Abstract
Autophagy, a catabolic process necessary for the maintenance of intracellular homeostasis, has recently been the focus of numerous human diseases and conditions, such as aging, cancer, development, immunity, longevity, and neurodegeneration. However, the continued presence of autophagy is essential for cell survival and dysfunctional autophagy is thought to speed up the progression of neurodegeneration. The actual molecular mechanism behind the progression of dysfunctional autophagy is not yet fully understood. Emerging evidence suggests that basal autophagy is necessary for the removal of misfolded, aggregated proteins and damaged cellular organelles through lysosomal mediated degradation. Physiologically, neurodegenerative disorders are related to the accumulation of amyloid β peptide and α-synuclein protein aggregation, as seen in patients with Alzheimer’s disease and Parkinson’s disease, respectively. Even though autophagy could impact several facets of human biology and disease, it generally functions as a clearance for toxic proteins in the brain, which contributes novel insight into the pathophysiological understanding of neurodegenerative disorders. In particular, several studies demonstrate that natural compounds or small molecule autophagy enhancer stimuli are essential in the clearance of amyloid β and α-synuclein deposits. Therefore, this review briefly deliberates on the recent implications of autophagy in neurodegenerative disorder control, and emphasizes the opportunities and potential therapeutic application of applied autophagy.
Keywords: Aggregate-prone proteins, Amyloid beta peptide, Autophagic flux, Clearance of toxic compounds, Neurodegenerative disorders
INTRODUCTION
Autophagy is a cellular process which can have a wide range of effects on pathological and physiological conditions, frequently leading to the removal of damaged/injured organelles and aggregated proteins (1). In more general terms, basal autophagy is important for removing misfolded proteins and damaged organelles, and therefore plays a crucial role in maintaining cellular homeostasis. Autophagy participates in a variety of normal physiological processes such as glucose homeostasis (2), lipid metabolism (3), aging (4) and many more. However, failure of autophagy causes cellular dysfunctions which render them unable to remove defective proteins or damaged organelles. Knockdown/inhibition of autophagy often causes premature aging in Caenorhabditis elegans (5). Interestingly, research in Drosophila melanogaster suggests that lifespan extends after enhanced autophagy (6). Several other studies have shown that deletion/mutation of the autophagy-related gene (Atg) provokes an imbalance of body homeostasis, resulting in obesity (7), insulin resistance (8), diabetes (9), and some neurodegenerative disease as well (10). In addition, neurodegenerative disease occurs when aggregated proteins are not accurately degraded by autophagy; for example, accumulation of aggregated forms of amyloid-β (Aβ) peptide (11) and tau protein (12) in Alzheimer’s Disease (AD), α-synuclein in Parkinson’s Disease (PD) (13), and that of huntingtin in Huntington’s Disease (HD) (14). Therefore, autophagy pathways could be a potential therapeutic target for the treatment of neurodegenerative diseases. Indeed, neuronal cells are easily damaged through impaired autophagy among all types of cells in physiological systems (15). Under-stimulated autophagy has been correlated with difficulty to maintain synaptic activity (16) and axonal homeostasis (17). Several studies have managed to demonstrate a relationship between neurodegeneration and the autophagy signaling pathway (18). This review would like to emphasize the understanding of the basic molecular mechanisms and regulation of autophagy in neurodegenerative disease control.
MOLECULAR MECHANISMS OF AUTOPHAGY
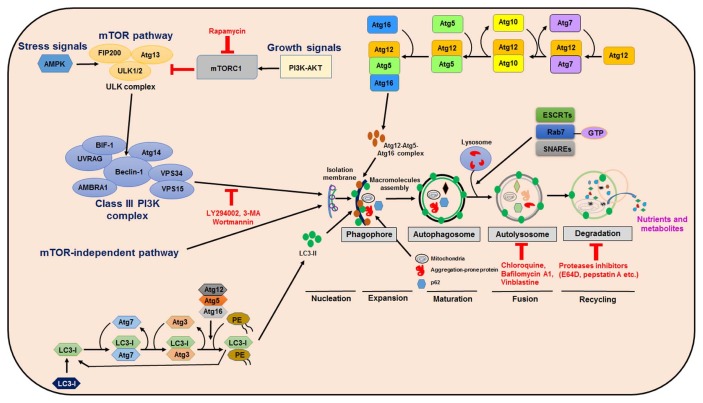
Autophagy occurs through consecutive steps in the molecular biological pathway, such as sequestration, delivery of autophagosome to lysosomes (autolysosomes), degradation and utilization of toxic components, and collectively every step demonstrates the diverse role of autophagy. Importantly, formation of phagophores are initiated during the early stages of autophagy, the production components of which are likely derived from numerous sources, including the endoplasmic reticulum (19), the endoplasmic reticulum-golgi intermediate compartment (20), the plasma membrane (21), recycling endosomes (22), the golgi complex (23), and lipid droplets (24). In fact, multiple autophagy-related genes are involved in membrane rearrangements and autophagosome formations that consequently become attached to the lysosomes. Generally, there are two ubiquitin-like conjugation reactions involved during autophagy. Firstly, there is the Atg12 conjugation to Atg5. Secondly, Atg16L1 binds to conjugate Atg12-Atg5 to form a complex which is essential for phagophore maturation (25). However, the second conjugation of Atg8 and LC3 is necessary to complete the autolysosome formation, in that the process of LC3 being cleaved through Atg4 to obtain cytosolic LC3-I. Finally, LC3-I is coupled to phosphatidylethanolamine (PE) during the macromolecular assembly of phagophores, which appears in the form of microtubule-associated protein light chain 3 (LC3-II) proteins (Fig. 1). It has been found that LC3-II localizes to the autophagosomal membrane, which is the most important marker of autophagy in mammal cells (26). Whereas with several adaptor proteins, such as the neighbor of BRCA1 gene 1 (NBR1), nuclear dot protein 52 (NDP52), and p62/sequestosome 1 proteins are directly bind to LC3-II (27, 28). Subsequently, misfolded proteins (29) or abnormal cellular organelles (as well as mitochondria (30), peroxisome (31) and the endoplasmic reticulum (32) are engulfed by double-membrane autophagosomes. Strikingly, the autophagosomes are ultimately combined with lysosomes to form an autolysosome, where the internal substrates are degraded to produce nutrients and metabolites (33).
Fig. 1.
Regulation of autophagy signaling pathway. Autophagy may initiate deprivation of nutrients or growth factors which activate AMPK and/or inhibition of mTORC1, leading to stimulation of ULK complex (FIP200 and ATG13). Beclin-1 become phosphorylated, leading to VPS34 activation and then initiation of phagophore formation. VPS34 complex function comprises a regulatory subunit like VPS15 (p150) and Beclin-1, their connection being with other regulatory factors e.g. AMBRA, ATG14, UVRAG, and BIF-1. Atg5–Atg12 conjugation involves Atg7 and Atg10 to form a complex, Atg12-atg5-Atg16 influences the formation of phagophores. Atg5 and Atg12 forms a complex with Atg16, which acts like an E3-function towards LC3-PE assembly (LC3-II). This has a role in the initiation of phagophore formation. LC3-II is a particular autophagy indicator marker which is eventually disrupted by autolysosomes. Autophagosome maturation also involves fusion with lysosomes which are mediated by Rab7, ESCRT and SNARE proteins, eventually leading to cargo degradation and recycling of nutrients and metabolites.
CLEARENCE OF AGGREGATE-PRONE PROTEINS THROUGH AUTOPHAGY IN NEURODEGENERATION
Recently, several studies revealed that intracellular protein aggregation and misfoldings are a common phenomenon of many neurodegenerative diseases, such as AD, PD, HD, and spinocerebellar ataxias (SCA) (34). To date, there are no efficient therapeutic approaches that can cure or prevent neurodegeneration diseases in humans. Thus, the need to understand the precise molecular mechanisms of neurodegeneration is urgent. To be studied are factors that regulate expression levels of protein misfolding and aggregate-prone proteins during the process of neurodegeneration. Usually, there are many methods to treat neurodegenerative diseases that might enhance degradation of prone-proteins. Among the most possible routes of misfolded protein clearance are the autophagy-lysosome and ubiquitin-proteasome pathways (Fig. 2). Although the ubiquitin proteasome system primarily degrades small-molecule proteins, its mechanism is not clear and it is unknown whether it is a possible therapeutic target for the clearance of aggregate-prone proteins. In addition, autophagic degradation of aggregate-prone proteins are related to decreasing protein aggregation and toxicity. Therefore, enhancing autophagy may be a promising therapeutic approach for neurodegenerative diseases, in that aggregate-prone proteins are used as autophagy substrates, such as tau (35), α-synuclein (36), mutant huntingtin (14), and mutant ataxin 3 (35), for their clearance of toxic products and having an effect on neurodegeneration control.
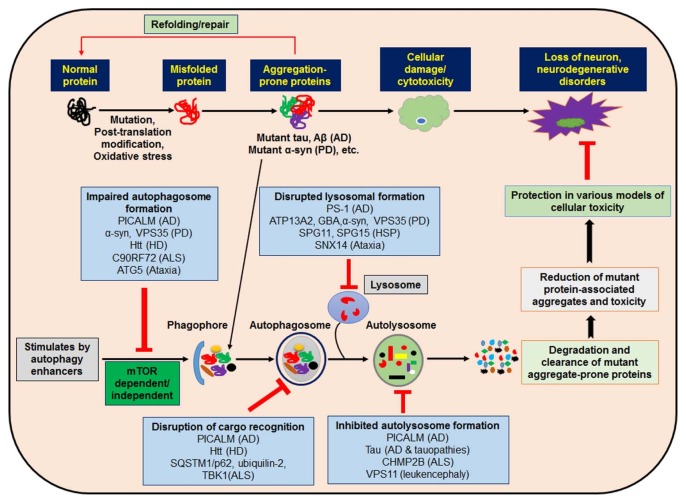
Fig. 2.
Modulation of autophagy and intersections between autophagy and disease-associated genes as a therapeutic approach for neurodegenerative disorders. Impaired autophagy is involved in the pathogeneses of several neurodevelopmental and neurodegenerative disorders. Different cytosolic mutant and aggregate proteins are linked with neurodegenerative symptoms, which may predominantly be removed through autophagy. Pharmacological stimulation of small molecule autophagy enhancers may initiate autophagy via mTOR dependent/independent pathways that reduce mutant protein-associated aggregates and toxicity, leading to cytoprotection and rescue against neurodegeneration. Several genes connected with neurodegenerative disorders have now been associated with autophagy function. These genes perform a number of diverse steps throughout the autophagic cycle, from early steps of autophagosome development through autolysosome formation. Their proposed sites of action are now designated, along with the neurodegenerative disease with which they are associated.
Autophagy in Alzheimer’s disease
AD is a multifactorial, irreversible, progressive and the most common type of dementia in elderly people worldwide. It is characterized by the gradual diminishment of memory and thoughts, and ultimately the capability to carry out any cognitive function (37). Recently it has been shown that the main pathological feather of AD is comprised of intracellular neurofibrillary tangles containing hyperphosphorylated tau proteins, self-aggregating extracellular Aβ plaques, the weakening of cholinergic function, and eventually autophagy dysfunction (38, 39). However, the Aβ peptide derived from the successive splitting of amyloid precursor protein (APP) through the β-site APP cleavage enzyme 1 (BACE1) and γ-secretase also has been linked to neurodegeneration. Accumulation of this peptide has been shown to cause cognitive deficits and neuronal dysfunction in the cortex and hippocampus during AD pathogenesis (40). Particularly, Aβ may also be produced in autophagosomes, which seem to comprise APP and presenilin-1 enzymes that participate in the cleavage of APP to Aβ (41). Additionally, autophagy conditions may play an important role in Aβ secretion into extracellular space, where it contributes to the accumulation of plaques. Numerous studies shows that deletion of Atg7 in transgenic mice models of APP results in fewer Aβ extracellular secretions as well as plaque formations; therefore autophagy up–regulation would simply lead to degraded Aβ secretion (42). Furthermore, other research has suggested that autophagy is involved in the removal of Aβ under physiological states, suggesting that maintenance of Aβ homeostasis in a healthy brain is essential (43). In correlation with these findings, it has been indicated that autophagy activation is decreased in AD patient brains and animal models (44). More importantly, the direct link between autophagy reduction activity and promotion of accumulation of Aβ in the brain consequently demonstrates their contribution to AD pathogenesis (45). Therefore, targeting Aβ and tau protein should be considered as an important therapeutic approach against AD pathogenesis via upregulation of the autophagy process.
Perhaps most importantly, class III PI3K/beclin-1 signaling has been found to be a constructive and significant regulator in autophagy (46). For that reason, it seems that modulation of autophagy represents a feasible method for Aβ clearance in the brain (45). Moreover, substantial efforts have been made to recognize safe and effective pharmacological stimulation of autophagy for AD treatment (47). In practical terms, the existing outcomes of ongoing research have provided biochemical, epidemiological, neuropathological and genetic evidence to further motivate the design of viably therapeutic approaches against AD. Incidentally, most anti-AD research groups are now embracing the discovery of BACE1, a rate-limiting enzyme in Aβ generation, which acts as an enzymatic inhibitor (48). To understand the molecular basis of AD in autophagy, recent studies have been shown that elevated protein levels of BACE1 found in AD patients, as well as mouse models, suggest the down-regulation of BACE1 expression as a novel target for AD treatment (49, 50). Even though this approach may offer some improvement, an effective therapy to halt the development of neural damage in AD patients is still missing. Meanwhile, medications for AD have already been approved by the FDA, such as an NMDA receptor antagonist and four other acetylcholinesterase (AChE) inhibitors (51). It is possible that combined therapy with NMDA receptor antagonists along with AChE inhibitors could facilitate greater treatment efficacy. Otherwise, multifunctional agents are also being considered as possible drug candidates for AD (52). For that reason, research for multi-target compounds has been constantly growing for the treatment of AD (53).
For treatment purposes, autophagy enhancers treated in various AD mouse models have produced useful evidence related to a decrease in Aβ levels. For example, carbamazepine, an mTOR-independent autophagy stimulator, improves memory deficits by enhancing autophagy via decreasing Aβ formation in APP/PS1 double-transgenic mice (54). Notably, Aβ plaque formation diminishes by rapamycin in triple-transgenic PS1/APP/Tau mice and improves cognitive defection through inducing autophagy (55). Lithium treatment also suppresses tau protein in aged APP/PS1 mutant mice pathology via GSK3β inhibition, found to disturb tau phosphorylation (56). Interestingly, latrepirdine, an antihistamine drug, shows to increase autophagy by reducing the Aβ peptide through inactivating the mTOR complex 1, which has been found to recover cognition function in an AD mice models (57). Correspondingly, trehalose, a disaccharide sugar, reduces aggregation of insoluble tau, which enhances neuronal brainstem survival in the cerebral cortex via influences of autophagy (58). Moreover, rapamycin and lithium have been demonstrated to decrease the aggregation of p62 in cortical and hyperphosphorylated tau proteins (59, 60). Taken together, all of this evidence indicates that autophagy stimulation is a promising therapeutic approach for “tauopathies,” as well as for Aβ clearance in AD mouse models.
Autophagy in Parkinson’s disease
PD is a progressively degenerative disorder of the nervous system and the second most common neurodegeneration in humans. PD is a long-term degenerative brain disorder that mostly affects motor function of the central nervous system, due to the progressive degeneration of dopamine neurons in an area of the brain called the substantia nigra, where clumps of α-synuclein protein known as Lewy Bodies are formed (61). Dementia becomes more common in advanced phases of the disease, with depression and anxiety generally occurring in more than one third of people with PD (62). PD is also related with A53T and A30P point mutations of α-synuclein or leucine-rich repeat kinase 2 mutations, and sometimes other gene multiplication. PTEN-induced kinase 1 or Protein deglycase mutations in parkin have also c been associated with recessive, early-onset PD (63). The pathological feature of early-onset PD is Lewy body formation via α-synuclein aggregation (64). It has been found that α-synuclein point mutations are sometimes autophagy substrates and their excessive expression does not distress autophagy (65). For that reason, autophagy influences by lithium, trehalose, carbamazepine, calpastatin, and rilmenidine promote the removal of mutant α-synuclein (66). In contrast, wild-type α-synuclein overexpression reduces autophagosome synthesis in transgenic mice and mammalian cells through down regulation of Ras-related protein Rab-1A (65). Additionally, overexpression of α-synuclein has been proposed to prevent autophagy in the rat midbrain by repossessing transcription factor EB into combinations, thereby affecting cytoplasmic retention. Conversely, transcription factor EB overexpression activates autophagy and prevents neurotoxicity of α-synuclein-induction (67). Hence, autophagy activator protein beclin-1 ameliorates neuronal pathology in lentiviral-overexpression transgenic mice models through a decrease in α-synuclein accumulation (36). Even though the effectiveness of autophagy stimulation in vivo attempts further warrant consideration, absence of strong neurodegeneration phenotypes, together with dopaminergic neuronal susceptibility in numerous transgenic PD models, have prevented treatment trials in PD (68).
In the preclinical approach to PD, autophagy’s significance has been increased by the promising effects of several potential autophagy enhancers, such as rapamycin, lithium, metformin and trehalose - routinely used in laboratory settings to increase clearance of α-synuclein – thereby protecting neuronal cells (69). To date, the most comprehensively-tested autophagy-enhancer is rapamycin, isolated from bacterial strain of Streptomyces hygroscopicus. In particular, rapamycin blocks the upstream signaling factor mTOR by binding to its intracellular small protein FK506 binding protein 12 receptor, thus disrupting mTOR function (70). Rapamycin also has the capability to decrease α-synuclein accumulation and block α-synuclein induction of neurodegeneration in wild-type and A53T α-synuclein expressing PC12 cells (71), as well as α-synuclein overexpressing mice (72) and rats (67). In addition, rapamycin decreases neuronal cell death induce via 6-OHDA (73), rotenone (74) and MPTP mouse models of PD (74). Furthermore, rapamycin exclusively improves motor function in A53T α-synuclein overexpressing mice (75) and also shows neuroprotective activity in Drosophila melanogaster mutated PTEN-induced putative kinase-1 as well as parkin (76); although it has been noted to accompany a widespread number of side-effects (77). By comparison, another preclinical trial drug, lithium, induces autophagy in an mTOR-independent manner and has a role in in vivo as well as in vitro models of neuroprotection (78). In preclinical models of PD, lithium has been shown to inhibit aggregation of α-synuclein in PC12 cells expressing A53T and A30P α-synuclein (79). In addition, lithium has also been demonstrated to prevent rotenone-induction neurotoxicity and cell death through the stimulation of autophagy (80). When taken together, these preclinical studies via rapamycin in addition with lithium shows valuable effects on diverse models of PD-related pathological progressions, such as α-synuclein aggregation, which is associated with autophagy stimulation. These pioneering studies have demonstrated that the prospective role of autophagy-enhancing approaches in the setting of experimental drugs and medical treatments lends credence to the furtherance of research in PD-related pathology.
Autophagy in other neurodegenerative diseases
Huntington disease (HD), an autosomal dominant mutation, is a progressive neuronal genetic disorder that impacts muscle coordination and manifests in the formation of cognitive decline and dementia. Symptoms include unsteady movements, lowering of mental and emotional abilities, and finally psychiatric and behavioral problems (81). Pathologically speaking, the polyglutamine disorder group of neurodegenerative diseases are caused by the expansion of cytosine-adenine-guanine repeats in DNA encoding, therefore rendering the protein aggregation-prone and pathogenic (82). For treatment of HD with CCI-779, a rapamycin analog drug improved pathology in mice models of HD by diminishing the aggregation of mutant huntingtin protein via stimulation of the autophagy pathway (14). Interestingly, numerous mTOR-independent autophagy activators, for example Ca2+ channel blockers (loperamide, verapamil, amiodarone), inositol-lowering agents (lithium), calpain inhibitor (calpastatin), and NOS inhibitor (L-NAME), improved the removal of huntingtin as well as lowered toxicity in cellular HD models (66). Recently, a natural compound, the quaternary ammonium salt berberine (an isoquinoline alkaloid), has been found to prevent mutant Htt protein accumulation, alleviate HD symptoms, as well as activate autophagy in HD cell and mouse models (83). Another natural product, onjisaponin B (derivative of Radix Polygalae), has been found to control autophagy in PC12 cells through Atg7 as well as the AMPK-mTOR signaling pathway to enhance clearance of the Htt mutant protein and α-synuclein mutant (84). Therefore, it has been suggested that modulation of autophagy in HD might be a favorable approach to the treatment of this neurodegeneration.
Spinocerebellar ataxia (SCA), a progressive, genetic neuronal disease, is described by means of gradually growing problems associated with muscle stiffness, speech and swallowing difficulties, which also leads to involuntary eye movements. In addition, SCA is caused by the aggregation of mutant ataxin-3, thereby impairing learning and cognition function (85). Interestingly, modulation of autophagy is also involved in the removal of the abnormal ataxin-3 protein, and impaired autophagy may affect the aggregation of the mutant ataxin-3 protein that initiates cellular toxicity as well as cell death. Accordingly, targeting SCA3 through directing autophagy could become a novel therapeutic strategy (86). Likewise, trehalose, a natural alpha-linked disaccharide, is an mTOR-independent autophagy inducer that could be used to control the ataxin-3 protein in SCA. Recently, it has also been found that trehalose analogs - such as lactulose and melibiose-significantly decreased aggregation of abnormal ataxin-3 protein in a cell model, which was also associated with the stimulation of autophagy, along with a reduction in free radical production (87). Therefore, autophagy induction could potentially be used as a therapeutic strategy to control SCA.
Amyotrophic lateral sclerosis (ALS), a rare group of neurological disorders, is the effect of gradual degeneration and death of nerve cells; mainly motor neurons which control voluntary muscle function, resulting in difficulty in speaking, walking, swallowing, chewing, and finally breathing (88). To control this disease, it is necessary to induce autophagosomes through a decreasing mTOR complex 1, using a Cu-Zn superoxide dismutase mutant mice model (89). It is suspected that p62 could bind to superoxide dismutase mutations in an ubiquitin-independent manner to target its removal by autophagy (90). Conversely, mTOR-independent autophagy stimulation by lithium eases removal of mutant superoxide dismutase and hinders the disease from the start. However, treatment with trehalose has been found to be more beneficial in superoxide dismutase mutant mice through upregulation of mTOR-independent autophagy, as is found in ALS (91). Therefore, autophagy stimulation is very essential issue for in vivo studies of ALS treatment.
THERAPEUTIC IMPLICATIONS OF AUTOPHAGY IN NEURODEGENERATION
For therapeutic purposes, it has been suggested that upregulation of autophagy through the mTOR complex 1-mediated pathway might help target the removal of aggregate protein molecules, as has been suggested by a decrease in cytotoxicity of mouse, zebrafish, and Drosophila studies as well (92). Tauopathies, α-synucleinopathies, SC 3, HD, and familial prion disease models, have all been implicated in the successful treatment of neurodegenerative disease through this strategy of stimulated autophagy. In particular, mTOR-independent autophagy inducers, rapamycin analogues such as rilmenidine and trehalose, haves been used in the treatment of those diseases (92). On the other hand, autophagy inhibitor activity increases the toxicity of proteins that lead to enhancement of neurodegeneration (92). Recently, it has been shown that the phosphatidylinositol-binding clathrin assembly protein, also known as PICALM, acts equally in the autophagosome formation and autophagosome degradation process, in AD genome-wide association studies (GWAS). Hence, altered PICALM activity in vitro as well as in vivo leads to the increase in toxicity and accumulation of tau in AD pathogenesis (93). Moreover, pharmacological inhibition of mTOR complex 1 is a demonstrated target in autophagy control in neurodegeneration (94). This also implies that rapamycin and its chemically synthesized analogues such as CCI-779, are widely used as potential activators of autophagy in yeast and mammalian cells in neurons, as well as in vivo in the mouse brain. Eventually, widespread preclinical animal model studies are required to explore the use of autophagy in the prevention of neurodegenerative disease.
Recent observations have found that inhibition or reduction of histone deacetylase 6 helps to rescue memory in 5XFAD AD mouse models and AD patients, which implies further therapeutic possibilities for AD, as well as other neurodegenerative diseases (95). In addition, it has been found that transplanted protein-induced pluripotent stem cells (iPSCs), derived from skin of mouse fibroblasts, differentiate into glial cells, resulting in the reduction of plaque depositions shown to alleviate cognitive dysfunction in 5XFAD transgenic AD mouse models. This suggests a favorable therapeutic application for AD (96). Similarly, insulin-degrading enzymes and major proteases of Aβ secreted from astrocytes have been shown to potentially alleviate AD symptoms through alternative, autophagy-based secretory pathways (97). Furthermore, statin, a class of lipid-lowering medications, has been demonstrated to induce autophagy in astrocyte cultures through AMPK-mTOR mediated pathways, and as it has been suggested that autophagy is essential in insulin-degrading enzyme secretion, modulation of autophagy could provide a possible therapeutic approach in Aβ pathology by increasing clearance of extracellular Aβ (98). Hence, accumulation of Aβ peptide participates in the pathological condition of AD, while inhibiting Aβ production or increasing Aβ removal may be implicated in slowing the progression of AD (40). In particular, the promotion of Aβ clearance is currently considered to be an additional therapeutic approach for AD. Thereby, autophagy has been found to have an important role in the clearance of Aβ under normal physiological conditions; for that reason it is essential to maintain Aβ homeostasis in a healthy brain (43). Most importantly, our current research is considerably directed to the identification of safe and more effective pharmacological inducers of autophagy in neurodegenerative diseases. Therefore, to target AD drug development, natural compound-mediated enhancement of autophagy might represent a sustainable strategy to Aβ clearance. To assess this question, alternatively a natural compound could be a potential drug candidate which facilities the enhancement of autophagy in AD patients. It has recently been found that several alkaloids, such as oxoisoaporphine, significantly decreased Aβ secretion in neuroblastoma cells when overexpressed in human APPsw Swedish mutants (99). Another recent report suggests that chronic administration of the isoquinoline alkaloid berberine significantly decreases Aβ deposits while promoting Aβ clearance. As a result, this was shown to improve cognitive impairments in 3XTg AD mice by enhancing activity of autophagy via the class III PI3K/beclin-1 pathway (100).
For the treatment of PD, several natural compounds have recently gained attention due to their ability to clear α-synuclein in animal and cell models. Specifically, studies have indicated that a natural compound, curcumin, derived from the curry spice of turmeric shows low toxicity in normal cells, and also significantly decreases the accumulation of α-synuclein in A53T cell via downregulation of the mTOR signaling via increasing autophagy (101). Interestingly, oxyresveratrol, a natural antioxidant, increases mTOR dependent autophagy in human neuroblastoma cell models independent of apoptosis (102). In addition, 18α-glycyrrhetinic acid, well known a gap-junction inhibitor, induces autophagy through upregulation of beclin-1, Atg5, Atg7 and LC3, as well as downregulation of p62 in neuroblastoma cells (103). Therefore, these reviews attempt to expound upon the important observation that the pharmacological targeting of autophagy-enhancing natural compounds hold a favorable implication for the development of PD therapies.
CONCLUSION
Even though a variety of autophagy-related proteins have been demonstrated to participate and control endogenous autophagy pathways (104), several studies have been performed to explore autophagy regulation through use of the active compounds of plants. Although numerous fundamental queries are essential for further address, many novel agents could one day be useful in a clinical approaches; thus the interest of research in autophagy is developing rapidly, and clinical applications might be anticipated in the near future. Furthermore, it is now important to characterize dysfunctional autophagy in diverse stages of genetic and molecular subtypes in order to better understand their roles in neurodegeneration. It is also necessary to produce clinical results demonstrating the efficacy of downstream autophagy regulation, which would help usher in an exciting new era for the development of neurodegenerative therapeutic strategies. Consequently, additional studies are required on the physiological roles of modulation of brain autophagy processes in neurodegenerative disease. Finally, we would like to screen novel, natural compounds that modulate autophagy and identify main therapeutic targets, such as the key molecular mechanisms that underlie the pathophysiological roles of neurodegeneration, with our main concern being the discovery of potential therapeutic drug targets.
ACKNOWLEDGEMENTS
This work was supported by “Korea Research Fellowship Program” (2016H1D3A1908615) and the NRF Research Program (2016M3C7A1913845), funded by the Ministry of Science, ICT and Future Planning, and also supported by the KIST Institutional Programs (Project No. 2E26820).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 5.Toth ML, Sigmond T, Borsos E, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 6.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 7.Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway. J Biol Chem. 2011;286:32324–32332. doi: 10.1074/jbc.M111.254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung HS, Lee M-S. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010;1201:79–83. doi: 10.1111/j.1749-6632.2010.05614.x. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 11.Spilman P, Podlutskaya N, Hart MJ, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39:423–438. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson’s disease. Brain. 2008;131:1969–1978. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- 14.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 15.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez D, Torres CA, Setlik W, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74:277–284. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 19.Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 20.Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pozueta J, Lefort R, Ribe EM, Troy CM, Arancio O, Shelanski M. Caspase-2 is required for dendritic spine and behavioural alterations in J20 APP transgenic mice. Nat Commun. 2013;4:1939. doi: 10.1038/ncomms2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shpilka T, Welter E, Borovsky N, et al. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015;34:2117–2131. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 26.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 28.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S, Holmstrom KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 31.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 33.Kirkin V, Lamark T, Sou Y-S, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Giacomelli C, Daniele S, Martini C. Potential biomarkers and novel pharmacological targets in protein aggregation-related neurodegenerative diseases. Biochem Pharmacol. 2017;131:1–15. doi: 10.1016/j.bcp.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Berger Z, Ravikumar B, Menzies FM, et al. Rapamycin alleviates toxicity of different aggregateprone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 36.Spencer B, Potkar R, Trejo M, et al. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson’s and Lewy body diseases. J Neurosci. 2009;29:13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Strooper B, Karran E. The Cellular Phase of Alzheimer’s Disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 38.Zare-Shahabadi A, Masliah E, Johnson GVW, Rezaei N. Autophagy in Alzheimer’s disease. Rev Neurosci. 2015;26:385–395. doi: 10.1515/revneuro-2014-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iqbal K, Liu F, Gong C-X. Tau and neurodegenerative disease: the story so far. Nat Rev Neurol. 2016;12:15–27. doi: 10.1038/nrneurol.2015.225. [DOI] [PubMed] [Google Scholar]
- 40.Lin L-F, Liao M-J, Xue X-Y, et al. Combination of Abeta clearance and neurotrophic factors as a potential treatment for Alzheimer’s disease. Neurosci Bull. 2013;29:111–120. doi: 10.1007/s12264-012-1287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boland B, Kumar A, Lee S, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson P, Loganathan K, Sekiguchi M, et al. Abeta secretion and plaque formation depend on autophagy. Cell Rep. 2013;5:61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 43.Xue Z, Guo Y, Zhang S, et al. Beta-asarone attenuates amyloid beta-induced autophagy via Akt/mTOR pathway in PC12 cells. Eur J Pharmacol. 2014;741:195–204. doi: 10.1016/j.ejphar.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Wolfe DM, Lee J-H, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37:1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan C-C, Yu J-T, Tan M-S, Jiang T, Zhu X-C, Tan L. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging. 2014;35:941–957. doi: 10.1016/j.neurobiolaging.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Jaber N, Zong W-X. Class III PI3K Vps34: essential roles in autophagy, endocytosis, and heart and liver function. Ann N Y Acad Sci. 2013;1280:48–51. doi: 10.1111/nyas.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Vicente M. Autophagy in neurodegenerative diseases: From pathogenic dysfunction to therapeutic modulation. Semin Cell Dev Biol. 2015;40:15–26. doi: 10.1016/j.semcdb.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Eketjall S, Janson J, Jeppsson F, et al. AZ-4217: a high potency BACE inhibitor displaying acute central efficacy in different in vivo models and reduced amyloid deposition in Tg2576 mice. J Neurosci. 2013;33:10075–10084. doi: 10.1523/JNEUROSCI.1165-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marwarha G, Raza S, Meiers C, Ghribi O. Leptin attenuates BACE1 expression and amyloid-beta genesis via the activation of SIRT1 signaling pathway. Biochim Biophys Acta. 20141842:1587–1595. doi: 10.1016/j.bbadis.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Murakami K, Watanabe T, Koike T, Kamata M, Igari T, Kondo S. Pharmacological properties of a novel and potent gamma-secretase modulator as a therapeutic option for the treatment of Alzheimer’s disease. Brain Res. 2016;1633:73–86. doi: 10.1016/j.brainres.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 51.Geldenhuys WJ, Darvesh AS. Pharmacotherapy of Alzheimer’s disease: current and future trends. Expert Rev Neurother. 2015;15:3–5. doi: 10.1586/14737175.2015.990884. [DOI] [PubMed] [Google Scholar]
- 52.Gauthier S, Molinuevo JL. Benefits of combined cholinesterase inhibitor and memantine treatment in moderate-severe Alzheimer’s disease. Alzheimers Dement. 2013;9:326–331. doi: 10.1016/j.jalz.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Ismaili L, Refouvelet B, Benchekroun M, et al. Multitarget compounds bearing tacrine- and donepezil-like structural and functional motifs for the potential treatment of Alzheimer’s disease. Prog Neurobiol. 2017;151:4–34. doi: 10.1016/j.pneurobio.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Zhang S, Zhang X, et al. Autophagy enhancer carbamazepine alleviates memory deficits and cerebral amyloid-beta pathology in a mouse model of Alzheimer’s disease. Curr Alzheimer Res. 2013;10:433–441. doi: 10.2174/1567205011310040008. [DOI] [PubMed] [Google Scholar]
- 55.Majumder S, Richardson A, Strong R, Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS One. 2011;6:e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Heng X, Li T, et al. Long-term treatment with lithium alleviates memory deficits and reduces amyloid-beta production in an aged Alzheimer’s disease transgenic mouse model. J Alzheimers Dis. 2011;24:739–749. doi: 10.3233/JAD-2011-101875. [DOI] [PubMed] [Google Scholar]
- 57.Steele JW, Lachenmayer ML, Ju S, et al. Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer’s mouse model. Mol Psychiatry. 2013;18:889–897. doi: 10.1038/mp.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135:2169–2177. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozcelik S, Fraser G, Castets P, et al. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS One. 2013;8:e62459. doi: 10.1371/journal.pone.0062459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimada K, Motoi Y, Ishiguro K, et al. Long-term oral lithium treatment attenuates motor disturbance in tauopathy model mice: implications of autophagy promotion. Neurobiol Dis. 2012;46:101–108. doi: 10.1016/j.nbd.2011.12.050. [DOI] [PubMed] [Google Scholar]
- 61.Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet (London, England) 373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 62.Sveinbjornsdottir S. The clinical symptoms of Parkinson’s disease. J Neurochem. 2016;139:318–324. doi: 10.1111/jnc.13691. [DOI] [PubMed] [Google Scholar]
- 63.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 65.Winslow AR, Chen C-W, Corrochano S, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams A, Sarkar S, Cuddon P, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110:E1817–1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kahle PJ. alpha-Synucleinopathy models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:87–95. doi: 10.1007/s00401-007-0302-x. [DOI] [PubMed] [Google Scholar]
- 69.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maiese K, Chong ZZ, Shang YC, Wang S. mTOR: on target for novel therapeutic strategies in the nervous system. Trends Mol Med. 2013;19:51–60. doi: 10.1016/j.molmed.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 72.Crews L, Spencer B, Desplats P, et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One. 2010;5:e9313. doi: 10.1371/journal.pone.0009313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Pan T, Rawal P, Wu Y, Xie W, Jankovic J, Le W. Rapamycin protects against rotenone-induced apoptosis through autophagy induction. Neuroscience. 2009;164:541–551. doi: 10.1016/j.neuroscience.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 74.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30:1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bai X, Wey MC-Y, Fernandez E, et al. Rapamycin improves motor function, reduces 4-hydroxynonenal adducted protein in brain, and attenuates synaptic injury in a mouse model of synucleinopathy. Pathobiol Aging Age Relat Dis. 2015;5:28743. doi: 10.3402/pba.v5.28743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 78.Forlenza OV, De-Paula VJR, Diniz BSO. Neuroprotective effects of lithium: implications for the treatment of Alzheimer’s disease and related neurodegenerative disorders. ACS Chem Neurosci. 2014;5:443–450. doi: 10.1021/cn5000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hou L, Xiong N, Liu L, et al. Lithium protects dopaminergic cells from rotenone toxicity via autophagy enhancement. BMC Neurosci. 2015;16:82. doi: 10.1186/s12868-015-0222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chao T-K, Hu J, Pringsheim T. Risk factors for the onset and progression of Huntington disease. Neurotoxicology. 2017;2130:21. doi: 10.1016/j.neuro.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Fan H-C, Ho L-I, Chi C-S, et al. Polyglutamine (PolyQ) diseases: genetics to treatments. Cell Transplant. 2014;23:441–458. doi: 10.3727/096368914X678454. [DOI] [PubMed] [Google Scholar]
- 83.Jiang W, Wei W, Gaertig MA, Li S, Li X-J. Therapeutic Effect of Berberine on Huntington’s Disease Transgenic Mouse Model. PLoS One. 2015;10:e0134142. doi: 10.1371/journal.pone.0134142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu A-G, Wong VK-W, Xu S-W, et al. Onjisaponin B derived from Radix Polygalae enhances autophagy and accelerates the degradation of mutant alpha-synuclein and huntingtin in PC-12 cells. Int J Mol Sci. 2013;14:22618–22641. doi: 10.3390/ijms141122618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Y-M, Lu C, Wu Z-Y. Spinocerebellar ataxia: relationship between phenotype and genotype - a review. Clin Genet. 2016;90:305–314. doi: 10.1111/cge.12808. [DOI] [PubMed] [Google Scholar]
- 86.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin C-H, Wu Y-R, Yang J-M, et al. Novel Lactulose and Melibiose Targeting Autophagy to Reduce PolyQ Aggregation in Cell Models of Spinocerebellar Ataxia 3. CNS Neurol Disord Drug Targets. 2016;15:351. doi: 10.2174/1871527314666150821101522. [DOI] [PubMed] [Google Scholar]
- 88.Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6:171. doi: 10.4103/2152-7806.169561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morimoto N, Nagai M, Ohta Y, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 90.Gal J, Strom A-L, Kwinter DM, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 92.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest. 2015;125:65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moreau K, Fleming A, Imarisio S, et al. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014;5:4998. doi: 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 95.Choi H, Kim HJ, Kim J, et al. Increased acetylation of Peroxiredoxin1 by HDAC6 inhibition leads to recovery of Abeta-induced impaired axonal transport. Mol Neurodegener. 2017;12:23. doi: 10.1186/s13024-017-0164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cha M-Y, Kwon Y-W, Ahn H-S, et al. Protein-Induced Pluripotent Stem Cells Ameliorate Cognitive Dysfunction and Reduce Abeta Deposition in a Mouse Model of Alzheimer’s Disease. Stem Cells Transl Med. 2017;6:293–305. doi: 10.5966/sctm.2016-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Son SM, Cha M-Y, Choi H, et al. Insulin-degrading enzyme secretion from astrocytes is mediated by an autophagy-based unconventional secretory pathway in Alzheimer disease. Autophagy. 2016;12:784–800. doi: 10.1080/15548627.2016.1159375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Son SM, Kang S, Choi H, Mook-Jung I. Statins induce insulin-degrading enzyme secretion from astrocytes via an autophagy-based unconventional secretory pathway. Mol Neurodegener. 2015;10:56. doi: 10.1186/s13024-015-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang L, Luo Y, Pu Z, et al. Oxoisoaporphine alkaloid derivative 8-1 reduces Abeta1- 42 secretion and toxicity in human cell and Caenorhabditis elegans models of Alzheimer’s disease. Neurochem Int. 2017;108:157–168. doi: 10.1016/j.neuint.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 100.Huang M, Jiang X, Liang Y, Liu Q, Chen S, Guo Y. Berberine improves cognitive impairment by promoting autophagic clearance and inhibiting production of beta-amyloid in APP/tau/PS1 mouse model of Alzheimer’s disease. Exp Gerontol. 2017;91:25–33. doi: 10.1016/j.exger.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 101.Jiang T-F, Zhang Y-J, Zhou H-Y, et al. Curcumin ameliorates the neurodegenerative pathology in A53T alpha-synuclein cell model of Parkinson’s disease through the downregulation of mTOR/p70S6K signaling and the recovery of macroautophagy. J Neuroimmune Pharmacol. 2013;8:356–369. doi: 10.1007/s11481-012-9431-7. [DOI] [PubMed] [Google Scholar]
- 102.Rahman MA, Bishayee K, Sadra A, Huh S-O. Oxyresveratrol activates parallel apoptotic and autophagic cell death pathways in neuroblastoma cells. Biochim Biophys Acta. 20171861:23–36. doi: 10.1016/j.bbagen.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 103.Rahman MA, Bishayee K, Habib K, Sadra A, Huh S-O. 18alpha-Glycyrrhetinic acid lethality for neuroblastoma cells via de-regulating the Beclin-1/Bcl-2 complex and inducing apoptosis. Biochem Pharmacol. 2016;117:97–112. doi: 10.1016/j.bcp.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 104.Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015;11:2114–2122. doi: 10.1080/15548627.2015.1099796. [DOI] [PMC free article] [PubMed] [Google Scholar]