Abstract
Neuroblastomas are associated with KIF1Bβ mutations within tumor suppressor region 1p36. In this issue of Developmental Cell, Li et al. (2016) show that KIF1Bβ binding releases calcineurin autoinhibition, leading to dephosphorylation of the DRP1 GTPase and subsequent mitochondrial fragmentation. KIF1Bβ impairment causes mitochondrial hyperfusion, impairing developmental apoptosis and promoting tumorigenesis.
During development of the sympathetic nervous system, neural progenitor cells depend on nerve growth factor (NGF) for regulation of differentiation and survival. Excess progenitors are normally limited through apoptosis when NGF becomes limiting during development, and tumors such as neuroblastoma and pheochromocytoma can arise when neural progenitors fail to die under these conditions. Analyses of familial forms of these cancers have identified alterations in the succinate dehydrogenase genes SDHB, SDHC, and SDHD, as well as in VHL, RET, and NF1. All of these genes encode products that regulate an NGF-dependent signaling pathway influencing neuronal survival. This pathway ultimately links the NGF receptor (TrkA) to the prolyl hydroxylase EGLN3, which is required for apoptosis upon NGF withdrawal (Lee et al., 2005). Thus, a failure to properly cull unwanted cells during development appears to predispose cells to oncogenic transformation. Suppression of neural crest-derived tumors such as neuroblastoma and pheochromocytoma has long been linked to a critical region in the distal short arm of chromosome 1 (1p36) that is deleted in a number of human malignancies. Understanding the molecular basis for this suppression has been an area of intense interest for decades.
Previous work scrutinizing 1p36 (Henrich et al., 2012) has highlighted the possible involvement of several genes in the region. The chromatin remodeler CHD5, which is crucial for neuronal differentiation via dual roles in facilitating gene expression and polycomb gene repression (Egan et al., 2013), is one candidate. More recently, particular interest has focused on loss-of-function mutations in KIF1B, a gene encoding members of the kinesin-3 family of motor proteins. KIF1B encodes two alternatively spliced iso-forms, KIF1Bα and KIF1Bβ. The isoforms share the same N-terminal motor domain but differ in their C-terminal cargo domains, and they are differentially involved in transport of mitochondria (KIF1Bα) and synaptic vesicle precursors (KIF1Bβ). KIF1Bβ, but not KIF1Bα, has also surprisingly been implicated in NGF-mediated neuronal apoptosis independent of its motor functions, prefiguring an unexpected role in sympathetic nerve development. In fact, though found in only a small percentage of neural crest tumors overall, a number of different KIF1B variants identified in tumors clearly impair KIF1Bβ function in studies in vitro, and one missense mutation segregated across three generations of a cancer-prone family with a number of neural and non-neural malignancies (Yeh et al., 2008). Recently, Chen et al. (2014) reported that KIF1Bβ interacts with RNA helicase A (DHX9), causing nuclear accumulation of DHX9 and subsequent induction of proapoptotic XIAP-associated factor 1 (XAF1), ultimately leading to apoptosis.
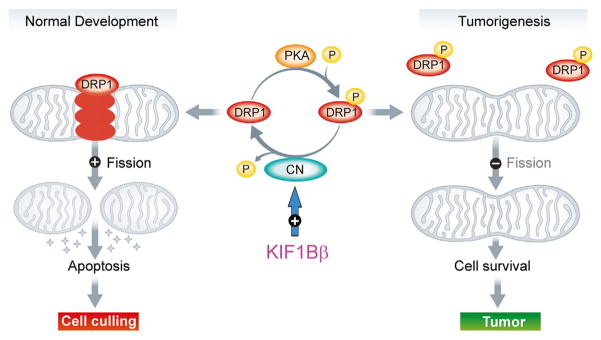
In this issue of Developmental Cell, Li et al. (2016) provide an additional mechanism to explain the effects of KIF1Bβ on tumorigenesis. These authors found that KIF1Bβ activates the Ca2+-calmodulin-dependent protein phosphatase calcineurin by binding and stabilizing the calmodulin-calcineurin complex, relieving enzymatic autoinhibition (more completely than Ca2+ alone) and permitting substrate recognition broadly. This result is quite daunting upon initial consider-since calcineurin is a ubiquitous mediator of cellular responses to Ca2+ signaling—with many substrates—and its dysregulation has already been implicated in a host of immune, neurodegenerative, and oncologic disorders. Importantly, out of the panoply of known calcineurin substrates, Li et al. (2016) focused on a critical role for calcineurin-dependent activation of DRP1, a dynamin-related, multimeric GTPase that mediates mitochondrial fission, a critical event during apoptosis. The authors found that KIF1Bβ affects mitochondrial morphology through calcineurin-dependent dephosphorylation of DRP1 Ser637, which is phosphorylated by protein kinase A (PKA). This regulated dephosphorylation is known to promote translocation of DRP1 to the mitochondrial surface, where it catalyzes mitochondrial fission (Cereghetti et al., 2008) (Figure 1). Under normal cellular conditions, over 95% of DRP1 is cytoplasmic, so this translocation is a key activation step. DRP1 opposes the activities of several intrinsic large mitochondrial GTPases (MFN1/2 and OPA1) that mediate mitochondrial fusion, and thus disruption of DRP1 function leads to mitochondrial hyperfusion. Imbalances in mitochondrial morphology have been seen in many disease models, and several of these GTPases regulating mitochondrial morphology (including DRP1) are mutated in neurologic disorders, emphasizing the importance of proper mitochondrial morphology for cellular health (Chan, 2012).
Figure 1. Model of KIF1Bβ-Dependent Effects on Mitochondrial Fission/Fusion Balance in NGF-Limiting Conditions.
KIF1Bβ activates calcineurin (CN) by relieving autoinhibition, resulting in dephosphorylation of DRP1 Ser637. This triggers DRP1 translocation to mitochondria and subsequent division, leading to cytochrome c release (stars), apoptosis, and cell death. In the absence of KIF1Bβ, DRP1 Ser637 is phosphorylated (P) by PKA and remains modified. Mitochondrial fission is impaired, resulting in mitochondrial hyperfusion, aberrant cell survival, and promotion of tumorigenesis. Figure adapted from Anderson and Blackstone (2013).
But how does mitochondrial hyperfusion relate specifically to tumor formation? Li et al. (2016) tested multiple known KIF1Bβ pathogenic mutations identified in neuroblastomas and pheochromocytomas, and all failed to activate calcineurin or stimulate DRP1 dephosphorylation. In addition, all seven hemizygous KIF1Bβ-deleted neuroblastomas examined had silencing of both DRP1 and KIF1Bβ—in stark contrast to 1p36 intact tumors—and low expression of these genes correlated with poor prognosis and reduced survival in neuroblastoma patients (Li et al., 2016). Moreover, depletion of DRP1 blocked KIF1Bβ-mediated apoptosis in NB1 neuroblastoma cells, and PKA gain-of-function tumorigenic mutations (expected to enhance phosphorylation at DRP1 Ser637 and inhibit mitochondrial fission) blocked KIF1Bβ-induced apoptosis as well. Together, these data provide compelling evidence for a critical role of impaired mitochondrial fission/fusion balance in high-risk and poor-prognosis neuroblastomas and further identify this process as a potential therapeutic target. However, it seems likely that other mechanisms may also contribute. Indeed, this calcineurin-dependent pathogenic mechanism has a clear link to the role of KIF1Bβ loss of function in impairing DHX9 nuclear localization (Chen et al., 2014), since the calcineurin docking motif (LXVP) in DRP1 (Slupe et al., 2013) is also present in DHX9, and calcineurin binds DHX9 only in the presence of KIF1Bβ (Li et al., 2016).
Since the first report of 1p36 deletions in neuroblastomas four decades ago, deletions in this region have been linked to wide range of tumors, extending well beyond those of the neural crest. The tumorigenic role of KIF1Bβ loss of function is becoming more and more compelling, but other nearby 1p36 genes such as CHD5, CAMTA1, CASZ1, and miR-34a may also be altered in dosage (Henrich et al., 2012), and disruptions in a number of these genes may work in synergy with one another, as well as with KIF1Bβ (in cells that express KIF1Bβ) to promote tumorigenesis. The non-motor roles played by KIF1Bβ in unexpected cellular locales continue to surprise.
References
- Anderson CA, Blackstone C. EMBO J. 2013;32:1496–1498. doi: 10.1038/emboj.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Wallis K, Fell SM, Sobrado VR, Hemmer MC, Ramsköld D, Hellman U, Sandberg R, Kenchappa RS, Martinson T, et al. Cancer Discov. 2014;4:434–451. doi: 10.1158/2159-8290.CD-13-0362. [DOI] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, et al. Dev Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Henrich KO, Schwab M, Westermann F. Cancer Res. 2012;72:6079–6088. doi: 10.1158/0008-5472.CAN-12-2230. [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG, Jr, Schlisio S. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Li S, Fell SM, Surova O, Smedler E, Wallis K, Chen ZX, Hellman U, Johnsen JI, Martinsson T, Kenchappa RS, et al. Dev Cell. 2016;36:164–178. doi: 10.1016/j.devcel.2015.12.029. this issue. [DOI] [PubMed] [Google Scholar]
- Slupe AM, Merrill RA, Flippo KH, Lobas MA, Houtman JCD, Strack S. J Biol Chem. 2013;288:12353–12365. doi: 10.1074/jbc.M113.459677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh IT, Lenci RE, Qin Y, Buddavarapu K, Ligon AH, Leteurtre E, Do Cao C, Cardot-Bauters C, Pigny P, Dahia PL. Hum Genet. 2008;124:279–285. doi: 10.1007/s00439-008-0553-1. [DOI] [PubMed] [Google Scholar]