ABSTRACT
Magnaporthe oryzae, the ascomycete fungus that causes rice blast disease, initiates conidiation in response to light when grown on Prune-Agar medium containing both carbon and nitrogen sources. Macroautophagy/autophagy was shown to be essential for M. oryzae conidiation and induced specifically upon exposure to light but is undetectable in the dark. Therefore, it is inferred that autophagy is naturally induced by light, rather than by starvation during M. oryzae conidiation. However, the signaling pathway(s) involved in such phototropic induction of autophagy remains unknown. We identified an M. oryzae ortholog of GCN5 (MGG_03677), encoding a histone acetyltransferase (HAT) that negatively regulates light- and nitrogen-starvation-induced autophagy, by acetylating the autophagy protein Atg7. Furthermore, we unveiled novel regulatory mechanisms on Gcn5 at both transcriptional and post-translational levels, governing its function associated with the unique phototropic response of autophagy in this pathogenic fungus. Thus, our study depicts a signaling network and regulatory mechanism underlying the autophagy induction by important environmental clues such as light and nutrients.
KEYWORDS: Atg7, autophagy, conidiation, Gcn5, histone acetyltransferase (HAT), Magnaporthe oryzae
Introduction
Rice-blast disease caused by the filamentous ascomycete M. oryzae is responsible for significant crop losses globally and annually.1 M. oryzae forms aerial hyphae (the hyphae that grow perpendicular into the air from the vegetative mycelial mat), when grown in darkness; whereas conidiation, formation of asexual spores, is initiated in response to light exposure.2 These fungal asexual spores known as conidia are the key determinant of the spread and severity of blast disease as they can be easily dispersed and transmitted by air.3 Functional analysis of several autophagy genes in M. oryzae, including MoATG1,4 MoATG4,5 MoATG5,6 MoATG8,7,8 MoATG9,9 and MoATG24,10 etc, has established the link between autophagy and fungal development. A systematic characterization of 16 genes essential for nonselective autophagy in M. oryzae demonstrates that autophagy is important for the establishment of rice blast disease.11 Overall these studies demonstrate a critical role of autophagy, including nonselective autophagy and some selective types of autophagy (e.g. mitophagy), in M. oryzae conidiation and/or infection. Autophagy likely serves diverse functions including programmed cell death, maintaining integrity of lipid bodies, and glycogen catabolism.4,7,8
Autophagy is a highly conserved catabolic process in eukaryotes, responsible for vacuolar (lysosomal) degradation of proteins, membranes and organelles. Autophagy is induced during several biological processes in response to environmental stress or pathogen invasion, and cellular remodeling during development and differentiation.12-14 The molecular basis of autophagy has been thoroughly investigated in yeasts and mammalian cells, by identification and functional characterization of 41 ATG genes (AuTophaGy) thus far.15-17 Among these genes, ATG8 has been established as the most reliable marker for autophagy induction and autophagy-associated vesicular compartments.18-20 MoATG8, the homolog of the yeast ATG8, is required for proper conidiation and pathogenicty in M. oryzae, and autophagy is naturally induced by light, the external stimulus for conidiation.8 Light-induced autophagy can be visualized as vesicular RFP-Atg8 signal distributed in vegetative mycelia as well as conidiation-related structures, including aerial hyphae, conidiophore and conidia.8 The findings pave the way to investigate the molecular mechanisms underlying such light-induced autophagy.
Recent studies unveil that histone acetyltransferase (HATs) and the histone deacetylases (HDACs) can directly modify autophagy proteins to regulate autophagy induction.21-23 Our previous study on phototropism in M. oryzae identifies a HAT-encoding gene, GCN5, as light responsive.24 HATs catalyze acetylation of histones on specific lysine residues,25 and thus epigenetically regulate global gene transcription, stress response, and metabolic flux in plants26-28 and yeasts.29,30 The family of GNATs (GCN5-related N-acetyltransferases) also has a function in drug resistance in bacterial pathogens.31,32 Recent clinical evidence suggests that Gcn5 may play a role in the pathology of cancer, asthma, COPD (chronic obstructive pulmonary disease) and Parkinson disease.33,34
In this study, we identified Gcn5 as an autophagy repressor in M. oryzae. The Gcn5 protein may shuttle between the cytosol and nucleus, and undergo post-translational cleavage and/or degradation in response to light. Our results suggest that Gcn5 acetylates Atg7 and thus represses autophagy in dark. Furthermore, Gcn5 may also regulate transcription of conidiation-related transcriptional regulator gene TFB5. Taken together, we propose that light induces TFB5 transcription via Gcn5, and meanwhile derepresses autophagy by removing the Gcn5-catalyzed acetylation on Atg7, to promote asexual reproduction in the rice blast fungus.
Results
Identification of the GCN5 genes in M. oryzae
We found 2 GCN5 genes, MGG_03677 and MGG_11716 in the M. oryzae genome. Sequence identity and similarity between these 2 Gcn5 proteins was 69.7% and 78.9%, respectively, as predicted by Needle (http://www.ebi.ac.uk/Tools/psa/emboss_needle/; Fig. S1). We named MGG_03677 as GCN5, and MGG_11716 as GCN5b. Given that our previous study on phototropism in M. oryzae identifies MGG_03677 as a light-inducible gene24 while MGG_11716 does not seem to respond to light exposure at the transcriptional level (data not shown), we focused here on GCN5 (MGG_03677) as a candidate gene to investigate phototropic induction of autophagy and conidiation in M. oryzae.
Gcn5 negatively regulates autophagy
To investigate the relationship between Gcn5 (which hereafter refers exclusively to MGG_03677) and autophagy during light-induced conidiation, we generated the gcn5Δ mutant by homologous recombination (Fig. S2A), and the GCN5OX strain (that overexpresses an N-terminal tagged GFP-Gcn5 fusion protein), both in an RFP-ATG8 background. The gcn5Δ mutant, and the GCN5OX strain were verified by Southern blot (Fig. S2B), and the transcriptional level of GCN5 in the gcn5Δ mutant or GCN5OX strain was examined by RT-PCR, with the wild-type (WT) RFP-ATG8 strain as control (Fig. S2C).
Next, we examined phototropic induction of autophagy in the gcn5Δ and GCN5OX strains. Autophagy was induced by light in the WT strain, visualized as punctate or vacuolar RFP-Atg8 signals (Fig. 1A), whereas in the gcn5Δ mutant, RFP-Atg8 appeared as small vesicles (autophagosomes) or vacuoles in both dark or light culture conditions (Fig. 1A). In contrast, RFP-Atg8 was undetectable in the GCN5OX mycelia grown either in presence or absence of light (Fig. 1A). The immunoblot analysis supported our interpretation that Gcn5 represses autophagy in M. oryzae. In WT, the amount of RFP peptide (cleaved from RFP-Atg8 fusion protein when autophagosomes fused with vacuoles; the band at the size of 26 kDa) was elevated/induced in response to light (Fig. 1B), suggesting that autophagy induction was enhanced. In the GCN5OX strain, however, little or no RFP band was detected in dark or light conditions (Fig. 1B). Autophagy activity was elevated upon loss of Gcn5, as RFP was detected in both dark and light conditions in the gcn5Δ mutant (Fig. 1B). Overall, we conclude that light-induced autophagy is repressed by Gcn5, although GCN5 was initially identified as a light-inducible gene in M. oryzae.
Figure 1.
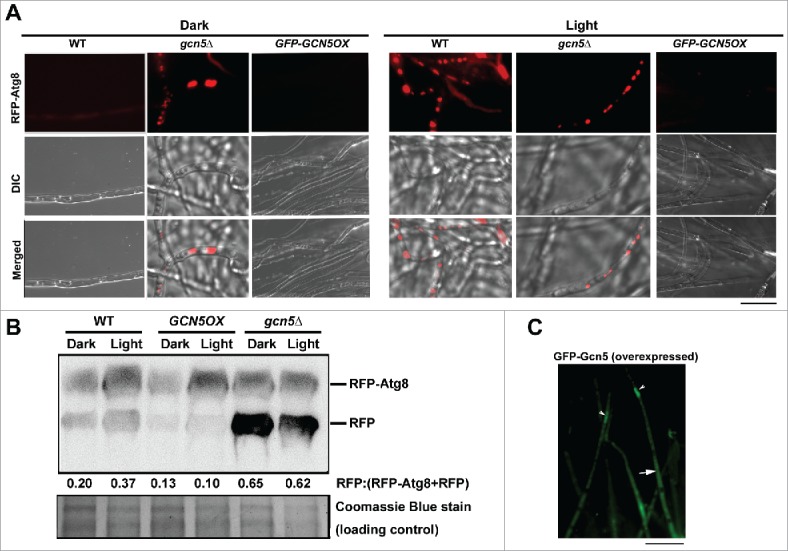
Gcn5 represses light-induced autophagy. (A) RFP-Atg8 in dark- or light- cultured mycelia of the WT, gcn5Δ and GCN5OX strains. Scale bar: 5 μm. (B) Total protein lysates from the indicated strains were analyzed by immunoblotting with anti-RFP antibodies, under light or dark conditions. The extent of autophagy was estimated by calculating the amount of free RFP compared with the total amount of intact RFP-Atg8 and free RFP (the numbers appear underneath the blot). Densitometric analysis was performed using ImageJ (https://imagej.nih.gov/ij/). (C) GFP-Gcn5 signal in the GCN5OX strain appears nuclear (arrowhead) as well as cytosolic (arrow). Scale bar: 5 μm. DIC, differential interference contrast.
We examined the subcellular localization of the Gcn5 protein in either light or dark conditions, by visualizing the overexpressed GFP-Gcn5 signal. GFP-Gcn5 appeared cytosolic (Fig. 1C, arrow) as well as nuclear (Fig. 1C, arrowhead). We costained the GFP-GCN5 mycelia with the fluorescent dye DAPI to verify the nuclear localization. Punctate GFP-Gcn5 colocalized well with the DAPI-stained nuclear compartment (Fig. S2D), thus confirming its nuclear localization. We infer that Gcn5, the histone modifier, likely moonlights as a cytosolic protein during asexual development in M. oryzae.
To test whether the repression on autophagy by Gcn5 is through transcriptional regulation of the ATG8 gene, we performed RT-PCR using total RNA from the mycelial cultures of WT, GCN5OX and gcn5Δ mutant exposed to light for 0, 2, 4, and 6 h. The input of total RNA was normalized and served as loading control (Fig. S2E). ATG8 transcripts at different time points of light exposure were in general comparable in the same strain (Fig. S2E), which seems to rule out the possibility that the light induced Gcn5 may regulate autophagy via repressing ATG8 transcription. However, we noticed that ATG8 transcripts were overall lower in the gcn5Δ mutant than that in the WT or the GCN5OX strain (Fig. S2E), indicating that Gcn5, the histone modifier and transcriptional activator, may at least partially play a role in activating ATG8 transcription. Nevertheless, given that autophagy was hyperinduced, instead of reduced, in the gcn5Δ mutant, compared with that in the WT (Fig. 1A and B), and that Gcn5 might be partially required for transcriptional expression of ATG8, we conclude that repression of autophagy by Gcn5 does not occur via transcriptional modulation of ATG8.
Considering that starvation is a well-established external stimulus to induce autophagy,35 although M. oryzae conidiation could not be induced solely by starvation in dark (our unpublished data), we next asked whether starvation-induced autophagy is repressed by Gcn5. The vegetative mycelia of WT, gcn5Δ and GCN5OX mutants were cultured in CM (nitrogen replete) and shifted to MM-N (nitrogen starvation) for further 6 h to induce autophagy. Interestingly, autophagy was induced in the gcn5Δ mutant even in CM, while induction of autophagy was repressed by overexpressed GCN5, under nitrogen starvation as shown by microscopic analysis (Fig. 2A). Under extended nitrogen starvation (12 h), RFP-Atg8 appeared as faint punctate structures in the cytosol, and was undetectable in the vacuoles in the GCN5OX mutant (Fig. S2F). In contrast, WT mycelia showed spherical vacuoles, with weak RFP signal in its lumen (Fig. S2F, arrowheads) under such extended starvation. We inferred that prolonged nitrogen starvation in the presence of continuous light, results in an incomplete or aberrant induction of autophagy, as visualized by the RFP-Atg8 puncta that failed to fuse with the vacuoles. Such RFP-Atg8 puncta likely represent incomplete PAS (phagophore assembly site) structures. Biochemical analysis using CM (nitrogen-replete) cultured and nitrogen starved (12 h) mycelia of WT, GCN5OX and gcn5Δ strains confirmed that nitrogen starvation based autophagy induction was also repressed by overexpressed GCN5 while hyperactivated with loss of the GCN5 gene (Fig. 2B). Cumulatively, the above results demonstrate that Gcn5 also represses autophagy in M. oryzae under nitrogen starvation conditions.
Figure 2.
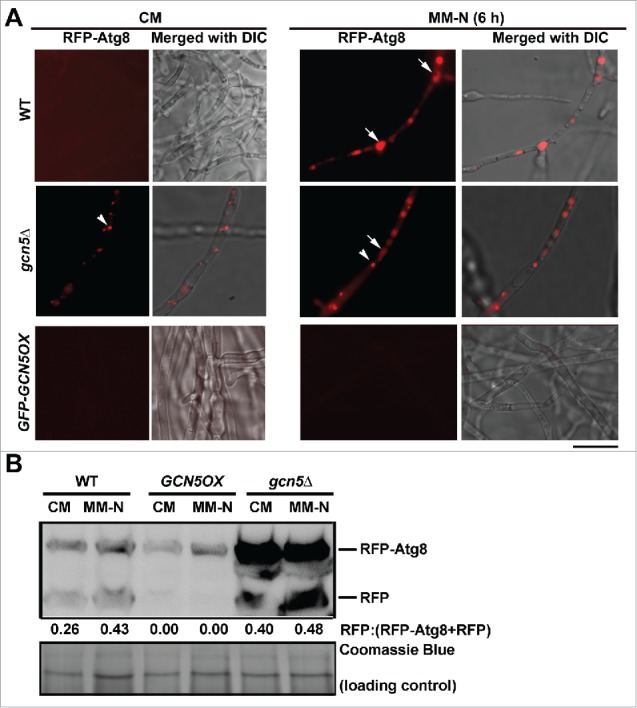
Gcn5 represses starvation-induced autophagy. (A) Gcn5 negatively regulates autophagy under nitrogen starvation conditions. RFP-Atg8 in CM- or MM-N-cultured mycelia of the WT, gcn5Δ and GCN5OX strains. Scale bar: 5 μm. (B) Total protein lysates from WT, gcn5Δ and GCN5OX strain were analyzed by immunoblotting with anti-RFP antibodies. The extent of autophagy was estimated by calculating the amount of free RFP compared with the total amount of intact RFP-Atg8 and free RFP (the numbers appear underneath the blot). Densitometric analysis was performed using ImageJ (https://imagej.nih.gov/ij/).
Gcn5 represses autophagy by acetylation of Atg7
We screened for potential regulator(s) of autophagy in response to light by immunoprecipitation using GFP-Gcn5 as bait (Fig. S2G), and identified the immunoprecipitated (IP) proteins by mass spectrometry. GFP and Gcn5 proteins were among the identified proteins thus further validating the reliability of our earlier results. As expected, histones 2B, 3 and 4 were all identified as the Gcn5-interacting proteins (data not shown), confirming the involvement of Gcn5 in acetylation of histones. More importantly, we identified several proteins with established functions in autophagy regulation, including autophagy proteins Atg8 and Atg27 (Table 1), which might also physically interact with Gcn5.
Table 1.
List of identified Gcn5-interacting proteins related to autophagy.
| Gene ID/name | Uniprot ID | MW | Mascot score* | E-value |
|---|---|---|---|---|
| MGG_00446/CK2 subunit β (beta) | L7IH74 | 38927.6 | 67.7 | 3.14E-06 |
| MGG_03696/CK2 subunit α (alpha) | L7J0C6 | 39488.7 | 66.54 | 1.15E-05 |
| MGG_05651/CK2 subunit β (beta)-2 | G4MNM6 | 31287.6 | 51.26 | 2.96E-05 |
| MGG_06962/Ypt1 | G4MNT9 | 70759.72 | 40.21 | 6.39E-03 |
| MGG_06154/Ras-like protein | G4MZY8 | 24097.9 | 46.72 | 4.73E-02 |
| MGG_07176/GTP-binding protein Rho1 | L7I1M4 | 21830 | 54.13 | 1.64E-04 |
| MGG_01607/Calnexin | L7IX88 | 63854.2 | 48.9 | 2.97E-04 |
| MGG_06860/Coatomer subunit β (beta) | L7J3Z0 | 106213 | 31.45 | 1.99E-02 |
| MGG_14666/SEC13 | L7JMJ6 | 31915.1 | 40.22 | 4.92E-03 |
| MGG_06910/SEC23 | A4R1J7 | 85917.3 | 31.25 | 5.29E-02 |
| MGG_01062/Atg8 | B6VCT7 | 14369.3 | 29.14 | 5.27E-02 |
| MGG_09565/Pmk1 | G4N0Z0 | 41303 | 27.08 | 6.40E-02 |
| MGG_09499/Ras-2 | G4N1S3 | 26891.3 | 30.03 | 2.54E-02 |
| MGG_07145/Cullin-1 | L7HTP0 | 88223.7 | 23.23 | 6.73E-02 |
| MGG_09564/SEC24 | L7I4V3 | 116723 | 20.02 | 1.38E-01 |
| MGG_04830Vps26B | L7IJI1 | 35801.6 | 22.41 | 1.04E-01 |
| MGG_02386/Atg27 | L7IPM7 | 38353.7 | 49.08 | 2.56E-04 |
| MGG_03511/Coatomer subunit α (alpha) | L7IXA5 | 136066 | 21.49 | 2.19E-01 |
| MGG_09294/Coatomer subunit ζ (Zeta) | L7J2Q7 | 22247.3 | 20.92 | 3.72E-02 |
| MGG_00345/Rim15 kinase | L7JKF9 | 212094 | 29.23 | 7.46E-03 |
Masct score ≥20 as threshold.
We further compared the acetylated proteins differentially present in GCN5OX and gcn5Δ mutants, and identified the possible acetylated lysine residue(s), by LC-MS/MS. Selected acetylated proteins and the predicted acetylated residues are listed in Table 2. As expected, histones 2A, 2B, 2A.Z, 3, 3-like and 4 were identified, indicating the reliability of the LC-MS/MS analyses. Noticeably, most of the acetylated lysine residues were present in both GCNOX and gcn5Δ samples, and at comparable levels (0.5<intensity ratio<2, Table 2). This suggests that acetylation of histone proteins did not depend entirely on the Gcn5 function. Among the acetylated proteins identified, we noticed that Atg7, encoded by MGG_07297, contained an acetylated lysine residue at 338, which was present only in the GCN5OX strain but absent in gcn5Δ (Fig. 3A; Table 2). This indicated that acetylation of Atg7 at K338 might be specifically catalyzed by Gcn5. Interestingly, another identified acetylated site, Lysine107, did not depend on Gcn5 function as it was acetylated in the gcn5Δ mutant as well as in the GCN5OX strain (Table 2).
Table 2.
List of predicted acetylated proteins and sites.
| Protein ID | Protein | Sequence window | Score | Modified sequence | Charge | m/z | Intensity OX/delt |
|---|---|---|---|---|---|---|---|
| Histone proteins | |||||||
| >tr|G5EHN4|G5EHN4_MAGO7 | Histone H4 | ___MTGRGKGGKGLGKGGAKRHRKILRDNIQ | 151.44 | _GLGK(ac)GGAK(ac)R_ | 2 | 464.272 | 0.65 |
| TGRGKGGKGLGKGGAKRHRKILRDNIQGITK | 151.44 | _GLGK(ac)GGAK(ac)R_ | 2 | 464.272 | 0.65 | ||
| _______MTGRGKGGKGLGKGGAKRHRKILR | 90.696 | _GK(ac)GGK(ac)GLGK_ | 2 | 443.261 | 0.78 | ||
| >tr|L7ICZ8|L7ICZ8_MAGOY | Histone H3 | RKSTGGKAPRKQLASKAARKSAPSTGGVKKP | 185.6 | _QLASK(ac)AAR_ | 2 | 443.759 | 0.55 |
| YKPGTVALREIRRYQKSTELLIRKLPFQRLV | 285.83 | _YQK(ac)STELLIR_ | 2 | 646.864 | 0.26 | ||
| TKQTARKSTGGKAPRKQLASKAARKSAPSTG | 185.6 | _K(ac)QLASK(ac)AAR_ | 2 | 528.812 | 0.53 | ||
| _MARTKQTARKSTGGKAPRKQLASKAARKSA | 138.08 | _STGGK(ac)APR_ | 2 | 408.222 | 1.18 | ||
| ______MARTKQTARKSTGGKAPRKQLASKA | 138.08 | _K(ac)STGGK(ac)APR_ | 2 | 493.275 | 0.77 | ||
| >tr|G4N793|G4N793_MAGO7 | H3-like centromeric protein cse-4 | YRPGTLALREIRRYQKSTDLLMRKLPFARLV | 110.66 | _RYQK(ac)STDLLMR_ | 3 | 484.926 | 0.00 |
| >sp|L7I1W3|H2B_MAGOY | Histone H2B | ___MPPKAADKKPASKAPATASKAPEKKDAG | 168.98 | _PASK(ac)APATASK(ac)APEK_ | 2 | 769.415 | 1.03 |
| ________MPPKAADKKPASKAPATASKAPE | 127.76 | _AADK(ac)K(ac)PASK_ | 2 | 500.277 | 1.22 | ||
| APATASKAPEKKDAGKKTAASGDKKKRTKTR | 135.02 | _DAGK(ac)K(ac)TAASGDK_ | 2 | 616.81 | 1.32 | ||
| >sp|L7HZV6|H2A_MAGOY | Histone H2A | __________MTGGGKSGGKASGSKNAQSRS | 113.4 | _(ac)TGGGK(ac)SGGK(ac)ASGSK_ | 2 | 652.826 | 0.74 |
| GGKASGSKNAQSRSSKAGLAFPVGRVHRLLR | 58.674 | _SSK(ac)AGLAFPVGR_ | 2 | 616.343 | 0.00 | ||
| ____MAGGKGKSSGGKSSGGKTSGEGPKKQQ | 121.28 | _SSGGK(ac)SSGGK(ac)TSGEGPK_ | 2 | 796.382 | 0.81 | ||
| >sp|A4QVR2|H2AZ_MAGO7 | Histone H2A.Z | AGGKGKSSGGKSSGGKTSGEGPKKQQSHSAR | 80.638 | _SSGGK(ac)SSGGK(ac)TSGEGPK_ | 2 | 796.382 | 1.05 |
| _________MAGGKGKSSGGKSSGGKTSGEG | 121.28 | _GK(ac)SSGGK(ac)SSGGK_ | 2 | 560.783 | 0.49 | ||
| Autophagy Proteins | |||||||
| >tr|L7HTU6|L7HTU6_MAGOY | Ubiquitin-like modifier-activating enzyme ATG7 | SEMPKVTGWERHPSSKLQARVISLAEYMDPT | 66.299 | _HPSSK(ac)LQAR_ | 2 | 533.294 | ∞ |
| RADGSIRNFNTIEDFKKADKGAILRQAGAQI | 881.865 | _NFNTIEDFK(ac)K_ | 2 | 649.325 | 0.35 | ||
Figure 3.
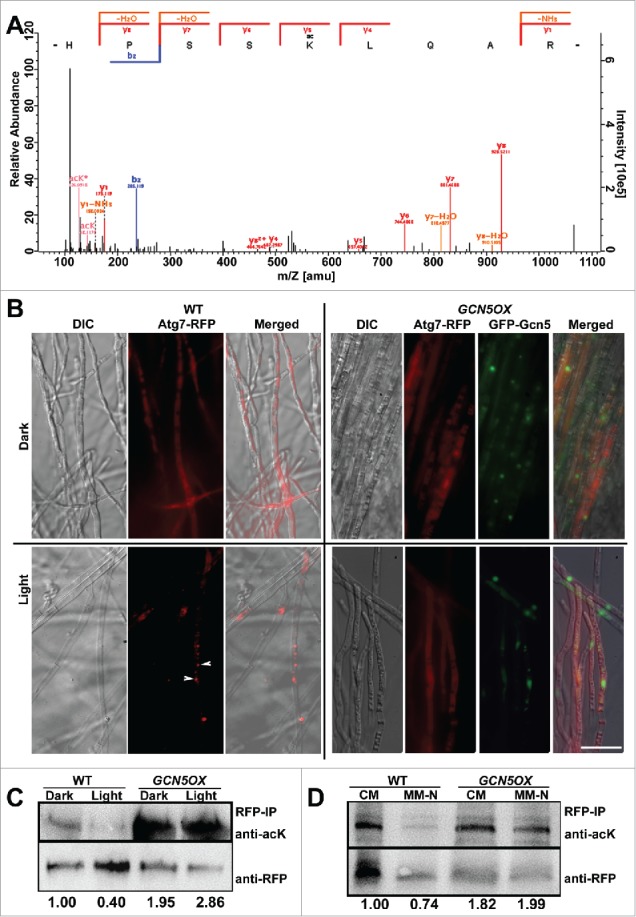
Gcn5 represses autophagy via acetylation on Atg7. (A) Identification of Atg7 K338 acetylation by means of liquid chromatography–mass spectrometry (LC-MS)/MS analysis. The MS/MS spectrum of a double-charged ion at mass/charge ratio (m/z) 533.294 for MH22+ corresponding to the mass of the acetylated peptide HPSSK(ac)LQAR. The labeled peaks correspond to masses of b, y ions of acetylated peptide fragments. (B) Subcellular localization of Atg7-RFP in WT and GCN5OX mycelia cultured in dark-light cycle. Arrowhead depicts punctate Atg7-RFP signal in WT mycelia exposed to light for 6 h. Arrow, vacuole. Scale bar: 10 μm. (C) RFP-IP (immunoprecipitated) proteins from total lysates of dark or light cultured WT or GCN5OX mutant were subjected to immunoblotting with anti-acK antibody. Detection with anti-RFP serves as loading control. Relative abundance of acetylated Atg7 was calculated as a ratio of acK/RFP band, with densitometric analysis performed by ImageJ software (https://imagej.nih.gov/ij/). The numbers under the blots are relative fold change, as normalized to the lane of WT dark, which was arbitrarily set as 1.00. (D) RFP-IP (immunoprecipitated) proteins from total lysates of liquid-cultured WT or GCN5OX mutant, in either rich (CM) or nitrogen-depleted (MM-N) medium, were subjected to immunoblotting with anti-acK antibody. Detection with anti-RFP serves as loading control. The relative fold changes of acetylated Atg7 were calculated following the same way as in Figure 3C, and labeled under the blots.
Atg7 is a ubiquitin-like modifier-activating enzyme that participates in post-translational processing of Atg8 for autophagy induction.36 Therefore, we speculated that acetylated Atg7 may be an inactive form and that light induces autophagy by removing such Atg7 acetylation. To test this hypothesis, we expressed an Atg7-RFP fusion protein in both the WT and GCN5OX backgrounds, and observed the subcellular localization of Atg7-RFP fusion protein in the dark-light cycles. Atg7-RFP appeared cytosolic in dark-grown mycelia in the WT, whereas it became punctate, presumably corresponding to the PAS, or vacuolar in response to light exposure (Fig. 3B). In contrast, Atg7-RFP remained cytosolic in GCN5OX mycelia in both dark and light culture conditions (Fig. 3B, arrowheads). We further detected Atg7 acetylation by immunoblot with RFP-Trapped proteins against the anti-acetyl lysine (anti-acK) antibody. Acetylation of Atg7 appeared higher in the GCN5OX mutant, both in dark or light conditions, compared with the WT (Fig. 3C). The acetylation level of Atg7-RFP protein decreased significantly in response to light exposure in the WT, while it was retained in the GCN5OX strain, likely due to prolonged activity and function of Gcn5 therein (Fig. 3C). Similarly, overexpression of GCN5 resulted in elevated levels of Atg7 acetylation even under nitrogen starvation (Fig. 3D). Nitrogen starvation also induced punctate Atg7-RFP in WT mycelia, whereas cytosolic Atg7-RFP was predominant in the GCN5OX mycelia cultured both in rich or nitrogen-depleted medium (Fig. 4A). These results indicate that starvation-induced autophagy could also be effectively repressed by continued Gcn5-catalyzed acetylation of Atg7.
Figure 4.
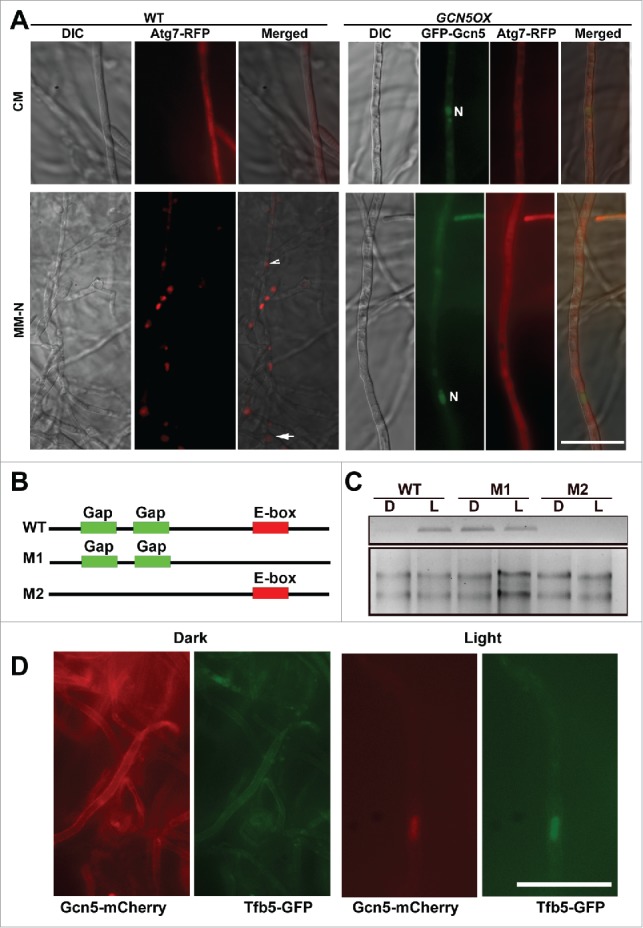
Transcriptional and post-translational regulation of Gcn5 during dark-light cycles. (A) Subcellular localization of Atg7-RFP in WT and GCN5OX mycelia cultured in rich (CM) medium or under nitrogen starvation for 6 h. Scale bar: 10 μm. “N” denotes nuclei visualized by GFP-Gcn5; arrow and arrowhead depict vacuolar and punctate Atg7-RFP signal, respectively. (B) Schematic representation of the GCN5 promoter, not drawn to scale. Green boxes represent Gap boxes and red box as E-box. (C) GFP transcription driven by WT, M1 or M2 variant of the GCN5 promoter was examined by RT-PCR. Primers used for RT-PCR are listed in Table S1. Total RNA (2 μg) serves as a loading control. (D) Subcellular localization of Gcn5-mCherry under dark or light conditions. Arrow denotes the colocalization of Gcn5-mCherry with nuclear Tfb5-GFP, upon light induction. Scale bar: 10 μm.
Light-dependent transcriptional regulation of GCN5
It was puzzling that light-inducible GCN5 could repress the photo-induced autophagy in M. oryzae. To explore possible light-dependent regulation of Gcn5 function, we attempted to screen for light responsive elements (LREs) in the promoter region of GCN5. Two Gap boxes as 5′-ATGAA(G/A)A-3′ repeats essential for light induction of the GapA and GapB genes in plants37 were found at −936 to −928 and −918 to −911 (Fig. 4B, green highlighted). In addition, a 5′-CACGTG-3′ E-box sequence, which is a negative regulatory element modulating light-dependent gene expression identified in flies,38 was also found at −179 to −173 (Fig. 4B, red highlighted). We generated the constructs carrying eGFP reporter gene under the control of WT, M1 or M2 mutated promoters of GCN5, as illustrated in Fig. 4B, and individually expressed them in the wild-type M. oryzae strain. The light-dependent regulation of GCN5 promoter activity was verified by RT-PCR with eGFP-specific primers (Fig. 4C, Table S1). In parallel, we tested the transcription of GCN5 during the time-course analysis of light exposure, using RT-PCR with GCN5-specific primers (Table S1), and found that the GCN5 transcripts were increased subtantially during the time-course of light exposure (Fig. 5C).
Figure 5.
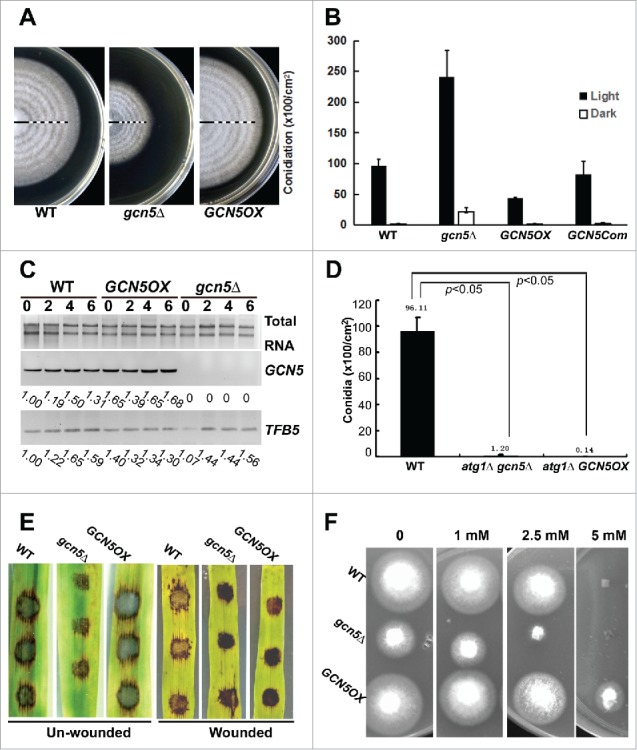
Biological functions of Gcn5. (A) Banding pattern of the WT, gcn5Δ and GCN5OX strains. Closed bar represents the growth phase in dark, and open bar for growth under light. Five dark-light (12 h-12 h) cycles were given following growth in constant dark for 3 d. The PA medium supporting mycelial growth was kept at 60°C for 3 to 5 h, for fast dehydration to achieve better contrast of the banding, before photography. (B) Bar chart depicting quantitatively assessed conidiation in the indicated strains, grown in dark or light conditions. Mean values ( ± SE) presented as percentage points were derived from 3 independent experiments (n = 15 colonies for each sample). (C) TFB5 transcripts in WT, gcn5Δ and GCN5OX cultures exposed to light for 0, 2, 4 and 6 h. Total RNA (2 μg) serves as a loading control. Primers used for RT-PCR are listed in Table S1. (D) Bar chart depicting quantitatively assessed conidiation in the indicated strains. Mean values ( ± SE) presented as percentage points were derived from 3 independent experiments (n = 15 colonies for each sample). (E) Barley leaf explants (intact or wounded) were inoculated with conidia from the indicated strains and disease symptoms assessed after 7 d. (F) Sensitivity toward oxidative stress was tested in the WT, gcn5Δ and GCN5OX strains, cultured on PA solid medium supplemented with H2O2 at concentration of 0 (control), 1, 2.5, 5, and 10 mM.
Under an epifluorescence microscope, WT-eGFP showed faint/weak signal in dark and was significantly enhanced under light (Fig. S3A). M1-eGFP showed a relatively higher eGFP signal in both dark and light conditions, compared with that in WT-eGFP (Fig. S3A), confirming that E-box serves as a negative element to repress gene expression in the dark. On the other hand, M2-eGFP was not induced by light (Fig. S3A), suggesting the key role of Gap boxes in light signaling. The light-dependent regulation of GCN5 promoter activity was further verified by immunoblot with anti-GFP antiserum (Fig. S3B).
Overall, we conclude that the Gap box acts as a positive element for light-induced expression of GCN5, while the E-box represses the GCN5 transcription in the dark and confers the light-dependent expression of GCN5 together with the Gap box.
Post-translational processing of Gcn5 in response to light
We generated a strain with GCN5-mCherry driven by its native promoter and at its genomic locus, as illustrated in Figure S3C. In the dark, Gcn5-mCherry signal was undetectable or appeared as dim, cytosolic signal (Fig. 4D). At 12h post-photo-induction, we observed weak nuclear localization of Gcn5-mCherry, which overlapped with Tfb5-GFP, a nuclear marker specific to light induction (Fig. 4D and S3D).24 In immunoblot analysis using anti-RFP antibody, 2 bands of approximately 36 and 20 kDa were detected, and were clearly more abundant in light-cultured mycelia (Fig. S3E, denoted by #). We infer that these were partial Gcn5-mCherry fusion protein variants, as the size was smaller than the calculated molecular mass of full-length Gcn5-mCherry (Gcn5 as approximately 44 kDa and mCherry as 26 kDa, Fig. S3C). The cleaved C-terminal region of Gcn5 tagged with mCherry was calculated as 9 to 10 kDa, corresponding to about 100 amino acids. As shown in the schematic representation of Gcn5 protein domain organization (Fig. S3C), Gcn5 contains an N-terminal acetyltransferase (NAT) domain that catalyzes histone acetylation, and a C-terminal bromodomain, that interacts specifically with acetylated lysine. The cleaved C-terminal 100-amino acid peptide of Gcn5 corresponds exactly to the bromodomain. We inferred that histone acetylation increases in response to light exposure, leading to increased association between acetylated histone and Gcn5 bromodomain, resulting in its nuclear retention. The subcellular localization of the NAT domain was not visualized by Gcn5-mCherry, but could be followed in the GFP-GCN5 OX strain, as both nuclear and cytosolic (Fig. 1C). Immunoblot with anti-GFP antibody against total lysates from dark- and light-cultured GFP-GCN5 OX strain confirmed that a cleavage occurs between the NAT domain and the C-terminal bromodomain, as a band corresponding to 60 kDa was detected (Fig. S3F, triangle) and assumed as GFP-NAT. The full-length GFP-Gcn5 fusion protein (70 kDa) was also detected (Fig. S3F, asterisk), and it appeared more abundant in the dark-cultured mycelia than in the light-cultured mycelia, likely due to light-triggered cleavage of Gcn5. The cleaved NAT domain may be unstable, as multiple bands of various sizes were detected, likely as intermediates of protein degradation (Fig. S3F, double triangles). A 26-kDa fragment, corresponding to GFP peptide alone, was also detected (Fig. S3F, double asterisk). Overall, we propose that the NAT domain and the bromodomain of Gcn5 protein are likely cleaved and separated upon light induction. The bromodomain was retained in the nucleus, likely by associating with the acetylated histones; while the cytosolic NAT domain was subjected to subsequent degradation. This could partially explain why light-inducible GCN5 represses light-induction of autophagy in M. oryzae: as light likely destabilizes the Gcn5 protein and thus relieves its repression on autophagy via Atg7 acetylation. In the dark, although the GCN5 transcription is low, the basal level of Gcn5 protein may be intact and stable, and it localizes in the cytosol to keep Atg7 acetylated and inactive.
Gcn5 regulates M. oryzae conidiation
We further investigated the biological function of Gcn5 by analyzing vegetative growth, circadian banding of vegetative colony in dark-light cycles, conidiation and pathogenicity of the gcn5Δ, GCN5-complemented strain and GCN5OX mutants, with WT strain as a control. Radial growth was significantly slower in the gcn5Δ mutant (radius = 1.53 ± 0.02 cm; P<0.01), while marginally faster in the GCN5OX mutant (radius = 2.35 ± 0.03 cm; 0.05<P<0.1), compared with WT (radius = 2.28 ± 0.02 cm), after growth in dark for 7 d. The reduction of radial growth could be fully restored in the GCN5-complemented strain (radius = 2.20 ± 0.04 cm; P = 0.18), verifying that such growth defect was indeed caused by the loss of GCN5 function. The banding of gcn5Δ colony was comparable to that of WT, although the mutant colony was smaller than WT (Fig. 5A). Similarly, the GCN5OX strain showed a banding pattern comparable to that of WT (Fig. 5A), suggesting that Gcn5 may not be a core component or regulator of the circadian clock in M. oryzae.
Under the light, conidiation was largely increased in the gcn5Δ mutant, while reduced approximately 50% in the GCN5OX strain, compared with the WT (Fig. 5B, P<0.05). In the dark, the WT and the GCN5OX mutant produced hardly any conidia while the gcn5Δ mutant was able to produce significantly more conidia than the WT (Fig. 5B, P<0.05). The conidiation of the GCN5-complemented strain was comparable to that of the wild-type strain (P>0.1), but distinct from either the gcn5Δ mutant or the GCN5OX mutant (P<0.05; Fig. 5B). We previously showed that Tfb5 is a light-inducible transcriptional factor and essential for M. oryzae conidiation.23 Here, we further verified whether overexpressed GCN5 could repress conidiation via transcriptional repression of the TFB5 gene. As shown in Fig. 5C, TFB5 was induced during the time-course of light exposure in the WT strain as well as in the gcn5Δ mutant, while it was constantly induced in the GCN5OX strain in either dark or light conditions (Fig. 5C). Therefore, we conclude that GCN5 is involved but not essential for TFB5 transcription, and the reduced conidiation in the GCN5OX strain is not caused by repression of the TFB5 transcription.
We inferred that Gcn5 may repress conidiation in the dark, likely by repressing autophagy. To further confirm this hypothesis, we deleted the ATG1 gene in both gcn5Δ and GCN5OX mutants (Fig. S4A and B), as ATG1 has been reported to be essential for autophagy in M. oryzae as the Moatg1Δ mutant showed significant reduction in conidia production.4 The resultant atg1Δ gcn5Δ double mutant was significantly reduced in conidiation (Fig. 5D, P<0.05), which was similar to the reported phenotypic defects of the Moatg1Δ mutant.4 Conidiation was also reduced in the atg1Δ GCN5OX mutant (Fig. 5D, P < 0.05). This suggests that conidiation phenotype observed in gcn5Δ and GCN5OX mutants were indeed related to autophagy activity.
Although the gcn5Δ mutant produced abundant conidia, such conidia were unable to infect the host successfully. Infection assays with barley leaf explants showed that the gcn5Δ conidia were significantly reduced in pathogenicity, while WT or GCN5OX conidia were able to cause typical blast disease lesions (Fig. 5E). The gcn5Δ conidia could better infect the wounded leaves than the intact leaves, however, the infection was still weaker than the wild-type conidia did, under the same conditions (Fig. 5E). Furthermore, regulation of autophagy as well as pathogenicity by the GCN5 gene was verified (Fig. S4C and D).
The reduced pathogenicity upon GCN5 deletion may result from the oxidative stress response mediated by Gcn5 as reported for its yeast, fungal or mammalian orthologs.30,39,40 The sensitivity assay toward hydrogen peroxide further confirmed that gcn5Δ was hypersensitive to oxidative stress, compared with the WT or the GCN5OX strain (Fig. 5F). Pathogenicity assays with mycelial plugs of the atg1Δ, atg1Δ gcn5Δ, or atg1Δ GCN5OX strains were also performed, with intact or wounded barley leaves, and the result showed that these 3 strains were incapable of causing blast disease (Fig. S4E), consistent with the previous reports on the essential role of autophagy in M. oryzae infection.4,7
Discussion
In eukaryotic organisms, HAT complexes are coactivators important for transcriptional activation by modifying chromatin.41-43 In this study, we identified an ortholog of GCN5 from M. oryzae, encoding a component of the HAT complex important for M. oryzae conidiation and pathogenicity. We also identified a GCN5 paralog, namely GCN5b, in M. oryzae genome. GCN5b is not required for M. oryzae conidiation or pathogenicity and hence was not characterized further in this study. Recently, it has been reported that Hat1 regulates the stress response and virulence in Candida albicans.44 The GCN5 ortholog is involved in morphological transition and virulence in C. albicans45 and Ustilago maydis.46 But overall, the knowledge on physiological functions of HAT or HDAC components and associated regulatory mechanisms in pathogenic fungi is really limited, and potential phototropic regulation of these Gcn5 HATs was not predicted or verified. To our knowledge, involvement of HAT in fungal asexual production has not been documented previously, therefore, the findings from this study add not only a new member to the list of HATs that are essential for fungal development and pathogenicity, but also further expand the biological functions modulated by HATs, and importantly, for the first time, reports a phototropic signaling pathway and a regulatory mechanism mediated by protein acetylation and deacetylation cycles.
A yeast HAT Esa1 and its mammalian ortholog KAT5/TIP60 acetylate Atg3/ATG3 or Atg1/(mammalian ULK1/2) respectively, to induce autophagy in response to physiological cues.21,22 In M. oryzae, the Esa1 ortholog is not involved in regulating autophagy or conidiation (our unpublished data). Instead, Gcn5 represses autophagy, in contrast to yeast Esa1 or mammalian KAT5, without light. We found that Gcn5 directly acetylates Atg7 to repress autophagy induction, which is consistent with the observation that acetylation of ATG7 by EP300/p300 acetyltransferase in mammalian cells inhibits autophagy,47,48 and that SIRT1-catalyzed deacetylation of autophagy protein is required for autophagy induction in mouse.49 A recent study shows that anacardic acid, a nonspecific HAT inhibitor, is effective in preventing M. oryzae pathogenicity.50 Anacardic acid has been demonstrated to directly inhibit histone acetyltransferases (HATs) like EP300/p300, KAT2B/PCAF and KAT5/Tip6051 and to induce autophagy in human lung carcinoma A549 cells.52 Our results are consistent with these reports in that the gcn5Δ mutant was reduced in pathogenicity, and showed elevated autophagy activity. Besides direct modification on cytosolic autophagy proteins, HATs including Gcn5 may also regulate autophagy via histone modification. It has been reported that histone H3 hyperacetylation in Drosophila melanogaster, is associated with transcriptional downregulation of several autophagy-essential ATG genes, including ATG5, ATG7, and ATG14, which may restrict autophagic activities.53 Our study also proved that ATG8 transcription was not repressed upon GCN5 overexpression, instead, it depended on GCN5 for its transcription (Fig. S2E). However, we did not rule out the possibility that overexpressed GCN5 may also lead to hyperacetylation of target histones and likely downregulation of ATG genes, thus resulting in the observed repression of autophagy pathway.
Our results showed that Gcn5 is subject to regulation by a dark-light cycle via the positive and negative LREs located in its promoter region. Interestingly, we also found same LREs in GCN5 orthologs in C. albicans and Ustilago maydis, as well as in the Ep300/p300 acetyltransferase gene in Mus musculus (Fig. S4F), although whether these GCN5 orthologs were also subjected to light modulation through LREs, remains to be functionally verified. Overall, our study demonstrates a novel light-responsive pathway mediated by Gcn5 to induce autophagy and conidiation in the rice-blast fungus M. oryzae. Such phototropic regulation on autophagy induction might be conserved in eukaryotic organisms, given that the LREs are conserved in the promoter regions of several HAT-encoding genes in other organisms. We are still unclear about the biological significance of light-induction of GCN5 transcription, as light subsequently destabilizes this gene product. We attempted to examine the dependence of conidiation-related gene TFB5 transcription on GCN5, as yeast Gcn5 is known to be a TFB5 activator,54 in response to carbon homeostasis and/or hypoxia,55,56 but not to light. Our result showed that TFB5 transcripts accumulated in the WT strain during light exposure (Fig. 5C). Such light-induction of TFB5 transcription was not dependent on GCN5 as the level of TFB5 transcription in the gcn5Δ mutant was comparable to that of WT (Fig. 5C). TFB5 transcription was induced in the dark in the GCN5OX strain but showed no obvious increase in the light condition (Fig. 5C). This result indicates that Gcn5 is involved but not critical for TFB5 activation, and the reduced conidiation in the GCN5OX strain was not caused by TFB5 repression. We further infer that transcriptional induction of GCN5 by light may be transient, to activate conidiation-related genes including TFB5.
In summary, we propose that light regulates GCN5 expression, as well as stability and subcellular localization of Gcn5 protein during the dark-light cycles. Gcn5 represses autophagy via acetylation on Atg7 in the dark; while it releases repression of autophagy upon photo-induction and subsequent translocation into the nucleus and/or upon degradation. Phototropic induction of GCN5 may contribute to transcriptional regulation of TFB5, which encodes a conidiation-specific transcription factor in M. oryzae. Such phototropic regulation of GCN5 transcription and the stability of the subcellular localization of the Gcn5 protein promote robust light-induced autophagy as well as conidiation in M. Oryzae. Future studies should certainly aim to identify the other targets of the Gcn5 transcriptional activator module, and further investigate the regulatory mode of GCN5 function in M. oryzae.
Materials and methods
Fungal strains and growth conditions
The M. oryzae wild-type strain B157 (Field isolate, mat1–2) was originally obtained from the Directorate of Rice Research, India. M. oryzae strains were propagated on Prune-agar (PA) medium or complete medium (CM) as described.57 The composition of MM-N (used for nitrogen starvation) was as reported previously.58 To assess the growth and colony characteristics, M. oryzae isolates were cultivated on CM agar or PA medium, at 28°C for 1 wk. Mycelia used for total protein extraction were obtained by growing the relevant strains in liquid CM for 2 to 3 d, with gentle shaking, followed by inoculation in CM or MM-N for about 6 h. For quantitative analysis of conidiation and testing the pathogenicity, standard procedures were followed.59
Plasmid constructs and fungal transformants
To generate gcn5Δ, the 5′ UTR and 3′ UTR were amplified and ligated sequentially into the Agrobacterium Transfer-DNA vector pFGL820 to flank the ILV1 cassette.23 The resultant plasmid was then transferred into the M. oryzae strain carrying an ectopic RFP-ATG8,8 to induce a locus-specific knockout. To construct overexpressed GFP-GCN5, GFP was fused in frame to the N-terminus of the GCN5 ORF, and under the constitutive promoter of RP27.23 The transformed M. oryzae was selected by bialaphos resistance, and with ectopic GFP-GCN5. To construct Gcn5-mCherry, mCherry was fused in frame to the last 0.5 kb of the GCN5 ORF and the plasmid transformed into M. oryzae, such that the mCherry was fused to the GCN5 coding sequence at its genomics locus. For GCN5 complementation, the GCN5 locus, including promoter region (1 Kb), ORF and terminator (0.3 Kb), was PCR-amplified and ligated into pFGL821 (addgene, 58223) WT, M1 and M2 varieties of the GCN5 promoter were synthesized by Thermo-Fisher (Shanghai) and fused with the eGFP coding sequence respectively, on the vector pFGL93224 (for M1 GCN5Prom-eGFP) or pFGL821 (for WT GCN5Prom-eGFP and M2 GCN5Prom-eGFP). To generate atg1Δ, the 5′ UTR and 3′ UTR were amplified and ligated sequentially into the vector pFGL821 to flank the HPH1 cassette. The resultant plasmid was then transferred into the WT, gcn5Δ or GCN5OX strains, respectively, to induce a locus-specific knockout. For C-terminal tagging of Atg7 with dsRed, ATG7 ORF was PCR amplified and ligated into pFGL932, under the RP27 promoter. The fragment containing PR27 promoter-ATG7 was released by BamHI/EcoRI double digestion and ligated to pFGL821, followed by ligation of PCR-amplified dsRed CDS into EcoRI site. The RP27 Promoter-ATG7-dsRed sequence was introduced into the WT and GCN5OX strains, as an ecotopic copy. The primers used for gene deletion, complementation and GFP, mCherry or dsRed tagging are listed in Table 3.
Table 3.
Oligonucleotide primers used for plasmid construction in this study.
| Gene (Locus) | Description | Enzyme sites | Primer sequence |
|---|---|---|---|
| GCN5(MGG__03677) | Deletion construct | — | 5′-AATGTGAATTCGAACACAAAACGTTCAG-3′ |
| — | 5′-GGAACGGGATCCGTGCTATCTCGTTGGG-3′ | ||
| PstI | 5′-GACTGTTCTGCAGTTTGCTGACCCATTTGTACATGAT-3′ | ||
| HindIII | 5′-GAGTGTTAAGCTTCTAGGCGTCGGATCACAGTGCGCAT-3′ | ||
| GFP-tagging (N-terminal) under RP27 promoter | BamHI | 5′-GAGAGTGGATCCATAAATGTAGGTATTACCTGTA-3′ | |
| NcoI | 5′-GAGAGTGACCATGGTTTGAAGATTGGGTTCCTACGA-3′ | ||
| NcoI | 5′-GAGAGTGACCATGGTGAGCAAGGGCGAGGAGCTGT-3′ | ||
| KpnI | 5′-GAGAGTGAGGTACCCTTGTACAGCTCGTCCATGCCGAG-3′ | ||
| KpnI | 5′-GAGAGTGAGGTACCTCTACAGCAACAGAAGATAGTACAAATGGC-3′ | ||
| EcoRI | 5′-GTGTGAATTCTGCGCATTAAAGTCATGGTTCAAGATGTGA-3′ | ||
| mCherry-tagging (C-terminal) | EcoRI | 5′-GTGTGAATTCGAGGGCGGCACCATAATGCAGTGCA-3′ | |
| NdeI | 5′-GGAATTCCATATGTGGTTCAAGATGTGACCACTCGGGG-3′ | ||
| NdeI | 5′-GGAATTCCATATGAAGGGCGAGGAGGATAACATGG-3′ | ||
| BamHI | 5′-GTGTGGATCCCTACTTGTACAGCTCGTCCATGCCG-3′ | ||
| Complementation | EcoRI | 5′-GAGTGAGAATTCGTTCAGTTTCGGGGAATCTTACGCC-3′ | |
| KpnI | 5′-AGAGTGAGGTACCCAAACCAACTATATGCATGTTCTCAAAAGCCGAC-3′ | ||
| ATG1 (MGG_06393) | Deletion construct | EcoRI | 5′-GTGTGAATTCAAGCCTCCAGATACAGAGGTTCTGT-3′ |
| BamHI | 5′-GTGTGGATCCCTCAACCGGGTATTGTTTCATCTT-3′ | ||
| PstI | 5′-GTGTCTGCAGAAGCGAGACGGTTGGCCCCCTAA-3′ | ||
| HindIII | 5′-GTGTAAGCTTAAGCCTCCAGATACAGAGGTTCTGT-3′ | ||
| ATG7 (MGG_07297) | dsRed-tagging (C-terminal), under RP27 promoter | NcoI | 5′-AACCCAATCTTCAAACCATGGGAAATGATGAGGCGGCCG-3′ |
| EcoRI | 5′-CTATGACATGATTACGAATTCAAGCATCTCACCATCCCCTTC-3′ | ||
| EcoRI | 5′-CCGGAATTCATGGACAACACCGAGGACGTCATCAAG-3′ | ||
| EcoRI | 5′-CCGGAATTCGAAACGCGTTTTATTCTTGTTGACATGGAGCTATTAAATCACTACTGGGAGCCGGAGTGGCGGG-3′ |
Epifluorescence microscopy
M. oryzae cells expressing fluorescent protein-fused chimera were grown under requisite conditions. Epifluorescence microscopy was performed using an Axio Observer Z1 microscope (Zeiss, Jena, Germany) equipped with an sCMOS camera (PCO Edge, Kelheim, Germany). To visualize the nucleus, mycelia were fixed with 0.1% TritonX and stained with 1 μg/ml DAPI (Sigma-Aldrich, D9542) at room temperature for 10 min, followed by 3 washes with PBS.
Immunoblotting and immunoprecipitation
For total protein extraction, mycelia grown under requisite conditions were ground into a fine powder in liquid nitrogen and resuspended in 0.3 to 0.5 ml extraction buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NONIDET P-40 Substitute (Sigma-Aldrich, IGEPAL® CA-630, I3021), with 2 mM PMSF and proteinase inhibitor cocktail (Sigma-Aldrich, cOmplete™, 11836170001). Lysates were cleared by centrifugation at 13,000 g for 30 min at 4°C. Total protein concentration was measured using the Bio-Rad Protein Assay (500–0006). Samples were resolved by 12% SDS-PAGE followed by western blotting with anti-RFP (rabbit; 1:1,000; Clontech, R10367), or anti-GFP (rabbit; 1:5,000; Invetrogen Molecular Probes, A6455). Secondary antibody was anti-rabbit (1:20,000; HiSec™HRP-conjugated, Ab202) followed by detection using the SuperSignal West Pico Chemiluminescent substrate (Pierce, 34080). Atg7 acetylation was assessed by immunoblot with RFP-Trapped (Chromotek RFP-Trap®_A, rta-100) proteins, against the anti-acetyl lysine (anti-acK) (Abcam, ab61257) antibody. Total lysates from the GFP-Gcn5 strain was subject to immunoprecipitation with GFP-Trap (Chromo Tek, gta-20). The IP proteins were then identified by mass spectrometry (Q Exactive, Thermo Finnigan, US).
Mass spectrometry
Gcn5-IP proteins (about 30 μg) were in-solution digested using trypsin (Sigma-Aldrich, T7409) for 20 h at 37°C, and the digested peptides were subject to Nano-HPLC/ESI-ion trap-MS/MS analysis with Q exactive (Thermo Finnigan, US). Raw MS data files were processed and analyzed using Mascot2.2 (Matrix Science, UK). The database search was performed with the following parameters: Database: uniprot; Taxonomy: Magnaporthe oryzae (39372); Enzyme: Trypsin; Dynamical modifications: Oxidation (M); Fixed modifications: Carbamidomethyl (C); Max Missed Cleavages: 2; ProteomicsTools: 3.1.6; Filter by score > = 20.
For identification of acetylated proteins and sites from GCN5OX and gcn5Δ mutants, total lysates (10 mg) from each strain were digested with trypsin for 20 h at 37°C, followed by enrichment of acetylated peptides with PTMScan Acetyl-Lysine Motif (acK) Kit (Cell Signaling Technology, 13416S). The peptides were separated on Easy-nLC1000 (Thermo Fisher Scientific Inc., Waltham, MA, US) with trap columns (EASY column SC001 traps 150 μm*20 mm [RP-C18]) and analysis column (EASY column SC200 150 μm*100 mm [RP-C18]). The peptides analyzed with Q exactive (Thermo Finnigan, US), eluted from the column with a linear solvent gradients (A: 0.1% formic acid [FA] 2% ACN; B: 84% ACN/0.1% FA) for 120 min at a flow rate of 300 nL/min (110 min of gradient from 0 to 45% buffer B; 8 min gradient from 45 to 100% buffer B; 2 min of 100% buffer B). This LC gradient was used for all mobile phase compositions. The mass spectrometer was operated in the positive ion mode at 2 kV and the capillary temperature was set to 180°C. The full scan was performed with enhanced mode, 350 to 1800 m/z. Raw LCMS/MS data files were processed with Maxquant software (version 1.3.0.5) for database searching, using uniprot_Magnaporthe_oryzae_39372_20160315.fasta, and with the following parameters: Main search ppm: 6; Missed cleavage: 4; MS/MS tolerance ppm: 20; De-Isotopic: TRUE; enzyme: Trypsin; Fixed modification: Carbamidomethyl (C); Variable modification: Oxidation (M), Acetyl (Protein N-term), Avetyl (K); Decoy database pattern: reverse; iBAQ: TRUE; Match between runs: 2 min; Peptide FDR: 0.01; Protein FDR: 0.01. Protein or peptide matching and annotation were performed with Perseus (version 1.3.0.4). The Mass Spectrometry analysis was performed by Shanghai Applied Protein Technology Co. Ltd. (http://www.aptbiotech.com/).
Statistical analysis
For quantification of radial growth or conidia production, 3 biological repeats were perform for culculation of average value and standard deviation. P value was culculated with TTEST function in Excel (Microsoft office 2007) with the setting as Two-Sample Assuming Unequal Variances.
Supplementary Material
Abbreviations
- ATG genes
autophagy-related genes
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- HAT
histone acetyltransferase
- IP
immunoprecipitation or immunoprecipitated
- LREs
light responsive elements
- NAT
N-terminal acetyltransferase
- PAS
phagophore assembly site
Disclosure of potential financial interests
The authors declare they have no competing financial interests.
Acknowledgments
We are grateful to Ziwei Qu (Temasek Life Sciences Laboratory, Singapore) for construction of the plasmids for GCN5 knockout and overexpression, and Fan Yang for construction of the vector pFGL820 and pFGL821. We thank the Shanghai Applied Protein Technology Co. Ltd for technology support in mass spectrometry identification of immunoprecipitated proteins and acetylated peptides/residues.
Funding
This work was supported by the National Basic Research Program of China (973 Program, grant number 2015CB150600) and the National Research Foundation, Singapore (Prime Minister's Office, NRF-CRP7–2010–02).
References
- [1].Talbot NJ. On the trail of a cereal killer: Exploring the biology of magnaporthe grisea. Annu Rev Microbiol 2003; 57:177-202; PMID:14527276; https://doi.org/ 10.1146/annurev.micro.57.030502.090957 [DOI] [PubMed] [Google Scholar]
- [2].Lee K, Singh P, Chung WC, Ash J, Kim TS, Hang L, Park S. Light regulation of asexual development in the rice blast fungus, magnaporthe oryzae. Fungal Genet Biol 2006; 43:694-706; PMID:16765070; https://doi.org/ 10.1016/j.fgb.2006.04.005 [DOI] [PubMed] [Google Scholar]
- [3].Talbot NJ. Having a blast: Exploring the pathogenicity of magnaporthe grisea. Trends Microbiol 1995; 3:9-16; PMID:7719639; https://doi.org/ 10.1016/S0966-842X(00)88862-9 [DOI] [PubMed] [Google Scholar]
- [4].Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 2007; 6:997-1005; PMID:17416896; https://doi.org/ 10.1128/EC.00011-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu TB, Liu XH, Lu JP, Zhang L, Min H, Lin FC. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010; 6:74-85; PMID:19923912; https://doi.org/ 10.4161/auto.6.1.10438 [DOI] [PubMed] [Google Scholar]
- [6].Lu JP, Liu XH, Feng XX, Min H, Lin FC. An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr Genet 2009; 55:461-73; PMID:19629489; https://doi.org/ 10.1007/s00294-009-0259-5 [DOI] [PubMed] [Google Scholar]
- [7].Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006; 312:580-3; PMID:16645096; https://doi.org/ 10.1126/science.1124550 [DOI] [PubMed] [Google Scholar]
- [8].Deng YZ, Ramos-Pamplona M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in magnaporthe oryzae. Autophagy 2009; 5:33-43; PMID:19115483; https://doi.org/ 10.4161/auto.5.1.7175 [DOI] [PubMed] [Google Scholar]
- [9].Dong B, Liu XH, Lu JP, Zhang FS, Gao HM, Wang HK, Lin FC. MgAtg9 trafficking in magnaporthe oryzae. Autophagy 2009; 5:946-53; PMID:19556868; https://doi.org/ 10.4161/auto.5.7.9161 [DOI] [PubMed] [Google Scholar]
- [10].He Y, Deng YZ, Naqvi NI. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2013; 9:1818-27; PMID:23958498; https://doi.org/ 10.4161/auto.26057 [DOI] [PubMed] [Google Scholar]
- [11].He M, Kershaw MJ, Soanes DM, Xia Y, Talbot NJ. Infection-associated nuclear degeneration in the rice blast fungus magnaporthe oryzae requires non-selective macro-autophagy. PLoS One 2012; 7(3):e33270; PMID:22448240; https://doi.org/ 10.1371/journal.pone.0033270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005; 121:567-77; PMID:15907470; https://doi.org/ 10.1016/j.cell.2005.03.007 [DOI] [PubMed] [Google Scholar]
- [13].Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963-6; PMID:9461583; https://doi.org/ 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- [14].Dementhon K, Saupe SJ, Clave C. Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus podospora anserina. Mol Microbiol 2004; 53:1625-40; PMID:15341644; https://doi.org/ 10.1111/j.1365-2958.2004.04235.x [DOI] [PubMed] [Google Scholar]
- [15].Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al.. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539-45; PMID:14536056; https://doi.org/ 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- [16].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931-7; PMID:17712358; https://doi.org/ 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- [17].Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy 2015; 11:2288-99; PMID:26565778; https://doi.org/ 10.1080/15548627.2015.1107692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al.. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488-92; PMID:11100732; https://doi.org/ 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- [19].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; https://doi.org/ 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435-46; PMID:10525546; https://doi.org/ 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al.. Function and molecular mechanism of acetylation in autophagy regulation. Science 2012; 336:474-7; PMID:22539722; https://doi.org/ 10.1126/science.1216990 [DOI] [PubMed] [Google Scholar]
- [22].Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, et al.. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 2012; 336:477-81; PMID:22539723; https://doi.org/ 10.1126/science.1217032 [DOI] [PubMed] [Google Scholar]
- [23].Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, et al.. Protein phosphorylation-acetylation cascade connects growth factor deprivation to autophagy. Autophagy 2012; 8:1385-6; PMID:22717509; https://doi.org/ 10.4161/auto.20959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deng YZ, Qu Z, Naqvi NI. Twilight, a novel circadian-regulated gene, integrates phototropism with nutrient and redox homeostasis during fungal development. PLoS Pathog 2015; 11:e1004972; PMID:26102503; https://doi.org/ 10.1371/journal.ppat.1004972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol 1997; 7:689-92; PMID:9285713; https://doi.org/ 10.1016/S0960-9822(06)00296-X [DOI] [PubMed] [Google Scholar]
- [26].Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, et al.. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J 2015; 84:1178-91; PMID:26576681; https://doi.org/ 10.1111/tpj.13076 [DOI] [PubMed] [Google Scholar]
- [27].Moraga F, Aquea F. Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front Plant Sci 2015; 6:865; PMID:26528322; https://doi.org/ 10.3389/fpls.2015.00865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Z, Cao H, Chen F, Liu Y. The roles of histone acetylation in seed performance and plant development. Plant Physiol Biochem 2014; 84:125-33; PMID:25270163; https://doi.org/ 10.1016/j.plaphy.2014.09.010 [DOI] [PubMed] [Google Scholar]
- [29].Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 2016; 26:211-25; PMID:26602697; https://doi.org/ 10.1101/gr.196337.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gaupel AC, Begley TJ, Tenniswood M. Gcn5 modulates the cellular response to oxidative stress and histone deacetylase inhibition. J Cell Biochem 2015; 116:1982-92; PMID:25755069; https://doi.org/ 10.1002/jcb.25153 [DOI] [PubMed] [Google Scholar]
- [31].Xie L, Zeng J, Luo H, Pan W, Xie J. The roles of bacterial GCN5-related N-acetyltransferases. Crit Rev Eukaryot Gene Expr 2014; 24:77-87; PMID:24579671; https://doi.org/ 10.1615/CritRevEukaryotGeneExpr.2014007988 [DOI] [PubMed] [Google Scholar]
- [32].Greene NP, Crow A, Hughes C, Koronakis V. Structure of a bacterial toxin-activating acyltransferase. Proc Natl Acad Sci U S A 2015; 112:E3058-66; PMID:26016525; https://doi.org/ 10.1073/pnas.1503832112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yin YW, Jin HJ, Zhao W, Gao B, Fang J, Wei J, Zhang DD, Zhang J, Fang D. The histone acetyltransferase GCN5 expression is elevated and regulated by c-Myc and E2F1 transcription factors in human colon cancer. Gene Expr 2015; 16:187-96; PMID:26637399; https://doi.org/ 10.3727/105221615X14399878166230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today 2009; 14:942-8; PMID:19577000; https://doi.org/ 10.1016/j.drudis.2009.06.008 [DOI] [PubMed] [Google Scholar]
- [35].Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of saccharomyces cerevisiae. FEBS Lett 1993; 333:169-74; PMID:8224160; https://doi.org/ 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- [36].Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004; 36:2503-18; PMID:15325588; https://doi.org/ 10.1016/j.biocel.2004.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Park SC, Kwon HB, Shih MC. Cis-acting elements essential for light regulation of the nuclear gene encoding the a subunit of chloroplast glyceraldehyde 3-phosphate dehydrogenase in arabidopsis thaliana. Plant Physiol 1996; 112:1563-71; PMID:8972600; https://doi.org/ 10.1104/pp.112.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Takanaka Y, Okano T, Yamamoto K, Fukada Y. A negative regulatory element required for light-dependent pinopsin gene expression. J Neurosci 2002; 22:4357-63; PMID:12040041; https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kikuchi H, Kuribayashi F, Takami Y, Imajoh-Ohmi S, Nakayama T. GCN5 regulates the activation of PI3K/Akt survival pathway in B cells exposed to oxidative stress via controlling gene expressions of Syk and Btk. Biochem Biophys Res Commun 2011; 405:657-61; PMID:21281601; https://doi.org/ 10.1016/j.bbrc.2011.01.088 [DOI] [PubMed] [Google Scholar]
- [40].O'Meara TR, Hay C, Price MS, Giles S, Alspaugh JA. Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot Cell 2010; 9:1193-1202; PMID:20581290; https://doi.org/ 10.1128/EC.00098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 2011; 39:1390-7; PMID:20972224; https://doi.org/ 10.1093/nar/gkq983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J Biol Chem 2001; 276:14773-83; PMID:11279013; https://doi.org/ 10.1074/jbc.M100125200 [DOI] [PubMed] [Google Scholar]
- [43].Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in chlamydomonas reinhardtii. Plant Cell 2011; 23:2285-301; PMID:21705643; https://doi.org/ 10.1105/tpc.111.085266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tscherner M, Zwolanek F, Jenull S, Sedlazeck FJ, Petryshyn A, Frohner IE, Mavrianos J, Chauhan N, von Haeseler A, Kuchler K. The candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog 2015; 11:e1005218; PMID:26473952; https://doi.org/ 10.1371/journal.ppat.1005218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chang P, Fan X, Chen J. Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet Biol 2015; 81:132-41; PMID:25656079; https://doi.org/ 10.1016/j.fgb.2015.01.011 [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Prieto JM, Rosas-Quijano R, Dominguez A, Ruiz-Herrera J. The UmGcn5 gene encoding histone acetyltransferase from ustilago maydis is involved in dimorphism and virulence. Fungal Genet Biol 2014; 71:86-95; PMID:25242418; https://doi.org/ 10.1016/j.fgb.2014.09.002 [DOI] [PubMed] [Google Scholar]
- [47].Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 2009; 284:6322-8; PMID:19124466; https://doi.org/ 10.1074/jbc.M807135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sebti S, Prébois C, Pérez-Gracia E, Bauvy C, Desmots F, Pirot N, Gongora C, Bach AS, Hubberstey AV, Palissot V, et al.. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci U S A 2014; 111:4115-20; PMID:24591579; https://doi.org/ 10.1073/pnas.1313618111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008; 105:3374-9; PMID:18296641; https://doi.org/ 10.1073/pnas.0712145105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Muzaffar S, Bose C, Banerji A, Nair BG, Chattoo BB. Anacardic acid induces apoptosis-like cell death in the rice blast fungus magnaporthe oryzae. Appl Microbiol Biotechnol 2016; 100:323-35; PMID:26381667; https://doi.org/ 10.1007/s00253-015-6915-4 [DOI] [PubMed] [Google Scholar]
- [51].Sun Y, Jiang X, Chen S, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett 2006; 580:4353-6; PMID:16844118; https://doi.org/ 10.1016/j.febslet.2006.06.092 [DOI] [PubMed] [Google Scholar]
- [52].Seong YA, Shin PG, Yoon JS, Yadunandam AK, Kim GD. Induction of the endoplasmic reticulum stress and autophagy in human lung carcinoma A549 cells by anacardic acid. Cell Biochem Biophys 2014; 68:369-77; PMID:23955513; https://doi.org/ 10.1007/s12013-013-9717-2 [DOI] [PubMed] [Google Scholar]
- [53].Eisenberg T, Schroeder S, Büttner S, Carmona-Gutierrez D, Pendl T, Andryushkova A, Mariño G, Pietrocola F, Harger A, Zimmermann A, et al.. A histone point mutation that switches on autophagy. Autophagy 2014; 10:1143-5; PMID:24879160; https://doi.org/ 10.4161/auto.28767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al.. A comprehensive genomic binding map of gene and chromatin regulatory proteins in saccharomyces. Mol Cell 2011; 41:480-92; PMID:21329885; https://doi.org/ 10.1016/j.molcel.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Abate G, Bastonini E, Braun KA, Verdone L, Young ET, Caserta M. Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochim Biophys Acta 2012; 1819:419-27; PMID:22306658; https://doi.org/ 10.1016/j.bbagrm.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hickman MJ, Spatt D, Winston F. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in saccharomyces cerevisiae. Genetics 2011; 188:325-38; PMID:21467572; https://doi.org/ 10.1534/genetics.111.128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol 2006; 61:61-75; PMID:16824095; https://doi.org/ 10.1111/j.1365-2958.2006.05194.x [DOI] [PubMed] [Google Scholar]
- [58].Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus magnaporthe grisea. Plant Cell 1993; 5:1575-90; PMID:8312740; https://doi.org/ 10.1105/tpc.5.11.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Deng YZ, Qu Z, He Y, Naqvi NI. Sorting nexin Snx41 is essential for conidiation and mediates glutathione-based antioxidant defense during invasive growth in magnaporthe oryzae. Autophagy 2012; 8:1058-70; PMID:22561104; https://doi.org/ 10.4161/auto.20217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


