Abstract
Type I natural killer T cells (NKT cells) are characterized by an invariant variable region 14–joining region 18 (Vα14-Vα18) T cell antigen receptor (TCR) α-chain and recognition of the glycolipid α-galactosylceramide (α-GalCer) restricted to the antigen-presenting molecule CD1d. Here we describe a population of α-GalCer-reactive NKT cells that expressed a canonical Vα10-Jα50 TCR α-chain, which showed a preference for α-glucosylceramide (α-GlcCer) and bacterial α-glucuronic acid–containing glycolipid antigens. Structurally, despite very limited TCRα sequence identity, the Vα10 TCR–CD1d–α-GlcCer complex had a docking mode similar to that of type I TCR–CD1d–α-GalCer complexes, although differences at the antigen-binding interface accounted for the altered antigen specificity. Our findings provide new insight into the structural basis and evolution of glycolipid antigen recognition and have notable implications for the scope and immunological role of glycolipid-specific T cell responses.
Natural killer T cells (NKT cells) arise in the thymus through random T cell antigen receptor (TCR) gene-recombination events that result in the expression of TCRs that preferentially interact with lipid-based antigens presented by CD1d on cortical thymocytes1. This unusual form of positive selection diverts developing thymocytes into the NKT cell lineage, which is characterized by rapid and diverse cytokine production and multipotent immunoregulatory abilities. NKT cells are traditionally categorized into two distinct populations: type I and type II (ref. 1). Type I NKT cells express an invariant α-chain variable region 14–α-chain joining region 18 (Vα14-Jα18; TRAV11-TRAJ18) TCR α-chain in mice or Vα24-Jα18 (TRAV10-TRAJ18) in humans and can recognize the prototypical glycolipid antigen α-galactosylceramide (α-GalCer). Type II NKT cells are also CD1d restricted but do not express this invariant TCR α-chain or recognize α-GalCer. Instead, type II NKT cells express a range of TCRs2, although they seem to be enriched for TCRs that incorporate Vα3.2-Jα9 and Vα8 (ref. 3) and recognize distinct glycolipid antigens such as sulfatide4. Mainly because CD1d–α-GalCer tetramers are the tool of choice for the study of NKT cells, most studies of glycolipid-reactive T cells have focused on type I NKT cells. The invariant Vα14-Jα18 TCR α-chain is a defining feature of type I NKT cells to the extent that these cells are not detected in mice with deletion of the gene encoding Jα18 (Tcra-Jtm1Tgi mice; called ‘Jα18−/− mice’ here)5, now commonly used as a model of deficiency in type I NKT cells. Furthermore, differences between Jα18−/− mice and mice with deletion of the gene encoding CD1d (Cd1d−/− mice) are presumed to indicate a role for type II NKT cells, which are still present in the former strain but not the latter strain6–9. Although human Vα24− NKT cells reactive to the CD1d–α-GalCer tetramer have been described10–12, these cells retain the Jα18 junctional region that characterizes type I NKT cells10,12.
Type I NKT cells are stimulated by an array of microbial and self-derived lipid-based antigens13. Given the invariant nature of the NKT cell TCR α-chain, this suggests that the NKT cell TCR β-chain has a role in determining differences in antigen specificity. Indeed, mouse NKT cells, which have a more diverse Vβ repertoire than do humans, frequently use three Vβ genes (Vβ8, Vβ7 and Vβ2; TRBV13, TRBV29 and TRBV1, respectively). The structures of various type I NKT cell TCR–CD1d–antigen complexes14–18 have provided insight into the basis of type I NKT cell recognition and some clues about the role of differences in Vβ use. In all NKT cell TCR–CD1d–antigen structures solved so far, a conserved, tilted and parallel docking mode relative to the CD1d antigen-binding cleft has been observed. In this common framework, the invariant NKT cell TCR α-chain, and in particular the Jα18 complementarity-determining region 3 (CDR3) α-loop, dominates the interaction with CD1d. Further, NKT cell TCR-mutagenesis experiments have demonstrated the importance of the Vα14 and Vα24 CDR1α and Jα18 CDR3α loops in interacting with CD1d when they are bound to a diverse array of lipid-based antigens19–22. Although differences are observed in the way Vβ8.2 and Vβ7 NKT cell TCRs interact with CD1d, the overall docking mode is largely conserved. Collectively, these studies suggest that the invariant Vα14-Jα18 α-chain has the principal role in determining the conserved NKT cell TCR-CD1d docking topology.
Here we define a previously unknown population of Vα10-Jα50+ NKT cells (called ‘Vα10 NKT cells’ here) that fits neither the type I nor the type II NKT cell category. Analogous to classical type I NKT cells, the Vα10 NKT cells were defined by a previously unknown canonical TCR α-chain rearrangement and shared some functional attributes with type I cells, including CD1d-mediated recognition of α-GalCer, but recognized other glycolipids differently, including self and bacterial lipids. Here we provide an analysis of several key attributes of this NKT cell subset, including TCR use, antigen reactivity, functional responses and the structural basis of antigen recognition by Vα10 NKT cells.
RESULTS
Identification of T cells reactive to CD1d-α–GalCer in Jα18−/− mice
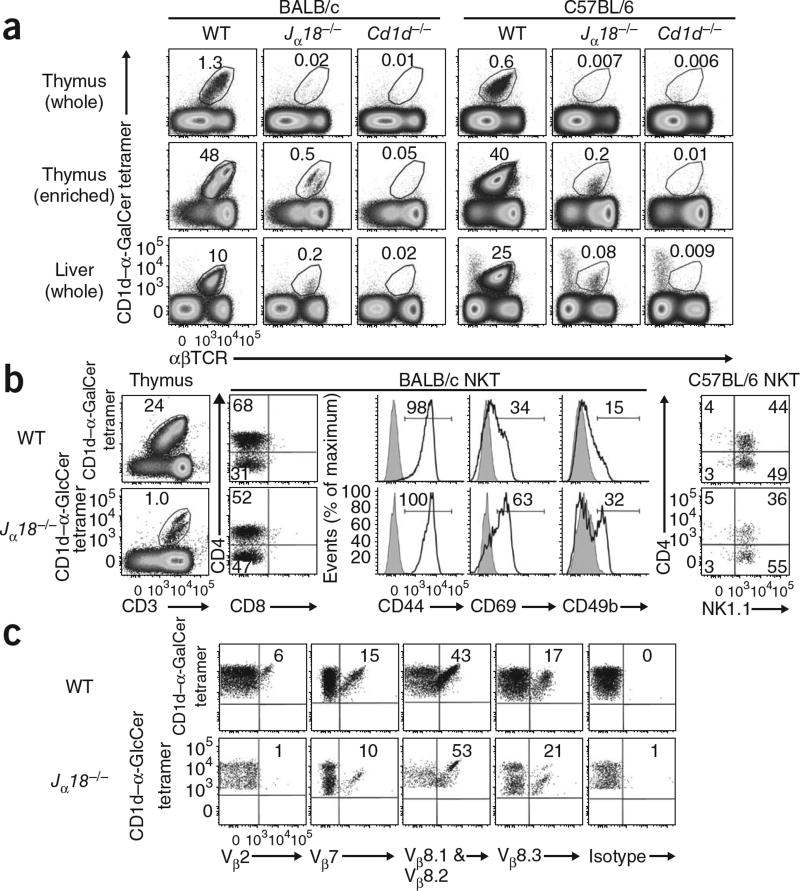
Jα18−/− mice are considered to completely lack type I NKT cells reactive to CD1d–α-GalCer1; however, we observed a small population of cells reactive to the CD1d–α-GalCer tetramer in the thymus and liver of Jα18−/− mice that was absent from Cd1d−/− mice (Fig. 1a). This population was most obvious on the BALB/c background, although it was also detectable in C57BL/6 mice, and was most prominent after enrichment by depletion of CD8+ and CD24+ thymocytes. Similar to BALB/c type I NKT cells23 (Fig. 1a), these cells could often be categorized into two subsets based on the amount of TCR expression; they also included CD4+ and CD4− subsets, were CD44hi and CD69int (Fig. 1b) and expressed a TCR Vβ repertoire enriched for Vβ8 and Vβ7 (Fig. 1c). In C57BL/6 mice, these cells expressed the activating NK cell receptor NK1.1, similar to C57BL/6 type I NKT cells (Fig. 1b). Accordingly, these cells, which lacked the invariant Vα14-Jα18 TCRα yet recognized CD1d–α-GalCer, were representative of neither classical type I NKT cells nor type II NKT cells and represent a previously unrecognized population of NKT cells.
Figure 1.
Identification of Jα18−/− T cells reactive to CD1d–α-GalCer. (a) Flow cytometry of thymocytes and liver lymphocytes isolated from BALB/c and C57BL/6 wild-type (WT), Jα18−/− and Cd1d−/− mice (n = 7–9 mice per genotype) and stained with tetramer loaded with CD1d–α-GalCer and monoclonal antibody to αβTCR; thymocyte populations were also enriched for NKT cells by complement-mediated depletion of CD24+ and CD8+ thymocytes. Numbers above outlined areas indicate percent tetramer-positive αβTCR+ cells. (b) Expression of CD4 versus CD3, CD8, CD44, CD69 and CD49b (black lines) and isotype-matched control antibody staining (gray shading) on NKT cells reactive to CD1d–α-GalCer (BALB/c wild-type cells) or CD1d–α-GlcCer (BALB/c Jα18−/− cells); right, expression of CD4 and NK1.1 in NKT cells from mice on the C57BL/6 background. Numbers above outlined areas, in quadrants or above bracketed lines, indicate percent positive cells in each area. (c) Expression of various TCRβ Vβ regions by NKT cells from BALB/c wild-type and Jα18−/− mice. Numbers in quadrants indicate percent positive cells in each. Data are representative of three (a) or two (b,c) separate experiments.
Jα18−/− NKT cells express a Vα10-Jα50 TCR α-chain
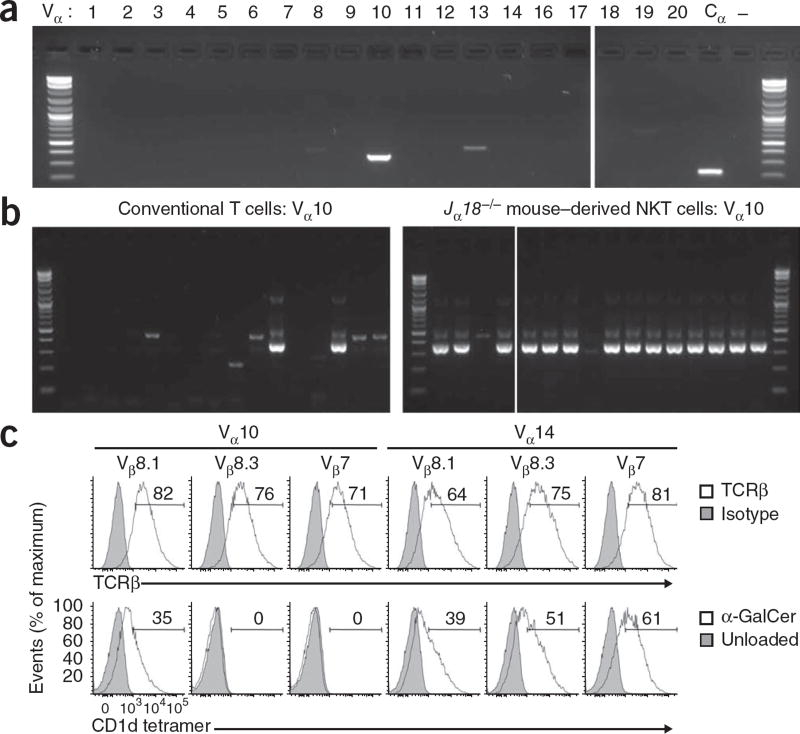
We next determined whether NKT cells derived from Jα18−/− mice expressed the gene encoding Vα14 or, if not, which genes encoding TCR Vα were used by these cells. RT-PCR-based analysis with a panel of primers for each gene encoding TCR Vα showed a strong band corresponding to Vα10 (TRAV13), but little expression of other Vα genes, apart from a faint band corresponding to Vα13 that sequencing confirmed was nonfunctional (Fig. 2a and data not shown). We then used single-cell RT-PCR analysis to determine the frequency of Vα10 NKT cells. As expected, most conventional T cells (negative for the CD1d–α-GalCer tetramer) did not express Vα10; however, most (14 of 16) of the NKT cells from Jα18−/− mice reactive to the CD1d–α-GalCer tetramer were Vα10+ (Fig. 2b). Furthermore, sequence analysis of the PCR products showed that all Vα10+ NKT cells expressed the gene encoding Jα50 (Table 1). Of 33 Vα10+ sequences from four independent cell sorts, all included Jα50 and one of five similar CDR3α sequences, each with the same length. Moreover, Vα10 has nine subtypes (TRAV13-1 through TRAV13-5, and TRAV13D-1 through TRAV13D-4), and all sequences were TRAV13-3 or TRAV13D-3 (which differ by a single amino acid in the framework region), which suggested a potential role for both the CDR1α and CDR2α regions of the Vα10 NKT cell TCR in antigen recognition. Sequence analysis of the TCR β-chain with Vβ8.1- and Vβ8.2-specific primers showed diverse expression of TCR Jβ and diverse length and composition of CDR3β (Table 1). Thus, similar to the Vα14 sequence of type I NKT cells, the TCR α-chain sequence of Vα10 NKT cells was nearly invariant, whereas the TCR β-chain was heavily biased toward Vβ8 use but was diverse in the CDR3β region.
Figure 2.
Jα18−/− CD1d–α-GalCer+ NKT cells express a semi-invariant Vα10-Jα50–Vβ8+ TCR. (a) PCR analysis of cDNA isolated from CD1d–α-GalCer-reactive cells sorted from BALB/c Jα18−/− thymuses and amplified with a panel of primers specific for each TCRα V-gene segment or the α-chain constant region (Cα). – (far right), Cα primers with no cDNA. (b) Single-cell PCR analysis for Vα10 on cDNA isolated from Vβ8.1 and Vβ8.2+ cells positive for CD1d–α-GalCer tetramer (right; Jα18−/− mouse–derived) or Vβ8.1 and Vβ8.2+ CD4+ αβTCR+ cells negative for the CD1d–α-GalCer tetramer (left; conventional T cells) sorted from BALB/c Jα18−/− thymuses; n = 16 cells per panel. (c) Staining of surface TCRβ (top row) and unloaded or α-GalCer-loaded CD1d tetramer (bottom row) on green fluorescent protein–gated human epithelial 293T cells transfected to express full-length rearranged Vα10-Jα50 or Vα14-Jα18 TCR α-chain, plus Vβ8.1, Vβ8.3 or Vβ7 TCR β-chain, and CD3 complex. Isotype, isotype-matched control antibody. Numbers above bracketed lines indicate percent-positive cells. Data are from one experiment (a,b; one for each) or are representative of one (Vβ8.1) or two (Vβ8.3 and Vβ7) experiments (c).
Table 1.
Sequence analysis of Vα10 NKT cells
| TCRα | Gene segment | ||
|
| |||
| Sequence | Vα | V-N | Jα |
|
| |||
| 1 | Vα10 | 104CAIAS | SSFSKLVFGQGTSLSVVP (Jα50) |
| 2 | Vα10 | 104CAIRS | SSFSKLVFGQGTSLSVVP (Jα50) |
| 3 | Vα10 | 104CAMKA | SSFSKLVFGQGTSLSVVP (Jα50) |
| 4 | Vα10 | 104CAMRA | SSFSKLVFGQGTSLSVVP (Jα50) |
| 5 | Vα10 | 104CAMQF | SSFSKLVFGQGTSLSVVP (Jα50) |
|
| |||
| TCRβ | |||
|
| |||
| Sequence | Vβ | V-NDN | Jβ |
|
| |||
| 1 | Vβ8.1 | 104CASRLGG | YEQYFGPGTRLTVL (Jβ2.7) |
| 2 | Vβ8.2 | 104CASGDWGA | NTGQLYFGEGSKLTVL (Jβ2.2) |
| 3 | Vβ8.2 | 104CASGGTGGR | EQYFGPGTRLTVL (Jβ2.7) |
| 4 | Vβ8.2 | 104CASGDWGA | EQYFGPGTRLTVL (Jβ2.7) |
| 5 | Vβ8.1 | 104CASRGQG | TEVFFGKGTKLTVV (Jβ1.1) |
| 6 | Vβ8.2 | 104CASGAGLGGRD | NYAEQFFGPGTRLTVL (Jβ2.1) |
| 7 | Vβ8.2 | 104CASGGRLGG | YAEQFFGPGTRLTVL (Jβ2.1) |
| 8 | Vβ8.2 | 104CASSDIWGGH | EQYFGPGTRLTVL (Jβ2.7) |
|
| |||
| NKT cell subset | CDR1α | CDR2α | CDR3α |
|
| |||
| Vα10 (Vα10-Jα50) | TTLNS | SPSWA | CAIASSSFSKLV |
| Type I (Vα14-Jα18) | VTPDNH | LVHEND | CVVGDRGSALGR |
PCR analysis of unique sequences for TCR α-chains (33 total sequences) and β-chains (13 total sequences) of Vα10 NKT cells sorted from thymus or liver, amplified with primers specific for Vα10, and Vβ8.1 and Vβ8.2 (top); and CDR1α, CDR2α and CDR3α sequences from Vα10 and type I NKT cell TCR α-chains (below). Superscripted numbers adjacent to sequences (top) indicate position of first amino acid in sequence; bolding (below) indicates type I NKT cell TCR residues critical for CD1d–α-GalCer interactions. N, nucleotide additions to TCR sequences; D, diversity region.
Data are representative of three (TCRα) or two (TCRβ) separate experiments.
The Vα10-Jα50 TCRα facilitates recognition of CD1d–α-GalCer
Alignment of the sequences of Vα10-Jα50 and Vα14-Jα18 showed little homology; moreover, none of the residues with a key role in CD1d-antigen recognition in the Vα14-Jα18 NKT cell TCR19–21 were conserved in the Vα10-Jα50 TCR (Table 1). Therefore, we directly determined whether this TCRα facilitated recognition of CD1d–α-GalCer. We transfected human epithelial 293T cells with rearranged Vα10-Jα50 or Vα14-Jα18 TCR α-chains, plus Vβ8.1 TCR β-chain with a sequence derived from Vα10 NKT cells (Table 1, sequence 1) or with Vβ8.3 or Vβ7 TCR β-chains derived from irrelevant T cell lines (H-2Db restricted and influenza A specific), and CD3 complex. The Vα10-Jα50+ TCR paired with each TCRβ to support surface expression of αβTCR, and when paired with the Vβ8.1+ TCRβ, it conferred recognition of CD1d–α-GalCer (Fig. 2c). The reactivity of the Vα14-Jα18 TCR α-chain to CD1d–α-GalCer was more permissive, with TCR β-chains Vβ8.1, Vβ8.3 and Vβ7 all conferring binding. Thus, we have identified a previously unknown canonical NKT cell TCR α-chain distinct from Vα14-Jα18 yet still able to confer recognition of CD1d–α-GalCer when paired with a permissive TCR β-chain. The finding that the irrelevant Vβ8.3 and Vβ7 TCR β-chains did not support binding of the CD1d-α-GalCer tetramer suggested that CDR3β is also important for the reactivity of the Vα10 NKT cell TCR to the CD1d-α-GalCer tetramer.
Glycolipid recognition by Vα10 NKT cells
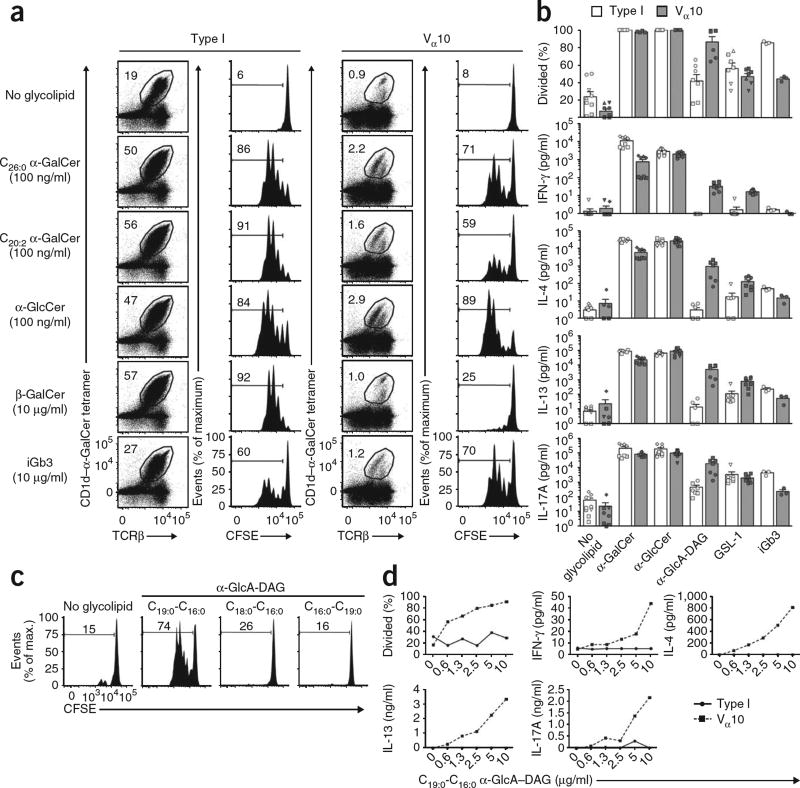
In addition to recognizing α-GalCer, type I NKT cells can recognize a range of glycosphingolipid antigens. Given the very different amino acid sequences of the Vα10-Jα50 TCR and the Vα14-Jα18 TCR of type I NKT cells, we labeled thymocyte samples enriched for Vα10 and type I NKT cells with the fluorescent dye CFSE and assessed their proliferative responses in vitro to a panel of lipid-based CD1d ligands that included bacterial and self lipid antigens (Fig. 3, Supplementary Fig. 1 and data not shown). Although both types of NKT cells proliferated considerably in response to α-GalCer with a saturated 26-carbon (C26:0) acyl chain (KRN7000) and a α-GalCer analog with a di-unsaturated 20-carbon (C20:2) acyl chain24, the strongest proliferative response of Vα10 NKT cells was in response to C20:2 α-glucosylceramide (α-GlcCer)25. The mammalian glycolipids iGb3 and β-GalCer stimulated both types of NKT cells, although the response of Vα10 NKT cells to β-GalCer was more variable among experiments (Fig. 3a and data not shown).
Figure 3.
Vα10 NKT cells have a unique hierarchy of antigen recognition. (a) Frequency of NKT cells among thymocyte populations obtained from BALB/c wild-type and Jα18−/− mice, depleted of CD8+ and CD24+ cells and cultured for 72 h with glycolipid-pulsed antigen-presenting cells (Jα18−/− splenocytes), and proliferation of CFSE-labeled type I and Vα10 NKT cells (gated on CD1d–α-GalCer tetramer, with an additional gate to exclude CFSE spectral overlap (not shown)). Numbers above outlined areas or brackets indicate percent positive cells in each; numbers above bracketed lines indicate percent divided cells. Data are from one of two similar experiments. (b) Proliferation (top) and cytokines in supernatants (below) of NKT cells positive for the CD1d–α-GalCer tetramer sorted from BALB/c wild-type and Jα18−/− mice, then labeled with CFSE and cultured for 72 h (4 × 103 cells per well) in the presence of no glycolipid, α-GalCer (C26:0; 500 ng/ml), α-GlcCer (C20:2; 500 ng/ml), α-GlcA–DAG(mixture of variants; 10 µg/ml), GSL-1 (1 µg/ml) or iGb3 (10 µg/ml), plus 20 × 103 sorted splenic CD11c+ dendritic cells. Each symbol shape represents a different experiment. IFN-γ, interferon-γ. Data are from up to four independent experiments (mean and s.e.m. of three to twelve replicates). (c) Proliferation of gated Vα10 NKT cells positive for the CD1d–α-GalCer tetramer, sorted from BALB/c Jα18−/− thymus, labeled with CFSE and cultured for 72 h in the presence (1 × 103 cells per well) or absence (5 × 103 cells per well) of α-GlcA-DAG (10 µg/ml), plus sorted CD11c+ dendritic cells (20 × 103 per well). Numbers above bracketed lines indicate percent divided cells. Data are from one experiment with two replicates. (d) Proliferation and cytokine concentrations in supernatants of NKT cells positive for the CD1d–α-GalCer tetramer, sorted from BALB/c wild-type thymus (type I) or Jα18−/− thymus (Vα10), labeled with CFSE and cultured for 72 h (2 × 103 cells per well) with 20 × 103 sorted CD11c+ dendritic cells and doubling dilutions of C19:0-C16:0 α-GlcA-DAG. Data are from one of two similar experiments (mean of duplicate cultures).
To determine if Vα10 NKT cells recognize glucose-containing microbial glycolipid ligands, we assessed the proliferation and cytokine production (interferon-γ, interleukin 4 (IL-4), IL-13 and IL-17A) in additional experiments involving coculture of purified CFSE-labeled Vα10 and type I NKT cells in the presence of purified CD11c+ dendritic cells plus the following antigens: α-glucuronosyl diacylglycerol (α-GlcA-DAG) derived from Mycobacterium smegmatis (a mixture of three variants (C19:0-C16:0, C18:0-C16:0 and C16:0-C19:0) at a ratio of 1:1:1 (wt/wt/wt), with one being the naturally occurring glycolipid; Supplementary Fig. 1), α-glucuronosyl ceramide (GSL-1) derived from Sphingomonas species, α-GalCer, α-GlcCer and iGb3 (Fig. 3b). Although most Vα10 and type I NKT cells proliferated in response to α-GalCer and α-GlcCer, α-GlcA-DAG elicited a stronger proliferative response from Vα10 NKT cells than from type I NKT cells. Furthermore, Vα10 NKT cells produced 10- to 100-fold more IL-4, IL-13 and IL-17A than did type I NKT cells in response to α-GlcA–DAG. Compared with the response of the control cells without glycolipid, the response of type I NKT cells to this antigen ranged from weak to undetectable. The proliferative response to GSL-1 was similar for Vα10 and type I NKT cells, although we also observed more production of IL-4 and IL-13 by Vα10 NKT cells in response to this glycolipid (albeit to a lesser extent than in response to α-GlcA–DAG). We also detected low concentrations of interferon-γ (<50 pg/ml) in cultures of Vα10 NKT cells in response to these microbial glycolipids. Conversely, type I NKT cells seemed to produce stronger responses to α-GalCer and iGb3.
To determine which of the three α-GlcA-DAG variants stimulated the Vα10 NKT cells, we tested each ligand separately (Fig. 3c). The naturally occurring compound C19:0-C16:0 α-GlcA-DAG26 elicited the strongest proliferative response. Dose-response experiments with C19:0-C16:0 α-GlcA-DAG showed greater stimulation of Vα10 NKT cells at a range of doses from 10 µg/ml down to 0.6 µg/ml (Fig. 3d). Type I NKT cells showed little response at any dose but responded well to the positive control (α-GlcCer; data not shown). These data suggest that although there is some overlap in specificity, Vα10 NKT cells are functionally distinct from type I NKT cells and tend to recognize glucose- and glucuronic acid–containing glycolipids.
Binding affinity of the Vα10 NKT cell TCR
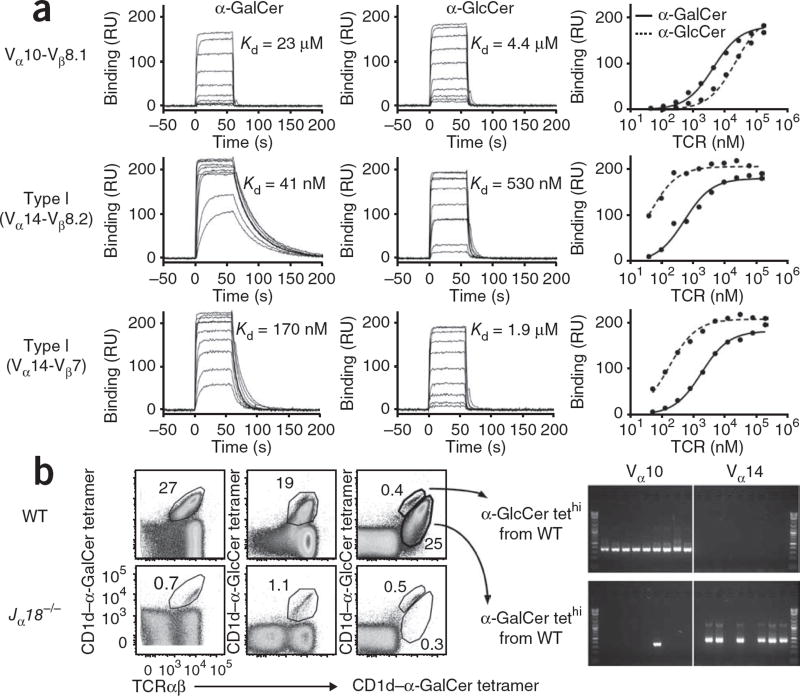
We used a soluble Vα10-Jα50 NKT cell TCR composed of paired TCR α- and β-chains (Table 1, sequence 1) from a Vα10 NKT cell clone carrying a TCR α-chain sequence present in both thymus and liver and measured affinity by surface plasmon resonance (Fig. 4a). As expected, control type I TCRs (Vα14-Jα18–Vβ8.2 and Vα14-Jα18–Vβ7; derived from published work14) bound to CD1d–α-GalCer with very high affinity (dissociation constants of 41 nM and 170 nM, respectively) and to CD1d–α-GlcCer with approximately 10% as much affinity (dissociation constants of 530 nM and 1.9 µM, respectively). Consistent with the results of the proliferation experiments (Fig. 3a), the Vα10 NKT cell TCR bound to CD1d–α-GlcCer with approximately fivefold higher affinity than it showed for CD1d–α-GalCer (dissociation constants of 4.4 µM and 23 µM, respectively). Thus, Vα10 NKT cell TCRs seemed to preferentially recognize CD1d–α-GlcCer better than CD1d–α-GalCer.
Figure 4.
Vα10 NKT cells have a higher affinity for α-GlcCer and are present in wild-type mice. (a) Binding of graded concentrations of Vα10 soluble TCR (Vα10–Vβ8.1 (175–0.05 µM)) or type I soluble TCR (Vα14–Vβ8.2 (150–0.04 µM) or Vα14–Vβ7 (200–0.05 µM)) to CD1d–α-GalCer or CD1d–α-GlcCer, after subtraction of results from those of a control flow cell (unloaded CD1d). Far right, saturation plots showing equilibrium binding to immobilized CD1d–α-GalCer or CD1d–α-GlcCer and the equilibrium dissociation constant (Kd) derived by equilibrium analysis. RU, response units. Data are representative of two independent experiments. (b) Staining profiles of α-GalCer tetramer (left) and α-GlcCer tetramer (middle) and dual tetramer labeling (right) in BALB/c wild-type or Jα18−/− thymocyte populations depleted of CD8+ and CD24+ cells, then simultaneously costained with CD1d tetramers loaded with α-GalCer and α-GlcCer. Numbers adjacent to outlined areas indicate percent cells in each. Far right, single-cell PCR analysis of Vα10 and Vα14 on wild-type NKT cells with high expression of the α-GlcCer tetramer (α-GlcCer tethi) or α-GalCer tetramer (α-GalCer tethi). Data are representative of three independent experiments.
On the basis of those differences in recognition, we determined whether costaining with CD1d tetramers loaded with either α-GlcCer or α-GalCer would allow us to distinguish Vα10 NKT cells from type I NKT cells in wild-type mice. Costaining of thymocytes showed a minor population that stained more brightly with the α-GlcCer tetramer (Fig. 4b), and a major population was stained more brightly with the α-GalCer tetramer. We assessed the TCR expression of these populations by single-cell PCR. All nine cells with high expression of the CD1d–α-GlcCer tetramer were Vα10+ (Fig. 4b), whereas none were Vα14+ and we successfully sequenced eight of nine as Vα10-Jα50+ (data not shown). In contrast, only one of nine cells with high expression of the CD1d–α-GalCer tetramer was Vα10+, and sequencing determined that this was a nonproductive Jα50− rearrangement (data not shown), whereas six of nine of these cells were Vα14+. Thus, Vα10+ NKT cells were present in wild-type mice and could be distinguished from type I NKT cells by costaining with CD1d tetramer loaded with either α-GalCer or α-GlcCer.
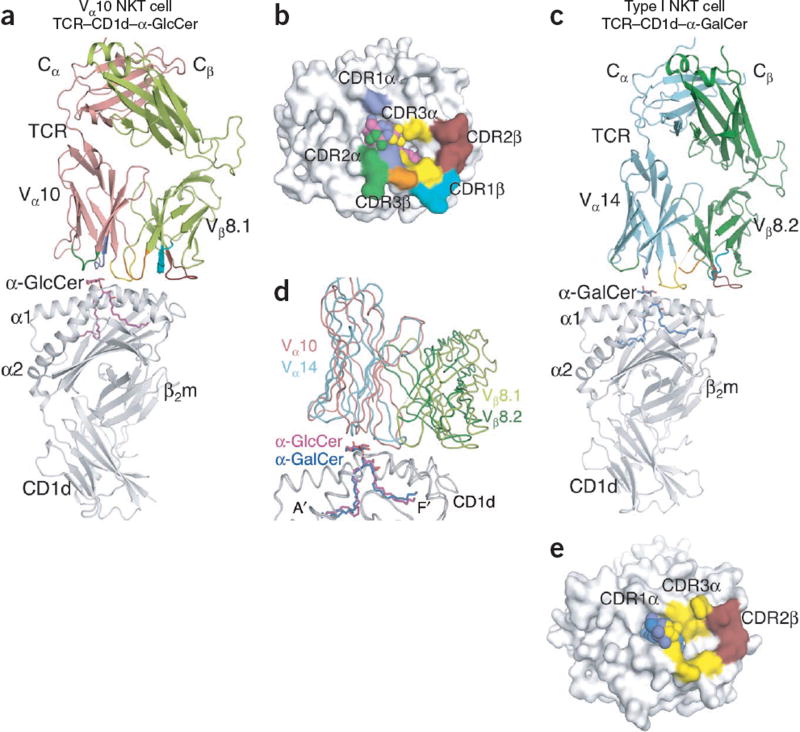
The Vα10 TCR–CD1d–α-GlcCer ternary complex
We determined the structure of the Vα10 TCR–CD1d–α-GlcCer complex at a resolution of 2.2Å (Fig. 5 and Supplementary Table 1). The Vα10 TCR adopted a parallel docking mode above the F′ pocket of the CD1d antigen-binding cleft (Fig. 5a,b) in a manner very similar to that of other type I NKT cell TCR–CD1d–α-GalCer complexes (Fig. 5c). However, there was a 14° difference in the docking angles, with the Vα10 TCR leaning more toward the F′ pocket than did the type I NKT cell TCR–CD1d–α-GalCer complex (Fig. 5d).
Figure 5.
Structural comparison of Vα10 NKT cell TCR–CD1d–α-GlcCer and type I NKT cell TCR–CD1d–α-GalCer. (a) NKT cell Vα10–Vβ8.1 TCR in complex with CD1d–α-GlcCer: magenta, α-GlcCer; gray, CD1d; salmon, Vα10; light green, Vβ8.1; purple, CDR1α; dark green, CDR2α; yellow, CDR3α; teal, CDR1β; ruby, CDR2β; orange, CDR3β. β2m, β2-microglobulin. (b) Footprint of the NKT cell Vα10–Vβ8.1 TCR on the surface of CD1d–α-GlcCer: spheres indicate α-GlcCer; colors of CD1d and CDR loops as in a. (c) Type I NKT cell Vα14–Vβ8.2 TCR in complex with CD1d–α-GalCer14: blue, α-GalCer; cyan, Vα14; dark green, Vβ8.2; colors of CD1d and CDR loops as in a. (d) Superposition of NKT cell Vα10–Vβ8.1 TCR–CD1d–α-GlcCer and type I NKT cell Vα14–Vβ8.2 TCR–CD1d–α-GalCer (colors as in a,c). (e) Footprint of the type I NKT cell Vα14–Vβ8.2 TCR on the surface of CD1d–α-GalCer: spheres indicate α-GalCer; colors of CD1d, α-GalCer and CDR loops as in a,c.
The interactions of the Vα10 TCR with CD1d spanned residues 72–87 and 145–154 of the α1 helix and α2 helix, respectively (Supplementary Table 2), which buried ~910Å2 after ligation; this was slightly greater than the buried surface area at the interface of type I NKT cell TCR–CD1d–α-GalCer (buried surface area, 760–860Å2 for Vβ8.2 and Vβ7 NKT cell TCRs14; Fig. 5b,e). At this interface, the TCR α- and β-chains contributed 59% and 41% of the buried surface area, respectively, with all CDRs contacting CD1d. CDR1α, CDR2α and CDR3α contributed 14%, 13% and 32%, respectively, whereas the CDR1β, CDR2β and CDR3β loops contributed 8%, 21% and 12%, respectively. Thus, whereas the CDR3α and CDR2β loops dominated interactions at the Vα10 TCR–CD1d–α-GlcCer interface, the involvement of CDR2α was unique to Vα10 NKT cells, in contrast to the type I NKT Vβ8+ TCR, in which only CDR1α, CDR3α and CDR2β mediated contact with CD1d–α-GalCer. Thus, the Vα10 and Vα14 NKT cell TCR–CD1d–antigen complexes showed similar docking modes, despite substantial differences in their Vα and Jα sequences.
Vα10 TCR–CD1d interactions
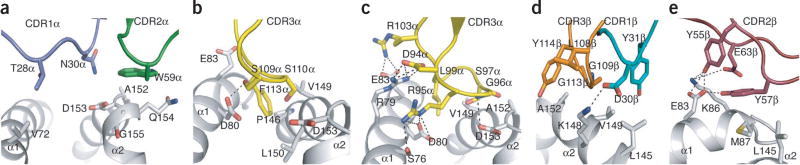
As Vα10 shares only 40% sequence identity with Vα14, we observed many differences in the interactions mediated by the respective TCR α-chains. In the Vα10 complex, Thr28α and Asn30α of CDR1α made van der Waals interactions with Val72 and Asp153 of CD1d, respectively (Fig. 6a), whereas in the type I complex, the CDR1α loop exclusively contacted α-GalCer14. The CDR2α loop of Vα10 TCR contained a bulky Trp59α residue that made extensive van der Waals interactions with residues spanning Ala152–Gly155 of the α2 helix of CD1d (Fig. 6a and Supplementary Table 2), whereas the corresponding loop in the type I TCR does not mediate any contacts with CD1d–α-GalCer14 or CD1d–α-GlcCer17. Jα50-encoded CDR3α interacted with residues from both the α1 and α2 helices of CD1d, including one hydrogen bond (between Ser109α and Asp80). Most notably, the bulky side chain of Phe113α protruded into the binding cleft between the α1 helix and α2 helix and made van der Waals interactions with Asp80 and Glu83 of the α1 helix and Pro146 and Val149 of the α2 helix (Fig. 6b, Supplementary Table 2). Accordingly, the CDR3α-CD1d interactions in the Vα10 complex were dominated by van der Waals contacts, whereas the type I NKT cell TCR CDR3α-CD1d interface was characterized by a large network of polar interactions14 (Fig. 6c).
Figure 6.
CD1d-mediated interactions with Vα10–Vβ8.1 NKT cell TCR. (a) Contacts of Vα10 NKT cell TCR CDR1α and CDR2α with CD1d. (b) Contacts of Vα10 NKT cell TCR CDR3α with CD1d. (c) Contacts of type I NKT cell TCR CDR3α with CD1d14. (d) Contacts of Vα10 NKT cell TCR CDR1β and CDR3β with CD1d. (e) Contacts of Vα10 NKT cell TCR CDR2β with CD1d. Purple, CDR1α; dark green, CDR2α; yellow, CDR3α; teal, CDR1β; ruby, CDR2β; orange, CDR3β; gray, CD1d; black dashed lines, hydrogen bonds and salt-bridge interactions.
Vβ8.1 shares 87% and 52% sequence identity with Vβ8.2 and Vβ7, respectively, and thus many contacts were conserved between the TCR β-chain of Vα10 and type I complexes. As noted before for type I NKT cell TCRs14, the TCR β-chain of the Vα10 NKT cell TCR contacted only CD1d (Supplementary Table 2). The interactions between Vα10 NKT cell TCR CDR1β and CD1d were mediated by Asp30β and Tyr31β, with Asp30β forming a salt bridge with Lys148, whereas the aromatic side chain of Tyr31β interacted with Val149 (Fig. 6d). These CDR1β-CD1d interactions were absent from the Vβ8.2 NKT cell TCR–CD1d–α-GalCer complex, whereas the Vβ7 complex included an equivalent salt bridge (Glu30β-Lys148)14.
In the CDR2β loop, Tyr55β, Tyr57β and Glu63β contacted CD1d, with Tyr55β and Tyr57β forming hydrogen bonds and van der Waals contacts with Glu83 and Lys86, which also formed a salt bridge with Glu63β. In addition, Tyr57β formed van der Waals interactions with Met87 and Leu145 of the CD1d α1 helix and α2 helix, respectively (Fig. 6e). These interactions were analogous to those mediated by the CDR2β loop of the Vβ8.2 type I NKT cell TCR14.
The hypervariable CDR3β loop ‘sat’ above the α2 helix, which allowed Leu108β, Gly109β, Gly113β and Tyr114β to make van der Waals interactions with CD1d (Fig. 6d). This suggests that the CDR3β loop has an important role in the Vα10 TCR–CD1d interaction and is consistent with the observation that Vβ8.3 and Vβ7 β-chains containing irrelevant CDR3β sequences failed to confer reactivity to the CD1d–α-GalCer tetramer when paired with Vα10 (Fig. 2c). Accordingly, despite the similar footprints of the Vα10 and type I NKT cell TCRs with CD1d, specific differences at the binding interface were readily apparent.
Plasticity of the Vα10 TCR interaction
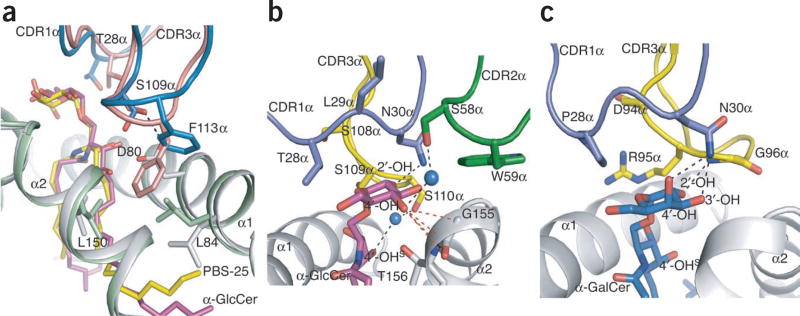
We also determined the structure of the Vα10 NKT cell TCR not bound to a ligand at a resolution of 2.9Å (Supplementary Table 1) and assessed the conformational changes that took place after ligation. In addition to small differences between the TCR alone and in complex in the Vα-Vβ domain positioning, some of the CDR loops had also moved to allow optimal binding to CD1d–α-GlcCer. In addition to the small change (root mean square deviation (r.m.s.d.), 1.1Å) in the CDR1α loop, we observed notable conformational changes in the CDR3β loop (r.m.s.d., 2.4Å, with maximal shift at Gly109β; data not shown) and CDR3α loop (r.m.s.d., 1.5Å, maximal shift of 2.8Å at Phe113α; Fig. 7a). In the Vα10 NKT cell TCR not bound to a ligand, the CDR3β interacted with the tip of CDR3α; however, in the complex, CDR3β moved away from CDR3α, making contacts with the α2 helix of CD1d instead (Fig. 6d). The movement in the CDR3α loop was particularly notable, as reorientation of side-chain residues enabled enhanced contacts with CD1d–α-GlcCer: Ser109α moved toward and formed a hydrogen bond with Asp80 of CD1d, and the Phe113α side chain was orientated to allow contacts with residues from both the α1 helix and α2 helix of CD1d (Fig. 7a). This Phe113α was in a position equivalent to that of Leu99α of the type I TCR (Fig. 6b,c), in which Leu99α was in a small hydrophobic niche formed by Leu84, Leu150 and Val149 of CD1d. Similarly, Phe113α was also in a hydrophobic niche formed by equivalent residues of CD1d. However, the longer side chain of Phe113α protruded further into the CD1d cleft, causing Leu84 and Leu150 to reorientate. The reorientation of Leu84 in turn pushed the tip of the sphingosine tail deeper into the F′ pocket of the binding groove compared with the CD1d-lipid binary complex (Fig. 7a). Thus, in contrast to the rigidity of the type I NKT cell TCR–CD1d–α-GalCer interaction, we observed a greater degree of plasticity for the Vα10 NKT cell TCR after interaction with CD1d–α-GlcCer.
Figure 7.
Lipid antigen specificity. (a) Overlay of Vα10 TCR not bound to a ligand and the binary complex of CD1d–PBS-25 (Protein Data Bank accession code, 1Z5L) on the Vα10 TCR–CD1d–α-GlcCer ternary complex: gray, CD1d in the Vα10 complex; magenta, α-GlcCer; salmon, Vα10 TCR in complex; blue, Vα10 TCR not bound to a ligand; light green, CD1d in binary complex; yellow, PBS-25 (α-GalCer analog with a shorter acyl chain). (b) Interactions with the Vα10 NKT cell TCR mediated by α-GlcCer: purple, CDR1α; dark green, CDR2α; yellow, CDR3α; magenta, α-GlcCer; gray, CD1d; blue, H2O molecules; −OHS, −OH on the sphingosine chain. (c) Interactions with the type I NKT cell TCR mediated by α-GalCer14: blue, α-GalCer; colors of CD1d and CDR loops as in b. Black dashed lines, hydrogen bonds; red dashed lines, van der Waals interactions.
Specificity of Vα10 TCR for α-GlcCer
The Vα10 NKT cell TCR interacted with α-GlcCer solely via the TCR α-chain (Supplementary Table 2), with the sugar directly under the CDR1α loop (Fig. 7b), which allowed the 2′ OH to form a hydrogen bond with Asn30α. In addition, Thr28α and Leu29α of the CDR1α loop made van der Waals interactions with α-GlcCer. Unexpectedly, CDR2α also made contact with the sugar group; Trp59α made van der Waals interactions with the 3′ OH, and there was an H2O-mediated hydrogen bond between the OH group of Ser58α and the 4′ OH (Fig. 7b). In the CDR3α loop, Ser108α, Ser109α and Ser110α made van der Waals contacts with the glucose as well as the OH groups of the sphingosine tail. Further, the OH group of Ser110α made an H2O-mediated hydrogen bond with 4′ OH of the sphingosine tail (Fig. 7b). These interactions, characterized by many van der Waals interactions and H2O-mediated hydrogen bonds were in contrast to the type I NKT cell TCR–α-GalCer interactions, in which many polar OH groups of the galactosyl ring were sequestered by hydrogen bonds with residues from the CDR1α and CDR3α loops (Fig. 7c).
The main difference in the glycosyl head groups of α-GlcCer and α-GalCer was in the orientation of the 4′ OH group. The ‘downward-pointing’ glucosyl 4′ OH group contacted the main chains of Gly155 and Thr156 of CD1d, with Gly155 and its neighboring residues interacting with the CDR2α loop. Accordingly, replacement of α-GlcCer with α-GalCer (Fig. 7b) may have resulted in a destabilizing loss of interactions with CD1d. Moreover, an upward-pointing galactosyl 4′ OH group may have sterically clashed with Ser58α of the CDR2α loop of Vα10, a residue that formed an H2O-mediated hydrogen bond with the glucosyl 4′ OH. Together these data provide a molecular basis for the differences between the type I NKT cell TCR and Vα10 NKT cell TCR in their fine specificity for glycolipid antigens.
DISCUSSION
In this study we have identified a unique population of NKT cells that fit neither the type I category nor the type II category; we have called these ‘Vα10 NKT cells’. These cells recognized α-GalCer presented by CD1d and were characterized by use of a semi-invariant α-chain (Vα10-Jα50) and a bias toward Vβ8 expression. Although the development of Vα10 NKT cells remains to be investigated, the very limited junctional diversity and the absence of these cells in Cd1d−/− mice suggest that they arise through TCR-mediated intrathymic selection events in a manner akin to that of type I NKT cells. In further support of that idea, we found that both CD4+ and CD4− subsets of Vα10 NKT cells existed; they were CD44hi, CD69lo−int and NK1.1+, they produced large amounts of a range of cytokines, and they were present at greater frequency in liver than spleen (data not shown), which suggests a developmental pathway that makes them more like type I NKT cells than conventional T cells.
The Vα10 NKT cells had a hierarchy of antigen reactivity different from that of type I NKT cells27, as demonstrated by their preferential recognition of α-GlcCer and other glucose-based glyco-lipids, including the microbial glucuronic acid–containing ligand α-GlcA-DAG from M. smegmatis26 and, to a lesser extent, GSL-1 from Sphingomonas species28–30. The finding that Vα10 NKT cells produced 10–100 times more interferon-γ, IL-4, IL-13 and IL-17 in response to α-GlcA-DAG than did type I NKT cells suggested that these cells are functionally distinct. Furthermore, the type of antigen may determine which population dominates the response and may also influence, through differences in cytokine production, the outcome of the response. Further studies are warranted comparing the response of Vα10 NKT cells with that of type I NKT cells in mouse-infection studies, especially with bacteria containing glucose-based α-linked glycolipids26,28–30. Notably, human Vα24−Jα18+ type I NKT cells respond less well to α-GlcCer than to α-GalCer12, which further discriminates between the Vα10 NKT cells defined here and human Vα24−Jα18+ NKT cells. Although CD1d-mediated intrathymic selection seems to impose limitations on TCR α-chain diversity, variations in αβTCR use nonetheless imbue CD1d-restricted T cells with a range of antigen specificities and cytokine-producing potential. The finding that these cells recognized and responded to the mammalian glycolipid antigen iGb3 also suggests that they have the potential for self-reactivity in a manner similar to that of iGb3-reactive type I NKT cells29,31,32.
Although type I and Vα10 NKT cells expressed highly disparate TCR α-chains, they docked onto CD1d-glycolipid antigen in a very similar manner. Nevertheless, we observed key differences in the type I and Vα10 NKT cell TCR-CD1d-antigen interfaces, which included the CDR2α loop of the Vα10 NKT cell TCR in contact with the lipid antigen. Moreover, we observed differences between the type I and Vα10 NKT cell TCRs in the degree of structural plasticity after they engaged their respective CD1d-antigen complex. Furthermore, the CDR3α loop of the Vα10 NKT cell TCR interacted with CD1d-antigen mainly through van der Waals interactions, in contrast to the polar bond–mediated network of the CDR3α-mediated interactions of the type I NKT cell TCR14,15. Collectively, the differences at the antigen-binding interface explain the differences between type I and Vα10 NKT cell TCRs in their antigenic specificity. The CDR3α loop is crucial for docking of type I NKT cell TCR–CD1d–antigen14,15,19–21,33. However, the Vα10 NKT cell TCR docked in a similar manner, which suggests that it may be the NKT cell TCR β-chain that dictates such a conserved NKT cell TCR-CD1d docking topology. In particular, the CDR2β loop of mouse Vβ8+ and human Vβ11+ NKT cell TCRs has two tyrosine residues that form a conserved set of interactions with a defined region on CD1d. These observations have resonance with the Vβ8.2-mediated interaction ‘codons’ observed in the major histocompatibility complex–restricted response, in which two conserved tyrosine residues are considered to dictate TCR–major histocompatibility complex bias34,35. However, the Vβ7 NKT cell TCR has only one tyrosine residue in the corresponding CDR2β loop yet docks onto CD1d in a conserved manner14. Similarly, mutagenesis studies of the Vβ2 NKT cell TCR, which has no tyrosine residues in the CDR2β loop, is also predicted to dock onto CD1d in a conserved manner21. Thus, the conserved docking topology may be a consequence of the fact that an innate-like TCR must recognize a relatively monomorphic antigen-presenting molecule.
In summary, we have identified a previously unknown population of CD1d-restricted NKT cells with a unique canonical Vα10-Jα50 TCRα chain that recognize a partially but not completely overlapping array of glycolipids, including self and bacterial antigens. Our findings have also identified a feature of the interaction of TCRs with antigen-presenting molecules: although the diverse repertoire of αβTCRs allows them to bind peptide–major histocompatibility complex molecules in a range of docking modes36, the diversity of the NKT cell TCR repertoire is associated with a conserved CD1d docking mode. Accordingly, our findings highlight a fundamental difference between peptide- and lipid-mediated recognition by the TCR.
ONLINE METHODS
Mice
C57BL/6 and BALB/c mice were derived from the Peter MacCallum Cancer Centre or were from the Walter and Eliza Hall Institute of Medical Research. The following mice were bred at Peter MacCallum Cancer Centre: BALB/c CD1d1.CD1d2-deficient mice (BALB/c Cd1d−/−; 11 backcrosses; from Jackson Laboratories)37; BALB/c Jα18-deficient mice (BALB/c Jα18−/−; 10 backcrosses; originally provided by M. Taniguchi)5; C57BL/6 Jα18-deficient mice (C57BL/6 Jα18−/−; 12 backcrosses; originally provided by M. Taniguchi)5; C57BL/6 CD1d-deficient mice (C57BL/6 Cd1d−/−; 11 backcrosses; originally provided by Jackson Laboratories)38. Mice over 6 weeks of age were used in all experiments, which were done in accordance with the Animal Ethics and Experimentation Committee of Peter MacCallum Cancer Centre.
Flow cytometry
Single-cell suspensions of thymus, spleen and liver were made as described39. Where indicated, thymocyte samples were enriched for NKT cells by depletion of CD8+ and CD24+ thymocytes as described39. Monoclonal antibody to CD16-CD32 (FcR block; 2.4G2; grown in-house) was included in cell-labeling experiments. Cells were stained with the following antibodies (all from BD Pharmingen): antibody to CD3 (anti-CD3; 145-2C11) anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD44 (IM7), anti-CD49b (HMα2), anti-CD69 (H1.2F3), anti-NK1.1 (PK136), anti-TCRβ (H57-597), anti-Vβ2 (B20.6), anti-Vβ7 (TR310), anti-Vβ8.1-Vβ8.2 (MR5-2), anti-Vβ8.3 (1B3.3) or rat IgG2b isotype-matched control antibody (A95-1). Mouse CD1d tetramers (produced in-house with a baculovirus-based CD1d expression system originally derived from M. Kronenberg) were loaded with α-GalCer (from Alexis Biochemicals or P. Savage (C24:1 PBS-44 analog)). In some cases, CD1d tetramer was loaded with α-GlcCer (C20:2 DB06-15 analog24,25). In some experiments, 7-amino-actinomycin D (Molecular Probes) and/or unloaded mouse CD1d tetramers were used for the exclusion of dead and nonspecifically stained cells. Cells were analyzed with an LSR II or FACSCanto (BD Biosciences) and data were processed with FlowJo (Tree Star), with bi-exponential scaling.
In vitro stimulation
For in vitro proliferation assays, two approaches were used. In one approach (Fig. 3a), thymocyte samples were depleted of CD8+ and CD24+ cells39 and labeled for 10 min at 37 °C with 2 µM CFSE (carboxyfluorescein diacetate succinimidyl ester; Molecular Probes), then the thymocytes (1.5 × 105) were cultured for 72 h with 3 × 105 BALB/c Jα18−/− splenocytes that had been pulsed the night before with α-GalCer (C26:0 or C20:2; 100 ng/ml), α-GlcCer (C20:2; 100 ng/ml)24,25, β-GalCer (C12; 10 µg/ml; Avanti Polar Lipids) or iGb3 (C26:0; 10 µg/ml; Alexis Biochemicals). Glycolipid stock solutions were dissolved in either dimethyl sulfoxide or a solution of 0.5% (vol/vol) Tween-20, sucrose (56 mg/ml) and l-histidine (7.5 mg/ml) and were sonicated for ~15 min immediately before use. For the second approach (Fig. 3b–d), purified αβTCR+ thymic NKT cells positive for CD1d–α-GalCer tetramer (1 × 103 to 4 × 103 cells) were labeled with 1 µM CFSE and were cultured for 72 h in 50 µl culture medium in the presence of 2 × 104 splenic CD11c+ dendritic cells (from BALB/c Jα18−/− mice) with or without the following synthetic glycolipids: C26:0 α-GalCer (500 ng/ml), α-GlcCer (500 ng/ml), M. smegmatis α-GlcA-DAG (0.6–10 µg/ml; either separate or a mixture of three analogs (C18:0-C16:0, C19:0-C16:0 and C16:0-C19:0) at a ratio of 1:1:1 (wt/wt/wt); structures, Supplementary Fig. 1), Sphingomonas species GSL-1 (C14:0 α-glucuronosyl ceramide40; 1 µg/ml; National Institutes of Health Tetramer Core Facility) or iGb3 (10 µg/ml; Alexis Biochemicals). Cells were cultured in RPMI-1640 medium (Invitrogen Life Technologies) supplemented with 10% (vol/vol) FCS (JRH Biosciences), penicillin (100 U/ml), streptomycin (100 µg/ml), Glutamax (2 mM), sodium pyruvate (1 mM) and nonessential amino acids (0.1 mM) and HEPES buffer (15 mM), pH 7.2–7.5 (all from Invitrogen Life Technologies), plus 50 µM 2-mercaptoethanol (Sigma-Aldrich). Culture supernatants were analyzed for cytokines with a Cytometric Bead Array Mouse Flex Set (BD Biosciences).
Synthesis of α-GlcA-DAG (C19:0-C16:0)
Three glycolipid variants derived from M. smegmatis26 were synthesized: 1-O-((R)-10-tuberculostearyl)-2-O-palmityl-sn-glyceryl α-D-glucopyranosiduronic acid (C19:0-C16:0;) was prepared from 1-O-tert-butyldiphenylsilyl-2-O-p-methoxybenzyl-sn-glycerol and 2,3,4-tri-O-benzyl-6-O-acetyl-α-d-glucopyranosyl iodide, followed by C6-oxidation and esterification (B.C. et al., personal communication). Hydrogenolysis of 1-O-((R)-10-tuberculostearyl)-2-O-palmityl-sn-glyceryl (benzyl 2,3,4-tri-O-benzyl-α-d-glucuronate) gave the title compound [α]D23 +39.2 (c 0.25 in dimethyl sulfoxide), δH (500 MHz; dimethyl sulfoxide) 0.81 (d, 3H), 0.85 (t, 6H), 1.03–1.23 (m, 51H), 1.50 (m, 4 H), 2.24–2.31 (m, 4 H), 3.22 (dd, 1H), 3.55 (dd, 1H), 3.69 (dd, 1H), 3.77 (d, 1H), 4.16 (dd, 1H), 4.32 (dd, 1H), 4.69 (d, 1H), 5.11 (m, 1H); δC (125 MHz; dimethyl sulfoxide) 13.91, 19.55, 22.09, 24.41, 24.44, 26.40, 26.44, 28.43, 28.45, 28.69, 28.71, 28.76, 28.96, 29.02, 29.07, 29.36, 29.37, 31.29, 32.06, 33.42, 33.53, 36.42, 36.44, 62.19, 65.48, 69.58, 71.38, 71.76, 72.61, 99.37, 171.06, 172.25, 172.53; high-resolution mass spectrometry–electrospray ionization [M + Na]+ calculated for C44H82O11, 809.5749; found a mass/charge ratio of 809.5739.
Single-cell PCR and RT-PCR
For analysis of genes encoding Vα regions, RNA was extracted from sorted NKT cells with an RNeasy kit (Qiagen) and cDNA was synthesized with oligo(dT15) primers (Promega) and AMV reverse transcriptase (Promega) in accordance with the manufacturers’ instructions. NKT cell cDNA was amplified by PCR with a panel of Vα-specific primers41 and GoTaq Master Mix (Promega). For single-cell PCR, cDNA from sorted αβTCR+ Vβ8+ thymic NKT cells positive for CD1d–α-GalCer tetramer was generated42 and then was amplified by two rounds of semi-nested PCR43 with sense primers for Vα10 (external, 5′-CCTTGGTTCTGCAGGAGGGGGAG-3′, and internal, 5′-AACGTCGCAGCTCTTTGCAC-3′), Vα14 (external: 5′-TTGTCCGTCAGGGAGAGAACTGC-3′, and internal: 5′-GACACAGGCAAAGGTCTTGTGTCC-3′) or Cα (5′-GAACCTGCTGTGTAC-3′), each used with the Cα antisense primer (both rounds; 5′-TGGCGTTGGTCTCTTTGAAG-3′). PCR products were separated by electrophoresis through a 1.5% agarose gel and were sequenced as described43. TCR residue numbering is in accordance with the International ImMunoGeneTics database44.
Cloning and expression of genes encoding mouse NKT cell TCRs
Paired TCRα and TCRβ sequences from a single Vα10 NKT cell (Table 1, sequence 1 for both TCRα and TCRβ) were used to design a chimeric transcript that also contained a modified human Cα domain to aid in refolding14. Sequences codon-optimized for Escherichia coli were synthesized (Genscript) and then transferred into a pET30 expression vector (Novagen). Soluble TRAV13D-3-TRAJ50–TRBV13-3-TRBJ2-7 (Vα10-Jα50–Vβ8.1-Jβ2.7) and TRAV11-TRAJ18–TRBV13-2 (Vα14-Jα18–Vβ8.2-Jβ2.1) NKT cell TCRs were expressed in E. coli strain BL21 and inclusion body proteins were prepared, refolded and purified by gel filtration as described45.
Transfection of 293T cells with TCR
Full-length TCRα transcripts TRAV13D-3 (Vα10; CDR3α sequence 1, Table 1) and TRAV11 (Vα14) were cloned from Vα10 and type I NKT cell cDNA, respectively, by RT-PCR. Full-length TCRβ (TRBV13-3-TRBJ2-7; Vβ8.1-Jβ2.7; CDR3β sequence 1, Table 1) was synthesized (Genscript), and TRBV13-1 (Vβ8.3) and TRBV29 (Vβ7) were generated from H-2Db-restricted influenza A–specific clones (E.B.D. et al., personal communication). TCR sequences were verified and inserted into the plasmid pMIGII, and 293T cells were transfected with hydrolase element P2A–linked genes encoding CD3, TCRα and TCRβ as described46. After 48 h of culture to allow expression of proteins, cells positive for the expression of green fluorescent protein were analyzed for surface expression of TCR by flow cytometry.
Surface plasmon resonance
A ProteOn XPR36 protein-interaction array system (Bio-Rad) was used for surface plasmon resonance as described14. Streptavidin was coupled to a GLC Sensor Chip (Bio-Rad) by amine coupling (~500 response units), and biotinylated mouse CD1d–α-GalCer, CD1d–α-GlcCer or unloaded CD1d was captured on a separate flow cell for each (~600 response units each). Soluble TCRs were serially diluted and simultaneously injected over test and control surfaces at a rate of 30 µl/min. After subtraction of data from the control flow cell (unloaded CD1d), interactions were analyzed with ProteOn Manager software version 2.1 (Bio-Rad) and the Scrubber 2.0a program (Prot version; BioLogic Software), and steady-state dissociation constants were derived at equilibrium.
Crystallization, structure determination and refinement
The Vα10 TCR–CD1d–α-GlcCer ternary complex and the Vα10 TCR not bound to ligand (both at a concentration of 10 mg/ml in 10 mM Tris, pH 8.0, and 150 mM NaCl) were crystallized at 18 °C in 20% (vol/vol) PEG 1500 and 0.1 M 2-(N-morpholino)ethanesulfonic acid, pH 6.5, and in 20% (vol/vol) PEG 3350, 0.2 M NaI and 0.1 M bis-Tris-propane, pH 6.5 (Hampton Research), respectively. Crystals were flash-frozen before data collection in crystallization solution containing 25% (vol/vol) glycerol as a cryoprotectant. Data for the ternary complex and the TCR were collected at 100 K on the MX1 beamline and MX2 beamline, respectively, at the Australian Synchrotron (Melbourne). The ternary complex crystals diffracted to 2.2Å and belonged to space group P21 with two ternary complexes in each asymmetric unit, with an r.m.s.d. of 0.57Å over 784 Cα atoms. Thus, structural analysis was restricted to one complex in the asymmetric unit. The crystals of the TCR not bound to a ligand diffracted to 2.9Å and belong to the space group C2, with two TCR molecules in each asymmetric unit (Supplementary Table 1). Data for the ternary complex were processed with the Mosflm program for integrating single-crystal-diffraction data from area detectors (version 7.0.5)47 and were scaled with the SCALA scaling and data-merging program from the CCP4 Suite (Collaborative Computational Project, number 4)48. The complex was solved by the molecular-replacement method with the MOLREP program in CCP4, with Vα14–Vα8.2 TCR minus the CDR loops (Protein Data Bank accession code, 3HE6) and mouse CD1d minus the lipid (Protein Data Bank accession code, 1Z5L) as the search models. The electron density at the TCR–CD1d–α-GlcCer interface was unambiguous and the initial experimental phases showed an unbiased density for α-GlcCer in the antigen-binding cleft of CD1d. The data for the TCR not bound to a ligand were processed and scaled with the HKL-2000 program package49. The structure of the TCR was solved by the molecular-replacement method with the Phaser crystallographic software in CCP4, with Vα14-Vα8.2 NKT cell TCR minus the CDR loops (Protein Data Bank accession code, 3HE6) as a search model. COOT (Crystallographic Object-Oriented Toolkit)50 was used for model building (Supplementary Table 1). The quality of both structures was confirmed at the Research Collaboratory for Structural Bioinformatics Protein Data Bank Data Validation and Deposition Services website. All presentations of molecular graphics were created with the PyMOL molecular visualization system. For the NKT cell TCR not bound to a ligand, all CDR loops were modeled except CDR2α, which was also involved in crystal contacts and was thus excluded from analysis. For structural analysis, although the structure of the binary complex of CD1d–α-GlcCer was not available, the degree of plasticity in CD1d after engagement of the Vα10 TCR was evaluated by comparison of the Vα10 TCR–CD1d–α-GlcCer in complex with CD1d linked to PBS-25, an analog of α-GalCer with a shorter (C8) acyl chain51.
Supplementary Material
Acknowledgments
We thank M. Taniguchi (Chiba University Graduate School of Medicine) for Jα18−/− mice; M. Kronenberg (La Jolla Institute for Allergy and Immunology) for the baculovirus-based CD1d expression system; P. Savage (Brigham Young University) for α-GalCer (C24:1 PBS-44 analog); the Australian Synchrotron staff at the MX1 and MX2 beamlines of the Australian synchrotron for assistance with data collection; S. Mattarollo, S. Doak, S. Berzins and A. Denton for discussions and assistance with some experiments; K. Field, N. Sanders and M. Reitsma for assistance with flow cytometry; and M. Stirling and the staff of the Peter MacCallum Cancer Centre Animal House and D. Maksel from the Protein Crystallography Unit at Monash University for technical assistance. Supported by the Cancer Council of Victoria, the National Health and Medical Research Council of Australia (A.P.U., L.C.S., M.J.S. and D.I.G.), the Australian Research Council (D.I.G., O.P. and J.R.), the Cancer Research Institute (G.C.) and the US National Institutes of Health (AI45889 to S.A.P.).
Footnotes
Accession codes. Protein Data Bank: Vα10 complex, 3RUG; Vα10 TCR not bound to a ligand, 3AXL.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
A.P.U. identified and carried out cellular and molecular characterization of Vα10 NKT cells and produced protein complexes for crystallographic studies; O.P. solved the crystal structures and did structural analysis; G.C. and K.K. carried out studies of glycolipid specificity and function; L.C.S. did surface plasmon resonance studies; D.G.P., E.B.D., L.K.-N., J.P.V., S.J.T., G.S.B., B.C., A.G.B., S.J.W., P.I., S.A.P., J.M., M.J.S., J.R. and D.I.G. provided intellectual input and key reagents and assisted with experimental design and interpretation and writing of the manuscript; and M.J.S., J.R. and D.I.G. led the investigation together and devised the project and contributed equally to this work.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/natureimmunology/.
References
- 1.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat. Rev. Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 2.Cardell S, et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J. Exp. Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SH, et al. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J. Exp. Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur. J. Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui JQ, et al. Requirement for Vα14 NKT cells in Il-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 6.Renukaradhya GJ, et al. Type I NKT cells protect (and type II NKT cells suppress) the host’s innate antitumor immune response to a B cell lymphoma. Blood. 2008;111:5637–5645. doi: 10.1182/blood-2007-05-092866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JH, Choi EY, Chung DH. Donor bone marrow type II (non-Vα14Jα18 CD1d-restricted) NKT cells suppress graft-versus-host disease by producing IFN-γ and IL-4. J. Immunol. 2007;179:6579–6587. doi: 10.4049/jimmunol.179.10.6579. [DOI] [PubMed] [Google Scholar]
- 8.Terabe M, et al. A nonclassical non- Vα14Jα18 CD1d-restricted (type II) NKT cell is sufficient for down-regulation of tumor immunosurveillance. J. Exp. Med. 2005;202:1627–1633. doi: 10.1084/jem.20051381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berzins SP, Smyth MJ, Godfrey DI. Working with NKT cells—pitfalls and practicalities. Curr. Opin. Immunol. 2005;17:448–454. doi: 10.1016/j.coi.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Gadola SD, et al. Structure and binding kinetics of three different human CD1d-α-galactosylceramide-specific T cell receptors. J. Exp. Med. 2006;203:699–710. doi: 10.1084/jem.20052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadola SD, Dulphy N, Salio M, Cerundolo V. V alpha 24-J alpha Q-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4+ and CD8αβ+ T lymphocytes. J. Immunol. 2002;168:5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- 12.Brigl M, et al. Conserved and heterogeneous lipid antigen specificities of CD1d-restricted NKT cell receptors. J. Immunol. 2006;176:3625–3634. doi: 10.4049/jimmunol.176.6.3625. [DOI] [PubMed] [Google Scholar]
- 13.Godfrey DI, et al. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin. Immunol. 2010;22:61–67. doi: 10.1016/j.smim.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the Vβ8.2 and Vβ7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J. Exp. Med. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wun KS, et al. A molecular basis for the exquisite CD1d-restricted Ag-specificity and functional responses of NKT cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallevaey T, et al. The molecular basis of NKT cell autoreactivity and recognition of self-CD1d. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wun KS, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J. Exp. Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 21.Mallevaey T, et al. T cell receptor CDR2β and CDR3β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florence WC, et al. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 2009;28:3579–3590. doi: 10.1038/emboj.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenstrom M, Skold M, Andersson A, Cardell SL. Natural killer T-cell populations in C57BL/6 and NK1.1 congenic BALB.NK mice-a novel thymic subset defined in BALB.NK mice. Immunology. 2005;114:336–345. doi: 10.1111/j.1365-2567.2004.02111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu KO, et al. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of α-galactosylceramides. Proc. Natl. Acad. Sci. USA. 2005;102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jervis PJ, et al. Synthesis and biological activity of α-glucosyl C24:0 and C20:2 ceramides. Bioorg. Med. Chem. Lett. 2010;20:3475–3478. doi: 10.1016/j.bmcl.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolucka BA, McNeil MR, Kalbe L, Cocito C, Brennan PJ. Isolation and characterization of a novel glucuronosyl diacylglycerol from Mycobacterium smegmatis. Biochim. Biophys. Acta. 1993;1170:131–136. doi: 10.1016/0005-2760(93)90062-e. [DOI] [PubMed] [Google Scholar]
- 27.Kawano T, et al. Cd1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 28.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 29.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 30.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur. J. Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 31.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 32.Dias BR, et al. Identification of iGb3 and iGb4 in melanoma B16F10-Nex2 cells and the iNKT cell-mediated antitumor effect of dendritic cells primed with iGb3. Mol. Cancer. 2009;8:116–129. doi: 10.1186/1476-4598-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kjer-Nielsen L, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J. Exp. Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 35.Dai S, et al. Crossreactive T Cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Smiley ST, Kaplan MH, Grusby MJ, Immunoglobulin E. Production in the absence of interleukin-4-secreting Cd1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda KH, Exley M, Snapper S, Balk SP, Stein-Streilein J. CD1-reactive natural killer T cells are required for development of systemic tolerance through an immune-privileged site. J. Exp. Med. 1999;190:1215–1226. doi: 10.1084/jem.190.9.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crowe NY, et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J. Exp. Med. 2005;202:1279–1288. doi: 10.1084/jem.20050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long X, et al. Synthesis and evaluation of stimulatory properties of Sphingomonadaceae glycolipids. Nat. Chem. Biol. 2007;3:559–564. doi: 10.1038/nchembio.2007.19. [DOI] [PubMed] [Google Scholar]
- 41.Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day EB, et al. The context of epitope presentation can influence functional quality of recalled influenza A virus-specific memory CD8+ T cells. J. Immunol. 2007;179:2187–2194. doi: 10.4049/jimmunol.179.4.2187. [DOI] [PubMed] [Google Scholar]
- 43.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc. Natl. Acad. Sci. USA. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefranc MP, et al. IMGT, the International ImMunoGeneTics database. Nucleic Acids Res. 1998;26:297–303. doi: 10.1093/nar/26.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garboczi DN, et al. Assembly, specific binding, and crystallization of a human TCR-αβ with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J. Immunol. 1996;157:5403–5410. [PubMed] [Google Scholar]
- 46.Holst J, et al. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- 47.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. Recent changes to the MOSFLM package for processing film and image plate data. 1992;26 [Google Scholar]
- 48.CCP4 (Collaborative Computational Project, 4) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Otwinoski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat. Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.