torque generation in the cortex (Naganathan et al., 2014). Although the microscopic mechanisms for chirality in bacteria are distinct, some conclusions of our work could generalize to other settings. In particular, we established an experimental approach to examine the role of surface structure and adhesion in chiral pattern formation.
Conclusions
Chirality is a general property of biological systems which is shared by very different species as the Gram-positive B. subtilis and the Gram-negative E. coli. Efficient adaptation of bacterial colonies to changing environmental conditions requires cooperative behavior and self-organization, which is based on the exchange of information. The different communication channels range from direct physical and chemical interaction to indirect interactions through chemotaxis and trails on the agar. Pattern formation in microbial colonies is a way to exchange information between the microscopic level and the macroscopic level. In other words, an exchange between the individual cells and the colony. Therefore, the full understanding of chiral growth, its origins and implications must focus on the relation between the individual cell and the colony organization. For E. coli colonies expanding on a substratum, the border between competing strains often exhibits chirality and always in a left-handed manner (when observed from the agar side). We find a correlation between the expression of surface structures involved in cell–cell and cell–surface adhesion or biofilm formation and the macroscopic degree of chirality; the more surface structures expressed, the less chiral the border between competing strains. This result was obtained by comparing pattern formation of MG1655 to pattern formation of MG1655 mutants with deletion of pili, curli fimbriae, colanic acid and antigen 43. Furthermore, we found chirality to be independent of flagella-mediated motility. The DH5α strain had more chiral borders than any of the MG1655 mutants, possibly because DH5α is a poorer biofilm former. We also find that substituting agar with agarose reduces chirality, suggesting that chirality largely depend on substratum composition. The exact mechanism by which the substratum mediates the chiral growth will be a subject of future investigation. Overall, intimate cell–substratum adhesion is necessary for chirality and extracellular features, such as fimbriae or biofilm, weakens this contact. Our results link macroscopic chiral colony formation to the microscopic biological features of E. coli’s cell wall and the substratum and suggest new ecological roles for several membrane-associated proteins during colony growth.
Acknowledgments
We thank G Thon and J van Gestel for providing us with S. pombe and B. subtilis strains, respectively. We also thank the anonymous referee for valuable comments. We are grateful to the Lundbeck foundation, the Danish National Research Foundation grant number DNRF116, the Carlsberg foundation and the Danish Research Councils for the financial support.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ali A, Grosskinsky S. (2010). Pattern formation through genetic drift at expanding population fronts. Adv Complex Syst 13: 349–366. [Google Scholar]
- Bachmann BJ. (1996). Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt FC editor, Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. ASM Press: Washington, DC, pp 2460–2488. [Google Scholar]
- Beloin C, Michaelis K, Lindner K, Hacker J, Ghigo J, Dobrindt U et al. (2006) The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J Bacteriol 188: 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Jacob E, Cohen I, Levine H. (2000). Cooperative self-organization of microorganisms. Adv Phys 49: 395–554. [Google Scholar]
- Ben-Jacob E, Cohen I, Shochet O, Tenenbaum A. (1995). Cooperative formation of chiral patterns during growth of bacterial colonies. Phys Rev Lett 75: 2899–2902. [DOI] [PubMed] [Google Scholar]
- Chang ACY, Cohen SN. (1978). Construction and characterization of amplifiable DNA cloning vectors derived from P15A cryptic plasmid. J Bacteriol 134: 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. (1996). Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA 93: 9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelis J-B, González-Morales N, Géminard C, Noselli S. (2014). Diversity and convergence in the mechanisms establishing L/R asymmetry in metazoa. EMBO Rep 15: 926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Dove SL, Kolter R. (2000). The outer membrane protein, Antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol 37: 424–432. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderichsen B. (1980). flu, A metastable gene controlling surface properties of Escherichia coli. J Bacteriol 141: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLuzio WR, Turner L, Mayer M, Garstecki P, Weibel DB, Berg HC et al. (2005). Escherichia coli swim on the right-hand side. Nature 435: 1271–1274. [DOI] [PubMed] [Google Scholar]
- Edie ES. (1914). On the resistance of trypsin solutions to heat. Biochem J 8: 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell FDC, Hallatschek O, Marenduzzo D, Waclaw B. (2013). Mechanically driven growth of quasi-two-dimensional microbial colonies. Phys Rev Lett 111: 168101. [DOI] [PubMed] [Google Scholar]
- Flack HD. (2009). Louis Pasteurs discovery of molecular chirality and spontaneous resolution in 1848, together with a complete review of his crystallographic and chemical work. Acta Crystallogr Sect A Found Crystallogr 65: 371–389. [DOI] [PubMed] [Google Scholar]
- Frank FC. (1953). On spontaneous asymmetric synthesis. Biochim Biophys Acta 11: 459–463. [DOI] [PubMed] [Google Scholar]
- Gayathri VS, Rao M. (2005). Fluctuation induced chiral symmetry breaking in autocatalytic reaction-diffusion systems. Europhys Lett 80: 4. [Google Scholar]
- de Gennes P-G, Prost J. (1995) The Physics of Liquid Crystals (International Series of Monographs on Physics). Oxford University Press: New York. [Google Scholar]
- van Gestel J, Weissing FJ, Kuipers OP, Kovács AT. (2014). Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J 8: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande C, Patel NH. (2009). Nodal signalling is involved in left-right asymmetry in snails. Nature 457: 1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi L, Tagliabue L, Bertagnoli S, Ieranò T, De Castro C, Landini P. (2008). Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 154: 2017–2024. [DOI] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. (2007). Genetic drift at expanding frontiers promotes gene segregation. Proc Nat Acad Sci USA 104: 19926–19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Nelson DR. (2010). Life at the front of an expanding population. Evolution 64: 193–206. [DOI] [PubMed] [Google Scholar]
- Hasman H, Chakraborty T, Klemm P. (1999). Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181: 4834–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H, Schembri MA, Klemm P. (2000). Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J Bacteriol 182: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley HE. (1970) The Divine Proportion: A Study in Mathematical Beauty. Courier Dover Publications: New York, NY, USA. [Google Scholar]
- Jafarpour F, Biancalani T, Goldenfeld N. (2015). Noise-induced mechanism for biological homochirality of early life self-replicators. Phys Rev Lett 115: 1–5. [DOI] [PubMed] [Google Scholar]
- Kjærgaard K, Schembri MA, Ramos C, Molin S, Klemm P. (2000). Antigen 43 facilitates formation of multispecies biofilms. Environ Microbiol 2: 695–702. [DOI] [PubMed] [Google Scholar]
- Klemm P, Krogfelt K. (1994) Type 1 fimbriae of Escherichia coli. In: Klemm P (ed). Fimbriae: Adhesion, Genetics, Biogenesis and Vaccines. CRC Press: Boca Raton, pp 9–26. [Google Scholar]
- Korolev K, Nelson D. (2011). Competition and cooperation in one-dimensional stepping-stone models. Phys Rev Lett 107: 1–5. [DOI] [PubMed] [Google Scholar]
- Korolev KS, Avlund M, Hallatschek O, Nelson DR. (2010). Genetic demixing and evolution in linear stepping stone models. Rev Mod Phys 82: 1691–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Müller MJI, Karahan N, Murray AW, Hallatschek O, Nelson DR et al. (2012). Selective sweeps in growing microbial colonies. Phys Biol 9: 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Xavier JB, Nelson DR, Foster KR. (2011). A quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am Nat 178: 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J-U. (2004). Conflicts of interest in biofilms. Biofilms 1: 265–276. [Google Scholar]
- Krogfelt KA, Bergmans H, Klemm P. (1990). Direct evidence that the FimH protein is the mannose-specific adhesion of Escherichia coli type 1 fimbriae. Infect Immun 58: 1995–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda R, Endo B, Abe M, Shimizu M. (2009). Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature 462: 790–794. [DOI] [PubMed] [Google Scholar]
- Mitri S, Clarke E, Foster KR. (2015). Resource limitation drives spatial organization in microbial groups. ISME J 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MJI, Neugeboren BI, Nelson DR, Murray AW. (2014). Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci USA 111: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganathan SR, Fürthauer S, Nishikawa M, Jülicher F, Grill SW. (2014). Active torque generation by the actomyosin cell cortex drives left–right symmetry breaking. Elife 3: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. (2000). Biofilm formation as microbial development. Annu Rev Microbiol 54: 49–79. [DOI] [PubMed] [Google Scholar]
- Oh YJ, Hubauer-Brenner M, Gruber HJ, Cui Y, Traxler L, Siligan C et al. (2016). Curli mediate bacterial adhesion to fibronectin via tensile multiple bonds. Sci Rep 6: 33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsén A, Jonsson A, Normark S. (1989). Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338: 652–655. [DOI] [PubMed] [Google Scholar]
- Owen P, Caffrey P, Josefsson LG. (1987). Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J Bacteriol 169: 3770–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30: 285–293. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. (2000). Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2: 450–464. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. (1999). Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol 181: 5993–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48: 933–946. [DOI] [PubMed] [Google Scholar]
- Rudge TJ, Steiner PJ, Phillips A, Haseloff J. (2012). Computational modeling of synthetic microbial biofilms. ACS Synth Biol 1: 345–352. [DOI] [PubMed] [Google Scholar]
- Saito Y, Sugimori T, Hyuga H. (2007). Stochastic approach to enantiomeric excess amplification and chiral symmetry breaking. J Phys Soc Japan 76: 1–14. [Google Scholar]
- Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL et al. (2009). Synergistic role of curli and cellulose in cell adherence and biofilm formation of aataching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Env Microbiol 11: 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri MA, Klemm P. (2001). Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J 20: 3074–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck G, Mika F, Hengge R. (2013). Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. (1995). The significance of bacterial colony patterns. BioEssays 17: 597–607. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, Dworkin M. (1988). Bacteria as multicellular organisms. Sci Am 6: 82–89. [Google Scholar]
- Shapiro JA, Hsut C. (1989). Escherichia coli K-12 cell-cell interactions seen by time-lapse video. J Bacteriol 171: 5963–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P-T, Yen P-W, Wang S-H, Lin C-H, Chiou A, Syu W-J. (2010). Factors affecting daughter cells’ arrangement during the early bacterial divisions. PLoS One 5: e9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Maeda R, Ando T, Okumura T, Nakazawa N, Hatori R et al. (2011). Chirality in planar cell shape. Science (80-) 339: 339–342. [DOI] [PubMed] [Google Scholar]
- Vejborg RM, Klemm P. (2009). Cellular chain formation in Escherichia coli biofilms. Microbiology 155: 1407–1417. [DOI] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. (1998). Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J Bacteriol 180: 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LQ, Ronaldson K, Park M, Taylor G, Zhang Y, Gimble JM et al. (2011). Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci USA 108: 12295–12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Furchtgott L, Huang KC, Shaevitz JW. (2012). Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA 109: E595–E604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. (1989). Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA 86: 6973–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Budrene EO, Othmer HG. (2011). Radial and spiral stream formation in proteus mirabilis colonies. PLoS Comput Biol 7: 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee E-C, Jenkins Na, Copeland NG, Court DL. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.