Abstract
Phylogenomics is extremely powerful but introduces new challenges as no agreement exists on “standards” for data selection, curation and tree inference. We use jawed vertebrates (Gnathostomata) as model to address these issues. Despite considerable efforts in resolving their evolutionary history and macroevolution, few studies have included a full phylogenetic diversity of gnathostomes and some relationships remain controversial. We tested a novel bioinformatic pipeline to assemble large and accurate phylogenomic datasets from RNA sequencing and find this phylotranscriptomic approach successful and highly cost-effective. Increased sequencing effort up to ca. 10Gbp allows recovering more genes, but shallower sequencing (1.5Gbp) is sufficient to obtain thousands of full-length orthologous transcripts. We reconstruct a robust and strongly supported timetree of jawed vertebrates using 7,189 nuclear genes from 100 taxa, including 23 new transcriptomes from previously unsampled key species. Gene jackknifing of genomic data corroborates the robustness of our tree and allows calculating genome-wide divergence times by overcoming gene sampling bias. Mitochondrial genomes prove insufficient to resolve the deepest relationships because of limited signal and among-lineage rate heterogeneity. Our analyses emphasize the importance of large curated nuclear datasets to increase the accuracy of phylogenomics and provide a reference framework for the evolutionary history of jawed vertebrates.
Keywords: cross-validation, jackknifing, Gnathostomata, molecular dating, phylogeny, RNA-Seq, substitution rates, transcriptome
Introduction
Understanding the evolutionary relationships among organisms is a prerequisite for any biological study aiming at explaining key processes such as adaptive radiations or evolutionary convergences. Evolutionary relatedness is generally represented with phylogenetic trees, which need to be robust and accurate if one aims at obtaining credible macroevolutionary inferences. In the last decade, genome-scale datasets (phylogenomics) have revolutionized molecular phylogenetics thanks to their ability to yield precise estimates of phylogeny and more precise divergence times by reducing sampling error, one of the major hurdles in the pre-genomics era. Several methodologies have been used to generate raw data for phylogenomics using high-throughput sequencing. But besides the obvious advantages, phylogenetic inference based on genomic data poses numerous challenges. For the assembly of genome-scale datasets, these include the removal of contaminants (from symbionts, pathogens or food items in the original sample or introduced during processing steps such as human DNA or sample cross-contaminations), misalignments due to erroneous sequence stretches (often produced by sequencing and annotation errors in low-coverage genome assemblies), the effective detection and removal of paralogs, and the presence of large amounts of missing data, often aggravated by the difficulty of identifying orthology in only partially assembled transcripts. Paralogy in particular can have dramatically detrimental effects on phylogenomic analyses1, but the robustness of tree topology to the inclusion of paralogs is generally not evaluated.
Because phylogenomics relies on hundreds or thousands of genes and taxa, manual data curation has become unfeasible and automatic solutions need to be devised. Phylogenomic analyses have generally relied on pooling evidence from multiple genes by concatenation or used “summary” coalescent-based species-tree methods. The size of genomic datasets also makes phylogenomics more sensitive to model misspecification (systematic error), which often translates into long-branch attraction problems2. Systematic error may be reduced with complex mixture models, but their application to large-scale phylogenomic matrices can sometimes become computationally intractable. In addition, phylogenomic alignments are known to inflate non-parametric bootstrap support values and Bayesian clade posterior probabilities, a precision not always accompanied by increased accuracy, thus rendering the interpretation of these support metrics difficult.
The above challenges regarding the quality of the data and the robustness of analytical approaches need to be carefully taken into account in order to produce reliable estimates of both phylogeny and divergence times. Jawed vertebrates (Gnathostomata) represent a good system to benchmark these challenges because of the availability of genomic data for many species but the remarkable absence of several species with key phylogenetic positions, and the relatively good knowledge of their phylogeny except for some nodes that were controversial. In addition, jawed vertebrates are among the best-studied organisms and include astonishing examples of convergent evolution (e.g., flight, echolocation, limb loss) and prominent instances of classic paraphyletic taxa such as "fishes" or "reptiles". Biologists have long been interested in understanding the evolutionary relationships among jawed vertebrates, first using morphological characters and later with sequence data. Molecular phylogenies have greatly contributed towards shaping the jawed vertebrate tree, in many instances corroborating classical morphology-based classifications, but sometimes establishing novel hypotheses such as the close relationship of turtles with crocodiles and birds. Studies relying on mitochondrial genomes (mitogenomes) have resolved several controversial issues3, but also recovered some unorthodox relationships4. Earlier molecular studies based on multiple genes obtained by classical Sanger-sequencing approaches have generally been limited by the number of genes or taxa, and were generally restricted to particular lineages such as ray-finned fishes5, amphibians6, squamate reptiles7, mammals8 or birds9. With the rise of genome-scale molecular datasets, it became possible to use ever larger datasets in an attempt at solving the relationships in the Tree of Life, and many nodes of the jawed vertebrate tree have been confirmed by phylogenomic analyses based on datasets obtained by second-generation sequencing and typically focusing on particular gnathostome clades10–17. Despite this growing consensus, some phylogenomic studies have also challenged important relationships, such as the monophyly and internal relationships of amphibians18 or the position of turtles19, demonstrating that crucial aspects of the jawed vertebrate tree still require careful attention. Further evolutionary relationships also remain controversial because of incongruence among molecular phylogenies or with morphological evidence, such as the close relationship of iguanian lizards with snakes20–22 or the relationships among tongueless frogs23,24. Convincingly resolving difficult nodes requires more than just a large number of genes, and instead a focus is needed on carefully avoiding and removing contaminations and errors in the data, and avoiding model misspecifications25.
Since their origin in the Ordovician (~470 Mya), jawed vertebrates have diversified into lineages with markedly different morphologies and life histories, including hyperdiverse radiations such as spiny-rayed fishes, birds, modern frogs (Neobatrachia) and placental mammals. As an appealing hypothesis, the main diversification bursts of these hyperdiverse radiations have been proposed to coincide with the Cretaceous-Paleogene boundary5,15, but due to uncertainties in timetree reconstruction and methodological disputes on molecular dating, this hypothesis remains contentious, especially for mammals26–28.
Here, we use a phylotranscriptomic approach to reconstruct the backbone of the jawed vertebrate tree based on a dataset of unprecedented size composed of 7,189 genes for 100 species representing all main gnathostome lineages (a total of 3,791,500 aligned amino acid positions). The dataset includes 23 newly generated transcriptomes from previously underrepresented clades occupying key phylogenetic positions, particularly early-branching ray- and lobe-finned fishes, lungfishes, amphibians and squamate reptiles. We devised a novel bioinformatic pipeline to assemble the largest and most informative dataset ever analysed for vertebrates (Supplementary Fig. 1) while focusing on the comprehensive removal of contaminants and paralogs. This dataset is subjected to thorough phylogenetic and molecular dating analyses. We present a strongly supported phylogenetic hypothesis, which is fossil-calibrated to yield robust divergence time estimations, thus providing a reference framework for the evolutionary history of jawed vertebrates.
Results and discussion
Phylotranscriptomic pipeline to assemble clean datasets
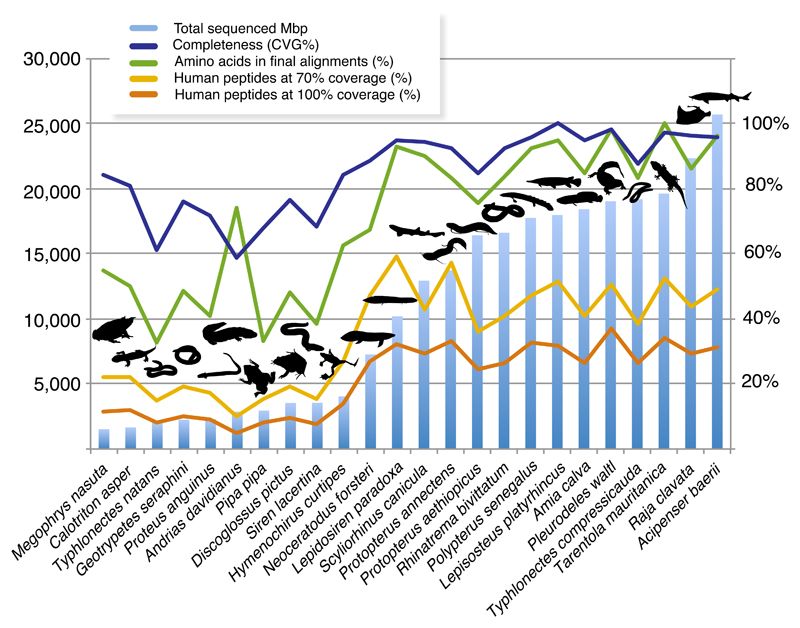
We developed a new bioinformatic pipeline (Supplementary Fig. 2) to assemble an informative and “clean” genome-scale dataset of jawed vertebrates using genome and transcriptome sequence data. For this study we collected RNA-Seq data for 23 previously unsampled gnathostome species representing key lineages. Sequencing effort for the new transcriptomes varied considerably among species (total sequenced base pairs ranged from 1.5 to 26 Gbp; Fig. 1 and Supplementary Table 1) and it correlated positively with (i) the average length of reconstructed transcripts (r=0.78; p=8.207x10-6), (ii) transcriptome completeness, measured as the proportion of recovered core vertebrate genes29 (r=0.78; p=6.173x10-6) and (iii) the total number of amino acids in final phylogenomic datasets (r=0.82 p=0.00066) (Supplementary Table 2). Despite considerable differences in sequencing effort, all transcriptomes were relatively complete (58.8 to 100% of the 233 core vertebrate genes were recovered; Fig. 1) and thousands of genes readily usable for phylogenomics were reconstructed (2,274 to 13,642 high-coverage genes per species, measured as human proteins at ≥70% length coverage; Fig. 1). Hence, deeper sequencing increased the completeness of transcriptome assemblies and the number of genes and amino acid positions in final alignments. Nevertheless, this tendency stabilized at approximately 10 Gbp of total data (for example, 50 million 100 bp-long read pairs), after which a higher sequencing effort did not significantly increase the above performance metrics (r<0.5 and p>0.05 in all correlations for transcriptomes with >10 Gbp of total data; Supplementary Table 2). Interestingly, genes missing in final phylogenomic matrices were essentially not different in species with shallow or deeply sequenced transcriptomes (assessed by GO enrichment tests with FDR<0.05 against the annotated set of 7,189 genes, run in Blast2GO, which suggests that sequencing effort does not significantly bias the types of genes present in final alignments.
Figure 1.
Transcriptome sequencing effort and performance in phylogenomic dataset assembly. Histogram represent sequencing effort as total number of sequenced (clean) Mbp (million bp). Transcriptome completeness is measured as the proportion of recovered core vertebrate genes (233 CVG; Hara et al.29). Genes effectively usable for phylogenomics are approximated by the proportion of human proteins reconstructed at full (100%) and nearly full (>70%) lengths (in proportion to a total of 22,964 human genes). The completeness the relevant species in our final phylogenomic dataset is shown as the proportion of amino acids across all 7,189 genes (3,791,500 aligned amino acids in total).
The new bioinformatic pipeline established herein warranted the high quality of alignments by addressing key issues in data integrity25, including several steps to minimize possible contaminations, resolution of paralogy, masking of misalignments, and minimizing missing data. During decontamination steps, BLAST similarity searches were used to identify potentially contaminant sequences from non-vertebrates and human sequences (in this latter case requiring high-identity at the nucleotide level). To remove any remaining contamination, we devised a sensitive protocol that identifies extremely long branches estimated on a fixed reference tree to flag possibly erroneous sequences, which were then removed. Per-sequence missing data was minimized by merging conspecific sequences (typically overlapping partially reconstructed transcripts) with SCaFoS30, and unreliably aligned regions discarded. A new tool based on profile hidden Markov models (HMM) was used to mask erroneous sequence stretches typically produced by frame shifts in ORFs or incorrect structural annotation. We implemented an innovative paralog-splitting pipeline that specifically targets distant paralogs (those particularly problematic for resolving the backbone of the tree) and further assessed the effect in the tree stability of including various levels of deep paralogy in the datasets. In order to do that, genes were classified into three sets that contained zero (NoDP), one (1DP) and two (2DP) deep paralogs (i.e., duplication events predating the origin of major jawed vertebrate lineages), which were then concatenated into three datasets that were separately analysed: NoDP (4,593 genes, 1,964,439 amino acids, 32% missing data), 1DP (1,162 genes, 668,132 amino acids, 36% missing data), and 2DP (1,434 genes, 1,158,929 amino acids, 39% missing data).
Backbone phylogeny of jawed vertebrates
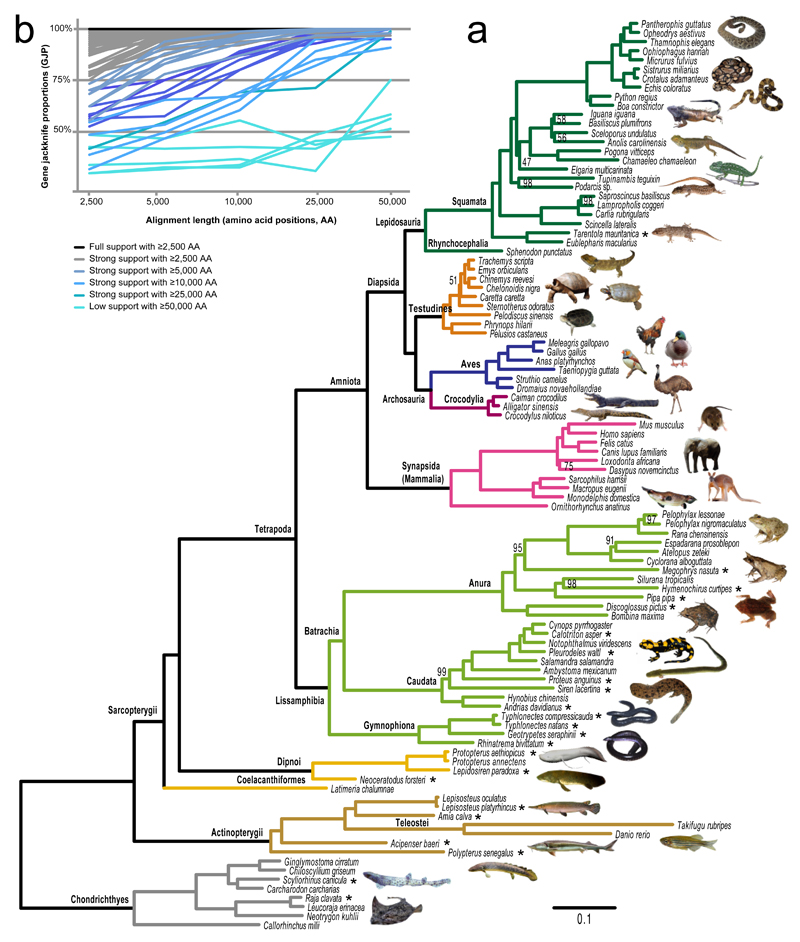
The phylogeny was estimated based on concatenated alignments by (i) maximum likelihood (ML) under the site-homogeneous LG+F+Γ and GTR+Γ models and 100 bootstrap replicates in RAxML and (ii) Bayesian inference (BI) under the more realistic site-heterogeneous CAT+Γ model in PhyloBayes. For computational tractability of large datasets under complex and computationally expensive models and to further assess the effect of gene sampling, BI analyses were performed on 100 gene jackknife replicates (~50,000 amino acids and ~180 genes per replicate), which were summarized in a final majority-rule consensus tree. Gene jackknifing measures the repeatability of the phylogenetic relationships across genes, which are randomly sampled without replacement from the total set of genes31. We employed gene jackknife proportions (GJP) as a stringent test for the robustness of the obtained relationships because they were estimated under the more realistic CAT model and based on virtually independent gene sets, each containing ~2.5% of the total alignment, as compared to the ~66% of the total alignment used in non-parametric bootstrapping. In addition, we carried out coalescent-based species tree analyses with ASTRAL-II with 100 replicates of multi-locus bootstrapping on the three nuclear datasets separately. All phylogenetic analyses of the paralog-free dataset (NoDP), including BI (Fig. 2a) and ML on the concatenated super-matrix and species tree analyses (Supplementary Figs. 3-5), reconstructed fully resolved and almost identical trees that were highly supported: 88% and 95% of the nodes in Fig. 2 received respectively full (100%) or high (>75%) GJP. All major uncontroversial vertebrate clades were recovered with full support: cartilaginous fishes (Chondrichthyes) were the sister group of bony fishes (Osteichthyes), including ray-finned (Actinopterygii) and lobe-finned (Sarcopterygii) fishes; within sarcopterygians, tetrapods (Tetrapoda) were monophyletic and encompassed amphibians (Lissamphibia), mammals (Mammalia), turtles (Testudines), birds (Aves), crocodiles (Crocodylia), lepidosaurian reptiles (Lepidosauria) and snakes (Serpentes). Even using relatively small alignments of ~5,000 amino acids (Fig. 2b, Supplementary Table 3), all the above nodes were recovered with strong support. In fact, these uncontroversial nodes were also recovered by a large proportion of single-gene trees (58-96% of the genes; Supplementary Table 4) though with varying levels of support.
Figure 2.
Backbone phylogeny of jawed vertebrates. (a) Bayesian majority-rule consensus tree from 100 independent MCMC chains derived from gene jackknife replicates (~50,000 amino acid positions each) of the NoDP nuclear dataset, estimated by PhyloBayes under the CAT+Γ model. All nodes received full gene jackknife support (100%), except those displaying the actual value. The scale bar corresponds to the expected number of substitutions per site. Asterisks denote new transcriptomic data generated in this study. (b) Effect of alignment length on the recovery of single nodes in the phylogeny assessed by gene jackknife proportions derived from the NoDP dataset.
In contrast, some of the relationships that remained hotly discussed during the past decades were not unambiguously recovered by single genes nor by relatively small-sized gene jackknife replicates (Fig. 2b and Supplementary Tables 3, 4). Thanks to the use of a larger dataset, our analyses however effectively resolved these controversial relationships with maximum support (Fig. 2a). (i) Lungfishes (Dipnoi) were the sister group of tetrapods, in agreement with the latest phylogenomic results12,32, and topology tests rejected the alternative hypothesis where coelacanth and tetrapods are sister taxa33 (Supplementary Table 5). (ii) Amphibians (Lissamphibia) were monophyletic and salamanders (Caudata) were the sister group of frogs (Anura) to the exclusion of caecilians (Gymnophiona) (Batrachia hypothesis34,35). Both the paraphyly of amphibians and the alternative sister group of caecilians and salamanders (Procera hypothesis18) were rejected by topological tests. (iii) Turtles were the sister group of crocodiles and birds (Archosauria), in agreement with the majority of previous phylogenomic studies10,11 and the latest morphological evidence36. Topology tests rejected the traditional view of turtles as primarily anapsids (early-branching within “reptiles”) as well as possible sister-group with either lepidosaurians or crocodiles18. (iv) The earliest offshoot within salamanders was Andrias (Cryptobranchidae) plus Hynobius (Hynobiidae)34 and the alternative position of Siren (Sirenidae) as the earliest-branching salamander clade37 was statistically rejected. (v) Lastly, our BI tree supports a close relationship between snakes and iguanian and anguimorph lizards (Elgaria) (Toxicofera7).
Only 4 out of 98 nodes in our phylogeny received relatively low support (<75% GJP; Fig. 2) and we consider these nodes in need of further confirmation. Besides relationships within crown-group iguanians and turtles, this applies to the sister-group between anguimorph (Elgaria) and iguanian lizards which was sensitive to the use of alternative models (GTR+Γ and LG+Γ+F in ML; Supplementary Figs. 3, 4) or the inclusion of deep paralogy (BI on the 1DP and 2DP datasets; Supplementary Figs. 6, 7), which recovered anguimorphs as the sister group of snakes (rejected however by topology tests; Supplementary Table 5). In agreement with Fig. 2a, coalescent-based analyses reconstructed an anguimorph + iguanian clade, which was robust to the inclusion of deep paralogy (Supplementary Figs. 5, 8, 9). In addition, only moderate support (75% GJP) was recovered for the controversial position14,17 of armadillo (Xenarthra) plus elephant (Afrotheria) sister to the remaining placental mammals (Atlantogenata13,16), in agreement with coalescent analyses (Supplementary Fig. 5), and the two alternative resolutions were rejected by topology tests (Supplementary Table 5). These problematic nodes correspond to fast radiations whose resolution requires extended taxon sampling in addition to accounting for incomplete lineage sorting. Our study minimized the possibility of model misspecification by using also complex evolutionary models and assessing the stability of tree topology to the effect of gene sampling and deep paralogy. For definitively resolving the above nodes, we argue for a careful exploration using suitable methodology and increased taxon sampling.
Robustness to gene sampling: size of gene jackknife replicates and gene trees
The use of gene jackknifing (100 replicates of ~50,000 amino acids each) allowed recovering an almost fully supported tree and resolving a number of controversial relationships. To explore the stability of the nodes in our tree and assess the amount of data required to recover them, we further analysed four sets of 100 gene jackknife replicates of increasing total length (~2,500, ~5,000, ~10,000 and ~25,000 amino acids) under ML. Relatively short replicates (~2,500 amino acids) recovered 33% and 76% of the nodes with full and high GJP, respectively (Fig. 2b). Increasing alignment length to 25,000 amino acids led to an increase of 47% of fully supported nodes (Fig. 2b; Supplementary Table 3). The relationships among the earliest-branching salamander lineages (Andrias, Hynobius, Siren) were particularly unstable and required long replicates (~50,000 amino acids) to be recovered with strong support. Gene length positively correlated with the proportion of final-tree bipartitions, more strongly for deep (>150 Ma; r=0.21, p<2.2 x10-16) than for recent (<150 Ma; r=0.13, p<2.2 x10-16) relationships, suggesting that longer genes correctly resolve more ancient nodes.
Mitogenomes and limits to phylogenetic resolution
To assess the phylogenetic resolution power of mitogenomes, we assembled a mitogenomic dataset matching the species in our nuclear datasets. Mitogenomic trees inferred by both ML and BI (Supplementary Figs. 10-17) correctly recovered some major clades with strong support, but failed to recover well-established relationships such as the monophyly of ray-finned and lobe-finned fishes or the sister-group position of platypus to all other mammals4, even after excluding the fastest evolving taxa and using complex mixture models to minimize long-branch attraction artefacts (Supplementary Figs. 14-15). Besides stochastic error due to limited alignment length, these incongruences most likely originate from long-branch attraction (despite using sophisticated models such as CAT-GTR), suggesting that mitogenomes are inadequate for resolving ancient divergences (>400 Ma) using currently available models of sequence evolution. The correlation between nuclear and mitochondrial rates is low (r=0.35; p<2.49·10-5; Supplementary Fig. 18) but still higher than expected from random datasets (r=0.13 ± 0.08; p> 0.05 averaged for 100 replicates). Hence, commonly assumed determinants of substitution rates, such as demography (population size changes, bottlenecks) or life history traits (body size, metabolic rate, generation time, genome size have to some extent influenced both genomes similarly, but other factors must be invoked to explain the observed rate disparity between the two genomes at many branches (Supplementary Fig. 18). These might include clade-specific variation in mitochondrial effective population sizes, genome-specific mutation rate, or acceleration of mitochondrial genes due to selection shifts in respiratory function.
No general relationship among evolutionary rates, species diversity and genome size
Comparing 44 main clades of jawed vertebrates of ages >150 Ma confirmed enormous differences in species diversity, from 1 to 31,826 species (Supplementary Table 6). Species diversity was not overall correlated with substitution rate (r=0.18, p=0.25) nor were higher rates significantly associated with higher species diversity in a sister group approach (Sign test, p=0.13). Our dataset includes the entire range of genome sizes in vertebrates (from 0.4 pg in pufferfish to 109 pg in lungfishes). Yet we found no association of genome size with evolutionary rate or species diversity (r=-0.28, p=0.061 and r=-0.13, p=0.44, respectively). Previous studies have also suggested that genome size might be associated with indels in coding regions38, but we detected no significant correlation, neither within conserved (r=0.1983, p=0.0722) nor within variable coding regions (r=0.0533, p=0.6325) as defined by the software BMGE (Block Mapping and Gathering with Entropy). The results of these correlation analyses were confirmed by a Bayesian joint modelling of the above traits with parameters of the evolutionary process at the sequence level (see Supplementary Table 7).
Divergence times of major lineages of jawed vertebrates
Genome-scale datasets have been shown to produce more precise and accurate divergence time estimates39, but this ultimately depends on the use of realistic evolutionary and clock models that appropriately account for among-lineage heterogeneities40 and multiple calibration intervals whose uncertainty and internal consistency is accounted for41,42. We applied an auto-correlated lognormal relaxed clock model and best-fitting sequence evolution model (CAT-GTR) to estimate genome-wide divergence times, averaged over 100 gene jackknife replicates. We used a conservative approach to setting calibrations, starting from multiple well-established calibrations with solid paleontological evidence and used conservative intervals to account for dating and phylogenetic uncertainty43 (Supplementary Table 8). On top of that, the internal congruence among these calibrations was verified through extensive cross-validation procedures in order to remove any poorly performing calibration, either examining the performance of single calibrations41 or removing one calibration at a time to check the congruence between estimated ages and priors42. The performance of each calibration scheme (named C16 and C30) derived from the above cross-validation strategies was assessed in independent dating analyses with a test dataset (a subset of the 14,352 most complete amino acid positions from the NoDP dataset that was computationally tractable with PhyloBayes). Both schemes produced largely congruent divergence times, but C16 yielded more reasonable dates within turtles, frogs, neoavian birds, modern frogs (overestimated in C30 if compared with previous data; www.timetree.org) or iguanian squamates and snakes (underestimated in C30) (Supplementary Table 9).
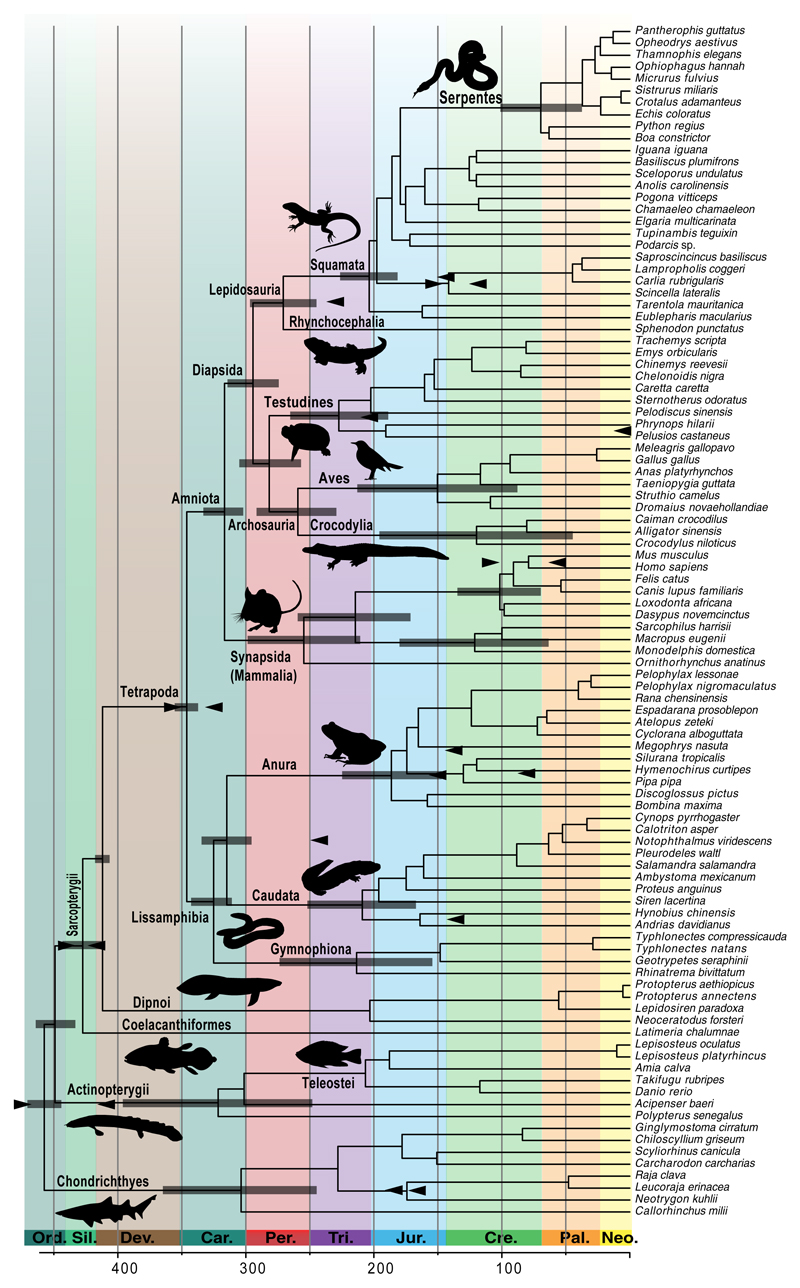
To estimate genome-wide divergence times, we calculated averaged divergence times (and conservative 95% credibility intervals; CrI) across 100 timetrees based of jackknife sampling of ~15,000 positions and the more stringently cross-validated C16 calibration scheme (Fig. 3). The genome-averaged timetree places the divergences among cartilaginous, ray-finned and lobe-finned fishes in the Ordovician, between 458 (CrI: 465–438) to 449 (462–431) Mya. The first split within lobe-finned fishes occurred in the Silurian ca. 427 (444–413) Mya and lungfishes separated from tetrapods in the early Devonian ca. 412 (419–408) Mya. The split between amphibians and amniotes occurred in the early Carboniferous ca. 346 (351–333) Mya and the three amphibian orders separated during the Carboniferous from 325 (338–307) to 315 (332–293) Mya, as did synapsids (mammals) and diapsids (turtles, archosaurs and lepidosaurs) ca. 317 (330–299) Mya. The origins of the main sauropsid groups, i.e., turtles, crocodiles, birds, squamates and tuatara (Sphenodon), took place in the Permian from 294 (313–273) to 259 (288–226) Mya. The crown diversification of extant frogs, salamanders and caecilians occurred in the late Triassic to early Jurassic between 213 (270–151) to 186 (231–153) Mya, almost simultaneously with the crown splits within squamates ca. 204 (228–183) Mya, cryptodiran turtles ca. 202 (243–159) Mya, pleurodiran turtles ca. 191 (248–116) Mya, and therian mammals ca. 214 (257–169) Mya.
Figure 3.
Time-calibrated phylogeny of jawed vertebrates. Divergences have been averaged across 100 timetrees estimated from independent gene jackknife replicates in PhyloBayes, using the subset of most congruent calibrations (C16; marked by arrows) and best-fit evolutionary (CAT-GTR+Γ) and relaxed clock (autocorrelated lognormal) models. Credibility intervals (CrI) are calculated as the absolute maximum and minimum values of 95% confidence intervals across 100 timetrees (only displayed for key nodes; see Supplementary Table 9 for detailed results). The dimensions of the scale is given in million years and main geological periods are highlighted.
Estimated divergences are generally in line with previous time-calibrated phylogenies using different dating methodologies, molecular data and calibrations, particularly for the deepest splits in the backbone44,45 as well as divergences within amphibians6, squamates46, snakes47 and placental mammals8. Estimated ages for crown-groups of cartilaginous and ray-finned fishes are younger compared to previous analyses48,49, which is likely caused by the removal of incongruent calibrations in the C16 scheme (the C30 scheme produced estimates more similar to previous studies for these groups; see Supplementary Table 9). The younger age of cartilaginous fishes, however, is consistent with recent paleontological analyses50. Compared to previous time-calibrated phylogenies, we obtain older divergences for turtles51 and birds15 but our estimates are in line with the ages of recently discovered fossils of stem turtles36 and an ornithuromorph bird that pushes back the origin of the group to at least 130.7 Mya52. The Cretaceous-Paleogene boundary (67 Mya) in our tree is not associated with a notable concentration of divergences, but our dataset does not capture the crown diversification of several species-rich taxa that might have occurred in this period, such as spiny-ray fishes, modern birds (Neoaves), boreoeutherian mammals, ranoid frogs, gekkonid geckos, or skinks. We support a diversification of placental mammals prior to the Cretaceous-Paleogene boundary ca. 102 (139–73) Mya, in agreement with most previous molecular and macroevolutionary studies8,39.
Reliability of phylogenomic analyses
Inferring phylogenies can be difficult, particularly in the presence of ancient or closely spaced speciation events, and the use of genome-scale datasets poses additional challenges related to poor data quality and more importantly systematic error25. In principle, the jawed vertebrate phylogeny is a solvable problem, being devoid of excessively old divergences and mostly long internal branches (Fig. 2a). It thus represents a good benchmark to test the abovementioned challenges. Yet, poor data quality18 (Supplementary Fig. 1) can lead to incorrect results (e.g., non-monophyly of amphibians, misplacement of turtles). We adopt a phylotranscriptomic approach to assemble an alignment of >7,000 genes for 100 species with rigorous quality controls. The quality and resolving power of our NoDP dataset are higher than those of previous studies, including Fong et al.18 and the most comprehensive dataset analysed to date12, with 70% vs. 17% and 61% mean congruence respectively for the two datasets, measured as the proportion of final-tree bipartitions recovered by single genes. The higher congruence of NoDP persisted after correcting for gene length (Supplementary Fig. 1). We thus confirm that RNA-Seq is a cost-effective method to anchor phylogenomic analyses, which can result in robust fossil-dated trees, provided that careful data curation and appropriate analytical methods are used. Moreover, we show that gene jackknifing allows stringently testing phylogenetic relationships and overcoming the limitations and possible biases of small datasets that aim to represent the entire genome, and that phylogenomics is resilient to limited levels of deep paralogy, as long as a large number of genes (>1,000) are used and internal branches are relatively long. In such cases, realistic models allow recovering correct phylogenetic hypotheses, even in the presence of extreme among-lineage evolutionary rate variation. In contrast, resolving closely spaced radiations, which were not targeted in this work, could require a detailed study with specific gene and taxon sampling14 and testing the robustness to model misspecification32. Overall, our results highlight the importance of data quality in phylogenomics, as well as the application of realistic evolutionary and clock models, and the validation of calibrations in timetree estimation, both a priori (based on paleontological data) and a posteriori (cross-validation).
Materials and Methods
An extended description of our bioinformatic pipeline and detailed Materials and Methods are available as Supplementary Information.
Assembly of phylogenomic datasets
New RNA-Seq data was generated for 23 gnathostome species using Illumina MiSeq (2x250 bp) and HiSeq2000 (2x50 bp, 2x100 bp) technologies. Available RNA-Seq data were downloaded from NCBI SRA. Transcriptomes were assembled de novo with Trinity or MIRA. Species names and accession numbers are available in Supplementary Table S10.
Nuclear datasets were assembled using a new pipeline summarized in Supplementary Fig. 2. Briefly, proteomes of 21 vertebrate genomes (ENSEMBL) were grouped into ortholog clusters and those not containing data for all major jawed vertebrate lineages were discarded. The resulting 11,656 protein clusters were aligned and positions of unreliable homology removed. To identify and resolve paralogy issues, we implemented a paralog-splitting pipeline based on gene trees. The obtained 9,852 ortholog clusters were complemented with new genomes and transcriptomes using the software Forty-Two (https://bitbucket.org/dbaurain/42/). Several decontamination steps were carried out. Any sequence contamination from non-vertebrates and human was detected by BLAST and eliminated. We searched for cross-contamination that can arise during library preparation using gene trees, and removed contaminants based on expression data. After eliminating overlapping redundant sequences that were too divergent, we filtered out incomplete or short sequences and alignments, leading to 7,687 genes. The paralogy splitting procedure was repeated to resolve any paralogy caused by the addition of new species, and gene alignments were classified into three datasets that contained zero (NoDP), one (1DP) and two or more (2DP) deep paralogs. Sequence stretches with unusually low similarity (usually due to frame shifts) were masked with HMM-cleaner (R. Poujol) and alignments were trimmed. For each gene, we used SCaFoS30 to merge conspecific sequences and resolve putative remaining paralogy. A third decontamination step used extremely long branches estimated on a fixed reference tree as proxy for contamination.
Mitochondrial datasets were assembled from mitogenomes available at NCBI with a taxon sampling mirroring the nuclear datasets plus a few additional species to reduce long-branch attraction artefacts expected in mitogenomic trees (Supplementary Table 11). The resulting alignments consisted of 106 species (2,773 amino acid positions) and 95 species (2,866 amino acid positions) after removing the fastest evolving species.
Phylogenetic inference
Concatenated nuclear gene sets (NoDP, 1DP and 2DP) were analysed separately using ML with RAxML v.853 under LG+F+Γ and GTR+Γ models and BI with PhyloBayes MPI v1.554 under the better fitting CAT+Γ model (selected after 10-fold cross-validation). The Bayesian consensus tree was calculated from 100 post-burnin tree collections, each from gene jackknife replicates of ~50,000 amino acid positions. Convergence was verified with the diagnostic tools of PhyloBayes. Branch support was computed from 100 bootstrap pseudo-replicates in ML, and from gene jackknife proportions (GJP) in BI. To assess the robustness to gene sampling, we analysed by ML gene jackknife replicates of ca. 2,500, 5,000, 10,000 and 25,000 aligned positions under the LG+Γ model. Coalescent analyses were run in ASTRAL-II v.4.10.12 using ML gene trees as input (estimated under best-fit models in RAxML) and node stability was assessed as local posterior support and 100 replicates of multi-locus bootstrapping.
The mitochondrial datasets were analysed by ML under MTREV+Γ and GTR+Γ models, and by BI under CAT+Γ and CAT-GTR+Γ models.
Molecular dating
Divergence times were estimated in PhyloBayes v.4.1 using best-fit CAT-GTR+Γ and auto-correlated lognormal clock models (selected after 10-fold cross-validation), a birth-death prior on divergence times and 30 calibration points with uniform priors and soft bounds (see Supplementary Table 8). After cross-validation procedures (see SI Materials and Methods), we applied the C16 and C30 calibration sets to compute timetrees based on a subset of 14,352 amino acid positions from NoDP (two independent chains). To estimate genome-wide divergence times, we estimated 100 timetrees from 100 gene jackknife replicates of ~15,000 amino acids from the NoDP dataset, using the most stringent C16 calibration scheme. Divergence times were averaged and conservative 95% credibility intervals (CrI) calculated as the absolute maximum and minimum values of 95% confidence intervals across 100 timetrees.
Nuclear and mitochondrial rates
Substitution rates were measured as branch lengths optimized under CAT+Γ and a reference tree (Fig. 2a) in PhyloBayes, independently for the nuclear (NoDP) and mitochondrial datasets, both pruned to a common subset of 78 species. Correlation between mitochondrial and nuclear rates was assessed by Pearson’s correlation among all pairs of internal and terminal branches. We simulated 100 random alignments characterized by the amino acid proportions of either mitochondrial or nuclear datasets, then branch lengths were optimized on a reference tree and rates correlated as above.
Association of life history traits and molecular features
We estimated Pearson’s correlation after correcting for phylogenetic non-independence among the following life history traits and molecular features: (i) genome size (retrieved from www.genomesize.com) versus number of gaps in either conserved or variable gene regions (defined by BMGE on untrimmed gene alignments), (ii) genome size versus nuclear substitution rate, and (iii) substitution rate versus species diversity (tabulated from the literature), for 44 lineages divided by an ad-hoc cut-off of >150 Ma defined to capture sister groups characterized by obvious differences in species diversity. Nuclear substitution rates and species diversity were also compared in a sister group approach, assessing by non-parametric Sign test whether higher substitution rates (tested by relative-rate tests) were associated with higher species diversity. We further used a Bayesian joint modelling to study the correlation between substitution rates, genome size and the number of gaps in conserved and variable gene regions.
Supplementary Material
Acknowledgements
We thank the following people for tissue samples: T. Ziegler (Andrias), M. Hasselmann (Neoceratodus) and O. Guillaume (Calotriton, Proteus) and V. Michael for technical assistance. N. Galtier kindly provided access to the Salamandra transcriptome. We acknowledge the use of computational resources from the University of Konstanz (HPC2), the Institute of Physics of Cantabria IFCA-CSIC (Altamira), the Fédération Wallonie-Bruxelles (Tier-1; funded by Walloon Region, grant #1117545), the Consortium des Équipements de Calcul Intensif (CÉCI; funded by the F.R.S.-FNRS, grant #2.5020.11) and the Réseau Québecois de Calcul de Haute Performance. II was supported by the Alexander von Humboldt Foundation (1150725) and the European Molecular Biology Organization (EMBO ALTF 440-2013). Further support came from the University of Konstanz (II), the Deutsche Forschungsgemeinschaft (DFG) (AM), the European Research Council (ERC Advanced Grant “Genadapt” No. 273900 to AM), and the Agence Nationale de la Recherche (TULIP Laboratory of Excellence ANR-10-LABX-41 to HP and ANR-12-BSV7-020 project “Jaws” to FD and JYS). This is contribution ISEM 2017-106 of the Institut des Sciences de l’Evolution de Montpellier.
Footnotes
Ethics statement and data availability
Animal experiments conformed to the European Parliament and council of 22/09/2010 (Directive 2010/63/EU) and the French Rural Code (Articles R214-87 to R214-137, decree No. 2013-118 of 01/02/2013). Experiments performed in France were authorized by the certificate No. 75-600. New RNA-Seq data are available at the SRA (Supplementary Table 10) and phylogenetic datasets, trees and custom scripts in Dryad (http://dx.doi.org/10.5061/dryad.r2n70).
Author contributions
MV, HP and II designed research. II, FD, JYS, AK, MJ, AM and MV contributed new data. II, DB, HB, MV and HP performed analyses. II, MV and HP drafted manuscript and all authors read and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Brown JM, Thomson RC. Bayes factors unmask highly variable information content, bias, and extreme influence in phylogenomic analyses. Syst Biol. 2017;66:517–530. doi: 10.1093/sysbio/syw101. [DOI] [PubMed] [Google Scholar]
- 2.Jeffroy O, Brinkmann H, Delsuc F, Hervé P. Phylogenomics: The beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Zardoya R, Meyer A. The evolutionary position of turtles revised. Naturwissenschaften. 2001;88:193–200. doi: 10.1007/s001140100228. [DOI] [PubMed] [Google Scholar]
- 4.Janke A, Erpenbeck D, Nilsson M, Arnason U. The mitochondrial genomes of the iguana (Iguana iguana) and the caiman (Caiman crocodylus): Implications for amniote phylogeny. Proc R Soc B-Biol Sci. 2001;268:623–631. doi: 10.1098/rspb.2000.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Near TJ, et al. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci USA. 2013;110:12738–12743. doi: 10.1073/pnas.1304661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irisarri I, et al. The origin of modern frogs (Neobatrachia) was accompanied by acceleration in mitochondrial and nuclear substitution rates. BMC Genomics. 2012;13:626. doi: 10.1186/1471-2164-13-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyron RA, Burbrink FT, Wiens JJ. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 2013;13:93. doi: 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meredith RW, et al. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 9.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 10.Chiari Y, Cahais V, Galtier N, Delsuc F. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria) BMC Biology. 2012;10:65. doi: 10.1186/1741-7007-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford NG, et al. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 2012;8:783–786. doi: 10.1098/rsbl.2012.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MY, Liang D, Zhang P. Selecting question-specific genes to reduce incongruence in phylogenomics: A case study of jawed vertebrate backbone phylogeny. Syst Biol. 2015;64:1104–1120. doi: 10.1093/sysbio/syv059. [DOI] [PubMed] [Google Scholar]
- 13.Tarver JE, et al. The interrelationships of placental mammals and the limits of phylogenetic inference. Genome Biol Evol. 2016;8:330–344. doi: 10.1093/gbe/evv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery EJP. Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol. 2013;30:2134–2144. doi: 10.1093/molbev/mst116. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song S, Liu L, Edwards SV, Wu S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci USA. 2012;109:14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan CC, et al. Heterogeneous models place the root of the placental mammal phylogeny. Mol Biol Evol. 2013;30:2145–2156. doi: 10.1093/molbev/mst117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong JJ, Brown JM, Fujita MK, Boussau B. A phylogenomic approach to vertebrate phylogeny supports a turtle-archosaur affinity and a possible paraphyletic Lissamphibia. PLoS ONE. 2012;7:e48990. doi: 10.1371/journal.pone.0048990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyson TR, et al. MicroRNAs support a turtle + lizard clade. Biol Lett. 2012;8:104–107. doi: 10.1098/rsbl.2011.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier JA, Kearney M, Maisano JA, Rieppel O, Behlke ADB. Assembling the squamate tree of life: Perspectives from the phenotype and the fossil record. Bull Peabody Mus Nat Hist. 2012;53:3–308. [Google Scholar]
- 21.Townsend TM, Larson A, Louis E, Macey JR, Sites J. Molecular phylogenetics of Squamata: The position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst Biol. 2004;53:735–757. doi: 10.1080/10635150490522340. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Wiens JJ. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol Part B. 2016;94:537–547. doi: 10.1016/j.ympev.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Irisarri I, Vences M, San Mauro D, Glaw F, Zardoya R. Reversal to air-driven sound production revealed by a molecular phylogeny of tongueless frogs, family Pipidae. BMC Evol Biol. 2011;11:114. doi: 10.1186/1471-2148-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bewick AJ, Chain FJJ, Heled J, Evans BJ. The pipid root. Syst Biol. 2012;61:913–926. doi: 10.1093/sysbio/sys039. [DOI] [PubMed] [Google Scholar]
- 25.Philippe H, et al. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 27.Springer MS, et al. Waking the undead: Implications of a soft explosive model for the timing of placental mammal diversification. Mol Phylogenet Evol. 2017;106:86–102. doi: 10.1016/j.ympev.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Phillips MJ. Geomolecular dating and the origin of placental mammals. Syst Biol. 2016;65:546–557. doi: 10.1093/sysbio/syv115. [DOI] [PubMed] [Google Scholar]
- 29.Hara Y, et al. Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics. 2015;16:977. doi: 10.1186/s12864-015-2007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: A tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 2007;7:S2. doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- 32.Irisarri I, Meyer A. The identification of the closest living relative(s) of tetrapods: Phylogenomic lessons for resolving short ancient internodes. Syst Biol. 2016;65:1057–1075. doi: 10.1093/sysbio/syw057. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, Schultze H-P. In: Major Events in Early Vertebrate Evolution: Paleontology, Phylogeny and Development. Ahlberg PE, editor. Taylor & Francis; 2001. pp. 289–314. [Google Scholar]
- 34.Pyron RA, Wiens JJ. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol Phylogenet Evol. 2011;61:543–583. doi: 10.1016/j.ympev.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 35.San Mauro D, Vences M, Alcobendas M, Zardoya R, Meyer A. Initial diversification of living amphibians predated the breakup of Pangaea. Am Nat. 2005;165:590–599. doi: 10.1086/429523. [DOI] [PubMed] [Google Scholar]
- 36.Schoch RR, Sues H-D. A Middle Triassic stem-turtle and the evolution of the turtle body plan. Nature. 2015;523:584–587. doi: 10.1038/nature14472. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Wake DB. Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2009;53:492–508. doi: 10.1016/j.ympev.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Gregory TR. Insertion–deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 39.Dos Reis M, et al. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc B-Biol Sci. 2012;279:3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepage T, Bryant D, Philippe H, Lartillot N. A general comparison of relaxed molecular clock models. Mol Biol Evol. 2007;24:2669–2680. doi: 10.1093/molbev/msm193. [DOI] [PubMed] [Google Scholar]
- 41.Near TJ, Meylan Peter A, Shaffer HB. Assessing concordance of fossil calibration points in molecular clock studies: An example using turtles. Am Nat. 2005;165:137–146. doi: 10.1086/427734. [DOI] [PubMed] [Google Scholar]
- 42.Sanders KL, Lee MSY. Evaluating molecular clock calibrations using Bayesian analyses with soft and hard bounds. Biol Lett. 2007;3:275–279. doi: 10.1098/rsbl.2007.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnock RCM, Parham JF, Joyce WG, Lyson TR, Donoghue PCJ. Calibration uncertainty in molecular dating analyses: There is no substitute for the prior evaluation of time priors. Proc R Soc B-Biol Sci. 2014;282 doi: 10.1098/rspb.2014.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King BL. Bayesian morphological clock methods resurrect placoderm monophyly and reveal rapid early evolution in jawed vertebrates. Syst Biol. 2017;66:499–516. doi: 10.1093/sysbio/syw107. [DOI] [PubMed] [Google Scholar]
- 45.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106:13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones M, et al. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara) BMC Evol Biol. 2013;13:208. doi: 10.1186/1471-2148-13-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsiang AY, et al. The origin of snakes: Revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol Biol. 2015;15:1–22. doi: 10.1186/s12862-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue JG, et al. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 2010;27:2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 49.Dornburg A, Townsend J, Friedman M, Near TJ. Phylogenetic informativeness reconciles ray-finned fish molecular divergence times. BMC Evol Biol. 2014;14:169. doi: 10.1186/s12862-014-0169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coates MI, Gess RW, Finarelli JA, Criswell KE, Tietjen K. A symmoriiform chondrichthyan braincase and the origin of chimaeroid fishes. Nature. 2017;541:208–211. doi: 10.1038/nature20806. [DOI] [PubMed] [Google Scholar]
- 51.Lourenço JM, Claude J, Galtier N, Chiari Y. Dating cryptodiran nodes: Origin and diversification of the turtle superfamily Testudinoidea. Mol Phylogenet Evol. 2012;62:496–507. doi: 10.1016/j.ympev.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Wang M, et al. The oldest record of Ornithuromorpha from the early cretaceous of China. Nat Commun. 2015;6:6987. doi: 10.1038/ncomms7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.