Abstract
The hormonal metabolite of vitamin D, 1,25-dihydroxyvitamin D3 (1,25D), binds to the vitamin D receptor (VDR) and promotes heterodimerization of VDR with a retinoid-X-receptor (RXR) to genomically regulate diverse cellular processes. Herein, it is revealed for the first time that VDR is post-translationally acetylated, and that VDR immunoprecipitated from human embryonic kidney (HEK293) cells displays a dramatic decrease in acetylated receptor in the presence of 1,25D-ligand, sirtuin-1 (SIRT1) deacetylase, or the resveratrol activator of SIRT1. To elucidate the functional significance of VDR deacetylation, vitamin-D-responsive-element (VDRE)-based transcriptional assays were performed to determine if deacetylase overexpression affects VDR/VDRE-driven transcription. In HEK293 kidney and TE85 bone cells, co-transfection of low amounts (1–5ng) of a SIRT1-expression vector elicits a reproducible and statistically significant enhancement (1.3- to 2.6-fold) in transcription mediated by VDREs from the CYP3A4 and cyp24a1 genes, where the magnitude of response to 1,25D-ligand is 6- to 30-fold. Inhibition of SIRT1 via EX-527, or utilization of a SIRT1 loss-of-function mutant (H363Y), resulted in abrogation of SIRT1-mediated VDR potentiation. Studies with a novel, non-acetylatable VDR mutant (K413R) showed that the mutant VDR possesses enhanced responsiveness to 1,25D, in conjunction with reduced, but still significant, sensitivity to exogenous SIRT1, indicating that acetylation of lysine 413 is relevant, but that other acetylated residues in VDR contribute to modulation of its activity. We conclude that the acetylation of VDR comprises a negative feedback loop that attenuates 1,25D-VDR signaling. This regulatory loop is reversed by SIRT1-catalyzed deacetylation of VDR to amplify VDR signaling and 1,25D actions.
Keywords: VDR, Vitamin D, Sirtuin-1, Resveratrol, Acetylation, Transactivation
1. INTRODUCTION1
Vitamin D can be acquired from intake of dietary sources/supplements or endogenously synthesized in the epidermis via UVB-stimulated photo-conversion of 7-dehydrocholesterol [1, 2]. Vitamin D is then transported to the liver and hydroxylated at the C-25 position by a mitochondrial cytochrome P450-containing (CYP) enzyme, CYP2R1, to generate 25-hydroxyvitamin D3 (25D), the primary circulating metabolite employed to quantitate clinical vitamin D status. The production of the active hormonal secosteroid occurs predominately in the kidneys via CYP27B1 which catalyzes the 1α-hydroxylation of 25-hydroxyvitamin D3 to 1α,25-dihydroxyvitamin D3 (1,25D). Circulating 1,25D bound to plasma vitamin D-binding proteins (DBPs) is systemically delivered to, and retained by, distal vitamin D target cells expressing the vitamin D receptor (VDR) [3]. Binding of 1,25D to VDR induces significant conformational alterations in the receptor, leading to nuclear accumulation and the recruitment of, and heterodimerization with its co-receptor, one of the retinoid X receptors (RXRs). By docking on vitamin D responsive elements (VDREs) in DNA, the ligand-activated VDR-RXR heterocomplex attracts co-regulatory transcription factors that influence the expression of target genes in tissues such as the small intestine, colon, kidney, bone, cardiac muscle, skin, and brain [4]. Classically, vitamin D regulates bone mineral homeostasis via signaling intestinal absorption, bone resorption, and renal reabsorption of calcium and phosphate to ensure proper remodeling of the mineralized skeleton [3, 5]. Extraosseous effects of vitamin D include oxidative stress reduction, immunoregulation, antimicrobial defense, xenobiotic detoxification, neuroprotection, cardiovascular maintenance, anti-inflammatory processes, insulin control, and nontoxic epithelial cell chemoprevention via regulation of cell proliferation and differentiation, in the many tissues where VDR and RXR are both expressed [5, 6].
1.1. 1,25D-VDR Signaling Modulators
In addition to 1,25D, several other nutritionally-derived compounds have been characterized recently as possible VDR agonists and/or potentiators capable of effecting receptor activation and downstream biological actions. We evaluated resveratrol (3,4',5-trihydroxy-trans-stilbene), a lipophilic compound, as a candidate for direct VDR binding and regulation [7]. Utilizing VDRE and mammalian 2-hybrid (M2H) transcriptional system technologies, we concluded that VDRE-mediated transcriptional activity and VDR-RXR heterodimerization were increased by resveratrol, as the combination of 1,25D and resveratrol yielded a cooperative effect [7]. However, radiolabeled-1,25D ligand displacement experiments revealed an increase in VDR-bound, radiolabeled 1,25D in the presence of resveratrol, indicating that resveratrol is not a VDR ligand and must potentiate VDR transactivation via an indirect pathway [7]. This Dampf Stone et al. study provided initial insights into the mechanism whereby resveratrol affects VDR activation; however, the precise molecular events underlying this interaction have yet to be determined. Consequentially, the aim of this study was to further investigate VDR potentiation by resveratrol through a focus on one of the target enzymes for this beneficial nutrient, namely the sirtuin-1 (SIRT1) deacetylase.
1.2. SIRT1 and Resveratrol
The silent information regulator-2 (Sir-2) family of proteins, commonly termed sirtuins, function as NAD+-dependent histone and non-histone protein deacetylases that couple the cleavage of NAD+ and deacetylation of lysine residues in histone/non-histone protein substrates to generate nicotinamide and the metabolite, O-acetyl-ADP-ribose [8]. Although most sirtuins retain deacetylase activity, SIRT4 has been shown to have only ADP-ribosyltransferase activity, whereas SIRT1 and SIRT6 have both deacetylation and weak ADP-ribosyltransferase activity [8–11]. Human nuclear sirtuin-1 (SIRT1), a class III HDAC, has been shown to regulate cell survival by inhibiting p53-dependent processes and modulating transcription, muscle cell differentiation, adipogenesis, preventing axonal degeneration, and increasing lifespan in a way that mimics caloric restriction [12–17].
Resveratrol, a phytoalexin constituent of grapes, cranberries, blueberries, and peanuts, is a natural polyphenolic antioxidant that has been proposed to possess putative anti-aging properties, presumably via its ability to scavenge oxidative free radicals [18, 19]. As expected, the compound has gained significant attention in the nutraceutical industry because of its proposed diverse, pro-health characteristics. Intriguingly, many of the observed health span bioactions of resveratrol correspond to benefits reported from elevated plasma levels of 25D. Resveratrol was found to be the most potent SIRT1 activator among a number of plant-derived phenols, eliciting a 13.4-fold increase in the catalytic rate of SIRT1, and enhancing the survival rate of cells stressed by irradiation [14]. Furthermore, resveratrol has been shown to enhance SIRT1-mediated cellular processes including protection from axonal degeneration, mobilization of adipose, and importantly, inhibition of NF-κB-regulated transcription [12, 17, 20].
1.3. Resveratrol-activated SIRT1 Deacetylation of Nuclear Receptors
Covalent modification (e.g., phosphorylation, ubiquitination/SUMOylation, glycosylation, acetylation) of nuclear hormone receptors has been reported to play an important role in the evolution of hormone responsiveness. Resveratrol-activated SIRT1 has been shown to result in the deacetylation of numerous nuclear receptors, thus leading to the potentiation of transcriptional activity. For example, the peroxisome proliferator-activated receptor gamma (PPARγ), found primarily in adipose tissue and the colon where it regulates fatty acid storage and glucose metabolism, was shown to be deacetylated at positions K268 and K293 in the presence of overexpressed SIRT1 or chemical activation of SIRT1 by resveratrol [21]. SIRT1-dependent deacetylation of PPARγ leads to the selective induction of genes responsible for producing brown adipose tissue and repression of white adipose tissue-generating genes associated with insulin resistance [21]. Additionally, SIRT1 has been shown to interact with steroid hormone receptor coregulator proteins including p300 and PGC-α with implications for muscle cell development, adipogenesis, and hepatic adipose storage/metabolism [13, 17, 22].
Furthermore, FXR, a regulator of lipid and glucose homeostasis, is deacetylated at K271 via overexpression of SIRT1 or resveratrol treatment, leading to increased heterodimerization with RXRα, DNA binding, and transcriptional activation of FXR-regulated genes [23]. Finally, thyroid hormone functioning via its nuclear receptor, thyroid hormone receptor β, regulates hepatic genes involved in mitochondrial fatty acid oxidation and gluconeogenesis. This receptor also has been shown to be activated by both resveratrol and SIRT1, leading to stimulation of fatty acid oxidation and inhibition of lipogenesis [24]. The liver X receptor (LXR) is regulated in a similar manner [25].
Predicated on the evolutionarily conserved nature of nuclear receptor mechanisms, in the present investigation the potential role of sirtuin-1 and resveratrol in modulating 1,25D-VDR signaling was analyzed via in vitro and intact human cultured cell experiments. VDR/VDRE-driven transcription is shown to be significantly potentiated by endogenous and exogenous SIRT1, suggesting that deacetylated VDR is more transcriptionally active than acetylated VDR. Direct comparison experiments reveal that VDR joins RXR, RAR, TR, PPARγ, and ER in a group of nuclear receptors for which the transcriptional activation function is augmented by SIRT1. With respect to resveratrol, we observe that this plant phenol is capable of mimicking both SIRT1-overexpression and 1,25D hormonal ligand treatment in augmenting VDRE-mediated transcriptional activation. Finally, novel mechanistic evidence is reported that the 1,25D-VDR hormone-receptor complex is enzymatically deacetylated by SIRT1, in part at human VDR residue K413, with resulting amplified transactivation of VDR target genes.
2. MATERIALS AND METHODS
2.1. Luciferase plasmid constructs
The DNA elements cloned upstream of the luciferase reporter vector, pLUC-MCS (Stratagene Corp., La Jolla, CA), included the vitamin D responsive elements termed XDR3, CYP24, and PER6; the estrogen responsive element (ERE), glucocorticoid responsive element (GRE), retinoid X receptor-activated responsive element (RXRE), farnesoid X receptor-activated responsive element (FXRE), liver X receptor-activated responsive element (LXRE), peroxisome-proliferator-activated receptor gamma responsive element (PPRE), retinoic acid responsive element (RARE), and thyroid hormone responsive element (TRE). Details concerning the construction of luciferase reporter plasmids containing the XDR3, CYP24, PER6, ERE, GRE, LXRE, and RARE responsive elements have been previously described [7]. XDR3 is the distal DR3 VDRE in the human CYP3A4 gene, CYP24 is a DNA fragment containing the two DR3 VDREs in the proximal promoter of the rat cyp24a1 gene, and PER6 is the everted repeat-6 VDRE in the proximal promoter of the human CYP3A4 gene. The sequences for these VDREs appear in tabular form in previously published work [26]. The RXRE reporter construct was based on a naturally occurring double-repeat RXR responsive element from the rat cellular retinol binding protein II gene with the sequence AAAATGAACTGTGACCTGTGACCTGTGACCTGTGAC (half sites are underlined) [27]. The FXRE reporter plasmid utilized for assaying FXR signaling was constructed by using the double-stranded oligonucleotide containing three copies of the FXRE from the human apolipoprotein E (ApoE) gene, CAGAGGTCAGAGACCTCTCCAGAGGTCAGAGACCTCTCCAGAGGTCAGAGACCTCTC [28–30]. The PPRE reporter construct utilized for evaluating PPARγ signaling was synthesized by using the double-stranded oligonucleotide containing three copies of the PPRE, also from the human ApoE gene, GGAGGGGAAGGGTCAGTGGAGGGGAAGGGTCAGTGGAGGGGAAGGGTCAG [28–30]. The reporter construct TRE utilized for assaying thyroid receptor (TR) signaling, was generated by inserting a double-stranded oligonucleotide containing two copies of the thyroid hormone responsive element from the rat myosin heavy chain gene, CTGGGAGGTGACAGGAGGACACGAGCTGGGAGGTGACAGGAGGACACGAG [31].
2.2. Expression plasmids
Synthesis of the K413R point mutation within the pSG5-VDR expression construct was accomplished using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, California) per the manufacturer’s protocol. Correct introduction of the mutation was confirmed by DNA sequencing. The SIRT1 deacetylase domain mutant plasmid, pECE-Flag-SIRT1 H363Y, was a gift from Michael Greenberg (Addgene plasmid #1792; Cambridge, MA) [32].
2.3. Mammalian cell culture
Three mammalian cell lines purchased from the American Type Culture Collection (ATCC, Manassas, VA) were employed in this study as representatives of classic 1,25D target tissues. Human osteosarcoma (HOS TE85, hereafter called TE85) cells were grown in Dulbecco’s modified eagle medium (DMEM) while human embryonic kidney (HEK293) cells were cultivated in minimal essential media (MEM) supplemented with 1 mM sodium pyruvate. Rat osteoblast/osteocyte-like (UMR-106) cells were cultured in DMEM/F12 (Hyclone Laboratories, Logan, UT, USA) supplemented with 2.5 mM-glutamine. All cell culture media were also supplemented with 10% fetal bovine serum (FBS) and 100 units/mL penicillin and 100 µg/mL streptomycin. The cells were passaged/plated in the indicated media and grown in a humidified atmosphere at 37°C and 5% carbon dioxide.
2.4. Transient transfection of cultured cells and luciferase assay
HEK293 and TE85 cells were plated in 24-well Falcon plates (Becton Dickinson, Franklin Lakes, NJ) at a density of 80,000 to 100,000 cells/well in the appropriate complete media 22 to 24 hours prior to transient transfection. Cells were transfected in individual wells using Express-In (Thermo Scientific, Waltham, MA) or PolyJet reagent (SignaGen Laboratories, Gaithersburg, MD) according to the specific manufacturer’s protocol. Briefly, each well received 2 µL Express-In or 1.25 µL PolyJet and DNA plasmids as specified in the figure legends, along with 20 ng of pRL-null. The pRL-null plasmid allows for the constitutive, low-level expression of Renilla reniformis luciferase to monitor DNA transfection efficiency. The luciferase-containing vectors (250 ng/well) were derived from the pLuc-MCS plasmid containing an oligonucleotide (cloned between the HindIII and BglII sites) with 2–3 copies of the appropriate DNA responsive element directly upstream of the Firefly (Photinus pyralis) luciferase gene (see section 2.1 for details). Twenty-two to twenty-four hours post-transfection, the cells were treated with either an ethanol (ETOH) vehicle control or the ligand for the nuclear receptor, with resveratrol (Enzo Life Sciences, Farmingdale, NY) added as a SIRT1-activator in select experiments as detailed in the figure legends. Subsequent to a 22–24 hour incubation, the cells were lysed in 1X passive lysis buffer (Promega, Madison, WI) and the whole-cell lysates were collected. Each lysate was then analyzed sequentially for Firefly and Renilla luciferase activity using a Dual Luciferase Assay Kit (Promega Corp., Madison, WI) and a Sirius Luminometer/software (Zylux Corp., Huntsville, AL) according to the manufacturers’ protocols. The luminescence value (relative light units, RLU) produced by the Firefly luciferase gene was then divided by the luminescence from the constitutively-active Renilla luciferase to obtain a normalized value that accounts for transfection efficiency, generalized cell death, and ligand-induced cellular toxicity. The Firefly/Renilla ratio was then multiplied by a scaling factor (usually 10,000) to streamline data presentation. The mean ratio of Firefly/Renilla for 3–24 biological replicates (wells) was calculated for each experimental treatment group and standard deviation values were computed and expressed as error bars. The data shown denote the average of all wells (biological replicates) within a single set of treatment groups. Thus, single sets of experiments with 3–24 biological replicates are depicted in all figure panels, with the n for number of biological replicates (individually treated and quantitated for transcriptional response) indicated in the appropriate figure legend.
2.5. Mammalian 2-Hybrid assays
HEK293 cells were transiently transfected and treated using the procedures outlined above. The pCMV-AD-VDR prey plasmid contains a VDR activation domain (AD) and the pCMV-BD-RXR bait vector contains the RXR “bait” DNA binding domain (BD) [33]. Each well received AD-VDR and BD-RXR, 20 ng of pRL-null, and a Firefly luciferase reporter construct (pFR-luciferase), as described in the figure legend. The negative controls were “empty” pCMV-AD and pCMV-BD vectors. After incubation with the appropriate ligands, transfected cells were assayed for luciferase activity and results were analyzed and graphically depicted as described above.
2.6. Total RNA isolation, cDNA synthesis, and mRNA expression analysis via quantitative real-time PCR
UMR106 cells were plated at 650,000 cells/well in a 6-well plate. The cells were transfected with 500 ng pSG5-VDR (expression plasmid for human VDR), using the PolyJet reagent. The cells were treated with either ethanol vehicle, 1, 10, or 100 nM 1,25D, or 25 µM Res. HEK293 cells were plated at 900,000 cells/well in a 6-well plate. The cells were transfected with 50 ng pSG5-VDR, 250 ng pTZ18U (non-specific, carrier DNA), and 5 ng pCMV-empty or pCMV-SIRT1 (parent plasmid construct or plasmid expressing human SIRT1, respectively) using the PolyJet reagent. The cells were treated with either ethanol vehicle, 10 nM 1,25D (Enzo Life Sciences), 10 nM 1,25D + 25 µM Res, or 10 nM 1,25D + 20 µM EX-527 (Sigma-Aldrich, St. Louis, MO) for 22 to 24 hours post-transfection. Note that the wells exposed to 10 nM 1,25D + 20 µM EX-527 was also pretreated with 20 µM EX-527 at the time of transfection to inhibit endogenous SIRT1. Total RNA was isolated from each well using the Aurum Total RNA Mini kit (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer’s instructions. The RNA yield was quantified via UV spectrophotometry and the RNA quality was estimated using the A260/A280 and A260/A230 ratios.
DNase-treated total RNA (1 µg) was reverse transcribed with an iScript cDNA Synthesis kit (Bio-Rad) and a DNA Engine™ thermocycler (Bio-Rad), to produce 20 µl of first-strand cDNA. For quantitative real-time PCR (qPCR), 1.5 µl of cDNA was used in a 10 µl reaction containing 5 µl FastStart Universal SYBR Green Master Mix with Rox (Roche Applied Science, Indianapolis, IN) and appropriate primers. Reactions were performed in 96-well PCR plates (Bio-Rad) in a Bio-Rad CFX96 instrument, with a 40-cycle profile. Data were analyzed by the comparative ΔΔCt method, as means of relative quantitation, normalized to an endogenous reference (GAPDH), relative to a calibrator (normalized Ct value from ETOH-treated cells), and expressed as 2−ΔΔCt according to the manufacturer’s protocol. For detection of human GAPDH and CYP24A1 transcripts, the following primer sets were utilized: human GAPDH, forward primer: 5'-ACAACTTTGGTATCGTGGAAGGAC-3' and reverse primer: 5'-CAGGGATGATGTTCTGGAGAGC-3' yielding a 129 bp product; and for CYP24A1, forward primer: 5'-CAGCGAACTGAACAAATGGTCG-3' and reverse primer: 5'-TCTCTTCTCATACAACACGAGGCAG-3', generating a 58 bp transcript. For detection of rat Nrf-2, the following primers were utilized: forward primer: 5′- CACGGATGATGCCAGCCAG-3′ and reverse primer: 5′- GCCCGCCCAGAAGTTCAGAGAG-3′ yielding a 164 bp transcript.
2.7. Immunoprecipitation and Western blotting
HEK293 cells were maintained as described above and plated at a density of 2×106 cells/100 mm dish, 24 hours prior to transfection. Cells were transfected with 2500 ng of pSG5-VDR or pSG5-VDR K413R in combination with 50 ng of pCMV-empty or pCMV-SIRT1 using 50 µL PolyJet reagent in serum/antibiotic-free media. After 24 hours, 800 µL FBS was added to each plate and cells were dosed with either ETOH vehicle, 10 nM 1,25D, or 25 µM Res.
The following day, the cell monolayers were washed with cold phosphate-buffered saline, and 500 µL of 1X lysis buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.3 mM zinc acetate, 300 mM KCl, 5 mM DTT, and 0.1% Tween-20) was added to each plate. Total cell lysates were generated using sonication (Fisher Scientific, Pittsburg, PA) with 3 × 20 second pulses on ice, and the samples were then cleared via centrifugation at 10,000 × g for 10 minutes at 4°C. Next, the supernatant was collected and immunoprecipitation assays were performed using 5 µL of rabbit polyclonal anti-VDR antibody generated using the oligopeptide antigen, EEHSKQYRCLSFQPECSMK (Affinity Bioreagents, Inc. Golden, CO); the samples were rocked at 4°C for 1 hour. Next, 40 µL of 5% BSA/lysis buffer equilibrated-50% Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology, Dallas, TX) were added to the lysate-antibody mixtures and the samples were then incubated overnight at 4°C with rocking. The beads were collected via centrifugation and washed three times with buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 300 mM zinc acetate, 150 mM KCl, 5 mM DTT, and 0.1% Tween-20). The immunoprecipitates were eluted from the beads by boiling for 5 minutes in SDS sample buffer (100 mM Tris-HCl, 10 mM dithiothreitol, and 4% SDS) and were then subjected to SDS-PAGE and Western blot analysis.
The protein samples were resolved on 4–15% SDS-PAGE gradient gels (Bio-Rad, Berkeley, CA) and transferred to nitrocellulose membranes using Tris-Glycine-SDS-20% methanol and electrophoresis. The membranes were blocked in 5% non-fat dry milk in Tris Buffered Saline-Tween-20 (TBST) for 1 hour. The membranes were washed with TBST and incubated in rabbit anti-acetyl-lysine antibody (Cell Signaling, Danvers, MA; 1:1000 dilution in 5% BSA in TBST) overnight. The blotted membranes were then washed with TBST and incubated with mouse anti-rabbit IgG (light-chain specific) peroxidase conjugate (Cell Signaling, Danvers, MA) diluted 1:5000 in 5% milk in TBST. The immunoreactive proteins were visualized using SuperSignal West Pico Chemiluminescent Substrate (Fisher Scientific, Pittsburg, PA) and x-ray film (Thermo Scientific) was employed for detection.
2.8. Statistical analysis
Data are expressed as means ± standard deviations. All data are expressed as fold-effects, with transcription in the presence of 1,25D set at 1.0-fold. Thus, all results are normalized to transcription driven by hormone, and presented graphically and numerically as fold-effect of the tested variable, e.g., SIRT1-overexpression, Res, EX-527, etc. Because the design of the experiments was a simple 2X2 motif in which the ability of SIRT1 to enhance the effectiveness of hormone-induced transcription was assessed, the Welch’s two-sample t-test was applied to the log transformed fold change to ascertain which specific experimental results differed from their controls. The Bonferroni method was used to correct for multiple comparisons in each experiment with the p-value significance threshold equal to 0.05 divided by the number of comparisons. For certain experiments an interaction term in a linear regression, with log fold change as the response, was used to test whether the effect of one factor was different in the presence and absence of another factor, and linear regression was also used to study dose-response when needed. Statistical analysis was conducted using Microsoft® Excel® 2013 and R version 3.3.2 (https://cran.r-project.org).
3. RESULTS
3.1. Acetylation Status of VDR
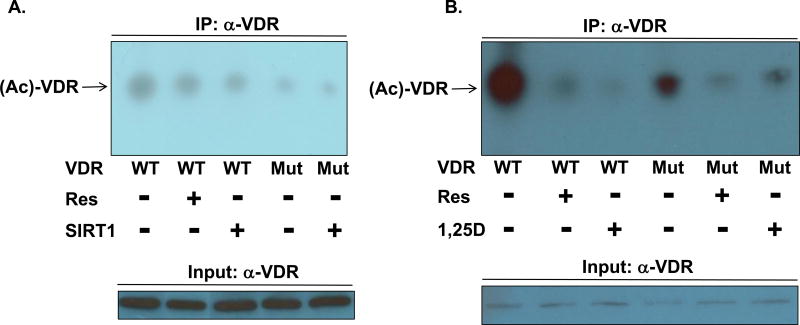
We hypothesized that SIRT1 elicits deacetylation of the vitamin D receptor, potentially affecting VDR transactivation capacity and possibly modulating the corresponding biological actions. Thus, VDR may serve as a mediator of certain sirtuin-1 bioeffects. To directly assess the acetylation status of VDR and the ability of SIRT1 to effect deacetylation of the receptor, we utilized a specific anti-acetyl-lysine antibody in Western blot assays. Wild-type (WT) or K413R mutant VDR were immunoprecipitated using an anti-VDR (α-VDR) antibody, followed by Western blotting and probing with the anti-acetyl-lysine antibody (Fig. 1). HEK293 cells transfected with WT VDR displayed robust expression of acetylated VDR (Ac-VDR; Figs. 1A and 1B, lane 1), whereas treatment with resveratrol or transfection of SIRT1 significantly decreased Ac-VDR (compare lanes 2 and 3 to lane 1 in panel A and lane 2 to 1 in panel B of Fig. 1), suggesting not only that SIRT1 targets VDR but that resveratrol promotes this process. In the context of HEK293 cells, K413R mutant VDR (Mut) displayed dramatically reduced lysine acetylation when compared to WT VDR (Figs. 1A and 1B, lane 4 versus 1), pinpointing K413 in VDR as a major site for acetylation/deacetylation. The observation that there is residual lysine acetylation in the K413R mutation that is further diminished in the presence of either SIRT1 (Fig. 1A, lane 5) or Res (Fig. 1B, lane 5) intimates that additional sites beyond K413 may exist as targets for lysine acetylation in VDR.
Figure 1. Assessment of VDR acetylation status in the presence or absence of SIRT1, 1,25D, or resveratrol.
(A) HEK293 cells were plated, transfected with pSG5-hVDR (wild-type/WT VDR) or pSG5-hVDR K413R (mutant/Mut VDR) alongside pCMV-SIRT1, and treated as indicated. Immunoprecipitation was conducted using a rabbit polyclonal anti-VDR antibody and Protein A/G PLUS-Agarose beads. A rabbit polyclonal anti-acetyl-lysine antibody was used to probe for acetylation levels of VDR. The identical rabbit polyclonal anti-VDR antibody was used for the immunoprecipitation and VDR input western blots (see Methods section for further details including chemiluminescent visualization of immunoreactive bands). (B) A similar experiment was performed in HEK293 cells treated with 10 nM 1,25D or 25 µM resveratrol. Each experiment was independently replicated (data not shown).
A surprising observation in the experiment illustrated in Fig. 1B is that the presence of 1,25D hormone also signals a drastic decrease in Ac-VDR in the wild-type background when compared to cells treated with ETOH vehicle only (Fig. 1B, lane 3 versus 1). Amazingly, 1,25D produces VDR-deacetylation in excess of that caused by the SIRT1-activator, resveratrol (Fig. 1B, lane 3 versus 2). This result suggests an action of 1,25D in driving VDR deacetylation, potentially via a conformational alteration in VDR which promulgates recruitment of endogenously activated SIRT1 to the 1,25D-VDR-RXR complex, possibly bound to VDREs across the genome. Interestingly, while this action of 1,25D ligand to promote VDR deacetylation also occurs in the K413R mutant, the magnitude of the reduction is less than that elicited by resveratrol, contrasting with the wild-type VDR where Res is more effective than 1,25D at diminishing this reduction (Fig. 1B, compare lane 6 versus 5 to lane 3 versus 2), indicating that when K413 is removed from the ensemble of VDR lysines, the receptor is less susceptible to 1,25D-induced, SIRT1-catalyzed deacetylation. This result implicates VDR K413 as a relevant site of acetylation in the modulation of VDR transactivity. Collectively, these results illuminate the significant capacity of 1,25D ligand, SIRT1, and its resveratrol activator to enhance deacetylation of VDR, likely to restore full transcriptional potency to acetylated VDR that has been feedback-attenuated in its activity by metabolic events within the intracellular milieu.
3.2. SIRT1 Enhances VDRE-mediated Transcription in Vitamin D Target Cells
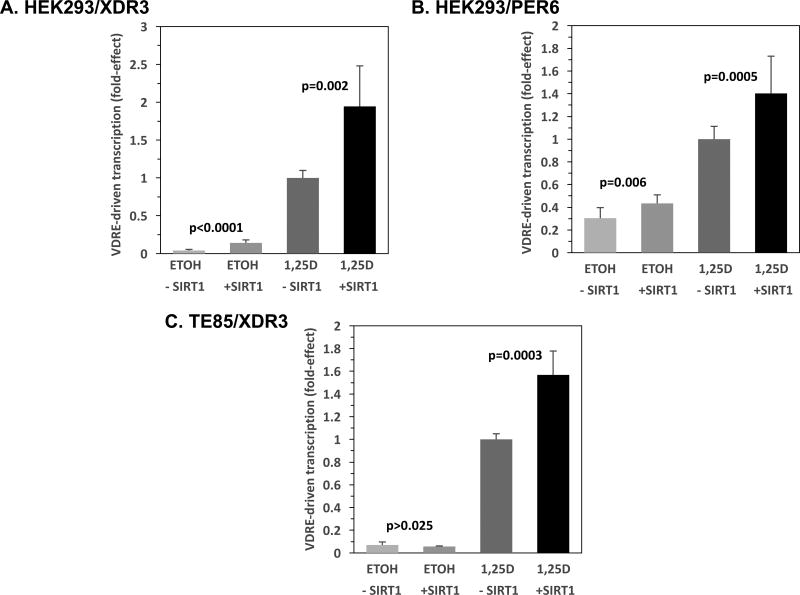
Several experiments were conducted to test the hypotheses generated from the data depicted in Fig. 1. The first approach was to evaluate the functional significance of VDR deacetylation by co-transfecting a SIRT1 expression plasmid into kidney and bone cells in culture. Figures 2A, B, and C depict VDRE-luciferase-based transcriptional experiments in human embryonic kidney (2A, 2B: HEK293) and osteosarcoma (2C: TE85) cells treated with ethanol (ETOH) vehicle or vitamin D hormone (1,25D), following co-transfection with SIRT1 expression vector, to evaluate the influence of the deacetylase on transcription. HEK293 cells transfected with 5 ng pCMV-SIRT1 (hereafter termed SIRT1; +SIRT1) and treated with 1,25D, displayed enhanced transcription of an XDR3-luciferase reporter plasmid 1.9-fold over the corresponding pCMV-empty (herein termed empty; -SIRT1), 1,25D-treated group (Fig. 2A, bar 4 versus 3; p=0.002). However, transfection of significantly higher levels of the SIRT1 expression plasmid (50 and 100 ng) decreased transcriptional activity (60% and 56%, respectively; p<0.01) in the presence of 1,25D (data not shown), perhaps caused by multi-target deacetylation via SIRT1 catalytic activity at these higher levels of expression. As a consequence of this observation, all further SIRT1 overexpression experiments were restricted to transfection of 1–5ng of SIRT1 plasmid. Somewhat surprisingly, the results pictured in Fig. 2A also reveal that SIRT1 overexpression significantly (3.4-fold) augments XDR3-driven transcription in the absence of 1,25D treatment (Fig. 2A, bar 2 versus 1; p<0.0001). Thus, the SIRT1 effect to potentiate VDRE-mediated transcription apparently does not require the 1,25D ligand, possibly the result of activation of unoccupied VDR. Note that the XDR3 VDRE employed in Fig. 2A is of the type consisting of a direct repeat (of the consensus half-element) with a spacer of three nucleotides found in the distal promoter of the human CYP3A4 gene.
Figure 2. SIRT1 Enhances VDRE-mediated Transcription in Vitamin D Target Cells.
(A) HEK293 cells were transfected with 5 ng pCMV-empty (-SIRT1) or 5 ng pCMV-SIRT1 (+SIRT1) in addition to 250 ng of XDR3 VDRE-luciferase containing two copies of the distal DR3 VDRE from the human CYP3A4 gene [56]. Twenty-four hours post-transfection, the cells were exposed to fresh medium containing ethanol (the vehicle control, ETOH) or 1 nM 1,25D in ethanol. After an additional 22–24 hours, the cells were lysed and analyzed sequentially for Firefly and Renilla luciferase activity as described in Methods. The Firefly/Renilla ratio was then multiplied by a scaling factor (usually 10,000), and converted to fold-effect of SIRT1 compared to 1,25D-treated cells transfected with empty vector control, which was set at 1.0-fold for standardization and to streamline data presentation. The fold-effect average for six biological replicates (n=6) was calculated for each experimental treatment group and standard deviation values were computed and expressed as error bars. (B) A similar experiment as in (A) was conducted with the PER6 VDRE-luciferase construct; however, the fold-effect average for twelve biological replicates (n=12) was calculated for each experimental treatment group. (C) A similar experiment as in (A) was conducted using TE85 osteoblast-like cells instead of HEK293 cells. P-values are noted in the figure panels.
Next, an alternative VDRE motif, the everted repeat (of the consensus half-element) with a spacer of six nucleotides, present in the proximal promoter of the human CYP3A4 gene (PER6), was utilized to construct the VDRE-luciferase reporter. Examination of this alternative VDRE motif in the context of a luciferase reporter construct is illustrated in Fig. 2B, showing that HEK293 cells transfected with 5 ng SIRT1-vector and treated with 1,25D, exhibited enhanced transcription of a PER6-luciferase reporter plasmid 1.4-fold over the corresponding empty vector, 1,25D-treated group (Fig. 2B, bar 4 versus 3; p=0.0005). Again, in the case of the PER6 VDRE motif, SIRT1 overexpression significantly (1.4-fold) augments XDR3-driven transcription in the absence of 1,25D treatment (Fig. 2B, bar 2 versus 1; p=0.006). Thus, the two most common VDRE motifs (DR3 and ER6) occurring in the genome are boosted in their ability to drive transcription when low amounts of SIRT1 expression vector are co-transfected into human kidney cells. To test another vitamin D target cell, namely osteoblast-like osteosarcoma cells, for the effects of SIRT1 overexpression on VDR function, we next transfected TE85 bone cells with SIRT1 and treated with 1,25D, again employing the XDR3-luciferase reporter plasmid. Transcription was amplified 1.6-fold by SIRT1 over the corresponding empty-vector, 1,25D-treated group (Fig. 2C, bar 4 versus 3; p=0.0003). However, distinct from kidney cells, in bone cells SIRT1 overexpression did not affect XDR3-driven transcription in the absence of 1,25D treatment (Fig. 2C, bar 2 versus 1; p>0.025, the Bonferroni adjusted threshold). In toto, the results illustrated in Fig. 2 are supportive of the hypothesis that SIRT1 elicits deacetylation of the vitamin D receptor, likely enhancing VDR transactivation capacity from the most prevalent VDRE motifs, at least in two major vitamin D target cells.
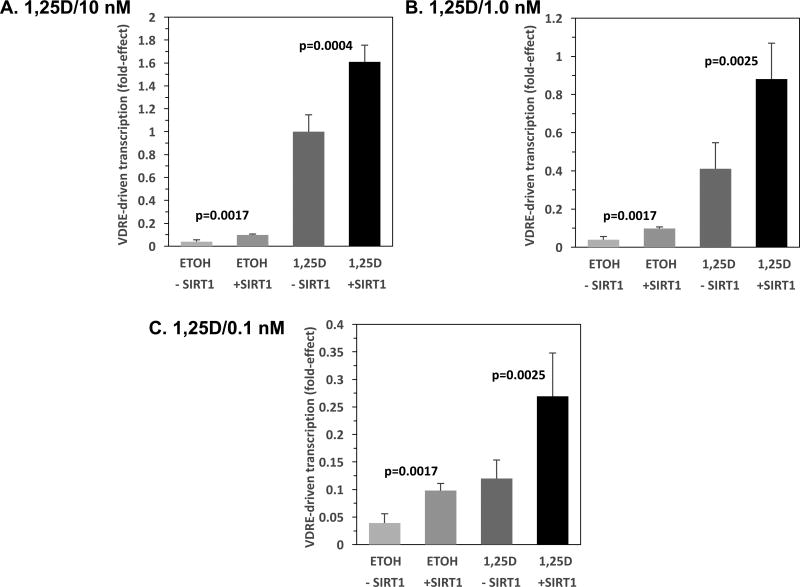
Because of the curious finding that the SIRT1 effect to increase VDRE-mediated transcriptional activation in HEK293 cells is apparently not dependent on the presence of 1,25D hormone, we conducted a dose-response experiment with respect to 1,25D treatment of HEK293 cells transfected with SIRT1 and the XDR3-luciferase reporter plasmid. Figures 3A, B and C depict XDR3-based transcriptional experiments in which HEK293 cells were transfected with 5 ng of the SIRT1 plasmid and treated with 10 nM, 1 nM and 0.1 nM concentrations of 1,25D, respectively. In HEK293 cells transfected with empty vector and exposed to 10 nM 1,25D, VDR/VDRE transcriptional activity increased 26-fold compared to ETOH vehicle (Fig. 3A, bars 3 versus 1). When SIRT1 was co-transfected, the 1,25D effect was 16.4-fold (Fig. 3A, bars 4 versus 2), and both +SIRT1 groups were significantly greater than their –SIRT1 controls (2.5-fold: p=0.0017 for ETOH and 1.6-fold: p=0.0004 for 1,25D-treated). In HEK293 cells transfected with empty vector and exposed to 1 nM 1,25D, VDR/VDRE transcriptional activity increased 10.5-fold compared to ETOH vehicle (Fig. 3B, bars 3 versus 1). When SIRT1 was cotransfected, the 1,25D effect was 9-fold (Fig. 3B, bars 4 versus 2), and both +SIRT1 groups were significantly greater than their –SIRT1 controls (2.5-fold: p=0.0017 for ETOH and 2.1-fold: p=0.0025 for 1,25D-treated). In HEK293 cells transfected with empty vector and exposed to 0.1 nM 1,25D, VDR/VDRE transcriptional activity increased 3.1-fold compared to ETOH vehicle (Fig. 3C, bars 3 versus 1). When SIRT1 was cotransfected, the 1,25D effect was 2.7-fold (Fig. 3C, bars 4 versus 2), and both +SIRT1 groups were significantly greater than their –SIRT1 controls (2.5-fold: p=0.0017 for ETOH and 2.2-fold: p=0.0025 for 1,25D-treated). Therefore, as the dose of 1,25D hormone is progressively titrated down from 10 to 1 to 0.1 nM, the fold effect of hormone descends from 26 to 10.5 to 3.1, respectively, whereas SIRT1-enhancement of VDRE-mediated transcription remains fairly constant in the 1.6- to 2.5-fold range. The 1,25D effect was positively associated with its dose in the absence or presence of SIRT1 (both p < 0.0001). This result confirms the dissociation of the 1,25D effects from SIRT1 effects on transcription, and is consistent with an allosteric influence of 1,25D contrasted with a catalytic action of SIRT1 on VDR deacetylation and associated transcriptional activation capacity.
Figure 3. Effect of 1,25D Concentration on SIRT1 Enhancement of VDRE-mediated Transcription in HEK293 Cells.
HEK293 cells were transfected with the XDR3 VDRE-reporter plasmid (250 ng/well) and 5 ng pCMV-empty (-SIRT1) or 5 ng pCMV-SIRT1 (+SIRT1). After 24 hours of transfection, the cells were refreshed with complete medium containing ETOH vehicle or the following final concentrations of 1,25D equal to: (A) 10 nM, (B) 1.0 nM, and (C) 0.1 nM. After an additional 22–24 hours, the cells were lysed and analyzed sequentially for Firefly and Renilla luciferase activity as described in Methods. The Firefly/Renilla ratio was then multiplied by a scaling factor (usually 10,000), and converted to fold-effect of SIRT1 compared to cells treated with 10 nM 1,25D and transfected with empty vector control, which was set at 1.0-fold in (A) for standardization and comparison of (B) and (C) to (A), and to streamline data presentation. The fold-effect average for six biological replicates (n=6) was calculated for each experimental treatment group and standard deviation values were computed and expressed as error bars. P-values are noted in the figure panels.
3.3. Resveratrol Mimics SIRT1 and 1,25D Effects on VDRE-mediated Transcription
As illustrated in Supplemental Figure 1 (Panel A), HEK293 cells exposed to only 25 µM Res displayed an increase (4.7-fold, p<0.0001) in XDR3-mediated transcription over the corresponding -Res control (bar 2 versus 1), whereas treatment with Res and 1 nM 1,25D elicited a 24-fold enhancement of transcription (bar 4 versus 2). In the same experiment, Res boosted 1,25D-activated transcription by 3.3-fold (Supplemental Figure 1 (Panel A; bar 4 versus 3, p=0.0007)), comparable to, and exceeding the 1.9-fold stimulatory effect exerted by SIRT1 for XDR3-driven transcription (see Fig. 2A). In Supplemental Figure 1 (Panel B), results are also depicted examining the influence of Res on VDRE-mediated transcription governed by the PER6 responsive element motif. In this case, 25 µM Res challenge exactly duplicates the action of a 1 nM dose of 1,25D (3.3-fold enhancement in transcription, p<0.0001, bar 2 versus 1). As with the XDR3-element, Res boosted 1,25D-activated transcription by 3.3-fold (Supplemental Figure 1 (Panel B, bar 4 versus 3, p<0.0001)), comparable to, and exceeding the 1.4-fold stimulatory effect exerted by SIRT1 for PER6-driven transcription (see Fig. 2B). Taken together, these experiments probing the effects of resveratrol and SIRT1 on VDR-VDRE-mediated transcription in HEK293 cells reveal that VDRE-directed transactivation is affected quite similarly by these two modulators, although challenge with the SIRT1-activator, Res, is slightly more effective than transfecting the SIRT1 overexpression plasmid. Thus, VDRE-mediated transcription is likely governed by the relative concentrations of catalytically active SIRT1 and occupied nuclear VDR.
To determine if this phenomenon of resveratrol mimicry persists in a natural cellular setting with endogenous SIRT1, plus an aging-relevant gene with VDREs in the native context of chromatin, we examined the induction of Nrf-2, a master anti-oxidant gene. As shown in Supplemental Figure 1 (Panel C), 1,25D dose-dependently induces Nrf-2 mRNA to a maximum of 3.4-fold over ETOH control in rat osteoblast-like, osteosarcoma cells, and 25 µM Res recapitulates the impact of 1 nM 1,25D, which is 65% of the maximal response for transcription. Thus, Res, which is not a VDR ligand, is nevertheless able to stimulate VDRE-driven transcription. We previously reported that Res enhances the binding of 1,25D to VDR, as well as the heterodimerization of VDR with RXR [7]. To determine one mechanism whereby Res activates VDR, probably via activation of its SIRT1 target enzyme, we performed mammalian-two-hybrid (M2H) protein-protein interaction studies in HEK293 cells. As previously published [7], the data in Supplemental Figure 1 (Panel D) show that Res accentuates VDR-RXR interaction 1.5-fold (p=0.0008, bar 2 versus 1). SIRT1-overexpression enhances VDR-RXR association 1.16-fold (p=0.009, bar 4 versus 3), indicating that at least part of the mechanism of Res function on transcription is through activation of SIRT1 deacetylase, followed by VDR-RXR heterodimeric binding to VDREs. This interpretation is consistent with the finding that both Res and SIRT1 trigger VDR-deacetylation (compare lanes 2 and 3 to lane 1 in panel A and lane 2 to 1 in panel B of Fig. 1). It is concluded that deacetylation of VDR by SIRT1, potentiated via its resveratrol activator, and facilitated by 1,25D ligand promulgated conformational change in VDR, all lead to augmented VDR attraction of its RXR heteropartner, as well as other transcriptional coregulators. Therefore, induction of vitamin D target gene expression is predicated on 1,25D ligand binding to confer a massive increase in RXR recruitment by VDR that produces VDRE-docking of the heterodimer to convey transcriptional activation of the gene in question. But the results herein reveal that this process can be modulated, subtly, through acetylation/deacetylation, apparently of un-liganded as well as liganded VDR, with SIRT1-catalyzed deacetylation of VDR rendering it a more potent ligand-binder, heterodimerizer, and trans-activator.
3.4. Inhibition or Mutational Inactivation of SIRT1 Decreases VDR Transactivation
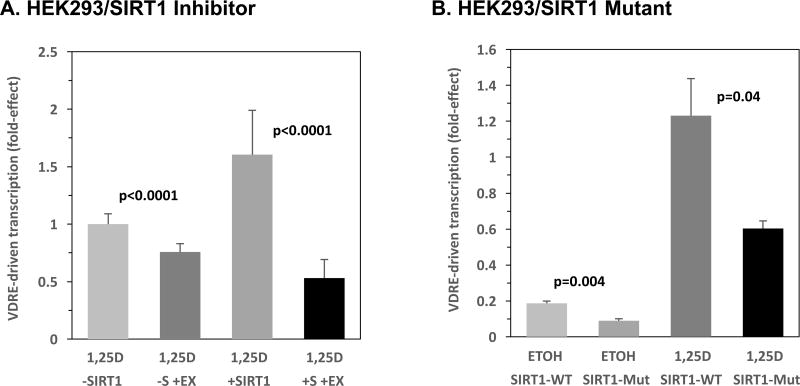
Initially, a small molecule SIRT1 inhibitor, EX-527, was used to attenuate the activity of endogenous SIRT1. The compound, 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide, known as EX-527, is characterized as a highly potent SIRT1 inhibitor with significant isoform selectivity [34]. A study conducted by Gertz et al. demonstrated that EX-527 occupies the nicotinamide site and a neighboring pocket of SIRT1 in addition to contacting the ribose of NAD+ or of the coproduct 2'-O-acetyl-ADP ribose [35]. Moreover, EX-527 stabilizes the closed SIRT1 conformation to prevent product release. HEK293 cells, containing endogenous VDR, were co-transfected with empty vector alongside the XDR3 VDRE reporter construct and dosed with 1,25D and EX-527. The results showed significantly decreased (24%; p<0.0001) VDR transactivation in the presence of 1,25D and EX-527 compared to 1,25D alone in the absence of SIRT1 co-transfection (Fig. 4A, bar 2 versus 1), revealing that EX-527 is inhibiting endogenous SIRT1 to attenuate VDRE-mediated transcription. Ectopic expression of SIRT1 induces a 1.6-fold (p=0.0005) increase in XDR3-driven transcription over the 1,25D empty (-SIRT1) vector group (Fig. 4A, bar 3 versus 1). Furthermore, treatment of HEK293 cells with 1,25D+SIRT1+EX-527 demonstrates transactivation levels that are decreased (67%; p<0.0001) compared to cells challenged with 1,25D and SIRT1 alone (Fig. 4A, bar 4 versus 3) indicating that EX-527 is inhibiting both endogenous and exogenous SIRT1.
Figure 4. Inhibition or Mutational Inactivation of SIRT1 Decreases VDR Transactivation.
(A) HEK293 cells were transfected with 1 ng pCMV-empty or 1 ng of pCMV-SIRT1, alongside 250 ng of the XDR3 VDRE-luciferase reporter. Twenty-four hours post-transfection, each of the two groups of cells (-SIRT1 (-S) and +SIRT1 (+S)) were treated with either 1 nM 1,25D or 1 nM 1,25D+20 µM EX-527 (+EX). After an additional 22–24 hours, the cells were lysed and analyzed sequentially for Firefly and Renilla luciferase activity as described in Methods. The fold-effect average for twelve biological replicates (n=12) was calculated for each experimental treatment group and standard deviation values were computed and expressed as error bars. (B) HEK293 cells were transfected with 1 ng pCMV-empty, pCMV-SIRT1 (WT SIRT1), or pECE-Flag-SIRT1 H363Y (SIRT1 deacetylase domain mutant, H363Y SIRT1, Mut) alongside 250 ng of the XDR3 VDRE-luciferase reporter. Twenty-four hours post-transfection, the cells were treated with either ETOH or 10 nM 1,25D, and then assayed for transcription. The fold-effect average for three biological replicates (n=3) was calculated for each experimental treatment group and standard deviation values were computed and expressed as error bars. P-values are noted in the figure panels.
Next, to establish whether SIRT1 activation of VDR requires the NAD-dependent deacetylase domain of SIRT1, a deacetylase-defective mutant (H363Y) was employed in XDR3 VDRE-based luciferase assays in HEK293 cells (Fig. 4B). The mutant SIRT1 plasmid (H363Y) encodes a protein that contains a point mutation at the highly conserved, catalytically active histidine residue located in the core domain of the functional protein, thus effectively abolishing deacetylase activity [15, 36]. When HEK293 cells were transfected with pCMV-empty, pCMV-SIRT1 (WT SIRT1), or pECE-Flag-SIRT1 H363Y (Mut SIRT1), and exposed to 1,25D, the 1,25D-treated cells expressing WT SIRT1 displayed a 1.2-fold increase in VDR transactivation (Fig. 4B, bar 3 versus SIRT1 empty vector+1,25D result normalized to 1.0-fold; data not shown, p<0.05), whereas the H363Y mutant SIRT1 exerted a diminution in VDRE-mediated transcription to 0.79-fold (Fig. 4B, bar 4) when compared to the empty vector control/1,25D-treated cells (Fig. 4B, –SIRT1 empty vector +1,25D result normalized to 1.0-fold; data not shown, p<0.05). Translated into relative effectiveness, the data depicted in Fig. 4B illuminate the H363Y mutant SIRT1 as being only 66% as effective as WT SIRT1 in boosting VDRE-mediated transcription, and the loss of function SIRT1 mutant actually represses VDRE-mediated transcription, mimicking the effects of the EX-527 inhibitor. These data highlight the importance of the NAD-dependent deacetylase domain of SIRT1 and, therefore, its significance in modulating VDRE-driven transcription. Interestingly, this same phenomenon of reversal of action on transcription by the SIRT1 mutant occurs in the absence of 1,25D hormone (Fig. 4B, ETOH controls, bar 2 versus 1, p=0.004), again dissociating 1,25D effects from SIRT1 effects on transcription, and pointing to an allosteric influence of 1,25D-ligand distinct from a catalytic action of SIRT1 on VDR deacetylation and associated transcriptional activation capacity. Thus, both selective inhibition of either endogenous or ectopic SIRT1 via EX-527, and expression of a catalytically-defective SIRT1 enzyme results in loss of the observed increase in VDR activity, indicating that the deacetylase domain of SIRT1 is required for potentiation of 1,25D-VDR signaling. One caveat to this conclusion is that the wild-type SIRT1 and SIRT1-mutant vectors contain different promoters, namely CV1 and SV40, respectively. Both are strong viral promoters, and we assume approximate equivalency of expression, but we have not proven this experimentally by quantitating the expressed SIRT1 enzymes. Nevertheless, in viewing Fig. 4, in toto, the data that the SIRT1 inhibitor produces an identical pattern/profile of results to that of the SIRT1 mutant (compare bars 1–4 in Fig. 4A with bars 1–4 in Fig. 4B), it is clear that the SIRT1 mutant and SIRT1 inhibitor elicit the same result. This observation offers compelling evidence for the role of SIRT1 catalytic activity in modulating 1,25D-VDR/RXR-mediated transcriptional control.
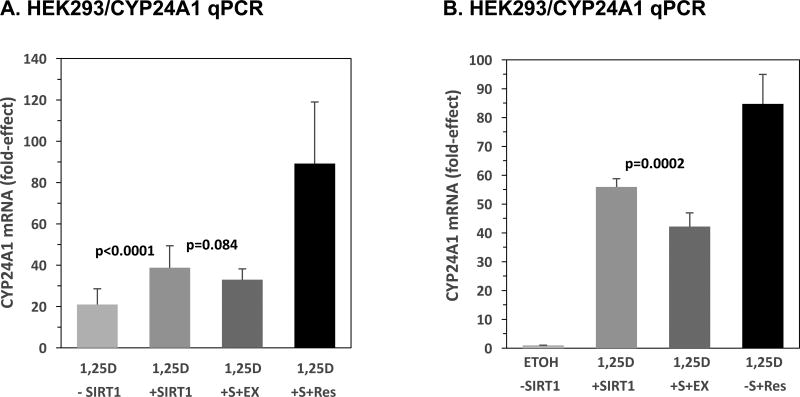
3.5. CYP24A1 mRNA Expression is Stimulated by SIRT1 and Resveratrol
Quantitative real time PCR (qPCR) was utilized to assess SIRT1-augmented induction of a bona fide 1,25D-target gene (CYP24A1) within the milieu of its natural chromatin context. The principal enzyme that exerts a negative feedback loop for homeostatic control of cellular and circulating 1,25D is CYP24A1, the protein catalyzing the initial step in 1,25D catabolism [37–39]. Briefly, the metabolic degradation of 1,25D via CYP24A1-catalyzed 24-hydroxylation of 1,25D to form 1,24,25-trihydroxyvitamin D3 (1,24,25D) serves to deactivate 1,25D, as 1,24,25D is approximately 10 times less potent than the active hormonal form [37, 39]. CYP24A1 is robustly induced by 1,25D-VDR in all 1,25D target tissues and can serve as a physiologically relevant and sensitive biomarker for VDR activation of endogenous cellular genes. It was therefore of interest to examine the regulation of endogenous CYP24A1 by SIRT1 and its resveratrol activator via qPCR, using the known regulation of CYP24A1 by 1,25D as a positive control. The results in Fig. 5A show that 1,25D alone boosts CYP24A1 mRNA levels by 21-fold in HEK293 cells in which VDR levels were amplified by the inclusion of an expression plasmid for VDR (bar 1); co-transfection with the pCMV-SIRT1 expression plasmid elicited a 39-fold increase, almost a doubling of 1,25D-induced CYP24A1 mRNA expression compared to cells receiving 1,25D and no exogenous SIRT1 (Fig. 5A, bar 2 versus 1, p<0.0001). Moreover, inclusion of the SIRT1-inhibitor, EX-527, apparently attenuates the response to 1,25D+SIRT1 to a 33-fold effect (Fig. 5A, bar 3 versus bar 2, p=0.084 > 0.025; not statistically significant). In contrast to EX-527, Res treatment amplifies the transcriptional action of 1,25D and SIRT1 to 89-fold (Fig. 5A, bar 4), supporting the concept of SIRT1 potentiation of VDRE-mediated transcription in a native chromatin environment. Because the impact of EX-527 was modest, and did not reach statistical significance, in the results illustrated in Fig. 5A, plus the larger variability of the resveratrol data than observed previously, we carried out another set of qPCR experiments to re-examine the effect of EX-527 and Res on the expression of CYP24A1 under conditions similar to those depicted in Fig. 5A. In this final qPCR experiment, HEK293 cells transfected with SIRT1 and treated with 1,25D+EX-527 display a 25% decrease in CYP24A1 mRNA expression when compared to 1,25D/SIRT1 in the absence of EX-527 (Fig. 5B, bar 3 versus 2, p=0.0002), and Res challenge produces a more consistent, 85-fold effect on transcription (Fig. 5B, bar 4). These results for endogenous CYP24A1 mRNA expression are in concert with the decreased VDR activity effected by EX-527 that was observed in the luciferase assay (see Fig. 4A), further confirming a role for SIRT1 in the regulation of 1,25D-VDR-mediated transcriptional activation of target genes in the setting of VDRE gene control in a natural chromatin context. Thus, the results pictured in Fig. 5A and B establish SIRT1 and resveratrol as authentic potentiators of the expression of a classical 1,25D target gene (CYP24A1), as assessed in the environment of the natural chromatin architecture.
Figure 5. CYP24A1 mRNA Expression is Stimulated by SIRT1 and Resveratrol.
(A) HEK293 cells were transfected with 5 ng pCMV-empty (-SIRT1, -S) or 5 ng pCMV-SIRT1 (+SIRT1, +S) in addition to 50 ng of pSG5-VDR and 250 ng of pTZ18U as carrier DNA. The cells were treated post-transfection with 10 nM 1,25D, 10 nM 1,25D+20 µM EX-527 (EX), or 10 nM 1,25D+25 µM Res. Quantitative PCR was performed on cell lysates and results are presented as a fold-induction after normalizing to human GAPDH. Error bars represent standard deviation. Results are the average of 15 biological replicates (n=15). (B) As in (A) except that cells were treated with ETOH, 10 nM 1,25D, 10 nM 1,25D+20 µM EX-527 (EX), or 10nM 1,25D+25 µM Res. Note that the group treated with EX-527 also received an additional dose of drug during time of transfection to inhibit endogenous SIRT1. Results shown are the average of nine biological replicates (n=9). P-values are noted in the figure panels.
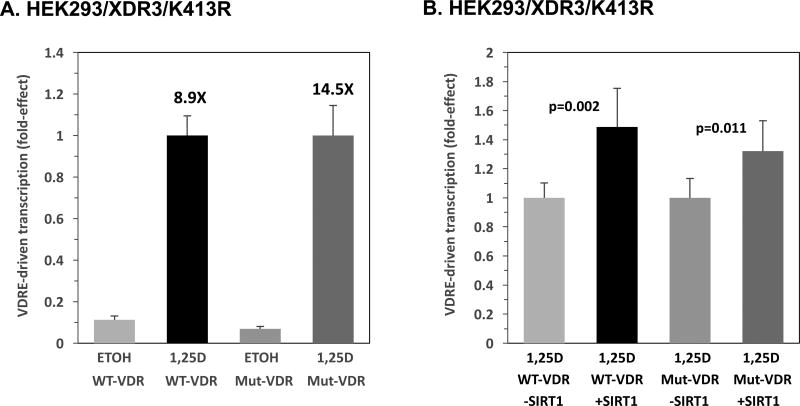
3.6. K413R Mutant VDR Possesses Increased Transactivation Capacity and is Less Sensitive to Transcriptional Enhancement by SIRT1
Given the catalytic role of SIRT1 as a deacetylase, we next examined the activity of a novel VDR mutant, K413R, that eliminates a conserved lysine within a candidate acetylation motif near the C-terminus of VDR. K413R was probed in the context of SIRT1-plasmid overexpression utilizing HEK293-based, XDR3-luciferase assays. The K413R VDR variant was generated via site-directed mutagenesis and constitutes alteration of a potential acetylation site in the ligand binding domain of VDR [40]. The substitution of arginine for lysine at amino acid position 413 of VDR renders this mutant non-acetylatable at this basic site while maintaining the conformational integrity and charge distribution of the receptor [25, 41]. It has already been demonstrated in the immunoprecipitation experiments pictured in Fig. 1A and B, that this VDR mutant is strikingly under-acetylated compared to wild-type VDR (e.g., lane 4 versus 1).
The activities of wild-type (WT) VDR and mutant (Mut) K413R VDR were evaluated in the absence and presence of 1,25D (Fig. 6A) as well as with the transfection of 5 ng SIRT1 in the 1,25D context (Fig. 6B). When either 25 ng of WT or mutant VDR expression plasmid were transfected into HEK293 cells, a greater (14.5-fold) increase in 1,25D-induced, XDR3-mediated transcription was observed in the mutant VDR group over that in WT VDR group (8.9-fold), as revealed in Fig. 6A, bars 4 and 2, and the difference in fold increase was statistically significant (p<0.0001), indicating that K413R is more responsive to 1,25D than wild-type VDR, likely because of the alteration of the transcriptionally desensitizing lysine residue at position 413 in VDR. These data are consistent with a significant role for acetylation of K413 in VDR. The transfection of SIRT1 does not appear to affect mutant VDR transcriptional activity to the degree that it influences WT VDR (Fig. 6B, bar 4 versus 2; p=0.27: not statistically significant). Specifically, SIRT1 co-transfection exerts a 1.3-fold increase (p=0.011) in mutant VDR/VDRE-mediated transcription in HEK293 cells, while SIRT1 co-transfection exerts a 1.5-fold increase (p=0.002) in wild-type VDR/VDRE-mediated transcription (Fig. 6B). However, these data also suggest that multiple post-translational modifications in VDR may occur at the K413 site in addition to acetylation, such as SUMOylation, which is postulated to repress VDR activity [41]. Finally, the fact that K413R VDR exhibited greater transactivation (1.3-fold) in the presence of SIRT1 compared to K413R VDR in the absence of SIRT1 (Fig. 6B, bar 4 versus 3, p = 0.011), suggests that multiple lysine sites in VDR may serve as targets of deacetylation by SIRT1, similar to SIRT1-induced deacetylation of PPARγ [21].
Figure 6. K413R Mutant VDR Possesses Increased Transactivation Capacity and is Less Sensitive to Transcriptional Enhancement by SIRT1.
(A) Embryonic kidney (HEK293) cells were transfected with either 25 ng of wild-type human VDR (WT) or 25 ng of mutant (Mut) K413R human VDR. The XDR3-mediated transcriptional activity of WT and Mut VDR in the presence and absence of 1 nM 1,25D was assessed. Results are the average of nine biological replicates (n=9). (B) A similar experiment to that in (A) was conducted with 25 ng of human WT VDR or K413R VDR, in this case testing the influence of 5 ng of SIRT1 vector versus the empty vector control only in the presence of 1,25D. Results are the average of 6 biological replicates (n=6). P-values are noted in the figure panels.
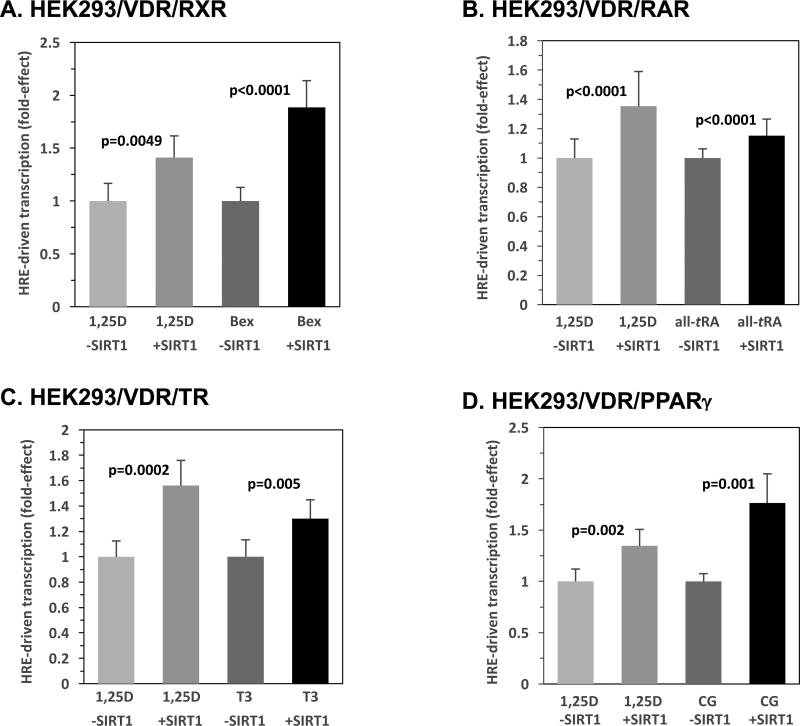
3.7. SIRT1 Modulates Hormone Responsive Element-mediated Transcription Driven by Multiple Nuclear Receptors
In order to investigate SIRT1 effects on transactivation by other nuclear receptors, select homodimerizing and RXR-heterodimerizing receptors were evaluated in the context of their cognate responsive element and ligand via transcriptional assays in HEK293 cells. Fig. 7A displays testing of the homodimeric retinoid X receptor using luciferase constructs containing the retinoid X receptor responsive element (RXRE) in the absence or presence of a low level of SIRT1 plasmid. VDR was included as a positive control for the effects of SIRT1 in this, and all subsequent, nuclear receptor analyses. RXRE-mediated transcriptional activity was escalated 1.9-fold (p<0.0001) in the presence of 100 nM Bexarotene (Bex) and SIRT1 over the empty vector group treated with Bex. The VDR positive control exhibited a 1.4-fold (p=0.0049) enhancement in VDRE-driven transcription. That both VDR and RXR activity are augmented by SIRT1 overexpression raises the question as to whether these two functions may cooperate in transactivation by VDR-RXR, as well as by additional nuclear receptor RXR-heterocomplexes. In the case of VDR, the magnitude of individual enhancement is smaller than that exerted by SIRT1 on RXR (Fig. 7A; p=0.03), implying that RXR and VDR are not cooperating, with only the primary receptor in the dimer, residing on the 3-prime half-element, being subjected to post-translational modulation.
Figure 7. SIRT1 Modulates Hormone Responsive Element-mediated Transcription Driven by RXR, RAR, TR and PPARγ.
HEK293 cells were transfected with 5 ng pCMV-empty or 5 ng of pCMV-SIRT1, and 5 ng of the expression plasmid for the following indicated nuclear receptors: RXR (A), RAR (B), TR (C), PPARγ (D), and 250 ng of the cognate responsive element (RXRE, RARE, TRE, and PPRE, respectively). After 24 hours, the cells were treated with either ETOH or the appropriate activating ligand (100 nM Bexarotene (Bex), 100 nM all trans retinoic acid (all-tRA), 100 nM triiodothyronine (T3), or 10 µM Ciglitizone (CG), respectively). VDR was also evaluated using the XDR3 VDRE-reporter and 1 nM 1,25D as a positive control in (A)–(D). The number of biological replicates was as follows: (A) n=6, (B) n=24, (C) n=6, (D) n=6. P-values are noted in the figure panels.
Figs. 7B, C, and D illustrate the influence of SIRT1 on a series of three receptors, like VDR, for which heterodimerization with RXR is an obligate step in the transactivation of target genes, namely retinoic acid receptor (RAR), thyroid hormone receptor (TR), and peroxisome proliferator-activated receptor gamma (PPARγ). These three close evolutionary relatives of VDR were also examined using a similar luciferase reporter system and reporter vectors containing the cognate RARE, TRE, and PPRE responsive elements, respectively (Figs. 7B, C, and D). Again, VDR was included as a positive control for SIRT1 action. HEK293 cells transfected with RARE, TRE, or PPRE luciferase constructs and challenged with the appropriate ligand (100 nM all trans retinoic acid (all-tRA), 100 nM triiodothyronine (T3), or 10 µM Ciglitizone (CG)), respectively, displayed 1.2-fold (Fig. 7B, p<0.0001), 1.3-fold (Fig. 7C, p=0.005), and 1.8-fold (Fig. 7D, p=0.001) increases in transactivation in the presence of exogenous SIRT1 compared with ligand treatment in the absence of SIRT1. Control VDR/VDRE transcription was augmented 1.4- to 1.6-fold by SIRT1 overexpression in Figs. 7B, C, and D, indicating that RAR is less sensitive than VDR to enhancement by SIRT1 (p=0.0028), TR is apparently less sensitive (difference not statistically significant; p=0.11), whereas PPARγ is the most responsive heterodimerizing nuclear receptor (p=0.02 compared to VDR). One explanation for this exquisite sensitivity of PPARγ could be that it is uniquely docked on the 5-prime half-element of the PPRE, and may be able to cooperatively employ its hetero-partner, RXR, residing on the 3-prime half-element, in SIRT1-bolstered transcription.
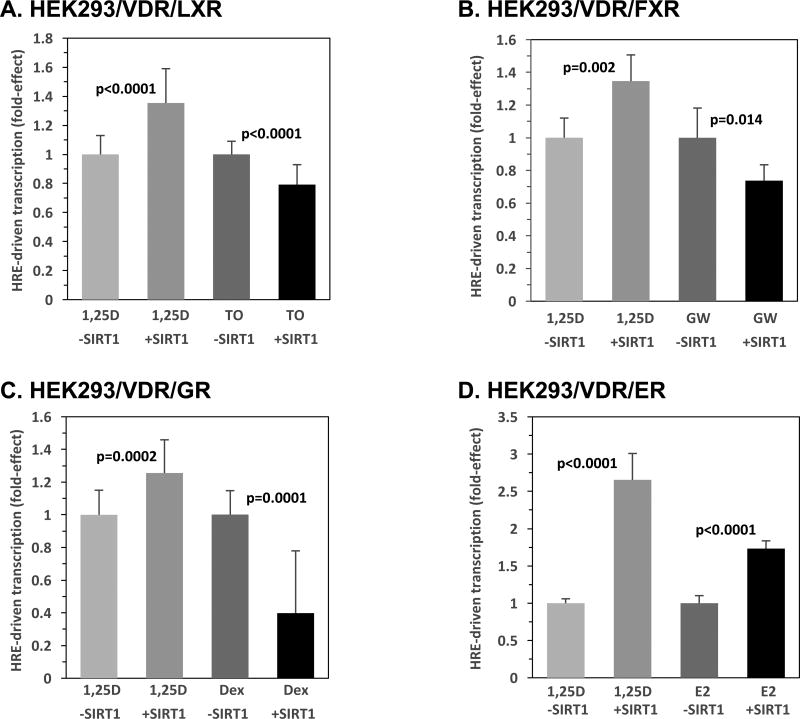
Lastly, as depicted in Fig. 8A–D, we evaluated the effect of SIRT1-overexpression on the liver X receptor (LXR), farnesoid X receptor (FXR), glucocorticoid receptor (GR), and estrogen receptor (ER), assessed in the context of their cognate responsive element and ligand via transcriptional assays in HEK293 cells. The respective responsive elements, LXRE, FXRE, GRE, and ERE, were linked to the luciferase reporter, and VDR served as a positive control in each set of experiments. As pictured in Fig. 8A and B for the RXR-heterodimerizing receptors LXR and FXR, along with their cognate ligands, 10 nM T0901317 (TO) and 100 nM GW-4064 (GW), respectively, the results differ markedly from those obtained with VDR, RAR, TR, and PPARγ, in that LXR/LXRE- and FXR/FXRE-driven transcription is decreased significantly. LXR and FXR display a 21% (p<0.0001) and 26% (p=0.014) respective reduction of transactivation in the presence of SIRT1 compared to the ligand-treated, empty vector group (Fig. 8A and B). It should be noted that the present results with FXR differ from those reported by Kemper et al.[23]; however, it is important to note the differences in cellular context, as Kemper et al. utilized a hepatic-derived line and in the present studies we employed embryonic kidney cells.
Figure 8. SIRT1 Modulates Hormone Responsive Element-mediated Transcription Driven by LXR, FXR, GR and ER.
HEK293 cells were transfected with 5 ng pCMV-empty or 5 ng of pCMV-SIRT1, and 5 ng of the expression plasmid for the following indicated nuclear receptors: LXR (A), FXR (B), GR (C), ER (D), and of the cognate responsive element (125 ng LXRE, 50 ng FXRE, 250 ng GRE, and 250 ng ERE, respectively). Note that in some cases, pTZ18U carrier DNA was added to achieve equal amounts of transfected DNA in each group. After 24 hours, the cells were treated with either ETOH or the appropriate activating ligand (10 nM T-0901317 (TO), 100 nM GW-4064 (GW), 1 µM Dexamethasone (Dex), and 0.5 µM estradiol (E2), respectively). VDR was also evaluated using the XDR3 VDRE-reporter and 1 nM 1,25D as a positive control in (A)–(D). The number of biological replicates was as follows: (A) n=24, (B) n=6, (C) n=18, (D) n=6. P-values are noted in the figure panels.
With respect to the homodimerizing receptors, GR and ER, Fig. 8C illustrates that GR/GRE activity in the presence of 1 µM Dexamethasone (Dex), a synthetic glucocorticoid, is decreased by 60% (p=0.0001) by SIRT1 overexpression when compared to the empty vector control treated in the presence of Dex. This SIRT1 suppression of transcription driven by GR/GRE occurs in the face of a 1.3-fold enhancement (p=0.0002) of VDR/VDRE-mediated transcription by SIRT1 (Fig. 8C). In contrast, in the case of ER/estrogen responsive element (ERE), in the presence of 0.5 µM estradiol (E2) and SIRT1, ERE-mediated luciferase activity was increased 1.7-fold (p<0.0001) by SIRT1 when compared to the empty vector control treated with E2. Thus, the functional effects of SIRT1 represent a broad spectrum of modulation in the setting of homodimerizing and heterodimerizing nuclear receptors, and the action of SIRT1 on transcription may be cell-selective. Nevertheless, at least for the vitamin D target cells in the kidney, VDR joins RXR, RAR, TR, PPARγ, and ER in a group of nuclear receptors for which the transcriptional activation function is augmented by SIRT1.
4. DISCUSSION
The potential of sirtuin-1 and resveratrol to modulate steroid and nuclear receptor superfamily signaling has been studied in the context of both homodimerizing and heterodimerizing receptors, and the results reveal differential SIRT1 effects in altering receptor activity [42]. Based on the evolutionarily-conserved nature of the steroid and nuclear receptor superfamily and the structural similarity that exists amongst resveratrol and other low-affinity, lipophilic VDR ligands, resveratrol was previously postulated and demonstrated experimentally by Guo and colleagues [43] to modulate intracellular VDR signaling. Independent investigations conducted by Jurutka and coworkers [26], as well as Dampf Stone and collaborators [7] revealed that resveratrol serves as an indirect modifier of the 1,25D-VDR signal transduction cascade, rather than serving as a direct VDR ligand.
In the present study, we investigated one resveratrol target enzyme, sirtuin-1, as a potential regulator of VDR signaling, examining both the cellular context and VDRE platform to determine if SIRT1 affected VDR function to control transcription. Experiments conducted in two distinct, cultured cell models (i.e., HEK293 and TE85) that represent VDR target tissues (i.e., kidney and bone, respectively) reveal that SIRT1 deacetylase overexpression statistically significantly potentiates VDR/VDRE-mediated transcriptional activation. Indeed, in kidney and bone cells in culture, low amounts (1–5 ng) of a SIRT1-expression vector elicit a reproducible and statistically significant enhancement (1.3- to 2.6-fold) in transcription mediated by VDREs from the CYP3A4 and cyp24a1 genes, where the magnitude of the transcriptional response to 1,25D-ligand is 6- to 30-fold (Figs. 2, 3, and 5). Furthermore, diverse VDRE platforms trigger VDRE-selective effects of the SIRT1 enzyme, including DR3 and ER6 responsive element motifs (Fig. 2). To further elucidate the effects of SIRT1 on 1,25D-VDR-mediated transactivation of target genes, we sought to determine which step(s) in the transactivation process was affected by SIRT1. Because the recruitment of and heterodimerization with RXR is fundamental to VDR signaling, the ability of SIRT1 to regulate VDR-RXR association was assessed. Heterodimerization of VDR-RXR increased modestly (16%) in the presence of SIRT1(Supplemental Fig. 1D), unveiling one potential molecular mechanism for the amplification of VDR activity by SIRT1, namely increased recruitment of RXR and/or stabilization of the ligand-bound VDR-RXR heterocomplex. This action of SIRT1 has a precedent with FXR, a heterodimerizing nuclear for which deacetylation by SIRT1 is also reported to facilitate heterodimerization with RXR [23]. It is acknowledged that some of the effects we observe for SIRT1-potentiation of VDR functions are modest in magnitude, ranging in magnitude from a low of 116% (Supplemental Fig. 1D) to a high of 260% (Fig. 8D), with an average transcription enhancement by SIRT1 of 1.68-fold obtained by considering 19 experiments with a total of 166 biological replicates. However, in the conceptualization of fine-tuning of ligand-controlled gene expression, the hormonal ligand must dominate in directing transcription (yielding a sizeable fold-effect), with environmental and other cell-specific fine-tuners conferring only measured/small perturbations that, in sum, contribute to appropriate gene expression control.
Moreover, experiments were conducted with an array of nuclear receptors that either homodimerize (i.e., ER, GR, RXR) or heterodimerize (i.e., FXR, LXR, PPARγ, RAR, and TR) with RXR, with the overall conclusion that VDR joins RXR, RAR, TR, PPARγ, and ER in a group of nuclear receptors for which the transcriptional activation function is augmented by SIRT1 (see Figs. 7 and 8). Except for the unique case of ER/ERE, the current results (Figs. 7 and 8) imply a potential RXR-dependence for the SIRT1 effect to enhance transcription, as also proposed for RAR by Singh and colleagues [44]. However, we observe herein that two RXR-heterodimerizing nuclear receptors, namely LXR (Fig. 8A) and FXR (Fig. 8B), are suppressed in their transactivation capacities by SIRT1 overexpression, at least in kidney cells. Therefore, it is not possible to generalize SIRT1 influence on nuclear receptor-mediated transcription, as it may be governed cell-specifically, perhaps to integrate the actions of nuclear receptors according to a variety of environmental challenges. It is tempting to speculate that the different effects of acetylation/deacetylation on the function of various nuclear receptors may be explainable, at least in part, by the location of the specific lysine residues that are acetylated and their role in the function of the receptor in question, in the physiologically relevant cell. Moreover, the stimulatory actions of SIRT1 on the activity of VDR, ER, TR and PPARγ could be employed therapeutically to potentiate cognate ligands to positively influence bone mineral homeostasis (VDR), inhibit bone resorption (ER), increase basal metabolic rate (TR), and boost insulin sensitivity (PPARγ).
The potential deacetylation of VDR was examined utilizing a K413R VDR mutant in the setting of VDRE-luciferase-based assays. The fact that the K413R mutant, which essentially represents a non-acetylatable variant of VDR, possesses enhanced transactivation capacity when compared to wild-type VDR (Fig. 6A), and is less sensitive to SIRT1-augmented transactivation than wild-type VDR (Fig. 6B), suggests that acetylation at this site is somehow inhibitory to VDR activity (including heterodimerization as cited above). The mechanism accounting for why this acetylation is inhibitory is not entirely clear from the position of this residue in VDR. K413 is located near the terminal VDR helix 12, but it neither appears to form part of the platform for interacting with coactivators, nor does it appear to be involved in the heterodimerization interface. Further, in the presence of SIRT1, K413R exhibited even greater transactivation, signifying the potential existence of multiple lysine sites in the VDR protein that may serve as target sites for acetylation/deacetylation and regulation by SIRT1, potentially including K45 in the DNA binding domain, K53/K55 in the nuclear localization domain, K302 in the ligand binding domain, and K321/K382 in the dimerization domain [40]. Regarding the additional candidate lysines in the VDR ligand binding domain, none of these residues (K302, K321, K382, nor K413) display any obvious participation in a functional interaction with ligand, coactivator, or the RXR heteropartner. Rather, each of these residues appears to interact with (a) residue(s) on an adjacent helix, suggesting that they may play a general role in VDR conformation. Interestingly, VDR residue K413 is conserved in mammals and birds, but not in fish and other lower species (e.g., lamprey), suggesting that its role as an acetylation/deacetylation modulator of VDR function is confined to homeostatic maintenance in higher organisms. Further experimentation will be necessary to investigate the functional significance of the additional internal lysines that are present in candidate acetylation motifs, and to explain why the ability of SIRT1 to enhance transactivation by VDR is potentially cell- and gene-specific. It should be noted that there exists the possibility that some effects observed for VDR in the present communication are the result of post-translational modification, specifically acetylation/deacetylation, of the RXR transcriptional partner for VDR. Finally, we also concede that we cannot exclude acetylation/deacetylation of another member(s) of the transcription meta-complex besides VDR and RXR impacting our results.
Importantly, immunoprecipitation and Western blot assays revealed a significant decrease in acetylated VDR in the presence of resveratrol and SIRT1, thus revealing the deacetylation of VDR by SIRT1 via a reaction enhanced by both the 1,25D VDR agonist, and resveratrol deacetylase activator. Based upon the authors’ experience in studying post-translation modifications of VDR, i.e., phosphorylation/dephosphorylation, we propose that acetylation influences protein-protein interactions between VDR and transcriptional factors/coregulators. The action of VDR deacetylation to promote heterodimerization with RXR is a viable starting point, but the magnitude of this effect is probably too small to account for the full amplitude of transcriptional enhancement. Consequently, the authors favor additional VDR-RXR interaction with known cofactors in 1,25D/VDR control of gene expression (e.g., CEBP, RUNX2, NURR1, CREBP, LEF-1, etc.), as well as novel candidate players. We propose that these acetylation-modulated VDR-inter-actors are possibly members of the transcription meta-complex, but also could generate chromatin looping to bring together VDR-RXR docked on VDREs with remote intra-gene sites of transcription factor localization.
To confirm the current findings by studying a gene in its native chromatin environment, experiments were conducted to assess the effects of both SIRT1 and resveratrol on the 1,25D-VDR-mediated induction of CYP24A1 mRNA expression. The results demonstrate enhanced induction of CYP24A1 by both SIRT1 and Res (Fig. 5A and B); however, resveratrol induced a more potent CYP24A1 activation, implying the existence of multiple resveratrol-regulated signaling mediators (e.g., FoxO, p53, and β-catenin) that may function to positively augment VDR bioeffects. Indeed, resveratrol has been shown to exert some of its actions in a SIRT1-independent manner, such as activation of the serine/threonine protein kinase ATM [45]. The significance of these potential actions of resveratrol, if any, to transactivation by VDR also awaits further investigation. Regardless of the breadth of resveratrol effects, we employed the specific SIRT1-inhibitor, EX-527 (Figs. 4A, 5A and B), and a SIRT1-inactivated mutant (Fig. 4B), to provide additional evidence for the crucial role for SIRT1-deacetylase activity in potentiating VDR/VDRE-mediated transcription. That Res is a classic SIRT1-activator is entirely consistent with the dramatic positive influence (Fig. 5A and B) of this putative anti-aging compound on SIRT1 enhancement of VDRE-driven transcription.
Of particular interest in the resveratrol-SIRT1/1,25D-VDR-partnership is the FoxO family of transcription factors that function to regulate cell growth/development, metabolism, and longevity, similar to the proposed actions of VDR [46–48]. SIRT1-catalyzed deacetylation has been linked to increased FoxO activity and DNA binding affinity [49, 50]. A study conducted by An and colleagues [51] demonstrated that VDR modulates the post-translational modification and function of FoxO proteins and serves as a selective regulator of SIRT1 action. The present study serves to unveil the crosstalk between VDR and SIRT1 as a positive feedback loop functioning to regulate gene products that coincide with FoxO and VDR anticancer effectors (e.g., repression of cyclins) [51, 52]. It will be of interest to examine the extent to which SIRT1 activity may be regulated by 1,25D-VDR, perhaps via multiple molecular mechanisms.
Intriguingly, experiments conducted by Firestein et al. [53] demonstrated that SIRT1 deacetylates β-catenin, a major etiological driver of colorectal malignancies, and suppresses the ability of β-catenin to trigger transcription and drive cell growth/division. Ligand-bound VDR is strongly implicated in the prevention of colon neoplasia and several studies have assessed the crosstalk between VDR and β-catenin signaling, including reports demonstrating that 1,25D attenuates beta-catenin-TCF-4 transcriptional activity, as well as inhibiting the expression of beta-catenin-TCF-4 target genes including c-myc [37, 54, 55]. Ultimately, a comprehensive understanding of the intersection of 1,25D-VDR, SIRT1, resveratrol, FoxO, and β-catenin signaling will be vital in the further elucidation of the molecular paradigms underlying SIRT1 and 1,25D-VDR action, as well as in their integration into the promotion of anticancer, anti-aging, and longevity pathways.
In conclusion, via immunoprecipitation experiments, we have demonstrated for the first time that VDR is acetylated/deacetylated (Fig. 1), and that this nuclear receptor displays a dramatic decrease in acetylation in the presence of 1,25D-ligand, SIRT1 deacetylase, or the resveratrol activator of SIRT1. The molecular basis and functional significance of VDR-deacetylation was characterized in a series of co-transfection experiments in kidney and bone cells. VDRE-based transcriptional assays revealed that low amounts (1–5 ng) of a SIRT1-expression vector elicit a reproducible and statistically significant enhancement (1.3- to 2.6-fold) in transcription mediated by VDREs from the CYP3A4 and cyp24a1 genes, where the magnitude of response to 1,25D-ligand is 6- to 30-fold. Inhibition of SIRT1 via EX-527, or utilization of a SIRT1 loss-of-function mutant (H363Y), resulted in abrogation of SIRT1-mediated VDR potentiation. Studies with a novel, non-acetylatable VDR mutant (K413R) showed that the mutant VDR possesses enhanced responsiveness to 1,25D, in conjunction with reduced, but still significant, sensitivity to exogenous SIRT1, indicating that acetylation of lysine 413 is relevant, but that other acetylated residues in VDR contribute to modulation of its activity. We conclude that the acetylation of VDR comprises a negative feedback loop that attenuates 1,25D-VDR signaling. This regulatory loop is reversed by SIRT1-catalyzed deacetylation of VDR to amplify VDR signaling and 1,25D actions.
Supplementary Material
HIGHLIGHTS.
Human VDR is post-translationally modified via acetylation, in part at lysine-413.
VDR deacetylation is mediated by SIRT1, and is enhanced by resveratrol, a SIRT1 activator, as well as by 1,25D ligand.
Deacetylation of VDR augments both ligand-dependent and apparent ligand-independent VDR activity.
SIRT1 and resveratrol amplify 1,25D-stimulated mRNA expression of CYP24A1 in embryonic, kidney-derived cells.
Deacetylation of other nuclear receptors by SIRT1 is stimulatory or inhibitory depending on the cellular/receptor context.
Acknowledgments
FUNDING
This work was supported by National Institutes of Health grants DK033351 to MRH and CA140285 to ETJ/PWJ. Arizona State University New College Undergraduate Inquiry and Research Experiences (NCUIRE) Program researcher grants were awarded to MSS and ZK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Nonstandard Abbreviations: 1α,25-dihydroxyvitamin D3, 1,25D; vitamin D receptor, VDR; retinoid X receptor, RXR; mammalian 2-hybrid, M2H; vitamin D responsive element, VDRE; sirtuin-1, SIRT1; resveratrol, Res
AUTHOR CONTRIBUTIONS
MSS contributed to study design, data collection, data analysis/interpretation, and drafting the manuscript. ZK, MAG, CMD, SM, and MCH contributed to data collection/analysis and revision of this manuscript. ADS, SFB, GKW, ETJ and CH contributed via critical revision of the article. ETJ and CH provided statistical expertise. MRH contributed via conception/design of work and a major final revision of the manuscript. PWJ, the corresponding author, contributed via conception/design of work, data interpretation, and critical revision of the article. All authors read and approved the final version of the manuscript.
DISCLOSURE STATEMENT
The authors disclose no actual or potential conflicts of interest.
References
- 1.Holick MF, Smith E, Pincus S. Skin as the site of vitamin D synthesis and target tissue for 1,25-dihydroxyvitamin D3. Use of calcitriol (1,25-dihydroxyvitamin D3) for treatment of psoriasis. Arch Dermatol. 1987;123:1677–1683a. [PubMed] [Google Scholar]
- 2.Webb AR, DeCosta BR, Holick MF. Sunlight regulates the cutaneous production of vitamin D3 by causing its photodegradation. Journal of Clinical Endocrinology and Metabolism. 1989;68:882–887. doi: 10.1210/jcem-68-5-882. [DOI] [PubMed] [Google Scholar]
- 3.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, Jurutka PW. Molecular mechanisms of vitamin D action. Calcified Tissue International. 2013;92:77–98. doi: 10.1007/s00223-012-9619-0. [DOI] [PubMed] [Google Scholar]
- 4.Bouillon R, Okamura WH, Norman AW. Structure-function relationships in the vitamin D endocrine system. Endocrine Reviews. 1995;16:200–257. doi: 10.1210/edrv-16-2-200. [DOI] [PubMed] [Google Scholar]
- 5.Haussler MR, Jurutka PW, Mizwicki M, Norman AW. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2vitamin D3: genomic and non-genomic mechanisms. Best practice & research. Clinical endocrinology & metabolism. 2011;25:543–559. doi: 10.1016/j.beem.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Bikle DD. Vitamin D metabolism, mechanism of action, and clinical applications. Chemistry & biology. 2014;21:319–329. doi: 10.1016/j.chembiol.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dampf Stone A, Batie SF, Sabir MS, Jacobs ET, Lee JH, Whitfield GK, Haussler MR, Jurutka PW. Resveratrol potentiates vitamin D and nuclear receptor signaling. Journal of Cellular Biochemistry. 2015;116:1130–1143. doi: 10.1002/jcb.25070. [DOI] [PubMed] [Google Scholar]
- 8.Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 9.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy Andrew J, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 Inhibits Glutamate Dehydrogenase and Opposes the Effects of Calorie Restriction in Pancreatic β Cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. Journal of Biological Chemistry. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 11.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 13.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Molecular Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 14.Howitz K, Bitterman J, Cohen H. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 15.Luo J, Nikolaev aY, Imai S, Chen D, Su F, Shiloh a, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 16.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 Represses Forkhead Transcription Factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 17.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Oliveira RM, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchal J, Pifferi F, Aujard F. Resveratrol in mammals: Effects on aging biomarkers, age-related diseases, and life span. Annals of the New York Academy of Sciences. 2013;1290:67–73. doi: 10.1111/nyas.12214. [DOI] [PubMed] [Google Scholar]
- 19.Raederstorff D, Kunz I, Schwager J. Resveratrol, from experimental data to nutritional evidence: The emergence of a new food ingredient. Annals of the New York Academy of Sciences. 2013;1290:136–141. doi: 10.1111/nyas.12147. [DOI] [PubMed] [Google Scholar]
- 20.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-jB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 23.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR Acetylation Is Normally Dynamically Regulated by p300 and SIRT1 but Constitutively Elevated in Metabolic Disease States. Cell Metabolism. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakran S, Sharma P, Attia RR, Hori RT, Deng X, Elam MB, Park Ea. Role of sirtuin 1 in the regulation of hepatic gene expression by thyroid hormone. The Journal of biological chemistry. 2013;288:807–818. doi: 10.1074/jbc.M112.437970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 Deacetylates and Positively Regulates the Nuclear Receptor LXR. Molecular Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 26.Jurutka PW, Whitfield GK, Forster R, Batie S, Lee J, Haussler MR. Vitamin D: A fountain of youth in gene regulation, Vitamin D: Oxidative Stress. Immunity, and Aging. 2013:3–35. [Google Scholar]
- 27.Mangelsdorf DJ, Umesono K, Kliewer SA, Borgmeyer U, Ong ES, Evans RM. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–561. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- 28.Galetto R, Albajar M, Polanco JI, Zakin MM, Rodriguez-Rey JC. Identification of a peroxisome-proliferator-activated-receptor response element in the apolipoprotein E gene control region. Biochem J. 2001;357:521–527. doi: 10.1042/0264-6021:3570521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kast HR, Nguyen CM, Sinal CJ, Jones Sa, Laffitte Ba, Reue K, Gonzalez FJ, Willson TM, Edwards Pa. Farnesoid X-activated receptor induces apolipoprotein C-II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Molecular endocrinology (Baltimore, Md.) 2001;15:1720–1728. doi: 10.1210/mend.15.10.0712. [DOI] [PubMed] [Google Scholar]
- 30.Mak Pa, Kast-Woelbern HR, Anisfeld AM, Edwards Pa. Identification of PLTP as an LXR target gene and apoE as an FXR target gene reveals overlapping targets for the two nuclear receptors. Journal of lipid research. 2002;43:2037–2041. doi: 10.1194/jlr.c200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 31.Tsika RW, Bahl JJ, Leinwand LA, Morkin E. Thyroid hormone regulates expression of a transfected human alpha-myosin heavy-chain fusion gene in fetal rat heart cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:379–383. doi: 10.1073/pnas.87.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng H-L, Jedrychowski MP, Gygi SP, Sinclair Da, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, N.Y.) 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 33.Jurutka PW, Thompson PD, Whitfield GK, Eichhorst KR, Hall N, Dominguez CE, Hsieh JC, Haussler CA, Haussler MR. Molecular and functional comparison of 1,25-dihydroxyvitamin D(3) and the novel vitamin D receptor ligand, lithocholic acid, in activating transcription of cytochrome P450 3A4. J Cell Biochem. 2005;94:917–943. doi: 10.1002/jcb.20359. [DOI] [PubMed] [Google Scholar]
- 34.Cen Y. Sirtuins inhibitors: The approach to affinity and selectivity. Biochimica et Biophysica Acta - Proteins and Proteomics. 2010:1635–1644. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Gertz M, Fischer F, Nguyen GTT, Lakshminarasimhan M, Schutkowski M, Weyand M, Steegborn C. Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2772–2781. doi: 10.1073/pnas.1303628110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. Journal of Biological Chemistry. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 37.Haussler MR, Haussler CA, Whitfield GK, Hsieh JC, Thompson PD, Barthel TK, Bartik L, Egan JB, Wu Y, Kubicek JL, Lowmiller CL, Moffet EW, Forster RE, Jurutka PW. The nuclear vitamin D receptor controls the expression of genes encoding factors which feed the "Fountain of Youth" to mediate healthful aging. Journal of Steroid Biochemistry and Molecular Biology. 2010;121:88–97. doi: 10.1016/j.jsbmb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs ET, Van Pelt C, Forster RE, Zaidi W, Hibler EA, Galligan MA, Haussler MR, Jurutka PW. CYP24A1 and CYP27B1 polymorphisms modulate vitamin D metabolism in colon cancer cells. Cancer Research. 2013;73:2563–2573. doi: 10.1158/0008-5472.CAN-12-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones G, Prosser DE, Kaufmann M. 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): Its important role in the degradation of vitamin D. Archives of Biochemistry and Biophysics. 2012:9–18. doi: 10.1016/j.abb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nature reviews. Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 41.Lee WP, Jena S, Doherty D, Ventakesh J, Schimdt J, Furmick J, Widener T, Lemau J, Jurutka PW, Thompson PD. Sentrin/SUMO specific proteases as novel tissue-selective modulators of vitamin D receptor-mediated signaling. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0089506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore RL, Dai Y, Faller DV. Sirtuin 1 (SIRT1) and steroid hormone receptor activity in cancer. The Journal of endocrinology. 2012;213:37–48. doi: 10.1530/JOE-11-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo C, Sinnott B, Niu B, Lowry MB, Fantacone ML, Gombart AF. Synergistic induction of human cathelicidin antimicrobial peptide gene expression by vitamin D and stilbenoids. Molecular Nutrition and Food Research. 2014;58:528–536. doi: 10.1002/mnfr.201300266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh CK, Kumar A, LaVoie Ha, DiPette DJ, Singh US. Resveratrol prevents impairment in activation of retinoic acid receptors and MAP kinases in the embryos of a rodent model of diabetic embryopathy. Reproductive sciences (Thousand Oaks, Calif.) 2012;19:949–961. doi: 10.1177/1933719112438972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH, Guo Z, Myler LR, Zheng S, Paull TT. Direct activation of ATM by resveratrol under oxidizing conditions. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0097969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 47.Bouchard C, Marquardt J, Brás A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. The EMBO journal. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim MJ, Ahn K, Park SH, Kang HJ, Jang BG, Oh SJ, Oh SM, Jeong YJ, Heo JI, Suh JG, Lim SS, Ko YJ, Huh SO, Kim SC, Park JB, Kim J, Kim JI, Jo SA, Lee JY. SIRT1 regulates tyrosine hydroxylase expression and differentiation of neuroblastoma cells via FOXO3a. FEBS Letters. 2009;583:1183–1188. doi: 10.1016/j.febslet.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 51.An B-S, Tavera-Mendoza LE, Dimitrov V, Wang X, Calderon MR, Wang H-J, White JH. Stimulation of Sirt1-regulated FoxO protein function by the ligand-bound vitamin D receptor. Molecular and cellular biology. 2010;30:4890–4900. doi: 10.1128/MCB.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 53.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan JB, Thompson PA, Vitanov MV, Bartik L, Jacobs ET, Haussler MR, Gerner EW, Jurutka PW. Vitamin D receptor ligands, adenomatous polyposis coli, and the vitamin D receptor FokI polymorphism collectively modulate beta-catenin activity in colon cancer cells. Molecular carcinogenesis. 2010;49:337–352. doi: 10.1002/mc.20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pálmer HG, González-Sancho JM, Espada J, Berciano MT, Puig I, Baulida J, Quintanilla M, Cano A, de Herreros AG, Lafarga M, Muñoz A. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. J Cell Biol. 2001;154:369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batie S, Lee JH, Jama RA, Browder DO, Montano LA, Huynh CC, Marcus LM, Tsosie DG, Mohammed Z, Trang V, Marshall PA, Jurutka PW, Wagner CE. Synthesis and biological evaluation of halogenated curcumin analogs as potential nuclear receptor selective agonists. Bioorganic and Medicinal Chemistry. 2013;21:693–702. doi: 10.1016/j.bmc.2012.11.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.