Abstract
The Group A Streptococcus remains a significant human pathogen causing a wide array of disease ranging from self-limiting to life-threatening invasive infections. Epithelium (skin or throat) colonization with progression to the subepithelial tissues is the common step in all GAS infections. Here, we used transposon-sequencing (Tn-seq) to define the GAS 5448 genetic requirements for in vivo fitness in subepithelial tissue. A near-saturation transposon library of the M1T1 GAS 5448 strain was injected subcutaneously into mice, producing suppurative inflammation at 24 h that progressed to prominent abscesses with tissue necrosis at 48 h. The library composition was monitored en masse by Tn-seq and ratios of mutant abundance comparing the output (12, 24 and 48 h) versus input (T0) mutant pools were calculated for each gene. We identified a total of 273 subcutaneous fitness (scf) genes with 147 genes (55 of unknown function) critical for the M1T1 GAS 5448 fitness in vivo; and 126 genes (53 of unknown function) potentially linked to in vivo fitness advantage. Selected scf genes were validated in competitive subcutaneous infection with parental 5448. Two uncharacterized genes, scfA and scfB, encoding putative membrane-associated proteins and conserved among Gram-positive pathogens, were further characterized. Defined scfAB mutants in GAS were outcompeted by wild type 5448 in vivo, attenuated for lesion formation in the soft tissue infection model and dissemination to the bloodstream. We hypothesize that scfAB play an integral role in enhancing adaptation and fitness of GAS during localized skin infection, and potentially in propagation to other deeper host environments.
Author summary
The WHO ranks the Group A Streptococcus (GAS) in the top 10 leading causes of morbidity and mortality from infectious diseases worldwide. GAS is a strict human pathogen causing both benign superficial infections as well as life-threatening invasive diseases. All GAS infections begin by colonization of an epithelium (throat or skin) followed by propagation into subepithelial tissues. The genetic requirements for M1T1 GAS 5448 within this niche were interrogated by in vivo transposon sequencing (Tn-seq), identifying 273 subcutaneous fitness (scf) genes with 108 of those previously of “unknown function”. Two yet uncharacterized genes, scfA and scfB, were shown to be critical during GAS 5448 soft tissue infection and dissemination into the bloodstream. Thus, this study improves the functional annotation of the GAS genome, providing new insights into GAS pathophysiology and enhancing the development of novel GAS therapeutics.
Introduction
The Group A Streptococcus (Streptococcus pyogenes, GAS) is a strict human pathogen of high prevalence worldwide [1–4]. The WHO ranks GAS in the top 10 leading causes of morbidity and mortality from infectious diseases, responsible for over 500,000 deaths annually [5]. Mucosal (throat) and epithelial (skin) surfaces represent GAS primary ecological niches, where GAS causes over 700 million reported cases of purulent, self-limiting infections (e.g., pharyngitis, impetigo) worldwide each year [4–6]. GAS can also gain access to normally sterile sites of the body (e.g., soft tissue, bloodstream) and produce life-threatening invasive diseases (necrotizing fasciitis and streptococcal toxic shock syndrome) [7–9]. GAS infections may also trigger the immune sequela acute rheumatic fever, a serious health threat in developing countries [6].
To successfully infect its human host, GAS must adapt to the different niches encountered during the infection process, including physicochemical environmental changes, fluctuating metabolic sources [10–13], as well as the immune response [14]. Molecular epidemiology studies have revealed that GAS pathogenesis is complex [for review, see [15]] with multiple GAS strains (over 230 distinct emm types) harboring distinct genetic determinants for tissue tropism and virulence potential [15–17]. Since the first complete genome of the M1 strain SF370 was released in 2001 [18], more than 53 GAS chromosomes have been completed and over 300 draft genomes are available as of June, 2017 (https://www.ncbi.nlm.nih.gov/genome/genomes/175). The GAS genome is genetically diverse with an average size of 1.85 Mb encoding ca. 1820 genes [15, 19]. Comparative genomics analyses established that the GAS pan-genome contains over 3900 genes with a set of ca. 1200 core genes [15]. A large proportion of GAS genes are still annotated as having no known or predicted function; and accurate functional annotation of GAS genes, particularly in the context of disease manifestations, is key to understanding GAS pathogenesis and improving diagnostics and therapeutics [20].
To investigate pathogenesis of the human-restricted GAS, in vivo infection models have been developed mostly using mice as the host and specific virulent GAS strains capable of producing human-like disease symptoms in these murine models [4, 21–26]. Historically, GAS isolates from the globally distributed M1T1 serotype have been commonly used to investigate GAS virulence potential in vivo. Among these, GAS strain 5448 is a clinical isolate representative of the M1T1 serotype that has been successfully employed in different mouse models of skin and tissue infections as well as necrotizing fasciitis [27–34]. GAS 5448 is also a model organism to study the increased virulence potential caused by mutations in the covS gene, i.e. the covS switch, during invasive infections by the M1T1 serotype [27, 34–36]. Furthermore, GAS 5448 genome sequence was recently released [37]. However, as found with many other GAS strains that are relevant to study virulence in in vivo mouse infection models, genetic manipulations of 5448 can prove difficult [38].
To accelerate functional genomics analyses of GAS pathogenesis, we developed a mariner transposon system (Oskar) for GAS to perform highly saturated mutagenesis [38] and subsequent high-throughput phenotype screens. Initially, we employed a transposon site hybridization (TraSH) screen to identify genes required for GAS 5448 fitness in an ex vivo human blood infection model [39–41]. To take advantage of increased sensitivity and resolution, we modified our mariner (Krmit) for transposon-sequencing (Tn-seq) [42], a method that uses massive parallel DNA sequencing to assay the frequency of transposon insertion sites within complex mutant libraries. Using Tn-seq, we were able to create highly saturated Krmit libraries in both M1T1 5448 and M49 NZ131 serotypes and define the GAS minimal core genome (essential, i.e., non-mutable, genes) for in vitro growth in rich media [43].
In this report, we present a genome-wide Tn-seq analysis to identify M1T1 GAS 5448 genetic determinants necessary for in vivo fitness using a murine model of skin and soft tissue infection. Immunocompetent hairless mice were subcutaneously inoculated with an M1T1 GAS 5448 Krmit library, and gene fitness was assessed from infected tissues to identify mutants differentially represented between the input (T0) and various output pools (12, 24 and 48 HPI) representing clinical progression from inflamed focus of infection to tissue destructive abscess. A total of 273 subcutaneous fitness (scf) genes related to M1T1 GAS 5448 fitness were identified (147 and 126 genes associated with decreased and increased fitness, respectively) in the subcutaneous environment, with 108 of those genes annotated as “of unknown function”. To validate our Tn-seq dataset, defined mutants were created in selected scf genes and tested using in vivo competition with parental GAS 5448 in the soft tissue of mice. Two such genes, scfA and scfB, encode putative membrane-associated proteins that are conserved among Gram-positive pathogens and were important for GAS fitness in the lesion at both 24 and 48 HPI. Defined mutants in scfAB were outcompeted by wild type GAS 5448 in vivo, and were attenuated for lesion formation in the soft tissue infection model as well as for subsequent dissemination into the bloodstream. Thus, ScfAB likely play an integral role in enhancing adaptation and fitness of GAS 5448 during localized skin infection, and potentially in other host environments. Our comprehensive Tn-seq-based dataset of scf genes in GAS will allow researchers to define critical pathways and assign functional attributes to the large number of GAS genes annotated as unknown function or hypothetical.
Results
Genetic determinants involved in GAS 5448 fitness in vitro
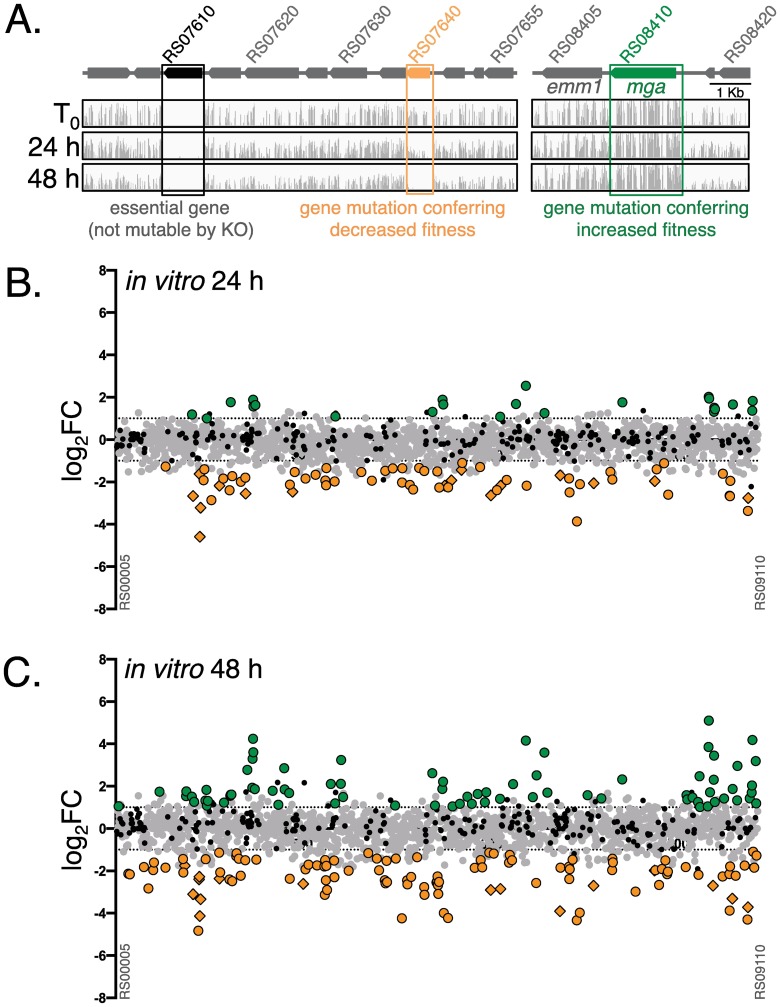
We recently constructed a near-saturation mariner transposon (Krmit) mutant library representing ca. 85,000 independent insertions in the M1T1 GAS strain 5448 genome that allowed us to combine Tn-seq with a Bayesian statistical model to predict the genes essential for in vitro growth in THY rich media [43]. Mutants lost from the input pool only after multiple passages in THY (termed critical) were proposed to represent genes important for fitness in vitro [43]. As these genes could alter the interpretation of the proposed in vivo Tn-seq screen, we re-analyzed our original Tn-seq datasets [43] obtained from two successive overnight passages (24 and 48 h) of the M1T1 5448 Krmit library in THY. Reads were summed for each GAS gene, and the ratios of mutant abundance comparing the output versus input mutant pools (fold change, FC) for each gene was calculated using the DEseq2 pipeline (see Material and methods). Results for 24 h and 48 h passages compared to the input (T0) were expressed using a log base 2 transformation of the FC (log2FC in S1 Table). Essential genes identified previously [43] (Fig 1, black) were removed from the current analyses. Genes with fewer insertions in the output pool (log2FC < -1, p < 0.05) were considered to confer decreased fitness (Fig 1, orange); those showing increased numbers of mutants in the output pools (log2FC > 1, p < 0.05) conferred a selective advantage or increased fitness (Fig 1, green), while genes with no significant change were considered neutral (Fig 1, grey).
Fig 1. Tn-seq analysis of M1T1 GAS 5448 fitness during in vitro growth in THY.
(A) Schematic of two representative chromosomal regions of M1T1 GAS 5448 (above) with the contribution (below) of GAS genes (arrows) during growth in THY determined by Tn-seq illustrated (IGV) with location (horizontal axis) and depth (vertical axis) of all Krmit transposon insertion sites identified in the GAS 5448 Krmit library initially (T0) and after 24-h and 48-h passages, respectively. Highlighted (boxed) are genes found to have a neutral effect (grey), decreased (orange), or increased (green) survival in the screen. Genes previously identified as essential [43] are shown in black and were not included in the fitness analyses. (B and C) Genome-scale summary of the ratios of mutant abundance (log2FC, Y axis) calculated using DEseq2 for each GAS 5448 gene (X axis), comparing the 24 h (B) and 48 h (C) output pools to the T0 input mutant pool. Gene mutations conferring decreased (log2FC < -1, p < 0.05) or increased fitness (log2FC > 1, p < 0.05) are indicated with orange and green circles, respectively. Neutral mutations (grey circles) and essential genes (black circles) are also indicated.
Analysis of M1T1 GAS 5448 in vitro fitness in THY identified 65 (24 h) and 112 (48 h) genes associated with decreased in vitro fitness, with 38 genes common to both time points (Fig 1B and 1C, orange circles; S1A Fig). Conversely, 23 (24 h) and 75 (48 h) genes were linked to increased in vitro fitness in THY, with 21 genes common to both time points (Fig 1B and 1C, green circles; S1B Fig). Clusters of Orthologous Genes (COG) enrichment analyses of genes identified during our in vitro Tn-seq analysis (n = 216) revealed that 65 (30%) encoded for proteins of unknown function, the highest COG for both increased and decreased fitness (S1C Fig). Additional genes linked to decreased in vitro fitness were primarily involved in the transport and metabolism of amino acids (~10%), inorganic ions (~8%), carbohydrates (~6%), and nucleotides (~6%) (S1C Fig). For increased in vitro fitness, gene products were also mostly related to the transport and metabolism of nucleotides (~10%), amino acids (~5%), and carbohydrates (~5%), as well as DNA replication (~8%) and signal transduction (~6%) (S1C Fig).
Validation of the in vitro Tn-seq screen
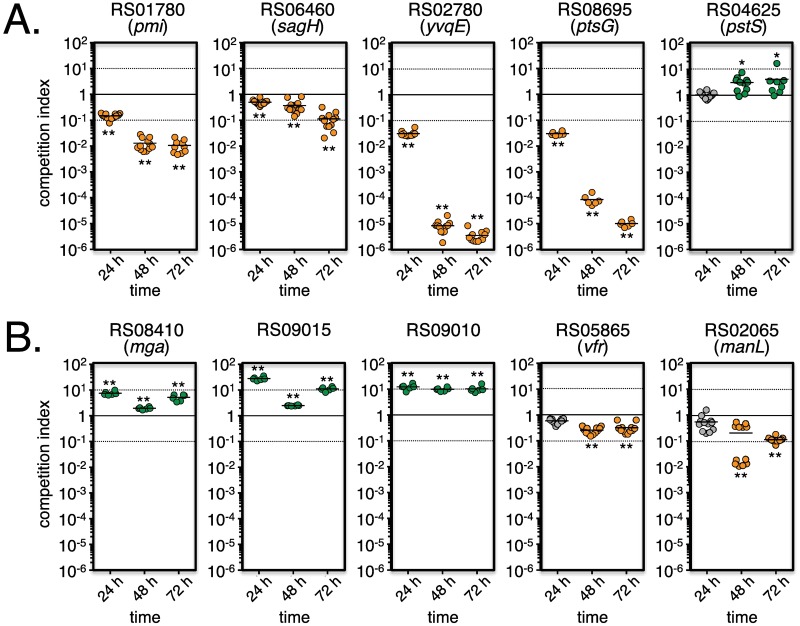
To provide experimental validation for our dataset, we selected 5 genes associated with decreased GAS fitness (RS01780/pmi, RS06455/sagH, RS02780/yvqE, RS08695/ptsG and RS04625/pstS) and 5 genes linked to increased GAS fitness (RS08410/mga, RS09015, RS09010, RS05865/vfr and RS02065/manL). Defined mutants were produced by insertional inactivation (see Material and methods) and growth assays using pure cultures in THY revealed growth parameters comparable to wild type (WT) 5448. To mimic the en masse Tn-seq screen, competition growth assays were performed in THY using similar CFU of WT and each mutant (see Material and methods). Population composition was monitored after plating serial dilutions after 3 successive overnight passages (i.e., 24, 48 and 72 h). Four of the five tested genes representing decreased in vitro fitness (pmi, sagH, yvqE, ptsG; Fig 2A) recapitulated the Tn-seq findings whereas pstS did not. For the genes showing increased fitness in Tn-seq, three (mga, RS09015 and RS09010) out of the 5 genes also demonstrated a positive competition index in direct competition with wild type 5448 in vitro (Fig 2B). Attempts to validate the remaining two genes (vfr and manL) were unsuccessful (Fig 2B). It is possible that the insertional mutagenesis strategy (pSinS, see Material and methods) employed for the pstS, vfr and manL genes could result in a partially functional truncated allele or have a polar effect on downstream genes. Overall, we were able to experimentally validate 70% of the tested genes, providing confidence in our fitness pipeline.
Fig 2. Validation of the in vitro Tn-seq screen.
Defined mutants (see Material and methods) in selected genes identified by Tn-seq as linked to decreased (A) or increased (B) fitness in vitro were grown in THY broth with GAS 5448 (ca. 1:1 ratio) in competitive growth assays. CFU counts were determined and strain ratios monitored after three successive 24-h passages (24 h, 48 h, 72 h) and expressed as competitive indexes. Results that validate the in vitro Tn-seq data are indicated as decreased (orange circles), increased (green circles), and neutral (grey circles) fitness in competitive growth assays. Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 (*) or <0.01 (**) was considered statistically significant.
An in vivo model of GAS skin and soft tissue infection suitable for Tn-seq
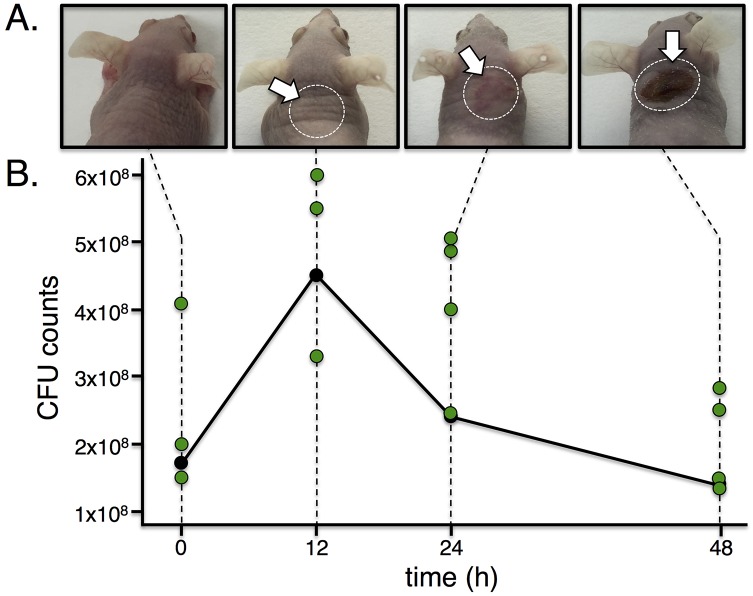
To interrogate GAS pathogenesis by Tn-seq, we selected a murine model of GAS subcutaneous infection using outbred immunocompetent hairless Crl:SKH1-hrBR mice [44–46]. In pilot experiments, mice were infected at the base of the neck with ca. 2×108 CFU of WT 5448 and progression of the disease followed over 48 h. Visual inspection of the infected tissues revealed the formation of a visible lesion 24 h post infection (HPI) that developed into a necrotic abscess by 48 HPI (Fig 3A). Histopathology of tissue samples from non-infected mice (S2A Fig) did not show any pathological alteration. At 12 HPI, inflammatory infiltrates (e.g., macrophages) were visible in the dermis, panniculus carnosus muscle (PCM) and hypodermis (S2B Fig, black arrows), with signs of suppurative inflammation associated with early necrosis (S2C Fig). However, the epidermis appeared asymptomatic and GAS cells were not visible (S2B Fig). At 24 HPI, epidermal thickening was observed, even though it was uninvolved in the infection process (S2D Fig). Ongoing inflammation of the hypodermis and necrotic PCM revealed prominent infiltration of macrophages (some intracellular GAS), lymphocytes and neutrophils (S2F Fig, blue, orange and green arrows, respectively); with bacterial growth in devitalized, edematous tissue (S2F Fig, black arrows). At 48 HPI, abscess formation was observed with extensive inflammation and tissue damage forming a pseudocapsule surrounding necrotic debris found in the hypodermis (S2E Fig). GAS chains were visible in the midst of necrotic tissues (S2G Fig, black arrows). Altogether, histopathology revealed that 5448 affected tissues in the deep reticular dermis, PCM and hypodermis, with inflammation visible at 12 and 24 HPI, leading to an abscess formation by 48 HPI in the mouse model of GAS skin and soft tissue infection as previously observed with other GAS strains [44, 45].
Fig 3. The murine soft tissue infection model and GAS M1T1 5448 are suitable for in vivo Tn-seq.
(A) Immunocompetent hairless Crl:SKH1-hrBR mice were infected subcutaneously with ca. 1–4 x 108 CFU of either wild-type GAS 5448 (shown) or the GAS 5448 Krmit library. Both produced marked lesions at 24 h that became necrotic and ulcerative by 48 h (white arrows, dotted circles). (B) Bacterial load within the excised lesion tissue was monitored by total CFU counts over time. The average counts for WT 5448 are displayed as black circles, and individual counts for the Krmit libraries in each lesion are indicated with green circles.
To verify that an adequate number of GAS cells could be retrieved from the M1T1 GAS 5448-infected tissues for Tn-seq, the tissue surrounding the site of infection was surgically extracted, homogenized by mechanical disruption and tissue lysates plated on blood agar plates for CFU counts. GAS cell numbers were recoverable in the range of 108 CFUs at the different time points tested (Fig 3B, black circles), a bacterial load that provides the complexity and numbers required for Tn-seq. We also observed that a proportion (15% on average) of the retrieved GAS cells from tissue lysates collected at 48 HPI produced mucoid colonies, a phenotype associated with GAS cells overproducing capsule. This is consistent with published reports showing that M1T1 GAS 5448 can acquire mutations in vivo functionally inactivating the covS gene (covS switch), leading to the derepression of the capsule-encoding hasABC operon [34, 36, 47–49].
Tn-seq analyses of M1T1 GAS 5448 during skin and soft tissue infection
Hairless Crl:SKH1-hrBR mice were infected with the M1T1 GAS 5448 Krmit mutant library [43], leading to lesions and tissue damage similar to those produced with WT 5448 (Fig 3, S2 Fig). Retrieval of M1T1 GAS 5448 Krmit mutants from excised lesions at 12, 24 and 48 HPI, yielded CFU counts in the 108 range (Fig 3B, green circles) comparable to those observed for wild type 5448 in the pilot experiments. Accumulation of mucoid colonies was also observed at the 24 and 48 HPI lesions, 5% and 25%, respectively. Analysis of selected mucoid colonies by AP-PCR to precisely identify transposon insertion sites revealed multiple independent Krmit mutations in the covS gene (S3 Fig) that were present in the input library prior to infection.
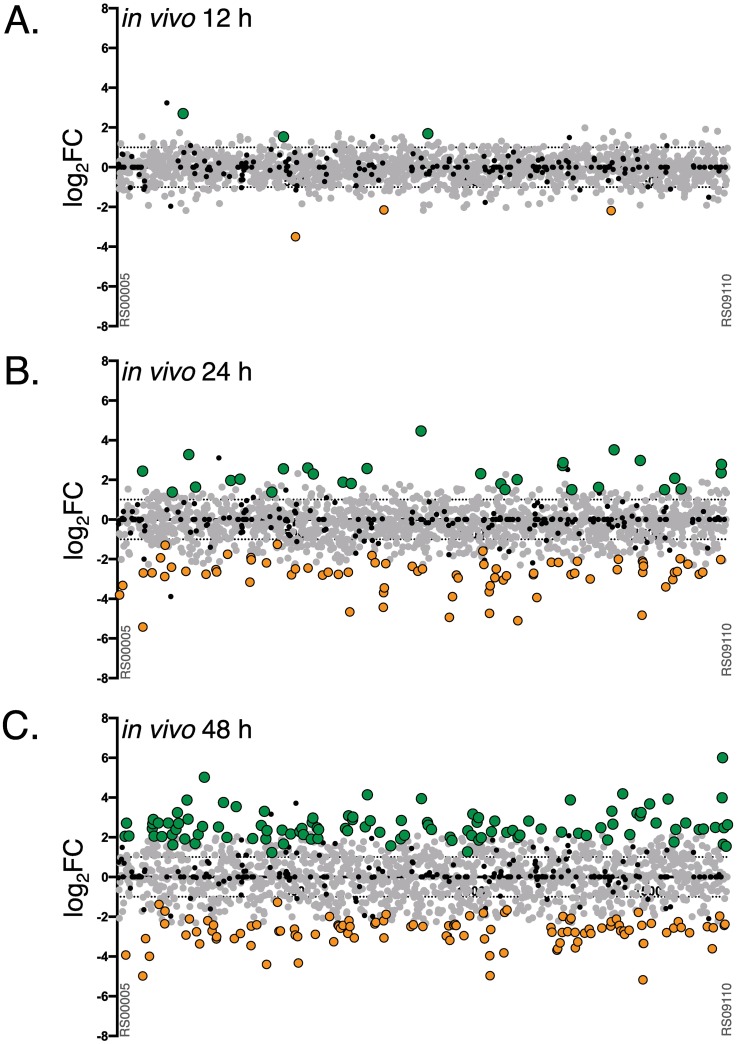
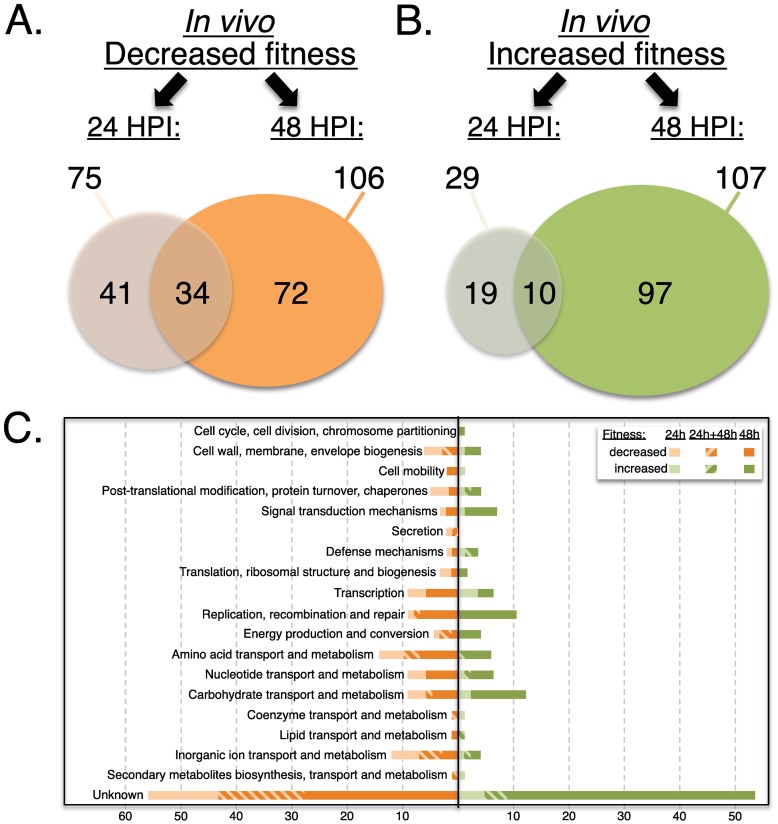
Initial attempts to produce Krmit insertion tags directly from homogenized infected tissues were unsuccessful due to the inability to extract non-sheared GAS gDNA required for Tn-seq. Consequently, homogenized tissue lysates were grown in THY broth for 4 h at 37°C to allow for a limited expansion of the mutant libraries (ca. 5–6 generations). GAS cells were collected after outgrowth, and Krmit insertion tags produced to allow for deep sequencing (see Material and methods). The ratios of mutant abundance were calculated for each GAS 5448 gene (log2FC) comparing the output at 12, 24 and 48 HPI to the input (T0) mutant pools to identify potential subcutaneous fitness genes (scf). At 12 HPI, the Krmit library composition remained relatively unchanged compared to the initial input library (Fig 4A, S2 Table), with only 6 genes identified (3 for increased, 3 for decreased fitness). Increasingly more substantial changes were observed at 24 then 48 HPI (Fig 4B and 4C, S2 Table). We found 75 (24 h) and 106 (48 h) genes associated with decreased in vivo fitness, with 34 common to both time points (Fig 5A). We also identified 29 (24 h) and 107 (48 h) genes linked to increased in vivo fitness, with 10 of those common to both time points (Fig 5B).
Fig 4. Tn-seq analyses of M1T1 GAS 5448 fitness during murine skin and soft tissue infection.
Genome-scale summary of the ratios of mutant abundance (log2FC, Y axis) calculated using DEseq2 for each gene (X axis) comparing the GAS 5448 Krmit library composition retrieved from in vivo lesions after 12, 24 and 48 HPI versus the T0 input pools (A, B and C, respectively). Gene mutations conferring decreased (log2FC < -1, p < 0.05) or increased fitness (log2FC > 1, p < 0.05) are indicated with orange and green circles, respectively. Neutral mutations (grey circles) and essential genes (black circles) are also indicated.
Fig 5. Comparison and classification of genes impacting M1T1 GAS 5448 fitness during in vivo lesion formation.
Venn diagrams comparing the number of genes showing either decreased (orange) or increased (green) fitness from the Tn-seq analyses of lesions 24 (A) and 48 (B) HPI. (C) Clusters of Orthologous Genes (COG) categories are indicated with their relative abundance for the same genes showing either decreased (orange) or increased (green) fitness at 24 h HPI (light shade), 48 h HPI (dark shade), and both (hatched).
COG enrichment analyses of the in vivo Tn-seq dataset revealed that the most represented category for genes linked to increased (55 of 147, 37%) or decreased (53 of 126, 42%) fitness at all time points was that of "unknown function" (Fig 5C). Of the remaining genes found at 24 and/or 48 HPI associated with decreased in vivo fitness, transport and metabolism of amino acids (~10%) and inorganic ions (~8%) were highly represented (Fig 5C). For genes linked to increased in vivo fitness, transport and metabolism of carbohydrates (~10%) and DNA replication (~8%) appeared to be limiting fitness in this environment (Fig 5C). When the Tn-seq dataset obtained for M1T1 GAS 5448 in vitro fitness in THY was compared to the in vivo results, 41 genes were found in both screens (S3 Table). This included 18 (decreased) and 9 (increased) genes with correlating phenotypes in both screens, while 14 genes presenting opposite phenotypes in vitro and in vivo.
Validation of the in vivo Tn-seq screen
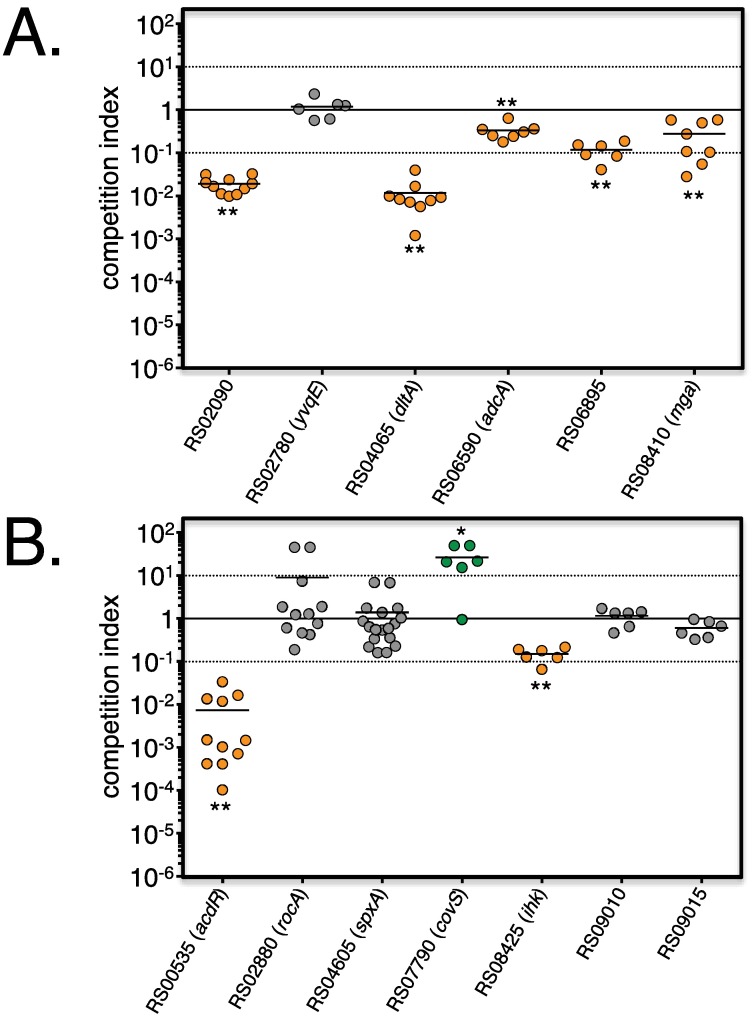
Insertional inactivation mutants were generated in 6 genes associated with decreased fitness in vivo (RS02090/lytR, RS02780 /yvqE, RS04065/dltA, RS06590/adcA), RS06895 and RS08410/mga) and tested individually against WT 5448 using an in vivo competitive infection assay in the murine skin and soft tissue model (see Material and methods). CD1 mice were subcutaneously infected with ca. 1:1 ratios of WT and each mutant, and population composition monitored after plating excised lesion lysates for CFU counts after 48 HPI.
Of the 6 tested mutants in genes linked to decreased fitness, 5 (lytR, dltA, adcA, RS06895 and mga) were outcompeted by 5448 in the lesions with a competitive index less than 1 (Fig 6A), strongly supporting the in vivo Tn-seq data. Only yvqE did not show any competitive defect in the assay, in contrast to the in vivo Tn-seq screen (Fig 6A). However, as mutants in yvqE showed an in vitro fitness defect (Fig 2A, S1 and S3 Tables), this likely represents a false positive in the in vivo Tn-seq screen due to the 4 h in vitro library expansion. In contrast, mga showed opposite phenotypes in vitro (increased fitness) and in vivo (decreased fitness) (S3 Table). Competitive assays confirmed that mga was critical during subcutaneous infection as seen in the in vivo Tn-seq (Fig 6A), but this is likely an underestimation given its potential recovery during in vitro expansion (Fig 2A).
Fig 6. Validation of the in vivo Tn-seq screen.
Insertional mutants in selected genes identified by Tn-seq as associated with decreased (A) or increased (B) fitness in vivo in lesions were mixed with GAS 5448 (ca. 1:1 ratio) and monitored during competitive infection during soft tissue infection of CD1 outbred mice. Total CFU were determined from homogenized lesions harvested at 48 HPI and ratios expressed as competitive indexes (mut/wt). Results correlating with the in vitro Tn-seq screen for decreased (orange circles) and increased (green circles) fitness are shown. Results not validating Tn-seq data are displayed as grey circles. Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 (*) or <0.01 (**) was considered statistically significant.
Selected mutants in genes associated with increased fitness in vivo (RS00535/adcR, RS02880/rocA, RS04605/spxA1, RS07790/covS, RS08425/ihk, RS09010 and RS09015) were also assayed by in vivo competition against WT 5448 (Fig 6B). Only 1 out of the 7 mutants tested (covS) recapitulated the in vivo Tn-seq findings, although rocA, which is associated with the covS genetic switch, trended upwards. Three of the tested genes (ihk, RS09010 and RS09015) were chosen as they were also found in the in vitro Tn-seq screen (S1 and S3 Tables) with 2 (RS09010 and RS09015) that were experimentally validated (Fig 2B); strongly suggesting that RS09010 and RS09015 correspond to false positive in the in vivo Tn-seq screen. Despite our pilot experiment finding pre-existing Krmit insertions in covS linked to mucoid colonies (S3 Fig), we cannot rule out that spontaneous mutations in covS and possibly rocA are occurring in vivo in mutants containing unlinked Krmit insertions. This suggests that mutations conferring a selective advantage for GAS in vivo or during the in vitro outgrowth step can bias the Tn-seq findings and emphasizes the importance of independent validation.
scfAB encode proteins of unknown function critical for M1T1 GAS 5448 pathophysiology
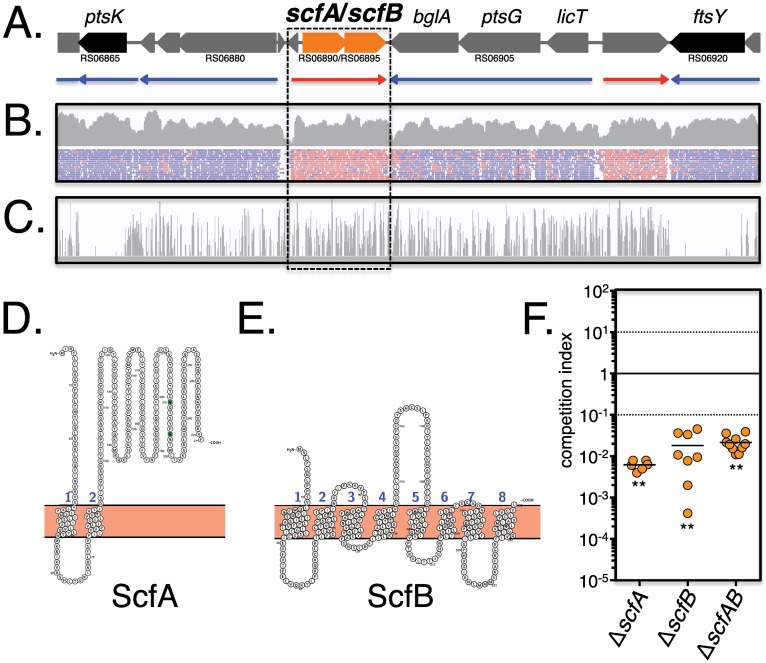
The in vivo Tn-seq screen identified RS06895 as critical during GAS 5448 fitness during subcutaneous infection at 24 h (S2 Table); and in vivo competitive infection assays conducted using a RS06895 Krmit mutant validated this finding (Fig 6A). Interestingly, the adjacent RS06890 gene was also shown to be necessary for in vivo fitness at 48 HPI (S2 Table). RNA-seq analyses performed on GAS 5448 grown in THY to exponential phase indicate that RS06890 and RS06895 likely form a bicistronic operon (Fig 7A and 7B). Importantly, both genes were dispensable during in vitro growth (Fig 7C, S1 Table), thus targets for knockout mutagenesis. RS06890 and RS06895 were renamed scfA and scfB, respectively, and subjected to further analysis.
Fig 7. The scfAB operon is dispensable in vitro, but is required for in vivo fitness.
(A) The M1T1 GAS 5448 genomic organization of scfAB (orange) with surrounding non-essential (grey) and essential (black) genes are shown with predicted transcriptional units from RNA-seq from (B) below with red (+ strand) and blue (- strand) arrows. (B) RNA-seq for the M1T1 GAS 5448 genomic region surrounding scfAB at late log phase in THY as displayed in IGV. RPKM (upper panel) and reads (lower panel) are shown with direction indicated as in (A). (C) Tn-seq analyses of the same genomic region after growth in THY and displayed in IGV. Height of the bars indicates the number of Krmit insertions mapping to that location. (D and E) Protein topology models of ScfA and ScfB (Protter), respectively, indicating membrane localization with 8 (ScfA) and 2 (ScfB) predicted transmembrane domains. (F) Non-polar mutants in the scfAB locus (ΔscfA, ΔscfB; and ΔscfAB) were mixed with parental GAS 5448 (ca. 1:1 ratio) and monitored during competitive soft tissue infection of CD1 outbred mice. Total CFU were determined from homogenized lesions harvested at 48 HPI and ratios expressed as competitive indexes (mut/wt). Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 (*) or <0.01 (**) was considered statistically significant.
The scfA and scfB genes are found within the GAS core genome [43] and both are annotated to encode for hypothetical membrane spanning proteins of unknown function. Using the topology prediction tool Protter [50] to analyze ScfA (300 AA; pI 8.20; Mw: ~33 kDa) identified eight putative transmembrane domains (Fig 7D, 1 to 8) and seven exposed domains (Fig 7D, a to g). Analyses of ScfB (271 AA; pI 7.77; Mw: ~31 kDa) indicated two possible transmembrane segments (Fig 7E, 1 and 2) and three projecting domains (Fig 7E, a to c). Homologs of GAS scfAB were found as linked genes of unknown function in the genomes of 21 Streptococcus, Enterococcus and Bacillus species, with the surrounding genetic context varying extensively (S4 Fig). In S. mutans, a transposon screen found that the homologs SMU.746 (scfA) and SMU.747 (scfB) were important for the acid stress response and biofilm formation [51]. Although the authors hypothesized that the genes encoded for an amino acid permease, that was not demonstrated [51]. In B. subtilis, expression of the uncharacterized ycgR (scfA) and ycgQ (scfB) is controlled by ECF σ factors involved in environmental stress response and cell envelope homeostasis [52]. In GAS, scfA was previously identified as required for M1T1 GAS 5448 survival in human blood in our TraSH screen [39], suggesting a role in disseminated GAS infection.
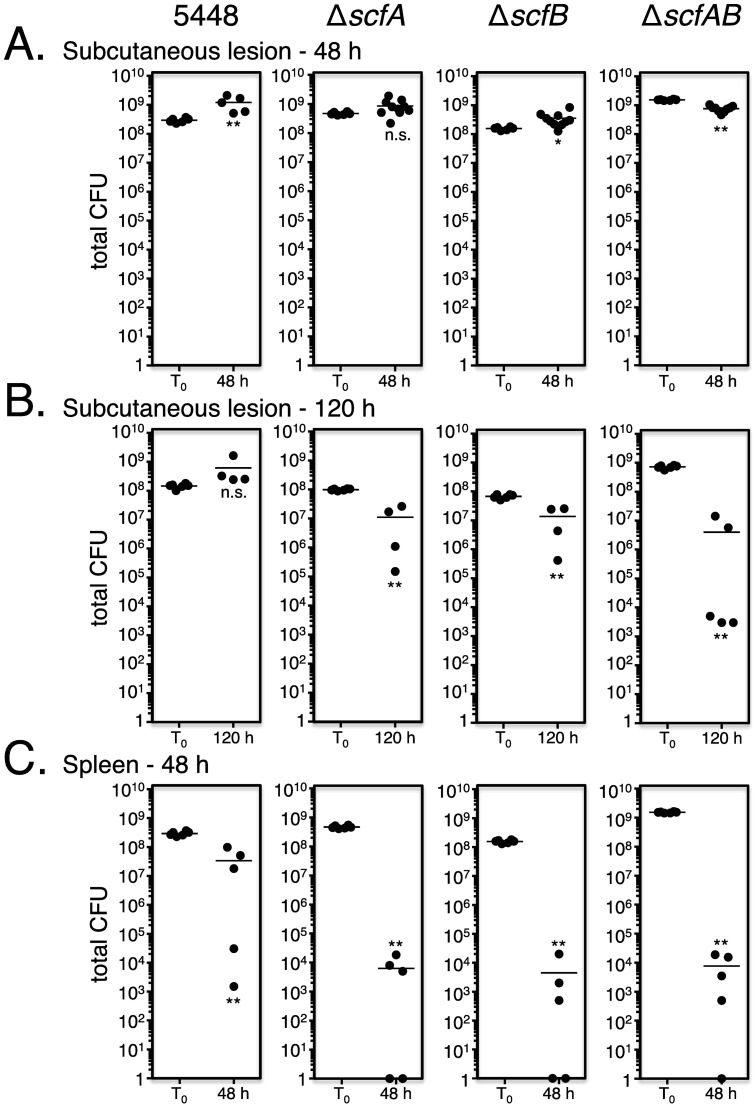
Non-polar mutations of scfA, scfB, and both scfAB in M1T1 GAS 5448 were constructed by allelic exchange and verified by qRT-PCR (S5 Fig). The resulting mutants (ΔscfA, ΔscfB and ΔscfAB) were tested against WT 5448 in in vivo competitive assays in the CD1 mouse subcutaneous infection model (see Material and methods). All three mutants were strongly outcompeted by WT 5448 in vivo after 48 HPI (Fig 7F). To assess whether the individual scfAB mutants were attenuated in vivo, each was used as pure culture to infect CD1 mice through the subcutaneous route and total bacterial burden (CFU) in lesions was compared to WT 5448 infection at 48 and 120 HPI. CFUs were similar to the infecting dose (108−109) after 48 HPI for all the tested GAS strains (Fig 8A). Although the cell counts at 120 HPI increased in the lesion infected with WT 5448, they decreased significantly for ΔscfA (1-log), ΔscfB (1-log), and ΔscfAB (2-log) (Fig 8B). Dissemination of GAS was also assessed by determining bacterial burden in the spleen at 48 HPI, and an approximate 5-log reduction in CFU was observed for ΔscfA, ΔscfB and ΔscfAB compared to WT 5448 (Fig 8C). Altogether, our results revealed that the conserved scfAB operon encodes putative membrane proteins that are critical for GAS 5448 during the initial subcutaneous infection as well as for dissemination into the bloodstream during invasive infection. Furthermore, our in vivo Tn-seq dataset allows us to assign functional information for M1T1 GAS genes previously annotated as "unknown function".
Fig 8. scfAB is required for soft tissue infection and dissemination.
Pure cultures of GAS 5448 and isogenic ΔscfA, ΔscfB and ΔscfAB mutants were individually inoculated (T0) subcutaneously into CD1 mice and total CFU counts were determined from lesions excised at 48 (A) and 120 (B) HPI to monitor infection, as well as from spleens harvested at 48 HPI (C) to assess dissemination compared to wild type GAS 5448. Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 (*) or <0.01 (**) was considered statistically significant.
Discussion
The Group A Streptococcus (GAS) remains a highly prevalent human pathogen capable of infecting multiple niches within its human host. Common to all GAS infections are the steps of initial colonization of an epithelium (throat or skin) and subsequent propagation to the subepithelial tissues [8]. In this report, we used the clinically relevant M1T1 GAS 5448 to perform the first in vivo Tn-seq screen to characterize, on a whole-genome level, those GAS genetic determinants (subcutaneous fitness genes, scf) functionally required within the subepithelial niche, as well as mutations that could potentially confer a selective advantage during the infection process. Although many were annotated with specific roles, we found that a substantial portion of the identified scf genes were of unknown function. Of these, we selected two undetermined genes, scfA and scfB, present in the GAS core genome and confirmed their role in vivo during soft tissue infection and GAS dissemination into the bloodstream.
Tn-seq to interrogate GAS pathophysiology en masse
High throughput Tn-seq screens (also TraDIS, HITS or IN-seq) [42, 53–55] take advantage of massive parallel DNA sequencing to monitor in a qualitative (nucleotide-level insertion location) and quantitative (mutant prevalence and gene fitness index) manner the composition of complex mutant pools [56–58]. These approaches permit biosystems-level analyses of bacterial genomes to accelerate functional genomics [20]. Depending on the experimental settings used (in vitro, ex vivo or in vivo), Tn-seq has helped define bacterial minimal genomes (i.e., essential genes) [42, 53–55], cell responses to environmental stresses [59, 60] or to antibiotics [61] and bacterial pathogenesis [62–65].
Tn-seq and other competition-based genetic screens (TraSH, STM) using complex mutant libraries do have certain limitations that prevent certain classes of genes from being identified. Classically, secreted and surface-associated molecules can be provided through trans-complementation by others in the library and mask the phenotype of that genes mutant in the library. For GAS virulence in soft tissue investigated here, important virulence factors such as toxins, superantigens, proteases, DNases, and capsule were not be expected in our screen likely due to this issue. In contrast, this approach may favor the discovery of important metabolic and regulatory pathways that are confined to the cytoplasm of GAS.
Our group has developed mariner-based genetic tools [38] to accelerate GAS functional genomic analyses through en masse TraSH [39] and Tn-seq [43] screens. Using two divergent GAS strains (M1T1 5448 and M49 NZ131), we recently conducted in vitro Tn-seq screens to characterize the GAS minimal core genome [43], defining potential targets for new therapeutics. GAS 5448 was isolated from a patient with necrotizing fasciitis (NF) and toxic shock syndrome [66] and is a representative of the globally disseminated M1T1 serotype that has been extensively used to study GAS pathogenesis in mice [39, 47, 67, 68]. Moreover, its genome sequence was recently completed [37]. For our essentiality studies, we generated a highly complex transposon mutant library in GAS 5448 containing over 85,000 unique Krmit insertions [43] that represented the baseline mutant pool used in this work. In our initial qualitative analyses of this 5448 Krmit library using a Bayesian statistical model [43], we predicted that 227 genes were essential (non mutable) and therefore indispensable in vitro. Such genes containing very few or no insertions are, by their nature, much more difficult to track during quantitative (i.e., fitness) Tn-seq analyses, and were not considered in our current study.
Here, the remaining dispensable genes were initially interrogated by Tn-seq for more subtle phenotypes conferring decreased or increased fitness in vitro. We monitored the composition of the GAS 5448 Krmit pool en masse to determine differential mutant abundance using DEseq2 [69] and EdgeR [70], two bioinformatics pipelines originally designed for RNAseq analyses. Other Tn-seq analysis tools are now available, such as MAGenTA [71, 72], which can track all individual insertions and generate fitness values for each insertion (nucleotide resolution). However, DEseq2 and EdgeR have recently gained acceptance for Tn-seq gene fitness analyses based on their availability and ease-of-use [58, 60, 73–75]. We compared the two pipelines by re-analyzing the datasets obtained for the GAS 5448 Krmit mutant libraries after two 24 h passages in THY rich medium [43] to compare mutant pool composition over time and selected DEseq2 for our analyses as it produced more conservative datasets.
A total of 139 genes associated with decreased GAS 5448 fitness in vitro were identified at the 24 h or 48 h time points using DEseq2, with 38 genes common to both (S1 Fig, S1 Table). Using the Bayesian statistical analysis, we had categorized genes as “critical” (n = 71) that were found essential only after 2 in vitro passages in THY and we proposed they might represent genes important for in vitro fitness. However, out of the 139 genes found linked to decreased fitness in vitro by DEseq2 (S1 Fig, S1 Table), only 22 were also found critical by Bayesian analysis [43] (Fig 1B and 1C, orange diamonds). This discrepancy is not surprising as the mode of data analyses between these two tools is quite different. Bayesian analysis takes into account the library coverage (Krmit positions) regardless of read depth (counts) and identifies significant gaps in the genome with no transposon insertions; therefore, a gene is essential if a portion of the gene (e.g., essential domain) lacks insertions. DEseq2 will miss this gene as it monitors the read counts for the entire gene length, and data integration would likely mask the results for the essential domain.
Data validation by in vitro competition growth assays confirmed 4 out of the 5 genes (80%) associated with decreased fitness that we tested (Fig 2A). Of the 98 mutations conferring increased in vitro fitness by DEseq2, validation experiments confirmed only 3 out of the 5 tested mutations (60%) (Fig 2B), revealing a slightly higher proportion of false-positive results with genes in this category. In general, false-positive results are not uncommon with Tn-seq screens [58, 64, 76] and may reflect a variety of reasons. The initial Tn-seq screen is a complex competitive environment involving thousands of mutants whereas validation studies tend to compare only a single mutant to the WT pathogen. Also, the defined mutant generated for validation are likely to differ genetically from the original transposon mutations found in the complex library. Genetic manipulation of GAS 5448 can be difficult and we used mutants produced by stable plasmid insertional inactivation instead of allelic exchange for our validation tests; a strategy that could create (i) a polar effect on adjacent genes when in operon or (ii) a truncated allele of the target gene that could still be functional. Overall, we were still able to validate 70% of the genes tested (Fig 2).
M1T1 GAS 5448 genetic determinants impacting fitness during subcutaneous infection
For our in vivo Tn-seq screen of GAS pathophysiology, we selected a subcutaneous infection model in immunocompetent Crl:SKH1-hrBR mice that displayed symptoms with the Krmit library similar to those observed with wild type GAS 5448 (S2 Fig) and supported high Krmit mutant counts (> 108 CFU) (Fig 3) allowing for pool complexity during infection. Our in vivo screen identified 147 genes (with 75 and 106 genes at 24 and 48 HPI, respectively) as potentially critical for the M1T1 GAS 5448 fitness during subepithelial infection. As observed for our in vitro fitness analysis, the most prevalent COG category (55 genes, 37%) was annotated as encoding proteins of unknown function.
Although we could validate 5 out the 6 genes (80%) identified as linked to decreased in vivo fitness, a defined mutant in RS02780/yvqE exhibited no defect when competed directly with wild type GAS 5448 in mice (Fig 6A). Since a yvqE mutant also displayed decreased competitive fitness in vitro (Fig 2A), it likely leads to a false positive finding in vivo. This result highlights that the 4 h in vitro library expansion step in our in vivo Tn-seq screen setting likely introduces some bias into our in vivo fitness datasets. Mutant pool expansion following in vivo Tn-seq screens, by mutant pool plating on agar [65, 76, 77] or culture in broth [64, 78], is often required when library DNA extraction directly from tissue represents a limiting step to the Tn-seq procedure. This extra step is particularly important when using the MmeI-based Tn-seq approach [72], as it requires high quality DNA (no shearing). In this case, our data revealed that it is important to take into account the competition phenotypes from both in vivo and in vitro validation experiments to distinguish phenotypes truly displayed in vivo.
Several established genes were found to be critical in the subepithelial environment, including a subunit of the D-alanine-activating enzyme DltA (RS04065/dltA) involved in the biosynthesis of D-alanylated lipoteichoic acid in GAS. A GAS 5448 dltA mutant was previously found to have diminished ability to adhere and invade human pharyngeal epithelial cells [79] and was attenuated for survival in whole human blood and neutrophils [80]. RS02090/cpsA, encoding a LytR-like transcriptional regulator known to regulate capsule expression, cell wall homeostasis and virulence in Streptococcus agalactiae (Group B Streptococcus, GBS) [81, 82], was also identified as required for GAS fitness in vivo. Our findings differ from published reports showing that a cpsA (lytR) mutant was more virulent than WT in subcutaneous infection [83]. However, these studies used an invasive M1 covS strain GAS 1529, suggesting that loss of LytR (CpsA) could increase GAS fitness in a covS- background.
RS06590/adcA, encoding a Zn-specific transporter [84], was also linked to in vivo fitness. Homologs of adcA have been found in other pathogenic streptococci, where it is typically found associated with an operon adcCBA. In GBS, AdcA was found to be required for Zn acquisition, proper cell morphology, and fitness in human amniotic fluid and cerebrospinal fluid [85]. In S. gordonii, adcA is important for manganese homeostasis, biofilm formation and natural competence [86, 87], while in S. pneumoniae adcA is critical for natural competence [88], fitness in human serum [89]; and virulence [89, 90]. Our study and others finding adcA important in vivo supports the hypothesis that zinc homeostasis is critical during multiple steps of GAS infection [39, 91].
The in vivo Tn-seq screen also identified mga as critical during GAS 5448 subepithelial infection, which is consistent with previous observations showing that a mga mutation led to virulence attenuation in different in vivo and ex vivo GAS infection models [11, 39, 92–95]. Interestingly, in vitro Tn-seq revealed two opposite phenotypes whereby inactivation of mga resulted in increased GAS 5448 fitness in THY (Fig 2B). This suggests that the mga-associated in vivo phenotype would be even stronger, except that the 4 h library expansion restored some of the in vivo reduction. Mga is an important PTS-regulatory domain (PRD)-containing virulence regulator (PCVR) in GAS that controls the expression of the virulence factor M protein [11]. However, the emm gene was not identified in the in vivo Tn-seq screen, suggesting that the mga phenotype observed in the subepithelial environment might not be related to emm expression. We are currently pursuing the exact role that Mga plays in M1T1 GAS 5448 fitness in this in vivo niche.
Our Tn-seq screen also identified 126 genes linked to increased in vivo fitness at 24 or 48 HPI, with 10 genes common to both. Interestingly, validation by in vivo competitive infection found that only 1 out of the 7 defined mutants tested (covS) recapitulated the in vivo Tn-seq findings (Fig 6B). Thus, in vivo Tn-seq demonstrated that covS mutants, and possibly rocA mutants through activation of covS, confer a fitness advantage in M1T1 GAS leading to selection in the subepithelial tissues. This reveals another potential technical bias that might be occurring during the in vivo Tn-seq whereby a covS mutation occurs spontaneously in the Krmit mutant pool independent from pre-existing covS Krmit insertions. This would result in genes found by Tn-seq associated with increased in vivo fitness that fail to validate. Moving forward, we will modify our in vivo Tn-seq screen and data analyses to take into account spontaneous covS mutations on M1T1 GAS 5448 fitness based on Tn-seq from near saturation Krmit libraries in the isogenic 5448AP (covS-) strain.
Of course, lack of validation may simply reflect differences between the defined and library mutants as for the in vitro validation or represent false positives (yvqE, RS09010, RS09015, and RS08425/ihk) based on the in vitro Tn-seq datasets. Although RS00535/adcR was initially identified as linked to increased in vivo fitness, our validation revealed that an adcR mutant was attenuated in vivo, which is consistent with observations by Sanson et al. [96]. Interestingly, AdcR can activate hasABC [96], the same capsule operon that is derepressed during the covS switch.
Role of ScfAB in subepithelial invasion and dissemination
Tn-seq screens represent a powerful approach to accelerate bacterial genomics, particularly by providing functional annotations for genes encoding proteins that are either poorly characterized or of unknown function [20, 58]. The most prevalent COG category of subcutaneous fitness (scf) genes found in our in vivo Tn-seq screen (108 genes), both for increased and decreased fitness, were those of "unknown function". We decided to focus on two adjacent genes, RS06890 (scfA) and RS06895 (scfB) that were critical for in vivo fitness as representative of this group. Analyses of defined mutants in this locus revealed that scfAB were in an operon that was important at the subepithelial site of infection as well as for GAS 5448 dissemination into the bloodstream (Figs 6–8). Both genes encode for putative transmembrane proteins and BLAST analyses did not reveal any homologs where a function was available. The scfAB operon is part of the GAS core genome and both genes appear conserved in GAS. Studies are underway to investigate the role of the scfAB operon during infection at different infection sites and in different GAS strains to see if their function is ubiquitous or strain-specific. Homologs of scfAB are found in other pathogenic streptococci and closely-related Gram-positive pathogens (S4 Fig); however, the locus has only been studied in one report. In S. mutans, scfA and scfB homologs were found in a transposon mutagenesis screen to impact acid stress response and biofilm formation [51, 52]. Although the authors proposed that they encoded amino-acid transporters, no further analysis was performed. Thus, scfA and scfB appear to be ubiquitous in other pathogenic streptococci and Gram-positive pathogens and further analyses are warranted to better understand their cellular function. Importantly, our in vivo Tn-seq was key in providing an in vivo phenotype for genes encoding "unknown function" proteins and has the potential to enhance our annotation of the GAS genome and the identification of novel therapeutic targets.
Concluding remarks
This work represents the first in vivo Tn-seq screen to functionally interrogate the GAS genome in the context of a disease-relevant environment. The identification of 147 genes potentially critical in vivo reveals that adaptation to in vivo niches is a complex process involving multiple aspects of the GAS cell physiology (metabolism, gene regulation, cell envelop). We are now investigating the genetic requirements of the M1T1 GAS 5448, and its counterpart 5448AP, using multiple infection models in vivo, ex vivo and in vitro. In addition, we have a rich dataset of genes with predicted function as well as those previously annotated as "of unknown function" that can be further mined to discover new aspects of GAS pathophysiology. We have also established complex Krmit libraries in additional GAS strains to help study the effect of genome variation on GAS pathogenesis.
Materials and methods
Bacterial strains and media
Bacterial strains used in this study are listed in S4 Table. GAS 5448 is a representative of the globally disseminated invasive serotype M1T1 clone isolated from a patient with necrotizing fasciitis and toxic shock [66]. The Krmit mutant library in GAS 5448 was produced by in vivo mariner mutagenesis [43] and contains over 85,000 independent mariner transposants (i.e., one Krmit TIS for every 22 nucleotides). GAS strains were routinely cultured in Todd-Hewitt medium (Alpha Biosciences) supplemented with 0.2% yeast extract (THY) as described [97]. Escherichia coli strain DH5α was routinely used as host for plasmid construction and preparation and cultured in Luria-Bertani (LB) medium (EMD Chemicals). Antibiotics (Gold Biotechnology) were used at the following concentrations: Spectinomycin (Sp) at 100 μg/ml for both E. coli and GAS, and Kanamycin (Km) at 50 μg/ml for E. coli and 300 μg/ml for GAS.
Molecular genetics
Oligonucleotides used in this study were synthesized by Integrated DNA Technologies, Inc. and are listed in S5 Table. Plasmids used in this study are presented in S4 Table. Plasmids were isolated using the Wizard Plus SV Minipreps kit (Promega) or the QIAGEN Plasmid Purification Midi Kit (QIAGEN). Restriction enzymes, Antarctic Phosphatase and T4 DNA ligase (New England Biolabs) were used according to the manufacturer’s instructions. PCR was performed using either Taq DNA polymerase (New England Biolabs) or High-Fidelity AccuPrime Pfx DNA polymerase (Life Technologies) with 1 μg of DNA template and 10 pmol of the appropriate primers (S5 Table). Arbitrary-primed-PCR (AP-PCR) experiments were conducted as previously described [38]. When necessary, PCR products were purified using Wizard SV Gel and PCR Clean-Up System kit (Promega). Transformations were performed with the Gene Pulser Xcell System apparatus (Bio-Rad) as recommended by the manufacturer, using electrocompetent cells of E. coli or GAS prepared as previously described [38, 98]. Genomic DNA (gDNA) from GAS was purified using the MasterPure Complete DNA Purification kit (Epicentre Biotechnologies). Sanger DNA sequencing was performed by Genewiz, Inc.
Gene insertional inactivation
When available, Krmit transposon mutants in GAS 5448 were selected (S4 Table). Mutants 5448KM07790, 5448KM08410, 5448KM09015 and 5448KM09010 present a Krmit integrated in the gene RS07790/covS [TA at position 211 within the ORF (TA211)], RS08410/mga (TA4), RS09015 (TA412) and RS09010 (TA471), respectively. Additional mutant strains in GAS 5448 (S4 Table) were produced using the pSinS/pHlpK system [43]: succinctly, an internal fragment of the gene of interest was amplified by PCR using the appropriate primers (S5 Table) and the resulting DNA fragment cloned into the BamHI site of the pSinS suicide vector. The resulting recombinant plasmid (S4 Table) was then transformed into GAS 5448 cells containing the pHlpK helper plasmid and mutagenesis carried out as previously described [43]. Plasmid integration within the targeted gene was verified by PCR using the appropriate primer(s) (S5 Table). For simplification in the Result section, insertional mutant strains were referred to by the name of the mutated gene (e.g., mutants 5448KM07790 and 5448ii01780 were designated RS07790 and RS01780, respectively).
In vitro competition growth assay
Competition growth assays in THY were carried after inoculation of 10 ml of broth with comparable amounts of exponentially-grown cells of GAS 5448 and the tested mutant strain (close to a 1:1 ratio). At different time intervals (24, 48, 72h), samples were collected, serial diluted (10-fold increments), cell suspension plated on either THY (whole population) or THY containing the appropriate antibiotic (mutant population) and cell counts determined. Competition index (CI) was calculated using the following formula: CI = (RM/RW)/(RM0/RW0), with RM0 and RW0 correspond to the ratio of the mutant and the ratio of the wild-type, respectively, in the initial inoculum (T0); and RM and RW correspond to the ratio of the mutant and the ratio of the wild-type, respectively, at the end of the competition growth assay. Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 was considered statistically significant.
Ethics Statement
Crl:SKH1-hrBR mice (Charles River Laboratories) were infected with GAS in AAALAC-accredited ABSL-2 facilities and following protocols approved by the University of Maryland IACUC (R-16-05) for humane treatment of animal subjects in accordance with guidelines set up by the Office of Laboratory Animal Welfare at NIH, Public Health Service, and the Guide for the Care and Use of Laboratory Animals; with every effort to limit distress and pain to animals taken.
Model of GAS 5448 subcutaneous infection suitable for Tn-seq
Overnight cultures of GAS 5448 or the GAS 5448 Krmit mutant library were diluted 1:20 to a final volume of 80 ml of THY broth and grown to late-logarithmic phase, GAS cell chains were disrupted by vortexing the culture for 10 min, and cells centrifuged at 6000 × g for 10 min and resuspended in saline to produce the infection inoculum (ca. 109 CFU/ml). Initial CFU counts of the infectious dose were confirmed by serial dilutions plated onto THY agar (THYA) plates. Five-week-old, outbred, immunocompetent, hairless female Crl:SKH1-hrBR mice (Charles River Laboratories) received subcutaneous injections of ca. 108 CFU (0.1 ml) of 5448 (or 5448 Krmit mutant library) at the base of the neck. At different times (t = 12, 24, 48 hours post infection), mice were euthanized by CO2 asphyxiation and skin lesions were excised.
For microscopy analyses, lesions were fixed with fresh 4% paraformaldehyde for 24 hours at 4°C, stored in 70% ethanol at room temperature, then paraffin embedded for sectioning and stained using a hematoxylin and eosin stain (H&E stain) for histology observations.
To quantify bacterial load, the lesion was placed into a sterile 2-ml screw-cap microtube containing 1.4 mm ceramic spheres (Lysing Matrix D, MP Biomedicals) and 1 ml of sterile saline; and skin tissues were homogenized using three successive 45-second bursts with a FastPrep FP120 BeadBeater (BioSpec Products) as recommended by the manufacturer. Tissue homogenates (3 ml final) were serially diluted (10-fold increments) in saline, plated onto THY agar, and CFU counts determined after overnight incubation at 37°C.
In vivo Tn-seq analyses
Tissue homogenates (3 ml final) were produced as described above using the skin lesions from mice infected with the GAS 5448Krmit mutant library and were transferred into 150 ml THY+Km and cultured at 37°C for 4 h. GAS cells were collected by centrifugation at 6000 × g for 10 min and GAS gDNA extracted as above. Tn-seq was carried using the MmeI protocol [42] with modifications described by Le Breton et al. [43]. In this study, twelve different MmeI adapters (S5 Table) were used to allow sample multiplexing on Illumina lanes. Libraries of Krmit insertion tags were sequenced (50-nt single end reads) on an Illumina HiSeq 1500 platform in the Institute for Bioscience and Biotechnology Research (IBBR) Sequencing Facility located at the University of Maryland, College Park. Tn-seq read datasets were analyzed (quality, filtering, trimming, alignment, visualization) as previously described [43] using the GAS 5448 genome [37] for read alignments. A more detailed description of the bioinformatics analyses is provided in S1 Text. The ratios of mutant abundance comparing the output to input mutant pools were calculated as a fold change (FC) for each GAS gene using the DEseq2 and EdgeR pipelines [69, 70].
Inactivation of scfA, scfB, and scfAB in GAS 5448
Nonpolar mutations in the scfAB locus were obtained by replacing the corresponding open reading frames (ORF) with the promoterless aphA3 gene using allelic exchange as previously described [38]. Primers, plasmid constructs and GAS strains are listed in S4 and S5 Tables. For the scfA mutation, DNA fragments flanking the scfA gene were amplified using the primer pairs oAX0478.1 and oAX0478.2 (before the 5′ end of scfA) (PCR 5’scfA) and oAX0478.3 and oAX0478.4 (within the 3′ end of scfA) (PCR 3’scfA) and subsequently ligated by PCR-splicing by overlap extension (SOE) to the aphA3 cassette. The resulting PCR product then was digested by BamHI and cloned into the BamHI-digested pCRS [38], creating the plasmid pAX0478K (S4 Table). Allelic replacement of the scfA by the aphA3 cassette was conducted as previously described [38], creating the GAS strain 5448ΔscfA. The same approach was carried out to mutate scfB using scfB flanking DNA fragments amplified with primers oAX0477.1 and oAX0477.2 (PCR 5’scfB) and oAX0478.3 and oAX0478.4 (PCR 3’scfB) to produce the pAX0477K plasmid and generate GAS strain 5448ΔscfB (S4 and S5 Tables). Expression of scfA and scfB in strains 5448ΔscfA and 5448ΔscfB was verified by qRT-PCR as previously described [40] using the appropriate primers (S2 Table). To generate the GAS strain 5448ΔscfAB, the aphA3 cassette was linked to PCR products PCR 5’scfA and PCR 3’scfB to create the plasmid pAX0478-77K (S4 and S5 Tables).
In vivo competition growth assay
Subcutaneous infections were carried out using 5- to 6-week-old female CD-1 mice (Charles River Laboratories). Cell suspensions of exponentially growing GAS cells were obtained by mixing equal amounts of GAS 5448 and the tested mutant strain (close to a 1:1 ratio) in saline (ca. 109 CFU/ml). Mice were anesthetized with ketamine, fur was removed from a ~3-cm2 area of the haunch with Nair (Carter Products), and 100 μl of a cell suspension in saline injected under the back skin. Mice were monitored twice daily for 2 days, euthanized by CO2 asphyxiation. Skin lesions lysates, CFU counts (in total population and mutant population) and competition indexes were obtained as described above. Unpaired student’s t-test was used to evaluate the significance of differences between groups; a p value of <0.05 was considered statistically significant.
RNA-Seq and data analysis
GAS 5448 cells were grown to exponential phase in 20 ml of THY broth and treated with the RNAlater reagent (Qiagen). RNA extraction, RNA-seq library preparation and massive parallel DNA sequencing were carried out as previously described [40]. Read analyses were conducted as previously described [40] using the GAS 5448 genome for alignment [37]. Visualizations of the sequencing mapping were performed using the Integrative Genomics Viewer (IGV) [99].
Infections with scfA, scfB and scfAB mutants
Cells suspensions in saline of pure cultures of GAS 5448, scfA, scfB and scfAB mutants were produced and injected subcutaneously in CD1 mice (108−109 CFUs) as described above. Mice were monitored twice daily for up to 5 days, euthanized by CO2 asphyxiation. Skin lesions and/or spleen were surgically harvested and cell lysates produced with the FastPrep FP120 BeadBeater and CFU counts determined as described above.
Accession number for public deposition of RNA-seq and Tn-seq data
Illumina sequencing reads from the RNA-seq and Tn-seq analyses were deposited in the NCBI Sequence Read Archive (SRA) under the accession number (PRJNA391181).
Supporting information
(DOCX)
Venn diagrams comparing the number of genes showing either decreased (orange) or increased (green) fitness from the Tn-seq analyses of M1T1 GAS 5448 grown in THY for 24 h (A) and 48 h (B). (C) Clusters of Orthologous Genes (COG) categories are indicated with their relative abundance for the same genes showing either decreased (orange) or increased (green) fitness in vitro at 24 (light shade), 48 h (dark shade), and both (hatched).
(EPS)
H&E stains of skin and associated soft tissues samples collected at 12, 24 and 48 HPI. Specific regions are indicated as follows: epidermis (EP), sebaceous gland (SG), hair shaft (HS), papillary dermis (PD), reticular dermis (RD), and panniculus carnosus muscle (PCM). (A) Non-infected tissues displaying normal epidermal thickness (ca. 12 μm) and features. (B) 12 HPI lesion with inflammatory infiltration (black arrows) in the dermis (both PD and RD) and PCM with early necrosis indicated. (C) Magnification of (B). (D) 24 HPI lesion with PCM inflammation and infiltration (black arrow); Hypo: hypodermis. (F) Magnification of (D) showing monocytes, macrophages, lymphocytes and neutrophils (blue, orange and green arrows, respectively) and visible GAS chains (black arrows). (E) 48 h HPI lesion showing extensive inflammation, tissue damage and abscess formation with pseudo-capsule (CAP) surrounding necrotic debris (Nec). (G) Magnification of (E) shows visible GAS chains (black arrows). Scale bars (in red) represent 100 μm on panels A, B, D and E; and 10 μm on panels C, F and G.
(EPS)
GAS 5448 Krmit libraries injected subcutaneously into immunocompetent Crl:SKH1-hrBR hairless mice were isolated from homogenized lesions and plated onto 5% TSA blood agar plates to look for mucoid (capsule-overproducing) colonies. (A) After 12 HPI, most colonies display a non-mucoid phenotype. (B) After 48 HPI, mucoid colonies (grey arrows) are distinguishable from the remaining colonies (white arrows). (C) AP-PCR analyses on gDNA isolated from selected mucoid mutants show Krmit insertions at different locations within the covS gene (green arrow heads) as indicated.
(EPS)
Genome alignment was generated using the Orthology Browser from the Microbes Online resource (www.microbesonline.org/) using the MGAS5005 genome as reference and the RS06890 homolog (M5005_Spy0477) as the query for the genome alignment. The graphic display presents genome alignments based on the 24 highest homologs to RS06890/M5005_Spy0477. Colored arrows depict homologous genes found within the display window of the different aligned genomes, while grey arrows represent genes that do not have a homolog in the displayed genome window.
(EPS)
Real-time quantitative PCR (qRT-PCR) analyses of relative transcript levels for scfA and scfB genes in the ΔscfA and ΔscfB mutants compared to wild-type 5448 grown in THY to late log phase. Error bars represent the standard errors from at least three biological replicates. Dashed lines indicate 2-fold significance. Significance was determined using comparisons of transcript levels relative to those of the gyrA gene.
(EPS)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
Acknowledgments
We would like to thank Rezia Era Dagting Braza and Shelby Foor for technical assistance and critical comments of the manuscript; Karen Tchuindjo for technical assistance; and Kristie Connolly and Slawomir Lukomski for helpful suggestions on the GAS subcutaneous infection model in Crl:SKH1-hrBR mice.
Data Availability
All data except for the raw sequencing reads are provided with the submitted materials. Illumina sequencing reads from the RNA-seq and Tn-seq analyses were deposited in the NCBI Sequence Read Archive (SRA) under the accession number (PRJNA391181).
Funding Statement
KSM, YLB, GSS, and EI were supported by grants (AI047928, AI134079) from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health. NMES and ATB were supported by an award (AI094773) from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health. Initial support was provided by a University of Maryland, Baltimore (UMB) and University of Maryland at College Park UMCP Seed Grant (2014/2015) awarded to YLB, KSM, and MES. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cunningham MW. Pathogenesis of Group A Streptococcal infections. Clin Microbiol Rev. 2000;13(3):470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reglinski M, Sriskandan S. The contribution of group A streptococcal virulence determinants to the pathogenesis of sepsis. Virulence. 2013;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan LK, Eccersley LR, Sriskandan S. Current views of haemolytic streptococcal pathogenesis. Curr Opin Infect Dis. 2014;27(2):155–64. doi: 10.1097/QCO.0000000000000047 [DOI] [PubMed] [Google Scholar]
- 4.Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 2007;15(7):318–25. doi: 10.1016/j.tim.2007.05.001 . [DOI] [PubMed] [Google Scholar]
- 5.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–94. doi: 10.1016/S1473-3099(05)70267-X . [DOI] [PubMed] [Google Scholar]
- 6.Cunningham MW. Pathogenesis of Group A Streptococcal infections and their sequelae. Adv Exp Med Biol. 2008;609:29–42. doi: 10.1007/978-0-387-73960-1_3 [DOI] [PubMed] [Google Scholar]
- 7.Weiss KA, Laverdiere M. Group A streptococcus invasive infections: a review. Can J Surg. 1997;40(1):18–25. [PMC free article] [PubMed] [Google Scholar]
- 8.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nature Reviews Microbiology. 2011;9(10):724–36. doi: 10.1038/nrmicro2648 [DOI] [PubMed] [Google Scholar]
- 9.Steer AC, Lamagni T, Curtis N, Carapetis JR. Invasive group A streptococcal disease: epidemiology, pathogenesis and management. Drugs. 2012;72(9):1213–27. doi: 10.2165/11634180-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalia A, Bessen DE. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J Bacteriol. 2004;186(1):110–21. doi: 10.1128/JB.186.1.110-121.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66(5):1056–65. doi: 10.1111/j.1365-2958.2007.06006.x . [DOI] [PubMed] [Google Scholar]
- 12.Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11(5):224–32. . [DOI] [PubMed] [Google Scholar]
- 13.McIver KS, Heath AS, Scott JR. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect Immun. 1995;63(11):4540–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voyich JM, Musser JM, DeLeo FR. Streptococcus pyogenes and human neutrophils: a paradigm for evasion of innate host defense by bacterial pathogens. Microb Infect. 2004;6:1117–23. [DOI] [PubMed] [Google Scholar]
- 15.Bessen DE, McShan WM, Nguyen SV, Shetty A, Agrawal S, Tettelin H. Molecular epidemiology and genomics of group A Streptococcus. Infect, Genet Evol. 2015;33:393–418. doi: 10.1016/j.meegid.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessen DE. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infection, Genetics, and Evolution. 2009;9(4):581–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessen DE, Hollingshead SK. Molecular epidemiology, ecology, and evolution of group A streptococcus In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. Washington, D.C.: ASM Press; 2000. p. 117–24. [Google Scholar]
- 18.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98(8):4658–63. doi: 10.1073/pnas.071559398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama F, Watanabe T, Nakagawa I. Streptococcus pyogenes Genomics In: Ferretti JJ, Stevens DL, Fischetti VA, editors Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2016. [Google Scholar]
- 20.Baric RS, Crosson S, Damania B, Miller SI, Rubin EJ. Next-generation high-throughput functional annotation of microbial genomes. mBio. 2016;7(5):e01245–16 doi: 10.1128/mBio.01245-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husmann LK, Dillehay DL, Jennings VM, Scott JR. Streptococcus pyogenes infection in mice. Microb Pathog. 1996;20(4):213–24. doi: 10.1006/mpat.1996.0020 [DOI] [PubMed] [Google Scholar]
- 22.Medina E, Lengeling A. Genetic regulation of host responses to group A streptococcus in mice. Briefings in Functional Genomics and Proteomics. 2005;4(3):248–57. Epub 2006/01/20. . [DOI] [PubMed] [Google Scholar]
- 23.Roberts S, Scott JR, Husmann LK, Zurawski CA. Murine models of Streptococcus pyogenes infection. Curr Protoc Microbiol. 2006;Chapter 9:Unit 9D 5. doi: 10.1002/9780471729259.mc09d05s02 . [DOI] [PubMed] [Google Scholar]
- 24.Scaramuzzino DA, McNiff JM, Bessen DE. Humanized in vivo model for streptococcal impetigo. Infect Immun. 2000;68(5):2880–7. Epub 2000/04/18. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson ME, Neely MN, Caparon MG. Animal Models of Streptococcus pyogenes Infection In: Ferretti JJ, Stevens DL, Fischetti VA, editors Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2016. [Google Scholar]
- 26.Watson ME Jr, Nielsen HV, Hultgren SJ, Caparon MG. Murine vaginal colonization model for investigating asymptomatic mucosal carriage of Streptococcus pyogenes. Infect Immun. 2013;81(5):1606–17. Epub 2013/03/06. doi: 10.1128/IAI.00021-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aziz RK, Kansal R, Aronow BJ, Taylor WL, Rowe SL, Kubal M, et al. Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS One. 2010;5(4):e9798 doi: 10.1371/journal.pone.0009798 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole JN, Pence MA, von Kockritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, et al. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. mBio. 2010;1(4). Epub 2010/09/10. doi: 10.1128/mBio.00191-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dohrmann S, Anik S, Olson J, Anderson EL, Etesami N, No H, et al. Role for streptococcal collagen-like protein 1 in M1T1 group A Streptococcus resistance to neutrophil extracellular traps. Infect Immun. 2014;82(10):4011–20. Epub 2014/07/16. doi: 10.1128/IAI.01921-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henningham A, Chiarot E, Gillen CM, Cole JN, Rohde M, Fulde M, et al. Conserved anchorless surface proteins as group A streptococcal vaccine candidates. Journal of Molecular Medicine (Berlin). 2012;90(10):1197–207. Epub 2012/04/25. doi: 10.1007/s00109-012-0897-9 . [DOI] [PubMed] [Google Scholar]
- 31.Hollands A, Pence MA, Timmer AM, Osvath SR, Turnbull L, Whitchurch CB, et al. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A Streptococcus M1T1 clone. J Infect Dis. 2010;202(1):11–9. doi: 10.1086/653124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kansal RG, Datta V, Aziz RK, Abdeltawab NF, Rowe S, Kotb M. Dissection of the molecular basis for hypervirulence of an in vivo-selected phenotype of the widely disseminated M1T1 strain of group A Streptococcus bacteria. Journal of Infectious Disease. 2010;201(6):855–65. doi: 10.1086/651019 . [DOI] [PubMed] [Google Scholar]
- 33.Kazmi SU, Kansal R, Aziz RK, Hooshdaran M, Norrby-Teglund A, Low DE, et al. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group A streptococcal isolates in vivo. Infect Immun. 2001;69(8):4988–95. doi: 10.1128/IAI.69.8.4988-4995.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13(8):981–5. doi: 10.1038/nm1612 . [DOI] [PubMed] [Google Scholar]
- 35.Aziz RK, Kotb M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerging Infect Dis. 2008;14(10):1511–7. doi: 10.3201/eid1410.071660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, et al. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 2004;51(1):123–34. [DOI] [PubMed] [Google Scholar]
- 37.Fiebig A, Loof TG, Babbar A, Itzek A, Koehorst JJ, Schaap PJ, et al. Comparative genomics of Streptococcus pyogenes M1 isolates differing in virulence and propensity to cause systemic infection in mice. Int J Med Microbiol. 2015;305(6):532–43. doi: 10.1016/j.ijmm.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 38.Le Breton Y, McIver KS. Genetic Manipulation of Streptococcus pyogenes (The Group A Streptococcus, GAS). Curr Prot Microbiol. 2013;30:Unit 9D.3. doi: 10.1002/9780471729259.mc09d03s30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, et al. Genome-wide identification of genes required for fitness of Group A Streptococcus in human blood. Infect Immun. 2013;81(3):862–75. doi: 10.1128/IAI.00837-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdes KM, Sundar GS, Vega LA, Belew AT, Islam E, Binet R, et al. The fruRBA operon is necessary for Group A Streptococcal growth in fructose and for resistance to neutrophil killing during growth in whole human blood. Infect Immun. 2016. doi: 10.1128/IAI.01296-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vega L, Valdes K, Sundar G, Belew A, Islam E, Berge J, et al. The transcriptional regulator CpsY is important for innate immune evasion in Streptococcus pyogenes. Infect Immun. 2016;85(3):pii: e00925–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–72. doi: 10.1038/nmeth.1377 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Breton Y, Belew AT, Valdes KM, Islam E, Curry P, Tettelin H, et al. Essential Genes in the Core Genome of the Human Pathogen Streptococcus pyogenes. Sci Rep. 2015;5:9838 Epub 2015/05/23. doi: 10.1038/srep09838 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connolly KL, Roberts AL, Holder RC, Reid SD. Dispersal of Group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS One. 2011;6(4):e18984 Epub 2011/05/07. doi: 10.1371/journal.pone.0018984 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukomski S, Montgomery CA, Rurangirwa J, Geske RS, Barrish JP, Adams GJ, et al. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67(4):1779–88. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller EW, Danger JL, Ramalinga AB, Horstmann N, Shelburne SA, Sumby P. Regulatory rewiring confers serotype-specific hyper-virulence in the human pathogen group A Streptococcus. Mol Microbiol. 2015;98(3):473–89. doi: 10.1111/mmi.13136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, et al. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20(10):1745–7. doi: 10.1096/fj.06-5804fje [DOI] [PubMed] [Google Scholar]
- 48.Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. American Journal of Pathology. 2006;169(3):927–42. doi: 10.2353/ajpath.2006.060112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192(5):771–82. doi: 10.1086/432514 . [DOI] [PubMed] [Google Scholar]
- 50.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30(6):884–6. https://doi.org/10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
- 51.Król JE, Biswas S, King C, Biswas I. SMU.746-SMU.747, a putative membrane permease complex, is involved in aciduricity, acidogenesis, and biofilm formation in Streptococcus mutans. J Bacteriol. 2014;196(1):129–39. doi: 10.1128/JB.00960-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asai K, Yamaguchi H, Kang CM, Yoshida K, Fujita Y, Sadaie Y. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol Lett. 2003;220(1):155–60. [DOI] [PubMed] [Google Scholar]
- 53.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106(38):16422–7. doi: 10.1073/pnas.0906627106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host and Microbe. 2009;6(3):279–89. doi: 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19(12):2308–16. doi: 10.1101/gr.097097.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barquist L, Boinett CJ, Cain AK. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biology. 2013;10(7):1161–9. doi: 10.4161/rna.24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature Reviews Microbiology. 2013;11(7):435–42. doi: 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burby PE, Nye TM, Schroeder JW, Simmons LA. Implementation and Data Analysis of Tn-seq, Whole-Genome Resequencing, and Single-Molecule Real-Time Sequencing for Bacterial Genetics. J Bacteriol. 2017;199(1): e00560–16 doi: 10.1128/JB.00560-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rau MH, Calero P, Lennen RM, Long KS, Nielsen AT. Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microbial Cell Factories. 2016;15(1):176 doi: 10.1186/s12934-016-0577-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci USA. 2016;113(48):13887–92. doi: 10.1073/pnas.1610856113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Opijnen T, Dedrick S, Bento J. Strain Dependent Genetic Networks for Antibiotic-Sensitivity in a Bacterial Pathogen with a Large Pan-Genome. PLoS Path. 2016;12(9):e1005869 doi: 10.1371/journal.ppat.1005869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Path. 2013;9(12):e1003800 doi: 10.1371/journal.ppat.1003800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2015;112(13):4110–5. doi: 10.1073/pnas.1419677112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria JJ, et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio. 2014;5(5):e01729–14. doi: 10.1128/mBio.01729-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22(12):2541–51. doi: 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, et al. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun. 2000;68(6):3523–34. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, et al. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16(4):396–400. Epub 2006/02/21. doi: 10.1016/j.cub.2005.12.039 . [DOI] [PubMed] [Google Scholar]
- 68.Zinkernagel AS, Hruz P, Uchiyama S, von Köckritz-Blickwede M, Schuepbach RA, Hayashi T, et al. Importance of Toll-like receptor 9 in host defense against M1T1 group A Streptococcus infections. Journal of Innate Immunity. 2012;4(2):213–8. doi: 10.1159/000329550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy KM, Antonio ML, van Opijnen T. MAGenTA; a Galaxy implemented tool for complete Tn-Seq analysis and data visualization. Bioinformatics. 2017; 1–3. doi: 10.1093/bioinformatics/btx320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Opijnen T, Lazinski DW, Camilli A. Genome-Wide Fitness and Genetic Interactions Determined by Tn-seq, a High-Throughput Massively Parallel Sequencing Method for Microorganisms. Curr Prot Microbiol. 2015;36:1E 3 1–24. doi: 10.1002/9780471729259.mc01e03s36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110(7):2623–8. doi: 10.1073/pnas.1222783110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Narayanan AM, Ramsey MM, Stacy A, Whiteley M. Defining genetic fitness determinants and creating genomic resources for an oral pathogen. Applied and Environmental Microbiology. 2017:AEM.00797-17. doi: 10.1128/AEM.00797-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Russell CW, Richards AC, Chang AS, Mulvey MA. The Rhomboid Protease GlpG Promotes the Persistence of Extraintestinal Pathogenic Escherichia coli within the Gut. Infect Immun. 2017:IAI.00866-16. doi: 10.1128/IAI.00866-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vries SPW, Gupta S, Baig A, Wright E, Wedley A, Jensen AN, et al. Genome-wide fitness analyses of the foodborne pathogen Campylobacter jejuni in in vitro and in vivo models. Sci Rep. 2017;7(1):1251 doi: 10.1038/s41598-017-01133-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host and Microbe. 2013;14(6):652–63. doi: 10.1016/j.chom.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gutierrez MG, Yoder-Himes DR, Warawa JM. Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front Cell Infect Microbiol. 2015;5:78 doi: 10.3389/fcimb.2015.00078 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, et al. D-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187(19):6719–25. doi: 10.1128/JB.187.19.6719-6725.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cox KH, Ruiz-Bustos E, Courtney HS, Dale JB, Pence MA, Nizet V, et al. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One. 2009;4(4):e5366 Epub 2009/04/30. doi: 10.1371/journal.pone.0005366 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol. 2012;194(7):1668–78. doi: 10.1128/JB.06373-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rowe HM, Hanson BR, Runft DL, Lin Q, Firestine SM, Neely MN. Modification of the CpsA protein reveals a role in alteration of the Streptococcus agalactiae cell envelope. Infect Immun. 2015;83:1497–506. doi: 10.1128/IAI.02656-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minami M, Ichikawa M, Ohta M, Hasegawa T. The cell envelope-associated protein, LytR, regulates the cysteine protease SpeB in Streptococcus pyogenes. Acta Pathologica, Microbiologica et Immunologica Scandinavica. 2012;120(5):417–26. doi: 10.1111/j.1600-0463.2011.02847.x [DOI] [PubMed] [Google Scholar]
- 84.Tedde V, Rosini R, Galeotti CL. Zn2+ Uptake in Streptococcus pyogenes: Characterization of adcA and lmb Null Mutants. PLoS One. 2016;11(3):e0152835 doi: 10.1371/journal.pone.0152835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moulin P, Patron K, Cano C, Zorgani MA, Camiade E, Borezée-Durant E, et al. The Adc/Lmb System Mediates Zinc Acquisition in Streptococcus agalactiae and Contributes to Bacterial Growth and Survival. J Bacteriol. 2016;198(24):3265–77. doi: 10.1128/JB.00614-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J Bacteriol. 2003;185(9):2887–900. doi: 10.1128/JB.185.9.2887-2900.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitrakul K, Loo CY, Gyurko C, Hughes CV, Ganeshkumar N. Mutational analysis of the adcCBA genes in Streptococcus gordonii biofilm formation. Oral Microbiol Immunol. 2005;20(2):122–7. doi: 10.1111/j.1399-302X.2004.00205.x [DOI] [PubMed] [Google Scholar]
- 88.Dintilhac A, Claverys JP. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148(2):119–31. doi: 10.1016/S0923-2508(97)87643-7 [DOI] [PubMed] [Google Scholar]
- 89.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol. 2011;82(4):904–16. doi: 10.1111/j.1365-2958.2011.07862.x [DOI] [PubMed] [Google Scholar]
- 90.Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25(4):727–39. [DOI] [PubMed] [Google Scholar]
- 91.Ong CL, Gillen CM, Barnett TC, Walker MJ, McEwan AG. An antimicrobial role for zinc in innate immune defense against group A streptococcus. J Infect Dis. 2014;209(10):1500–8. doi: 10.1093/infdis/jiu053 [DOI] [PubMed] [Google Scholar]
- 92.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol. 2013;88(6):1176–93. Epub 2013/05/09. doi: 10.1111/mmi.12250 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kihlberg BM, Cooney J, Caparon MG, Olsén A, Björck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19(5):299–315. [DOI] [PubMed] [Google Scholar]
- 94.Sanson M, Makthal N, Gavagan M, Cantu C, Olsen RJ, Musser JM, et al. Phosphorylation events in the multiple gene regulator of group A Streptococcus significantly influence global gene expression and virulence. Infect Immun. 2015;83(6):2382–95. Epub 2015/04/01. doi: 10.1128/IAI.03023-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Terao Y, Kawabata S, Kunitomo E, Murakami J, Nakagawa I, Hamada S. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol Microbiol. 2001;42(1):75–86. . [DOI] [PubMed] [Google Scholar]
- 96.Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015;43(1):418–32. doi: 10.1093/nar/gku1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gera K, McIver KS. Laboratory growth and maintenance of Streptococcus pyogenes (the Group A Streptococcus, GAS). Curr Prot Microbiol. 2013;30:Unit 9D.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, et al. , editors. Short Protocols in Molecular Biology. 3rd ed New York: John Wiley & Sons, Inc; 1997. [Google Scholar]
- 99.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011; 29(1):24–6. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Venn diagrams comparing the number of genes showing either decreased (orange) or increased (green) fitness from the Tn-seq analyses of M1T1 GAS 5448 grown in THY for 24 h (A) and 48 h (B). (C) Clusters of Orthologous Genes (COG) categories are indicated with their relative abundance for the same genes showing either decreased (orange) or increased (green) fitness in vitro at 24 (light shade), 48 h (dark shade), and both (hatched).
(EPS)
H&E stains of skin and associated soft tissues samples collected at 12, 24 and 48 HPI. Specific regions are indicated as follows: epidermis (EP), sebaceous gland (SG), hair shaft (HS), papillary dermis (PD), reticular dermis (RD), and panniculus carnosus muscle (PCM). (A) Non-infected tissues displaying normal epidermal thickness (ca. 12 μm) and features. (B) 12 HPI lesion with inflammatory infiltration (black arrows) in the dermis (both PD and RD) and PCM with early necrosis indicated. (C) Magnification of (B). (D) 24 HPI lesion with PCM inflammation and infiltration (black arrow); Hypo: hypodermis. (F) Magnification of (D) showing monocytes, macrophages, lymphocytes and neutrophils (blue, orange and green arrows, respectively) and visible GAS chains (black arrows). (E) 48 h HPI lesion showing extensive inflammation, tissue damage and abscess formation with pseudo-capsule (CAP) surrounding necrotic debris (Nec). (G) Magnification of (E) shows visible GAS chains (black arrows). Scale bars (in red) represent 100 μm on panels A, B, D and E; and 10 μm on panels C, F and G.
(EPS)
GAS 5448 Krmit libraries injected subcutaneously into immunocompetent Crl:SKH1-hrBR hairless mice were isolated from homogenized lesions and plated onto 5% TSA blood agar plates to look for mucoid (capsule-overproducing) colonies. (A) After 12 HPI, most colonies display a non-mucoid phenotype. (B) After 48 HPI, mucoid colonies (grey arrows) are distinguishable from the remaining colonies (white arrows). (C) AP-PCR analyses on gDNA isolated from selected mucoid mutants show Krmit insertions at different locations within the covS gene (green arrow heads) as indicated.
(EPS)
Genome alignment was generated using the Orthology Browser from the Microbes Online resource (www.microbesonline.org/) using the MGAS5005 genome as reference and the RS06890 homolog (M5005_Spy0477) as the query for the genome alignment. The graphic display presents genome alignments based on the 24 highest homologs to RS06890/M5005_Spy0477. Colored arrows depict homologous genes found within the display window of the different aligned genomes, while grey arrows represent genes that do not have a homolog in the displayed genome window.
(EPS)
Real-time quantitative PCR (qRT-PCR) analyses of relative transcript levels for scfA and scfB genes in the ΔscfA and ΔscfB mutants compared to wild-type 5448 grown in THY to late log phase. Error bars represent the standard errors from at least three biological replicates. Dashed lines indicate 2-fold significance. Significance was determined using comparisons of transcript levels relative to those of the gyrA gene.
(EPS)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
Data Availability Statement
All data except for the raw sequencing reads are provided with the submitted materials. Illumina sequencing reads from the RNA-seq and Tn-seq analyses were deposited in the NCBI Sequence Read Archive (SRA) under the accession number (PRJNA391181).