Abstract
Homeobox (HOX) proteins and the receptor tyrosine kinase FLT3 are frequently highly expressed and mutated in acute myeloid leukemia (AML). Aberrant HOX expression is found in nearly all AMLs that harbor a mutation in the Nucleophosmin (NPM1) gene, and FLT3 is concomitantly mutated in approximately 60% of these cases. Little is known how mutant NPM1 (NPM1mut) cells maintain aberrant gene expression. Here, we demonstrate that the histone modifiers MLL1 and DOT1L control HOX and FLT3 expression and differentiation in NPM1mut AML. Using a CRISPR-Cas9 genome editing domain screen, we show NPM1mut AML to be exceptionally dependent on the menin binding site in MLL1. Pharmacological small-molecule inhibition of the menin-MLL1 protein interaction had profound anti-leukemic activity in human and murine models of NPM1mut AML. Combined pharmacological inhibition of menin-MLL1 and DOT1L resulted in dramatic suppression of HOX and FLT3 expression, induction of differentiation, and superior activity against NPM1mut leukemia.
Keywords: Leukemia, MLL, DOT1L, NPM1, Homeobox Genes
Introduction
The clustered HOX genes are a highly conserved family of transcription factors that are expressed during early development and hematopoiesis(1-3). Specific members of the HOXA- and HOXB-cluster are required to maintain self-renewal properties of hematopoietic stem cells (HSCs)(3). Aberrant HOX gene expression is also a common feature of acute leukemias and can be activated by various oncogenes in murine leukemia models(3, 4). These genes are believed to play an important role during leukemogenesis since ectopic expression of specific members of the HOX-cluster induces leukemic transformation of murine HSCs(5). In human AML aberrant HOX expression is found in 40-60% of the cases, while the specific HOX expression pattern is heterogeneous and has been used to categorize AML cases into four main groups that correlate with the presence of specific molecular markers(4). Simultaneous HOXA- and HOXB-cluster expression is most commonly found and frequently associated with the presence of an NPM1 mutation(4, 6, 7), one of the two most frequently mutated genes in AML(8).
Mutant NPM1 co-occurs often with other mutations in AML, most commonly in the FLT3 (60%) and/or the DNMT3A (50%) gene(9-11). While NPM1mut AML lacking an internal tandem duplication (ITD) in the FLT3 gene is considered a relatively favorable genotype with overall survival rates of up to 60% in younger patients, the presence of a concurrent FLT3-ITD converts this genotype into a less favorable category(9, 11, 12). The receptor tyrosine kinase FLT3 is of particular interest because a) its mutation status is used as prognostic indicator guiding treatment decisions in AML patients and b) represents a molecular target for small-molecule inhibition. Clinical trials have shown activity of several FLT3 inhibitors against FLT3 mutated AML but resistance occurred frequently and quickly(13). Currently less than 50% of the (younger) NPM1mut FLT3-ITD positive AML patients achieve a sustainable remission and survival rates of NPM1mut AML patients >65 years of age are even more dismal(11, 14). These data highlight the need for novel therapeutic concepts, which may be less toxic and more efficacious.
NPM1 is an intracellular chaperone protein implicated in multiple cellular processes, such as proliferation, ribosome and nucleosome assembly(15, 16) but it has remained elusive how NPM1 mutations initiate and maintain AML thus hindering the development of targeted therapeutic approaches. It is also unknown what mechanisms drive aberrant HOX gene expression in these leukemias. Our current knowledge of HOX gene regulation in leukemias has so far come from studies of MLL1-rearranged leukemias. In these leukemias, chromosomal translocations cause a fusion of the mixed-lineage leukemia gene (MLL1, also known as MLL or KMT2A) with one of more than 60 different fusion partners resulting in the formation of an oncogenic fusion protein(17). MLL1-rearranged leukemias exhibit aberrant HOXA-cluster expression as a consequence of chromatin binding of the MLL1-fusion complex(18), which in turn requires the association with at least two other proteins, menin and LEDGF. Whereas menin facilitates LEDGF binding to the complex, the latter directly conveys chromatin binding of MLL1(19, 20). Menin, the protein encoded by the multiple endocrine neoplasia 1 gene (MEN1), is of particular interest as it is required by MLL1-fusion proteins for transformation and has been shown to be therapeutically targetable. Recently developed inhibitors of the menin-MLL1 interaction were demonstrated to have anti-leukemic activity in preclinical MLL1-rearranged leukemia models(21, 22). While NPM1mut AML lack MLL1-fusion proteins, studies of conditional MLL1 knock-out mice have shown that wildtype MLL1 (wt-MLL1) is required for maintenance of HOX gene expression during normal hematopoiesis(23, 24). In addition, it has been shown that wt-MLL1 also requires the interaction with menin to maintain HOX expression and self-renewal properties of hematopoietic progenitors(25, 26).
The histone 3 lysine 79 (H3K79) methyltransferase DOT1L is another protein of therapeutic interest involved in HOXA-cluster regulation of MLL1-rearranged leukemias(27-29). DOT1L is believed to be recruited to promoters of HOXA-cluster genes via the MLL1-fusion partner that is commonly part of the DOT1L binding complex(30). Small-molecule inhibitors of DOT1L were proven to have activity against preclinical models of MLL1-rearranged leukemias(31, 32), and are currently being tested in a clinical trial (NCT01684150) with promising first results(33). DOT1L has also been linked to HOX gene control during normal hematopoiesis. Higher states of H3K79 methylation, such as di- and trimethylation (me2 and me3) at the HOX locus are associated with high expression levels of these genes in lineage-, Sca-1+, c-Kit+ (LSK) murine hematopoietic progenitors and are converted into monomethylation (me1) as these cells mature into more committed progenitors lacking HOX expression(28). Whether Hoxb-cluster expression in LSKs is also associated with H3K79me2 and me3 is unknown to date.
We hypothesized that aberrant HOX and MEIS1 gene expression in NPM1mut AML might be driven by similar mechanisms that control these genes in normal HSCs and that high expression of FLT3, a reported downstream target of MEIS1, is indirectly driven via elevated MEIS1 transcript levels. In the current study, we use CRISPR-Cas9 genome editing and small-molecule inhibition to identify the chromatin regulators MLL1 and DOT1L as therapeutic targets that control HOX, MEIS1 and FLT3 expression and differentiation in NPM1mut leukemia.
Results
CRISPR-Cas9 mutagenesis demonstrates a requirement for MLL1 and the menin binding in NPM1mut leukemia
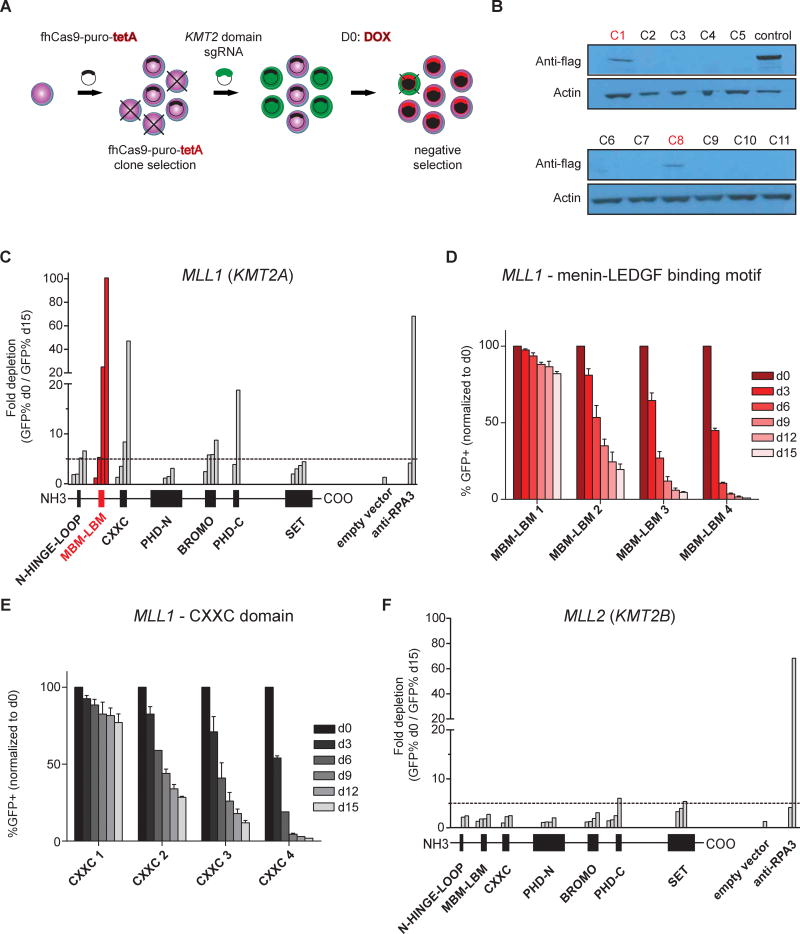
Since MLL1 has been shown to regulate critical gene expression programs, including HOX gene expression in normal hematopoiesis and MLL1-rearranged AML, we hypothesized that it might also regulate HOX gene expression in other settings. As an initial assessment we used an approach recently developed by Shi and co-workers that uses genome editing across exons encoding specific protein domains to determine the functional relevance of a given domain and the suitability for drug development(34). To assess potential dependencies of NPM1mut AML on specific protein domains of MLL1 and its most similar family member MLL2 (also known as KMT2B), we designed a negative selection CRISPR-Cas9 screen interrogating multiple domains of both proteins (Supplemental-Table 1+2). We generated a tetracycline-inducible Cas9-expressing OCI-AML3 cell line (OCI-AML3-pCW-Cas9, Fig.1A+B) as OCI-AML3 is the only commercially available human AML cell line harboring an NPM1mut. After transduction with 2-4 GFP+ sgRNAs interrogating each domain, Cas9 was induced and GFP+/GFP--ratio assessed over a period of 15 days. Cells expressing sgRNAs targeting exon 1 that encodes the menin-LEDGF binding motif of MLL1 as defined by Huang and co-workers(35) were rapidly outcompeted by their GFP- counterparts (Fig.1C+D; fold change of GFP+ d0/d15 up to 100). Similar results were obtained for one of two sgRNAs targeting the first exon of RPA3, a gene required for DNA replication that is used as a positive control, while there was no difference in GFP expression over time in an empty vector control guide (Fig.1C). Having found selection against the sgRNAs targeting the menin-LEDGF binding motif, we performed SURVEYOR assays of sgRNAs targeting the menin-LEDGF binding motif and the N-HINGE-LOOP motif. The latter is located just 5′ of the menin-LEDGF locus but is considered as non-essential for the menin-MLL1 interaction and showed less negative selection in our screen. We detected indel mutations at day 3 for both sgRNAs but at day 9 only for the N-HINGE-LOOP sgRNA (Supplemental-Fig.1). These results are consistent with the selection data from our screen that demonstrate outcompeting of the cells transduced with the menin-LEDGF sgRNAs. Interestingly, sgRNAs targeting exons that encode the CXXC domain of MLL1, also known to be involved in chromatin binding and required for MLL1-fusion leukemia, were also selected against in our screen (Fig.1C+E)(36, 37). Of interest, no significant phenotype was observed for sgRNAs targeting the C-terminal SET domain of MLL1 (Fig.1C). We validated these findings by performing a second screen using another independently engineered OCI-AML3-pCW-Cas9 clone (OCI-AML3-pCW-Cas9-C8, Fig.1B) and obtained similar results with sgRNAs targeting the menin-LEDGF binding motif being again the top hit (Supplemental-Fig.2A+B). We found no significant negative selection for any sgRNAs that interrogated the MLL2 protein domains (Fig.1F). These data suggest that the menin-LEDGF binding motif of MLL1 but not MLL2 is required for NPM1mut AML.
Figure 1. CRISPR-Cas9 mutagenesis of exons targeting MLL protein domains in NPM1mut AML cells.
(A) Experimental strategy for CRISPR-Cas9 negative selection screening: Engineering a clonal NPM1mut OCI-AML3 cell line that expresses a flag-tagged human codon-optimized Cas9 (fhCas9) vector containing a puro resistance gene (puro) and tetracycline-inducible transcriptional activator (tetA). GFP reporters of sgRNA constructs were used to track sgRNA negative selection after doxycycline induction of Cas9 (D0, day 0; DOX, doxycycline).
(B) Immunoblotting for flag-tagged hCas9 after doxycycline treatment in eleven OCI-AML3-Cas9 single cell clones. C1 and C8 clones were selected for two independent screens of MLL1 and MLL2.
(C and F) Summary of negative selection experiments with sgRNAs targeting exons encoding specific MLL1 and MLL2 protein domains. Negative selection is plotted as the fold depletion of GFP+ cells (d0 GFP% divided by d15 GFP%) during 18 days in culture. Each bar represents an independent sgRNA. The location of each sgRNA relative to the MLL1 or MLL2 protein is indicated along the x axis. The dashed line indicates a fivefold change. The data shown are the mean value of two independent replicates. Empty vector and anti-RPA3 sgRNA represent negative and positive controls.
(D and E) Negative selection competition assay that plots the percentage of GFP+ cells over time following transduction of OCI-AML3-Cas9 with the indicated sgRNAs. GFP+ percentage is normalized to the day 0 measurement following doxycycline induction of Cas9 (3 days after sgRNA transduction).
N-HINGE-LOOP, N-terminal hinge loop of the menin-LEDGF binding motif lacking many specific interactions; MBM-LBM, N-terminal fragment of MLL1 containing the menin and the LEDGF binding motif (as defined by Huang et al(35)); CXXC, CXXC-type zink finger domain; PHD-N, N-terminal plant homeodomains; BROMO, bromodomain; PHD-C , C-terminal plant homeodomain; SET, SET-domain. Data shown in panel C-F represent mean of biological duplicates.
Pharmacological disruption of the menin-MLL1 interaction suppresses HOX, MEIS1, and FLT3 expression and induces NPM1mut AML differentiation
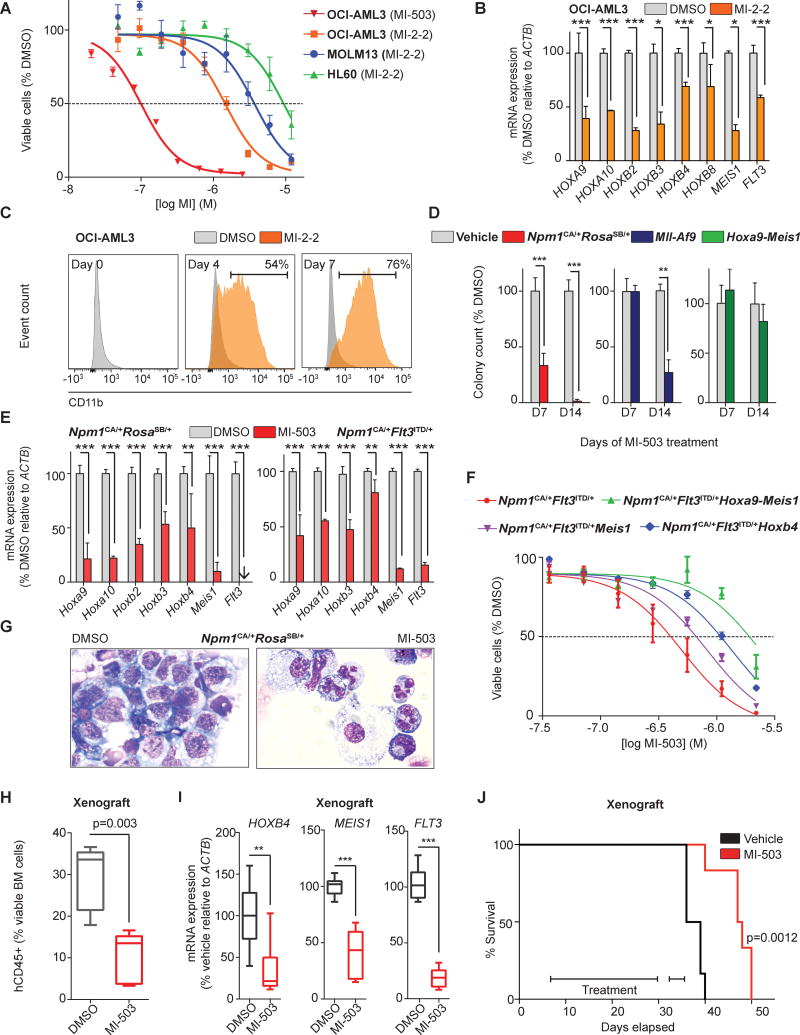
The genetic data presented above indicate that the menin-MLL1 interaction might be a therapeutic target in NPM1mut AML. We therefore assessed whether recently developed small-molecule inhibitors of the menin-MLL1 interaction might influence proliferation and gene expression in NPM1mut AML cells. First, we assessed the effects of MI-2-2, a small-molecule inhibitor of this protein interaction in the human NPM1mut AML cell line OCI-AML3(21). We observed a profound dose dependent reduction in cell proliferation that was even more pronounced than in the MLL1-rearranged MOLM13 cells that served as a positive control (Fig.2A). The HL60 AML cells lacking an NPM1mut or MLL1-fusion showed only a mild cell growth inhibitory effect at higher doses (Fig.2A). During the work on this study, MI-503, a novel menin-MLL1 inhibitor was described(22) that was synthesized by re-engineering the molecular scaffold of MI-2-2 resulting in enhanced drug-like properties and in vivo utility. To compare the two compounds and to define equivalent growth inhibitory concentrations, we generated dose response curves for both inhibitors confirming the higher potency of MI-503 over MI-2-2 (Fig.2A and Supplemental-Fig.3). MI-503 was used for most of the subsequent experiments.
Figure 2. Effects of menin-MLL11-i in human and murine NPM1mut leukemia cells in vitro and in vivo.
(A) Dose response curves from cell viability assays after 11 days of MI-2-2 or MI-503 treatment
(B) HOX gene expression in the human OCI-AML3 cells following 4 days of MI-2-2 [12μM] treatment.
(C) Cell differentiation upon menin-MLL1-i (MI-2-2: [12μM]) as determined by CD11b expression in OCI-AML3 cells (at day 0, 4, and 7 of treatment).
(D) MI-503 [2.5μM] treatment of murine Npm1CA/+RosaSB/+, Mll-Af9, and Hoxa9-Meis1-transformed cells in colony-forming assays assessed on day 7 and day 14 of treatment.
(E) Gene expression in murine Npm1CA/+Flt3ITD/+ and Npm1CA/+RosaSB/+ cells assessed on day 4 of MI-503 treatment [2.5μM].
(F) Dose response curves from cell viability assays after 11 days of MI-503 treatment comparing Npm1CA/+Flt3ITD/+ cells versus Npm1CA/+Flt3ITD/+ cells overexpressing Hoxb4, Meis1, or Hoxa9-Meis1.
(G) Morphological changes consistent with granulocytic monocytic differentiation in murine Npm1CA/+RosaSB/+ cells after 7 days of MI-503 [2.5μM] treatment.
(H) Assessment of leukemia burden in an OCI-AML3 xenotransplantation model after 7-12 days of MI-503 in vivo treatment as determined by human CD45 positive cells in the murine bone marrow.
(I) Gene expression changes after 12 days of MI-503 [50mg/kg bid IP] in vivo treatment.
(J) Survival of OCI-AML3 xenograft mice (n=6 mice/group) treated with MI-503 [50mg/kg bid IP].
Data represent averages of two independent experiments, each performed in three replicates (A, B, C, D, E, F) except for the dose response to MI-503 (red curve in A) that was once performed (in three replicates) to independently confirm sensitivity to menin-MLL1-i. Error bars represent standard error of the mean (SEM). The whiskers of box plots (H, I) represent minimal and maximal values of 5 (H) and 3 (I) mice per group, the box represents the SEM, the line represents the median.
We next assessed gene expression in the NPM1mut AML cells and found a significant down regulation of HOXA-, HOXB-cluster and MEIS1 gene expression upon 4 days of treatment with MI-2-2 (Fig.2B). Of these, MEIS1 appeared to be the most profoundly suppressed gene. This finding led us to explore a possible effect of menin-MLL1-inhibition (menin-MLL1-i) on FLT3 expression, as FLT3 is a reported downstream target of MEIS1(38). In fact, FLT3 transcript levels were also substantially and highly significantly suppressed upon MI-2-2 treatment (Fig.2B). To characterize the effects of pharmacological menin-MLL1-i in more detail, we assessed apoptosis and differentiation following MI-2-2 treatment. Whereas induction of apoptosis was moderate (Supplemental-Fig.4), we observed profound differentiation following menin-MLL1-i, as reflected by an increase in CD11b expression over time as determined by flow cytometry (Fig.2C; vehicle: 1.3%; MI-2-2: 76% (mean expression, day 7 of treatment)).
Next, we sought to validate our findings from the human OCI-AML3 cells on two independent murine conditional knock-in models of NPM1mut leukemia. One of the leukemias is engineered to possess both an Npm1 mutation and a Flt3-ITD (Npm1CA/+Flt3ITD/+), whereas in the other model secondary mutations were induced through use of a sleeping beauty transposon system (Npm1CA/+RosaSB/+)(39, 40). For in vitro drug testing leukemic blasts were harvested from moribund primarily transplanted mice and cultured. As Npm1CA/+RosaSB/+ cells did not grow in liquid culture medium but in methylcellulose only, we performed MI-503 treatment of these cells in colony-forming assays. Treatment for 7 and 14 days, respectively resulted in a profound inhibitory effect on colony-forming potential in the Npm1CA/+RosaSB/+ cells as well as in Mll-Af9 leukemia cells that served as a positive control, whereas Hoxa9-Meis1 transformed cells were unaffected (Fig.2D). Similar results were obtained for Npm1CA/+Flt3ITD/+ cells that grow in liquid culture. These leukemias exhibited profound sensitivity to MI-503 and MI-2-2 treatment in a dose dependent manner, which was even more pronounced than in the Mll-Af9 leukemia cells (Supplemental-Fig.3).
Next, we assessed the effects of MI-503 on Hox gene expression in both murine Npm1mut leukemia models. While baseline expression levels of individual Hox genes differed between the two murine leukemias, with for example lack of Hoxb2 expression in Npm1CA/+Flt3ITD/+ cells, we observed substantial suppression of Hoxa- and Hoxb-cluster genes as well as Meis1. Of these, Meis1 was the most profoundly suppressed gene and again accompanied by substantial down regulation of global Flt3 transcript levels (Fig.2E). To further explore whether the antiproliferative effects of MI-503 are causally related to Hox suppression in these leukemias, we expressed Meis1, Hoxb4, and Hoxa9-Meis1 ectopically in the Npm1CA/+FLt3ITD/+ murine leukemia cells. Of interest, all three scenarios rescued the Npm1CA/+FLt3ITD/+ cells from the menin-MLL1-i mediated antiproliferative effect when exposed to MI-503 treatment for 11 days as reflected by a shift of the dose response curves towards higher concentrations and increased IC50 values (Fig.2F).
As changes in Hox gene expression were followed by myeloid differentiation in the human cells, we then determined whether MI-503 also induces differentiation in the murine leukemias. In fact, cytological analysis of both Npm1CA/+RosaSB/+ and Npm1CA/+FLt3ITD/+ cells revealed morphological changes consistent with substantial myelo-monocytic differentiation (Fig.2G and Supplemental-Fig.5).
We next sought to investigate the therapeutic effects of menin-MLL1-i on leukemia burden in vivo using a disseminated human OCI-AML3 xenotransplantation model. OCI-AML3 cells were transplanted into NSG mice via tail vein injection and MI-503 treatment was initiated seven days later. Animals were sacrificed after seven (n=2 per group) and 12 days (n=3 per group) of MI-503 treatment (50mg/kg twice daily (bid) by intraperitoneal injection (IP)). Leukemia burden, as defined by the percentage of bone marrow cells expressing human CD45, was significantly reduced within the treated animal group compared to vehicle controls (Fig.2H). To assess effects of MI-503 on gene expression in vivo, we analyzed sorted human CD45 positive bone marrow cells harvested from these animals for HOXB4, MEIS1 and FLT3. 12 days of treatment resulted in dramatic suppression of these genes in the treated versus vehicle control animals (Fig.2I).
We then explored the effects of MI-503 treatment inhibition on survival in the OCI-AML3 xenotransplantation model in a separate experiment. Treatment was initiated 7 days post transplantation at 50mg/kg bid IP and continued for a total of 26 days excluding a 3-day break to allow recovery from local irritation at the injection site. MI-503 treatment resulted in a highly significant survival advantage compared to vehicle control animals (Fig.2J; p=0.0012; increase in median survival: 21%). Next, we sought to validate these findings in a murine model of NPM1mut leukemia. Mice transplanted with secondary Npm1CA/+RosaSB/+ leukemia cells were treated for 10 days with MI-503 versus vehicle control. Despite the aggressive character of this leukemia, with a disease latency of only 2-3 weeks, MI-503 significantly prolonged survival of leukemic animals (Supplemental-Fig.6; p=0.0045, increase in median survival: 31%). These data indicate that therapeutic menin-MLL1-i reverses a HOX gene dominated leukemogenic gene expression program in Npm1mut leukemias in vitro and in vivo thereby releasing the differentiation block in these cells and ultimately resulting in prolonged survival of mice suffering from NPM1mut AML.
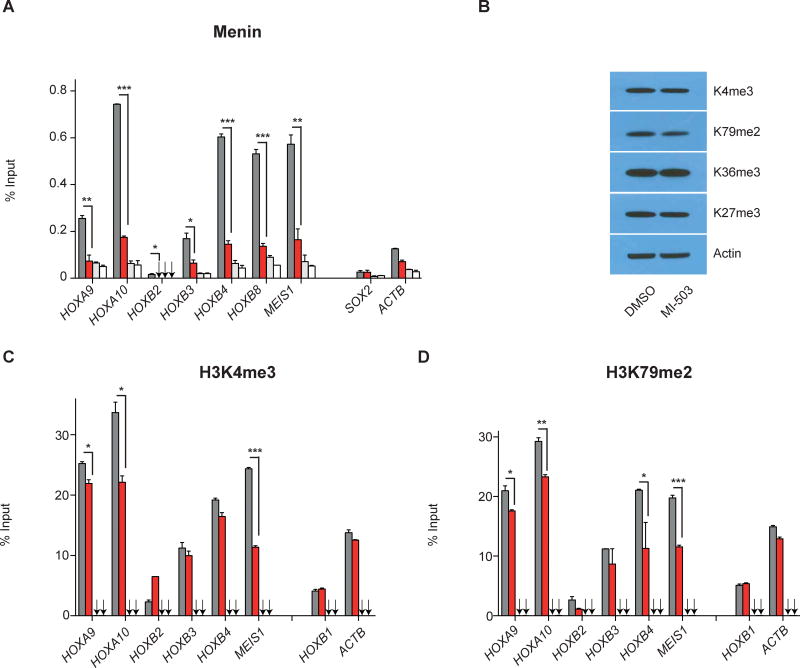
MI-503 treatment depletes menin from HOX and MEIS1 loci in NPM1mut leukemias
Menin associates with MLL1 at HOXA and MEIS1 promoters(19) and pharmacological interruption of the menin-MLL1 interaction diminishes menin occupancy at the HOXA locus in MLL1-rearranged leukemias(41). We assessed the abundance of menin at HOXA, HOXB and MEIS1 loci in the human NPM1mut leukemia cell line before and after pharmacological menin-MLL1-i. Chromatin Immunoprecipitation (ChIP) of menin followed by quantitative PCR demonstrated the presence of menin at expressed HOXA, HOXB and MEIS1 gene loci and menin was depleted from most of these loci after 5 days of MI-503 treatment (Fig.3A). Reduction of menin enrichment was particularly pronounced at the MEIS1 locus thereby correlating with findings from gene expression analysis (Fig.3A and Fig.2B).
Figure 3. Menin-MLL1-i depletes menin, H3K4me3, and H3K79me2 at HOX and MEIS1 gene loci in OCI-AML3 cells.
(A) Relative enrichment of menin at selected HOXA, HOXB, and MEIS1 gene loci in OCI-AML3 cells upon 5 days of MI-503 [2.5μM] treatment compared to drug vehicle as assessed by ChIP-PCR.
(B) Immunoblotting of indicated histone marks following 4 days of MI-503 treatment [2.5μM].
(C+D) H3K4me3 and H3K79me2 enrichment across the HOX gene locus and at MEIS1 following 6 days of MI-503 treatment [2.5μM] versus DMSO.
One representative experiment is shown, bar graphs represent averages of three replicates, arrows point to IgG control values, error bars represent SEM. Results were confirmed in two additional independent experiments.
While we did not detect global changes of several chromatin marks upon menin-MLL1-i (Fig.3B), we observed a locus-specific decrease of H3K4me3 at HOX and MEIS1 loci (Fig.3C), thus demonstrating the expected changes in histone methylation due to menin inhibition. Somewhat unexpectedly, we also found a significant reduction of H3K79me2 at those loci (Fig.3D). These data are consistent with decreased binding of the menin-MLL1 complex to HOX and MEIS1 loci following pharmacological menin-MLL1-i and also point to the H3K79 methyltransferase DOT1L as having a potential role in HOX gene regulation in NPM1mut leukemia cells.
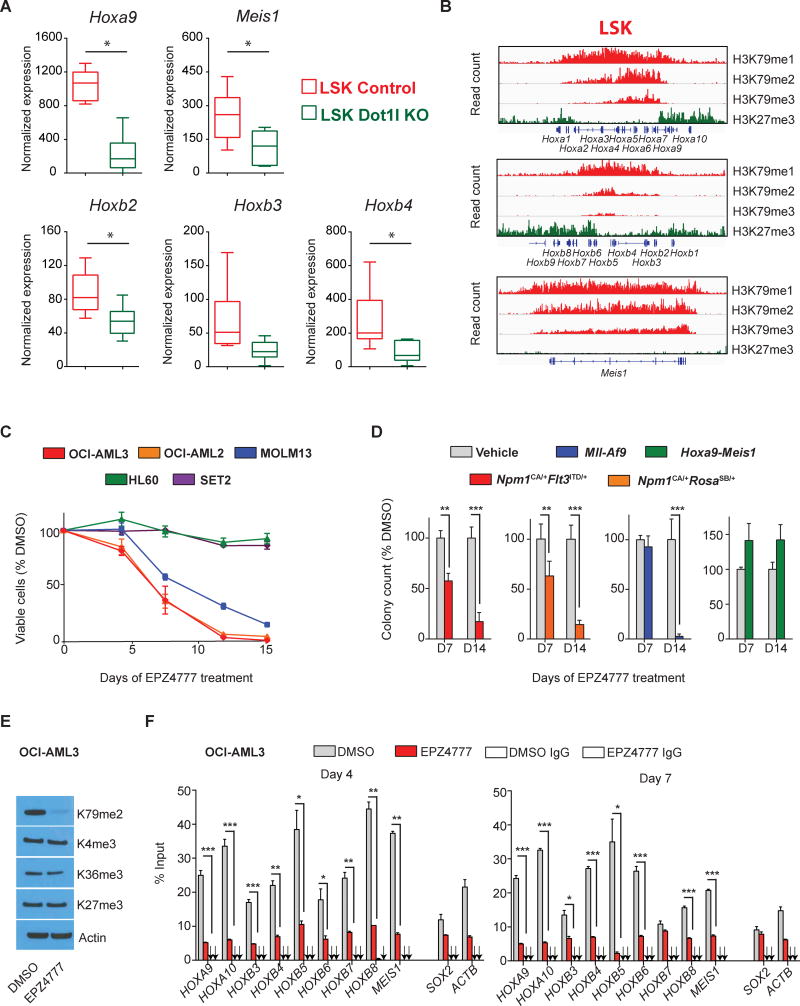
DOT1L is involved in HOXA- and HOXB-cluster regulation during normal hematopoiesis and in NPM1mut leukemia
The data described above suggest wt-MLL1 to be important for HOX gene control in NPM1mut AML, and also implicate DOT1L in HOX gene regulation in this disease. Recent studies of benign murine hematopoiesis demonstrated that Dot1l is another chromatin regulator critical for Hoxa-cluster regulation in early hematopoietic progenitors, whereas its role in Hoxb-cluster control is not well defined(28). To explore whether Dot1l might be involved in the control of Hoxb-cluster genes, we reanalyzed gene expression data from Dot1lfl/fl Mx1-Cre transgenic mice after polyinosinic-polycytidylic acid induced excision compared to their normal counterparts(27). In fact, we found the anterior Hoxb-cluster genes Hoxb2 and Hoxb4 to be significantly down regulated after homozygous deletion of Dot1l (Fig.4A), while there was also a trend for Hoxb3 that did not reach statistical significance. In addition, there was an association between high expression levels of early Hoxb-cluster genes and higher states of H3K79 methylation (H3K79me2 and me3) in normal murine LSKs (Fig.4B) mimicking the H3K79 profile across the expressed Hoxa-cluster genes in these cells. These findings extend the concept of Dot1l being critical for Hoxa-cluster regulation during normal hematopoiesis to the Hoxb-cluster, which together represent also the dominant expression pattern found in NPM1mut leukemias.
Figure 4. DOT1L is required for HOXB-cluster expression in early hematopoietic progenitors and a therapeutic target in NPM1mut leukemia.
(A) Expression of Hoxa9, Meis1, Hoxb2, Hoxb3, and Hoxb4 in LSK cells sorted from Dot1lfl/fl or Dot1lWT/WT mice crossed with Mx1-Cre mice after 10 days of pIpC treatment.
(B) Representative profiles for ChIP-seq using anti-H3K79me1, H3K79me2, H3K79me3, and H3K27me3 antibodies in LSK cells at the Hoxa- and Hoxb-cluster. The y axis scale represents read density per million sequenced reads.
(C) Growth of OCI-AML3, MOLM13, SET2, OCI-AML2, and HL60 cells exposed to EPZ4777 [10μM]. Viable cells were counted and replated at equal cell numbers in fresh media with fresh compound every 3-4 days. Results were plotted as percentage of split-adjusted viable cells in the presence of EPZ4777 [10μM] and normalized to DMSO control.
(D) Colony numbers of Npm1CA/+Flt3ITD/+, Npm1CA/+RosaSB/+, Mll-Af9, and Hoxa9-Meis1 cells exposed to 10μM EPZ4777 and compared to DMSO vehicle control. Data were obtained at day 7 (D7), when viable cells were harvested and replated in fresh methylcellulose with fresh compound and at day 14 (D14) of treatment.
(E) Immunoblotting of global histone marks in OCI-AML3 cells upon 4 days of EPZ4777 treatment [10μM].
(F) H3K79me2 levels across the HOXA- and HOXB-cluster locus and MEIS1 in OCI-AML3 cells after 4 and 7 days of EPZ4777 [10μM] treatment and compared to DMSO control as assessed by ChIP-PCR.
The box plots in (A) shows normalized expression values of 6 mice per group. Whiskers represent minimal and maximal values, the box represents the standard error of the mean, the line represents the median. Data in (C) and (D) represent averages of three independent experiments, each performed in three replicates. Error bars represent the SEM. For ChIP data in F, one representative experiment performed in three replicates is shown, arrows point to the IgG control values, error bars represent the SEM. Results were confirmed in two additional independent experiments.
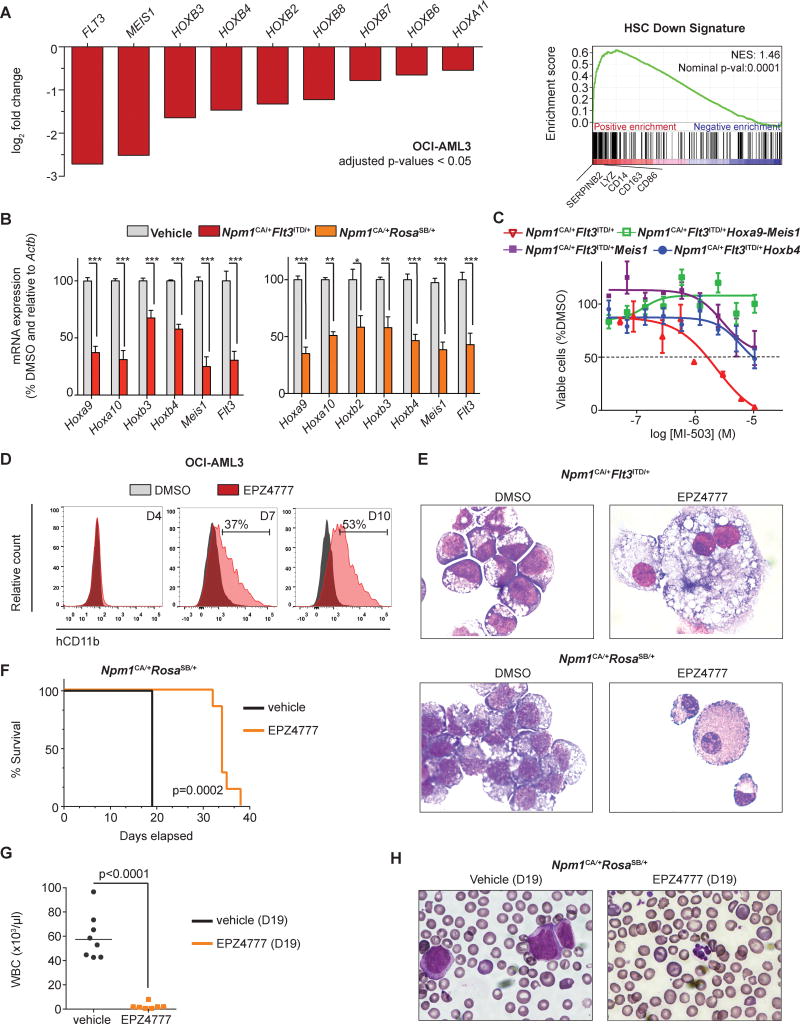
Based on these findings, we assessed the effect of EPZ4777, a small-molecule DOT1L inhibitor in NPM1mut leukemias. DOT1L-inhibition (DOT1L-i) led to a profound reduction in cell proliferation and colony-forming potential in the human and in both murine NPM1mut leukemia models. Similar results were obtained for MLL1-fusion leukemias whereas leukemias lacking an NPM1 mutation or MLL1-fusion such as human HL60 cells and murine Hoxa9-Meis1 in vitro transformed cells were unaffected (Fig.4C+D). Antiproliferative effects of DOT1L-i in the human cells were preceded by global and HOX and MEIS1 locus-specific reduction in H3K79me2 as determined by immunoblotting and ChIP-PCR, whereas global levels of other histone marks associated with transcriptional activation (H3K4me3, H3K36me3) and transcriptional repression (H3K27me2) were unchanged (Fig.4E+F). As hypothesized, DOT1L-i resulted in significant repression of HOX genes in human and murine leukemia cells while the pattern differed slightly among the models (Fig.5A, left panel and Fig.5B). While murine leukemias exhibited significant suppression of Hoxa, Hoxb and Meis1 genes (Fig.5B), the HOXB-cluster was suppressed in human cells but the HOXA-cluster was only mildly affected as assessed by RNA-sequencing (Fig.5A, left panel). Of note, MEIS1 was most profoundly down regulated across all NPM1mut models. This finding was accompanied by dramatic FLT3 suppression (Fig.5A, left panel and 5B). To control the results from the RNA-sequencing analysis for potential bias from standard normalization in the context of global transcriptional changes as they might occur with global changes of chromatin marks, we normalized on synthetic RNA spike-in controls as proposed by Loven and others and compared the results to standard normalization(42). No difference of global transcription levels was noted (data not shown), indicating that global reduction of H3K79me2 is not associated with global transcriptional down regulation in these cells but with suppression of a smaller HOX dominated gene subset. As EPZ4777 treatment resulted in profound suppression of Hox-cluster genes followed by substantial growth inhibition of Npm1mut leukemia cells, we next assessed whether retroviral overexpression of Hoxb4, Meis1, or Hoxa9-Meis1 influences the treatment response to EPZ4777 of these leukemias. As demonstrated for menin-MLL1-i, ectopic expression of these genes abolished sensitivity of Npm1CA/+FLt3ITD/+ cells to DOT1L-i (Fig.5C).
Figure 5. Effects of DOT1L-i on gene expression, cell differentiation, and leukemia initiating potential in NPM1mut AML cells.
(A) (Left panel) Log2 fold change of HOX genes, MEIS1, and FLT3 between OCI-AML3 cells treated for 7 days with 10μM EPZ4777 or DMSO vehicle control as assessed by RNA-sequencing. Only expressed HOXA- and HOXB-cluster genes are shown with a normalized read count of ≥100 reads within the vehicle control, ≥0.5 log2 fold change, and a p-value (adjusted for multiple testing) of < 0.05. (Right panel) GSEA of RNA-sequencing data showing enrichment of genes upregulated with EPZ4777 treatment for genes silenced in normal hematopoietic cord blood stem cells.
(B) Gene expression in murine Npm1CA/+Flt3ITD/+ and Npm1CA/+RosaSB/+ cells assessed on day 7 of EPZ4777 treatment [10μM] by quantitative PCR.
(C) Dose response curves from cell viability assays after 14 days of EPZ4777 treatment comparing Npm1CA/+Flt3ITD/+ cells versus Npm1CA/+Flt3ITD/+ cells overexpressing Hoxb4, Meis1, or Hoxa9-Meis1.
(D) Cell differentiation upon DOT1L-i (EPZ4777 [10μM]) as determined by flow cytometry for CD11b expression in OCI-AML3 cells (at day 0, 4, and 7 of treatment).
(E) Morphological changes in Npm1CA/+Flt3ITD/+ (upper panel) and Npm1CA/+RosaSB/+ (lower panel) cells consistent with monocytic differentiation in murine cells after 14 days of EPZ4777 [10μM] treatment.
(F) Kaplan-Meier survival curve of mice transplanted with pretreated Npm1CA/+RosaSB/+ leukemia cells (vehicle: n= 8 mice/group; EPZ4777: n=7 mice/group).
(G) White blood cell count and (H) morphology of mice transplanted with pretreated Npm1CA/+RosaSB/+ leukemia cells on day 19 post transplantation.
Data in (A) represent averages of three independently treated replicates per group, data in (B), (C), and (D) represent averages of three independent experiments, each performed in three replicates. Error bars represent the SEM.
Since gene expression changes observed in response to EPZ4777 treatment were also consistent with cell differentiation (Fig.5A, right panel), we next assessed the human NPM1mut AML cells for differentiation following DOT1L-i. As expected, we observed a strong increase of CD11b expression as determined by flow cytometry and morphological changes consistent with myeloid differentiation in the OCI-AML3 cells beginning at day 7 of treatment (Fig.5D). Npm1CA/+FLt3ITD/+ and Npm1CA/+RosaSB/+ cells also showed substantial monocytic cell differentiation after 14 days of pharmacological DOT1L-i (Fig.5E).
As HOX gene expression is associated with self-renewal properties, we next determined whether pharmacological DOT1L-i inhibits NPM1mut leukemia initiation in mice. Murine Npm1CA/+RosaSB/+ leukemia cells were treated ex vivo with EPZ4777 or drug vehicle for 10 days and equal numbers of viable cells were transplanted into sublethally irradiated recipient animals. A significant survival advantage was noted for the animals transplanted with treated cells compared to vehicle control in both NPM1mut leukemia models (Fig.5F and Supplemental-Fig.7) and animals transplanted with the EPZ4777-treated Npm1CA/+RosaSB/+ leukemia cells had normal WBC counts at disease onset of the vehicle control group (Fig.5F-H). Similar results were obtained for the Npm1CA/+FLt3ITD/+ cells but with longer disease onset within the treated group (Supplemental-Fig.7). These data are therefore consistent with DOT1L participating in HOX gene regulation and differentiation control of NPM1mut leukemia cells thereby reprogramming these cells towards a state with substantially decreased leukemia initiating and self-renewal properties.
DNMT3A mutations do not account for sensitivity of NPM1mut AML to DOT1L-i
In addition to the NPM1 mutation, OCI-AML3 cells harbor a DNMT3A mutation (R882C), which has been linked to aberrant HOX expression in AML in some studies(4) but was attributed to concomitant NPM1 mutations in others(43). To explore whether DNMT3A mutation status is associated with sensitivity to DOT1L-i, we included two additional cell lines with DNMT3A mutations in our study. While the JAK2 mutated SET2 cells (which harbor the most common AML-associated DNMT3A mutation R882H) did not show sensitivity to DOT1L-i, the OCI-AML2 cells (R635W reported in a single patient(44)) were found to be tremendously sensitive (Fig.4C). To understand these conflicting findings we searched for other genetic factors potentially influencing sensitivity to EPZ4777 in the OCI-AML2 cells. Unexpectedly, RNA-sequencing revealed the presence of an MLL-AF6 fusion transcript with breakpoints that aligned to the common breakpoint cluster region typically affected in MLL-AF6-rearranged leukemias that was confirmed by breakpoint spanning RT-PCR (Supplemental-Fig.8A). Whereas conventional cytogenetic analysis confirmed the normal 11q23 loci as reported, we detected a MLL1-split signal mapping to the derivative chromosome 1q using FISH (Supplemental-Fig.8B+C). These findings are consistent with a cryptic insertion of a functionally relevant MLL-AF6-fusion into the derivative chromosome 1. Also, we found the typical MLL1-fusion related gene signature with high-level HOXA expression that were profoundly down regulated upon DOT1L-i but lack of HOXB-cluster expression (Supplemental-Fig.9). Together, these data support a previous study reporting that DOT1L controls HOXA-cluster expression also in MLL1-fusion leukemia with no apparent DOT1L recruiting activity (such as MLL-AF6)(45). However, whether DNMT3A mutations are associated with sensitivity to DOT1L-i remains to be fully determined.
Combinatorial menin-MLL1-i and DOT1L-i synergistically suppresses HOX, MEIS1, and FLT3 expression and induces differentiation in NPM1mut leukemia
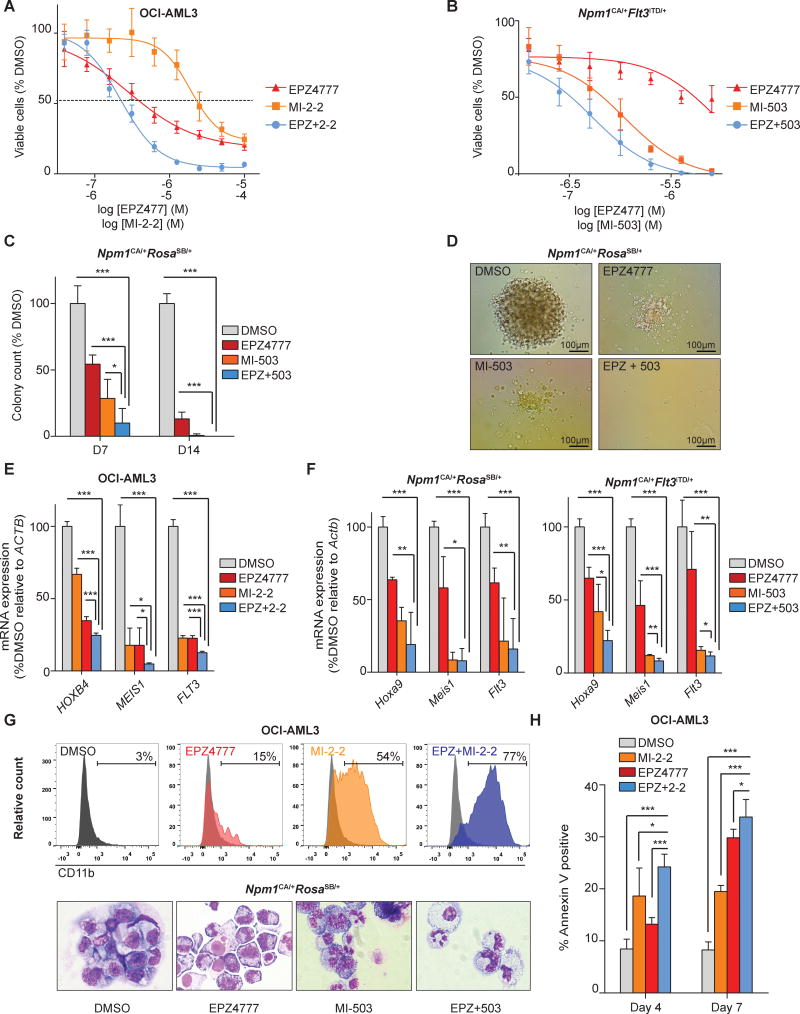
Our data are consistent with the menin-MLL1 interaction and DOT1L being both important regulators that control HOX, MEIS1, and FLT3 expression in NPM1mut AML. We therefore sought to investigate the effects of combinatorial menin-MLL1-i and DOT1L-i in the three models of NPM1mut AML. Combinatorial drug treatment in liquid culture resulted in improved growth inhibition as reflected by a shift of the dose response curve toward lower doses compared to each of the compounds alone in the human OCI-AML3 and the murine Npm1CA/+FLt3ITD/+ cells (Fig.6A+B). Of note, significant drug synergism was demonstrated for both the human and the murine NPM1mut leukemia using the Chou-Talalay algorithm(46) (Supplemental-Fig.10A+B; Supplemental-Table 3+4). Npm1CA/+RosaSB/+ leukemia cells that grow only in methylcellulose also exhibited substantially superior suppression of colony-forming potential when treated with both compounds compared to single drug treatment (Fig.6C+D).
Figure 6. Effects of combinatorial menin-MLL11-i and DOT1L-i in murine and NPM1mut leukemia cells.
(A) Dose response curves from cell viability assays of OCI-AML3 cells comparing 7 days of MI-2-2 [12μM], EPZ4777 [10μM], and combinatorial EPZ4777 [10μM] and MI-2-2 [12μM] (EPZ+2-2) treatment.
(B) Dose response curves from cell viability assays of Npm1CA/+Flt3ITD/+ leukemia cells comparing 11 days of MI-503 [2.5μM], EPZ4777 [10μM], or combinatorial EPZ4777 [10μM] and MI-503 [2.5μM] (EPZ + 503) treatment.
(C) Comparison of MI-503 [2.5μM], EPZ4777 [10μM], and combinatorial MI-503 [2.5μM] and EPZ4777 [10μM] treatment of murine Npm1CA/+RosaSB/+ leukemia cells in colony-forming assays assessed on day 7 (D7) and day 14 (D14) of treatment.
(D) Representative colony formation of Npm1CA/+RosaSB/+ leukemia cells following 7 days of EPZ4777 [10μM], MI-503 [2.5μM] or combinatorial EPZ4777 and MI-503 treatment.
(E) Gene expression changes of selected leukemogenic genes in human OCI-AML3 following 4 days of EPZ4777 [10μM], MI-2-2 [12μM] or combinatorial EPZ4777 and MI-2-2 treatment as assessed by quantitative PCR.
(F) Expression changes of Hoxa9, Meis1, and Flt3 in murine Npm1CA/+RosaSB/+ (left panel) and Npm1CA/+Flt3ITD/+ leukemia cells (right panel) following 4 days of EPZ4777 (10μM), MI-503 (2.5μM) or combinatorial EPZ4777 and MI-503 treatment as assessed by quantitative PCR.
(G) Cell differentiation of NPM1mut leukemia cells as assessed by flow cytometric analysis of CD11b in OCI-AML3 cells (upper panel) and cytology of Npm1CA/+ RosaSB/+ leukemia cells (lower panel). Representative pictures are shown in the lower panel and data were obtained after 4 days of treatment (EPZ4777: [10μM], MI-2-2: [12μM], MI-503: [2.5μM]).
(H) Apoptosis in OCI-AML3 cells following 4 days of EPZ4777 [10μM], MI-2-2 [12μM] or combinatorial EPZ4777 [10μM] and MI-2-2 [12μM] treatment as assessed by flow-cytometric staining for Annexin V.
Data represent averages of two independent experiments, each performed in three replicates (A, B, C, E, F, G, H). Error bars represent the SEM.
Gene expression analysis revealed that growth inhibition was preceded by enhanced suppression of leukemogenic gene expression after 4 days of treatment. While suppression levels of individual HOX genes differed across the human and murine NPM1mut leukemia models, we found that combinatorial treatment consistently suppressed MEIS1 transcript levels more than single drug treatment (Fig.6E+F). Although EPZ4777 reaches full suppression of gene expression not before seven days of treatment, we noted that combination treatment resulted in an additional relative reduction of MEIS1 expression compared to single menin inhibition already after four days (OCI-AML3, 73%, p=0.03; Npm1CA/+Flt3Itd/+, 31%, p=0.001; Npm1CA/+RosaSB/+, 8%, p=0.89). Similarly, we observed enhanced suppression of FLT3 in response to combinatorial treatment, that reached significance in the OCI-AML3 and the Npm1CA/+Flt3Itd/+cells (Fig.6E+F). Consistent with these findings, OCI-AML3 cells treated with the combination regimen exhibited less RNA-Polymerase II binding to the MEIS1 transcriptional start site and gene body region than single-treated cells as assessed by ChIP-PCR (Supplemental-Fig.11) pointing to a direct inhibitory effect of MEIS1 transcription.
Next, we assessed differentiation following combinatorial menin-MLL1-i and DOT1L-i. In OCI-AML3 cells differentiation was dramatically more pronounced with both compounds compared to single drug treatment alone and occurred earlier. After 4 days of treatment 77% of the co-treated OCI-AML3 cells expressed the monocytic differentiation marker CD11b compared to 54%, 15%, and 3% in the MI-2-2, EPZ4777, or vehicle control treated cells, respectively (Fig.6G, upper panel). Flow-cytometric data were validated by cyto-morphological analysis, which revealed more neutrophils or macrophages over blasts with 7 days of combinatorial versus single drug treatment, particularly pronounced in the murine Npm1CA/+RosaSB/+ cells (Fig.6G lower panel and Supplemental-Fig.12). We also detected a significant increase in apoptosis in the OCI-AML3 cells treated with both compounds compared to single treatment alone (Fig.6H). Overall, these data suggest that combinatorial menin-MLL1-i and DOT1L-i has synergistic therapeutic effects in NPM1mut leukemia likely conveyed by enhanced suppression of selected HOX genes, MEIS1, and FLT3 as well as induction of differentiation.
Combinatorial menin-MLL1 and DOT1L inhibitor treatment suppresses primary AML cells in vitro
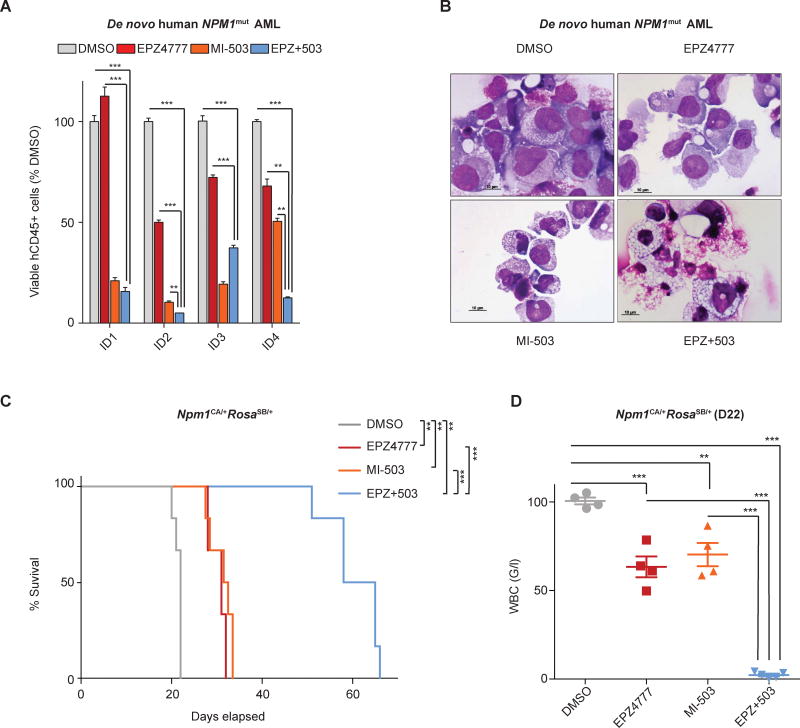
In order to assess the therapeutic potential of combinatorial menin-MLL1-i and DOT1L-i on primary AML patient samples, we used a human stromal cell co-culture assay to maintain and treat patient samples in vitro. Of 5 de novo NPM1mut AML patient samples (see patient details in Supplemental-Table 5), one was not maintainable in culture so the other 4 were used for experiments. 10 days of single drug treatment with MI-503 (2.5μM) or EPZ4777 (10μM) resulted in a profound reduction of viable cell counts compared to DMSO control in all 4 remaining primary AML samples and we observed an enhanced antiproliferative effect for the combinatorial compared to single drug treatment in 3 of 4 samples (Fig.7A). Cell differentiation was also more pronounced with combination therapy (Fig.7B and Supplemental-Fig.13). These data from primary NPM1mut AML patient samples further support single and combinatorial menin-MLL1-i and DOT1L-i as a potentially efficacious therapeutic concept for NPM1mut leukemia.
Figure 7. Effects of single and combinatorial menin-MLL11-i and DOT1L-i on primary NPM1mut AML patient samples and on leukemia initiating potential of murine NPM1mut leukemias.
(A) Viable cell numbers of four independent samples of de novo NPM1mut AML treated in co-culture assays with DMSO, EPZ4777 [10μM], MI-503 [2.5μM], or combinatorial EPZ4777 [10μM] and MI-503 [2.5μM].
(B) Representative pictures of cytospins from de novo NPM1mut AML blasts after 10 days of in vitro treatment with vehicle or EPZ4777 [10μM], MI-503 [2.5μM], or combinatorial EPZ4777+MI-503 [10μM+2.5μM] treatment.
(C) Kaplan-Meier survival curve of mice transplanted with pretreated Npm1CA/+RosaSB/+ leukemia cells comparing drug vehicle, EPZ4777, MI-503, or combinatorial EPZ4777 and MI-503 inhibition (n=6 mice/group).
(D) Engraftment values in the peripheral blood 22 days after transplantation of pretreated Npm1CA/+RosaSB/+ leukemia cells comparing drug vehicle, EPZ4777, MI-2-2, and combinatorial EPZ4777 and MI-503 inhibition (n=4 mice/group).
Bar graphs in (A) represent averages of three replicates assessing one of four independent AML patient samples. Each of the four samples was assessed independently. Error bars represent the SEM.
Combinatorial menin-MLL1-i and DOT1L-i synergistically abolishes NPM1mut leukemia initiating potential
Our in vitro data showed a synergistic drug effect for simultaneous menin-MLL1-i and DOT1L-i in NPM1mut leukemias. To assess the effects of combinatorial drug treatment on leukemia initiating potential, we transplanted equal numbers of viable Npm1CA/+RosaSB/+ leukemia cells pretreated ex vivo with drug vehicle, EPZ4777, MI-503, or the combination into sublethally irradiated recipient mice and assessed for leukemia onset and survival. Whereas both, EPZ4777 and MI-503 pretreatment led to significantly prolonged survival compared to vehicle control, we observed a significant survival advantage for the drug combination compared to either of the compounds alone (Fig.7C). Leukemia onset was also significantly delayed as reflected by significantly lower white cells count or CD45.2 engraftment at the time of death from leukemia within the control group (Fig.7D and Supplemental-Fig.14). These data confirm that both menin-MLL1-i and DOT1L-i diminishes leukemia initiation and indicate that simultaneous inhibition using both compounds shows enhanced effects on leukemia initiating (stem) cells.
Discussion
Despite being associated with high complete remission rates following standard chemotherapy, less than half of the NPM1mut AML patients achieve long-term disease-free survival due to older age at leukemia onset or concomitant adverse disease factors such as mutations in the FLT3 gene(9, 14). Research during the last decade has failed to establish mechanism-based targeted therapies specific for NPM1mut AML, but described distinctive biological features of this disease such as a HOX dominated gene expression signature(4, 6). Efforts to therapeutically target these genes are challenging as HOX and MEIS1 transcription factors are not amenable to direct pharmacological inhibition. Lack of knowledge how aberrant HOX expression is driven and maintained during NPM1mut leukemic transformation has hindered attempts to indirectly target these genes(3).
Here, we have identified the menin-MLL1 interaction and DOT1L as therapeutic targets that control expression of HOX genes, the HOX co-factor MEIS1, and FLT3 in NPM1mut AML. Using a CRISPR-Cas9 negative selection screen interrogating multiple MLL1 protein domains we discovered the menin-LEDGF binding site of MLL1 as a dependency in our screen. This is the same region that is critically involved in chromatin binding of the MLL1 complex(19, 20). Also, we detected a strong phenotype for at least one of the sgRNAs targeting the CXXC domain further pointing to chromatin binding of MLL1 as critically required for NPM1mut leukemias(37). Furthermore, pharmacological menin-MLL1-i diminishes menin enrichment at HOX and MEIS1 loci resulting in dramatic suppression of HOX and MEIS1 expression. Together these findings strongly support the concept that aberrant HOX/MEIS1 expression in NPM1mut AML is regulated via chromatin binding of the menin-wt-MLL1 complex. Whether HOX gene activation is a direct consequence of MLL1 binding to chromatin itself or dependent on other proteins that are indirectly recruited to chromatin via MLL1 remains to be determined. It is of interest to note that none of the sgRNAs targeting the C-terminal SET domain of MLL1 containing the H3K4 methyltransferase activity exhibited any significant phenotype. These data are in line with a recent study on MLL1-fusion leukemias where selective inactivation of the SET domain did not alter transcription of HOX genes or transformation potential(47) but future studies will need to assess the importance of this domain with more detailed experiments. In addition, we found no evidence that the SET domain of MLL2, the other MLL family member interacting with menin is involved in NPM1mut leukemogenesis.
We have also shown that DOT1L is another chromatin modifier important for the control of HOX gene expression in NPM1mut leukemias. We show that not only HOXA and MEIS1 but also HOXB expression is dependent on DOT1L in murine hematopoietic stem cell progenitor cells (LSK cells) and the high expression levels that are associated with higher states of H3K79 methylation at those gene loci point to an important role for DOT1L. While there is evidence that DOT1L is misdirected to HOXA-cluster loci in MLL1-fusion leukemias via the fusion partner protein(30), the data presented here suggest that regulatory mechanisms that normally control HOX gene expression in hematopoietic cells remain critical in NPM1mut AML. Just as we found with inhibition of the menin-MLL1 interaction, we found that pharmacological DOT1L-i resulted in HOX down regulation, cell growth inhibition, and profound differentiation of NPM1mut AML blasts. These data therefore extend the rationale for therapeutic DOT1L-i from MLL1-fusion leukemias to the much more prevalent NPM1mut AML and prompt further study as to the mechanisms by which HOX genes are controlled in various genetics subtypes of AML.
A particularly striking finding was that the profound down regulation of MEIS1 following menin-MLL1-i was accompanied by almost complete suppression of FLT3 expression. FLT3 has been previously shown to be a transcriptional target of MEIS1(38), and thus we believe that this suppression is most likely conveyed via MEIS1 rather than a direct effect caused by MLL1 or DOT1L-i. This observation might be of particular therapeutic importance as up to 66% of NPM1mut AMLs harbor concomitant activating FLT3 mutations associated with adverse outcome(8, 10, 11). Thus, menin-MLL1-i may particularly be attractive for patients exhibiting the unfavorable NPM1mut FLT3-ITD genotype.
Whereas our data provide evidence that HOX genes and MEIS1 in NPM1mut leukemias are regulated on a chromatin level via MLL1 and DOT1L, the specific role of the mutant NPM1 protein with regard to HOX and MEIS1 regulation remains elusive. However, the strong association between HOX expression in human NPM1mut AML or genetic knock-in models of NPM1mut leukemia in mice together with the above presented data suggests that the mutant NPM1 protein acts upstream of menin-MLL1 and DOT1L to deregulate gene expression, at least partially analogous to the functions of MLL1-fusion proteins in MLL-rearranged leukemias. The strong oncogenic transformation potential of HOX genes in general suggests that these genes are likely required for NPM1mut driven leukemogenesis. Whereas formal proof of this view was lacking, our data provide strong support for this concept as exogenous overexpression of selected HOX genes rescues antiproliferative effects of both DOT1L and menin-MLL1 inhibitors. We further show that combinatorial inhibition of menin-MLL1 and DOT1L has enhanced on-target activity as reflected by more profound HOX and MEIS1 suppression. These changes result in a synergistic antiproliferative and boosted differentiation effect, suggesting that therapeutic HOX targeting ultimately releases the differentiation block of NPM1mut AML blasts.
The availability of menin-MLL1 and DOT1L inhibitors should enable relatively quick translation of our findings into clinical testing. However, future studies will determine whether the potent anti-leukemic activity can be best harnessed when introduced into current standard chemotherapy regimens, thereby overcoming possible context-specific escape mechanisms that are frequently observed in AML. A proof of principle study on leukemia cell lines already demonstrated drug synergism of EPZ-5676 with standard chemotherapeutic agents in vitro(48). Another interesting combination partner might be dactinomycin that directly inhibits transcriptional elongation by inhibition of RNA Polymerases and was reported to have activity against NPM1mut leukemia in an anecdotal report on a single patient(49).
In summary, our data show that NPM1mut leukemogenesis is dependent on HOX and MEIS1 expression, which is controlled by specific chromatin regulatory complexes. We further demonstrate that these genes are therapeutically targetable via the menin-MLL1 interaction and the H3K79 methyltransferase DOT1L. Small-molecule inhibition of these two histone modifiers releases the differentiation block of NPM1mut leukemic blasts. Both compounds as single agents or in combination represent novel and possibly less toxic therapeutic opportunities for the relatively common NPM1mut leukemias and represent an attractive concept particularly for patients with the prognostically adverse genotype with concomitant FLT3-ITD and the difficult to treat elderly population.
Methods
Cell Culture and Cell Lines
The AML cell lines OCI-AML3, OCI-AML2, SET2, MOLM13, HL60, as well as 293T cells, and Hs27 cells were maintained under standard conditions. Cell line authentication testing (ATCC) was performed between March and November 2013 and verified identity and purity for all human AML cell lines used in this study. The murine leukemia models Npm1CA/+Flt3ITD/+ and Npm1CA/+RosaSB/+ were described previously39,40 and cells were cultured as previously described(29). With regard to the Npm1CA/+RosaSB/+ model, leukemias with two different sleeping beauty integration sites were used (A: target gene: Csf2, position: 54064482, and target gene: Mll1, position: 44649906; B: target gene: Csf2, position: 54250117; genomic positions are as per Ensemble assembly NCBIm37) for all in vitro studies. Experiments were performed using A as these cells were found to have a slightly higher colony-forming potential in vitro. Confirmation of the results was then performed with B.
CRISPR-Cas9 Screening
The pCW-Cas9 expression construct and the pLKO5.sgRNA.EFS.GFP vector were purchased from Addgene (#50661 and #57822). All sgRNAs in this study were designed using the Zhang laboratory's CRISPR design webpage at MIT and sequences are listed in Supplemental-Table 1+2. OCI-AML3-pCW-Cas9 cells were derived by retroviral transduction of OCI-AML3 cells with pCW-Cas9, followed by puromycin selection. Cells were plated in methylcellulose to obtain single cell-derived clones, and screened for Cas9 expression by immunoblotting using an anti-flag antibody. The MLL1 and MLL2 domain screen was performed in duplicates and repeated with two independent clones. Three days following sgRNA transduction Cas9 was induced and GFP+/GFP--ratio determined every three days for 15 days as described(34). For more details see supplemental methods.
In Vitro Studies
RNA purification, cDNA synthesis, qRT-PCR, immunoblotting, ChIP-PCR, flow cytometry, retroviral overexpression, and in vitro drug treatment in colony-forming or cell viability assays was performed using standard procedures. A detailed description is provided in the supplement. Drug synergy was assessed in viability assays comparing single versus constant ratios of combinatorial drug treatment using the Chou-Talalay method(46) and is also described in detail in the supplement.
RNA-sequencing and analysis
For gene expression analysis biological triplicates of OCI-AML3 cells were treated for 7 days with DMSO or 10μM EPZ4777. ERCC synthetic spike-in controls were added to cell lysis buffer of each sample based on cell number and prior to RNA isolation. For fusion detection triplicates of untreated OCI-AML2 cells were harvested for RNA purification. See supplemental methods for RNA purification, library construction, and sequencing analysis. Data were made publicly available at Gene Expression Omnibus (accession code: GSE85107).
OCI-AML3 xenograft model
For in vivo treatment experiments 7-10 week female NSG mice were injected via tail vein with 5×106 OCI-AML3 cells. Animals were randomized to vehicle (25% DMSO, 25% PEG 400, and 50% normal saline) or MI-503 (50mg/kg bid IP) starting on day 5 post transplantation. For assessment of leukemia burden, mice were sacrificed between 7 and 12 days of drug treatment and harvested bone marrow cells assessed for human CD45 expression using flow-cytometry. For gene expression analysis, RNA was isolated from MACS sorted human CD45+ cells. For survival analysis, treatment was initiated on day 7 for 26 days in total with a 3 day treatment break to allow partial recovery from local irritation at the injection sites. Mice were monitored daily for clinical symptoms. Moribund animals were euthanized when they displayed signs of terminal leukemic disease. All mouse experiments were approved by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center.
Murine Bone Marrow Transplantation Models
In vivo treatment studies
1×106 Npm1CA/+RosaSB/+ (model A) secondary leukemia cells, harvested from moribund animals were transplanted into sublethally irradiated (200cGy) female NSG mice via tail vein injection. Mice were randomized to vehicle or MI-503 (50mg/kg, bid, 10 days IP) and treatment initiated 7 days post transplantation.
Ex vivo treatment studies
a) Npm1CA/+Flt3ITD/+ and Npm1CA/+RosaSB/+ (model A) secondary leukemia cells were treated with DMSO or EPZ4777 (10μM) for 11 days in tissue culture or b) Npm1CA/+RosaSB/+ secondary leukemia cells for 10 days with either DMSO, EPZ4777 (10μM) or 4 days of MI-503 (2.5μM) plus 6 days of DMSO or 6 days of EPZ4777 (10μM) plus 4 days of combinatorial EPZ4777 (10μM) + MI-503 (2.5μM). 1×106 pretreated viable cells of each group were then transplanted into sublethally irradiated female NSG (Npm1CA/+RosaSB/+) or C57BL/6J (irradiated with 600cGy; Npm1CA/+Flt3ITD/+) recipient animals.
Human AML Co-Culture Assay
The Co-culture treatment assay of primary human AML samples was performed as previously reported(50) and is described in the supplement. Human AML samples were obtained under institutional review board-approved protocols from patients treated at Memorial Sloan Kettering Cancer Center following written informed consent.
Data Analysis and Statistical Methods
Statistical test computations were performed using Graph Pad Prism (v.6). Statistical significance for the in vitro assays was calculated using the Student's t-test. Survival analysis was estimated using the Kaplan-Meier method and p-values were calculated using the log-rank test. P-values were displayed as follows: *p < 0.05, **p < 0.005, ***p < 0.0005. In vitro assessment of menin-MLL1-i in the NPM1mut AML models was performed in at least two independent experiments each performed in three replicates using one of the two menin-MLL1 inhibitors (MI-2-2 or MI-503). In addition, results for most experiments were once confirmed with the other menin-MLL1 inhibitor. In vitro studies related to DOT1L-i were performed in three independent experiments each performed in three replicates.
Supplementary Material
Statement of Significance.
MLL1 and DOT1L are chromatin regulators that control HOX, MEIS1 and FLT3 expression and are therapeutic targets in NPM1mut AML. Combinatorial small-molecule inhibition has synergistic on target activity and constitutes a novel therapeutic concept for this common AML subtype.
Acknowledgments
We thank members of the Armstrong Laboratory for helpful comments on the manuscript; Rowena Eng, Huiyong Zhao, and Michael Hadler for excellent technical assistance; Olga Guryanova and Ross Levine for the SET2 and OCI-AML2 cell lines.
Financial Support: Supported by grants (to S.A.A.) from the National Institute of Health (CA66996, CA140575, and CA176745), the Leukemia and Lymphoma Society, E. d. S was supported by the NIH grant (P30 CA008748), and M.W.M.K. was supported by the German Research Foundation (DFG; KU 2688/1-1).
Footnotes
Disclosures: S.A.A is a consultant to Epizyme and Vitae Pharmaceuticals. All other authors declare no competing financial interests.
Author's Contribution: M.W.M.K. designed and conducted research, analyzed, and interpreted data, wrote the paper. E.S. performed in vivo experiments. Z.F. conducted research. A.S. performed computational RNA-sequencing analysis. C.W.C and A.D. designed experiments and developed methodology. M.C. conducted research. N.F. performed computational RNA-sequencing analysis. A.P. and C.G. provided NPM1mut leukemia models. J.E.B. synthesized small-molecule inhibitor. E. d. S. designed in vivo experiments and developed methodology. G.S.V. engineered and provided NPM1mut leukemia models. T.H. designed and cloned sgRNAs for MLL1 and MLL2 CRISPR domain screen, interpreted data. S.A.A. designed research, analyzed and interpreted data, wrote the paper.
References
- 1.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12223–7. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giampaolo A, Sterpetti P, Bulgarini D, Samoggia P, Pelosi E, Valtieri M, et al. Key functional role and lineage-specific expression of selected HOXB genes in purified hematopoietic progenitor differentiation. Blood. 1994 Dec 1;84(11):3637–47. [PubMed] [Google Scholar]
- 3.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013 Apr;27(5):1000–8. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 4.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015 Jun;29(6):1279–89. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007 Oct 15;26(47):6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007 Sep;21(9):2000–9. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 7.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005 Aug 1;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 8.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005 Jan 20;352(3):254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 9.Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005 Dec 1;106(12):3733–9. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 10.Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med. 2016 Feb 4;374(5):422–33. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 11.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008 May 1;358(18):1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 12.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005 Dec 1;106(12):3747–54. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 13.Daver N, Cortes J, Ravandi F, Patel KP, Burger JA, Konopleva M, et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015 May 21;125(21):3236–45. doi: 10.1182/blood-2014-10-605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostronoff F, Othus M, Lazenby M, Estey E, Appelbaum FR, Evans A, et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: a SWOG and UK National Cancer Research Institute/Medical Research Council report. J Clin Oncol. 2015 Apr 1;33(10):1157–64. doi: 10.1200/JCO.2014.58.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federici L, Falini B. Nucleophosmin mutations in acute myeloid leukemia: a tale of protein unfolding and mislocalization. Protein science : a publication of the Protein Society. 2013 May;22(5):545–56. doi: 10.1002/pro.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS letters. 2001 Oct 12;506(3):272–6. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 17.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007 Nov;7(11):823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 18.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006 Aug 17;442(7104):818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008 Jul 8;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005 Oct 21;123(2):207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Shi A, Murai MJ, He S, Lund G, Hartley T, Purohit T, et al. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 2012 Nov 29;120(23):4461–9. doi: 10.1182/blood-2012-05-429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015 Apr 13;27(4):589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995 Nov 30;378(6556):505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 24.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004 Mar;6(3):437–43. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 25.Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, Silva A, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci U S A. 2006 Jan 24;103(4):1018–23. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novotny E, Compton S, Liu PP, Collins FS, Chandrasekharappa SC. In vitro hematopoietic differentiation of mouse embryonic stem cells requires the tumor suppressor menin and is mediated by Hoxa9. Mech Dev. 2009 Jul;126(7):517–22. doi: 10.1016/j.mod.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011 Jul 12;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014 Dec 8;26(6):896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CW, Koche RP, Sinha AU, Deshpande AJ, Zhu N, Eng R, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med. 2015 Apr;21(4):335–43. doi: 10.1038/nm.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshpande AJ, Bradner J, Armstrong SA. Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. Trends in immunology. 2012 Nov;33(11):563–70. doi: 10.1016/j.it.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013 Aug 8;122(6):1017–25. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011 Jul 12;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein EM, Tallman MS. Mixed lineage rearranged leukaemia: pathogenesis and targeting DOT1L. Curr Opin Hematol. 2015 Mar;22(2):92–6. doi: 10.1097/MOH.0000000000000123. [DOI] [PubMed] [Google Scholar]
- 34.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology. 2015 Jun;33(6):661–7. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, Wan K, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012 Feb 23;482(7386):542–6. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010 Jan;17(1):62–8. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007 May 4;282(18):13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 38.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005 Jul 1;106(1):254–64. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011 May;43(5):470–5. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mupo A, Celani L, Dovey O, Cooper JL, Grove C, Rad R, et al. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia. 2013 Sep;27(9):1917–20. doi: 10.1038/leu.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL1 inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nature chemical biology. 2012 Mar;8(3):277–84. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, et al. Revisiting global gene expression analysis. Cell. 2012 Oct 26;151(3):476–82. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012 Jun 14;119(24):5824–31. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 44.Gaidzik VI, Schlenk RF, Paschka P, Stolzle A, Spath D, Kuendgen A, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013 Jun 6;121(23):4769–77. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 45.Deshpande AJ, Chen L, Fazio M, Sinha AU, Bernt KM, Banka D, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013 Jan 29; doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006 Sep;58(3):621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 47.Mishra BP, Zaffuto KM, Artinger EL, Org T, Mikkola HK, Cheng C, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell reports. 2014 May 22;7(4):1239–47. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klaus CR, Iwanowicz D, Johnston D, Campbell CA, Smith JJ, Moyer MP, et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. The Journal of pharmacology and experimental therapeutics. 2014 Sep;350(3):646–56. doi: 10.1124/jpet.114.214577. [DOI] [PubMed] [Google Scholar]
- 49.Falini B, Brunetti L, Martelli MP. Dactinomycin in NPM1-Mutated Acute Myeloid Leukemia. N Engl J Med. 2015 Sep 17;373(12):1180–2. doi: 10.1056/NEJMc1509584. [DOI] [PubMed] [Google Scholar]
- 50.Kühn MWM, Hadler MJ, Daigle SR, Koche RP, Krivtsov AV, Olhava EJ, et al. MLL partial tandem duplication leukemia cells are sensitive to small molecule DOT1L inhibition. Haematologica. 2015 May;100(5):e190–3. doi: 10.3324/haematol.2014.115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.