Abstract
Mobile sensing based on the integration of microfluidic device and smartphone, so-called MS2 technology, has enabled many applications over recent years, and continues to stimulate growing interest in both research communities and industries. In particular, it has been envisioned that MS2 technology can be developed for various cell functional assays to enable basic research and clinical applications. Toward this direction, in this paper, we describe the development of a MS2-based cell functional assay for testing cell migration (the Mkit). The system is constructed as an integrated test kit, which includes microfluidic chips, a smartphone-based imaging platform, the phone apps for image capturing and data analysis, and a set of reagent and accessories for performing the cell migration assay. We demonstrated that the Mkit can effectively measure purified neutrophil and cancer cell chemotaxis. Furthermore, neutrophil chemotaxis can be tested from a drop of whole blood using the Mkit with red blood cell (RBC) lysis. The effects of chemoattractant dose and gradient profile on neutrophil chemotaxis were also tested using the Mkit. In addition to research applications, we demonstrated the effective use of the Mkit for on-site test at the hospital and for testing clinical samples from chronic obstructive pulmonary disease patient. Thus, this developed Mkit provides an easy and integrated experimental platform for cell migration related research and potential medical diagnostic applications.
Keywords: Microfluidic device, Smartphone, Cell functional assay, Cell migration, Chemotaxis
1. Introduction
Mobile sensing based on the integration of microfluidic device and smartphone, so-called MS2 technology, is an emerging and fast developing research area in recent years (Erickson et al. 2014; Yang et al. 2016). It has been used as a mobile laboratory for a wide range of applications, which include biochemical detection and analysis such as water and food quality analysis, routine health test and disease diagnosis (Yang et al. 2016; Zhang and Liu 2016). The core components of MS2 are lab-on-chip (LoC) based analytical technologies in a portable and miniaturized manner, and the mobile sensing and data processing functions offered by the new generation of smartphone. Effective integration of the two key technologies critically empowers MS2 for many mobile sensing applications. Current applications of MS2 cover detection of various environmental and health indicators such as pH (Lopez-Ruiz et al. 2014), nitrite (Wang et al. 2015), heavy metal (Chen et al. 2014b; Wang et al. 2014), bacterial contamination (Hutchison et al. 2015; San Park et al. 2013; Zhu et al. 2012), blood glucose (Chun et al. 2014), proteins (Chan et al. 2015; Lillehoj et al. 2013; Preechaburana et al. 2012; You et al. 2013) and other pathogen-associated biomarkers (Fronczek et al. 2014; Stemple et al. 2014; Yeo et al. 2016). Some complicated assays such as enzyme-linked immunosorbent assay (ELISA) (Chen et al. 2014a; Wang et al. 2011) and polymerase chain reaction (PCR) (Jiang et al. 2014; Liao et al. 2016; Stedtfeld et al. 2012) were successfully implemented with the MS2 systems. Furthermore, MS2 systems offers advantages in test speed, self-containment and sample to result assay operation, which are required for in field test and point of care (PoC) diagnosis (D’Ambrosio et al. 2015; Hu et al. 2016; Laksanasopin et al. 2015; Mudanyali et al. 2012). Those MS2 applications integrated sophisticated assay control accessories and sample-chip-phone interfaces, which demonstrate the potential of MS2 to enable high-level biological applications.
Among the high-level biological applications, we envisioned that MS2 technology can be applied for various cell functional assays (Yang et al. 2016). To be more specific, here we refer cell functional assays to the in-vitro assays that can qualitatively or quantitatively measure the presence or level of functional activities of live biological cells (e.g. cell adhesion assay; cell migration assay). Indeed, growing efforts have been made to develop compact imaging systems so that cell functional assays can be performed without requiring specialized microscopy facilities. For example, various incubation microscopes were developed so the microscope can be placed inside a conventional incubator for cell imaging or directly control the temperature of the cell assay within the portable microscope (Jin et al. 2015; Pushkarsky et al. 2014; Walzik et al. 2015; Zhang et al. 2015). A highly integrated portable and robotically controlled live cell imaging system was employed for cell migration assay in a microfluidic device (Saito et al. 2016). In addition, USB microscopes or webcams were also used for functional cell and tissue imaging, which significantly lower the costs while maintaining adequate imaging performance (Isikman et al. 2012; Kim et al. 2011; Kim et al. 2012; Lynch et al. 2014; Walzik et al. 2015). We have previously performed cell chemotaxis test using a USB microscope based portable system and a smartphone for remote data monitoring (Wu et al. 2014). More recently, the new generation of smartphones with advanced hardware and software configurations led to growing development of smartphone-based microscopy applications (Wei et al. 2013; Zhu et al. 2013; Zhu et al. 2012). Collectively, these previous works support the concept of integrating mobile sensing devices with microfluidic chips and control systems to enable MS2-based cell functional assays. To our best knowledge, although smartphone-based imaging systems have been applied to image cells (Liu et al. 2014; Skandarajah et al. 2014; Smith et al. 2011; Zhu et al. 2011), they have not been used for cell functional assays.
In this direction, cell migration and chemotaxis assays represent an attractive target application to realize the potential of MS2 for advanced cell functional assays. Cell migration and chemotaxis are important for many biological and pathological processes such as host defense (de Oliveira et al. 2016; Kolaczkowska and Kubes 2013), tissue development (Laird et al. 2008), autoimmune disease (Luster et al. 2005) and cancers (Condeelis and Segall 2003; Friedl and Wolf 2003). In-vitro real-time visualization assays are widely used for cell migration and chemotaxis research, which typically require sophisticated chemical gradient generation, controlling incubation temperature, single cell imaging and quantitative data analysis (Funamoto et al. 2002; Muinonen-Martin et al. 2010). Over the past near two decades, microfluidic devices have become widely used research tools for quantitative cell migration and chemotaxis studies owing to their ability to better control microenvironments, low reagents consumption, simple and automated fluid handling systems (Huang et al. 2009; Jeong et al. 2010; Jeong et al. 2013; Li et al. 2016; Sackmann et al. 2014a; Vargas et al. 2014; Wu et al. 2013). Many of these devices can configure chemical gradients without the requirement of external fluid delivery instruments (Ge et al. 2015; Sackmann et al. 2012; Wu et al. 2016). Recently, new microfluidic methods have been developed to allow rapid immune cell chemotaxis test directly from a drop of whole blood by incorporating on-chip cell isolation module (Agrawal et al. 2008; Hamza and Irimia 2015; Jones et al. 2016; Sackmann et al. 2012; Sackmann et al. 2014b; Wu et al. 2016). Several studies have also employed microfluidic cell migration systems for disease orientated applications (Butler et al. 2010; Jones et al. 2014; Wu et al. 2015). These developments provide the general background of microfluidics technology for performing portable cell migration and chemotaxis assays on a smartphone platform. Such a platform will enable easy cell migration experiment for scientific research and rapid on-site cell migration test for potential clinical diagnostic applications (e.g. neutrophil chemotaxis test for lung disease diagnosis) without requiring specialized research facilities and skills.
Therefore, in this study we were motivated to construct a MS2-based cell migration test kit (we name it the Mkit) that integrates a new microfluidic device, a smartphone-based portable live cell imaging platform, reagents and accessories for cell migration assay and custom smartphone apps for image acquisition and data analysis. This Mkit is the first microfluidic platform coupled with smartphone for cell migration and chemotaxis test. We successfully validated this new Mkit by testing chemotaxis of both purified human blood neutrophils and breast cancer cell line. Moreover, the Mkit allows rapid chemotaxis test of neutrophils from a drop of whole blood with red blood cell (RBC) lysis. Finally, we demonstrated the effective use of the Mkit for on-site test at the hospital and testing patient samples for potential clinical diagnostic applications. Thus, this effective Mkit demonstrates the potential of MS2-based cell functional assays.
2. Materials and Methods
2.1 Assembly of Mkit
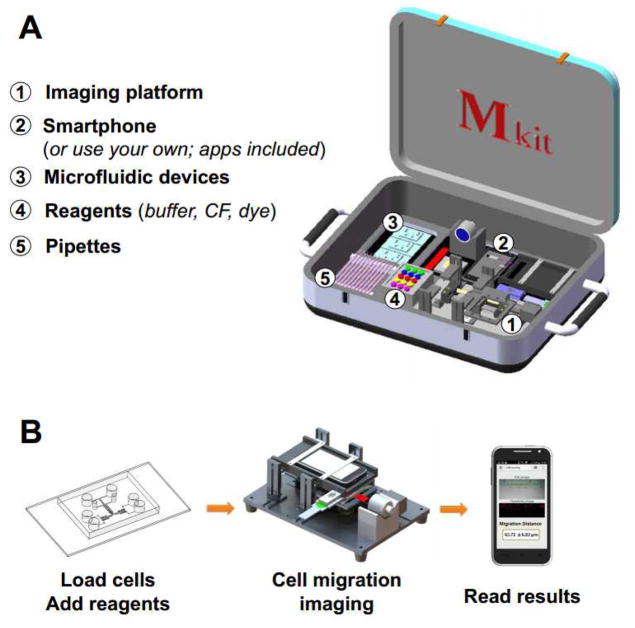
The components of the Mkit and the method of cell migration test are illustrated in Fig. 1.
Fig. 1. Illustration of the Mkit.
(A) Components of the Mkit; (B) Operation flow of the Mkit. CF indicates chemotactic factor.
2.1.1 Reagents and supplies
BSA, RBC lysis buffer, FITC-Dextran, N-Formyl-Met-Leu-Phe (fMLP), DME/F12, epidermal growth factor (EGF), Paraformaldehyde (PFA) and Rhodamine 6 G were purchased from Sigma-Aldrich. The EasySep Direct Human Neutrophil Isolation Kit was purchased from STEMCELL Technologies Inc. RPMI-1640, DMEM, Fetal Bovine Serum (FBS) and DPBS were purchased from Fisher Scientific. Pipettors or squeeze pipettes were used for adding reagents and samples to the microfluidic device.
2.1.2. Smartphone-based imaging platform
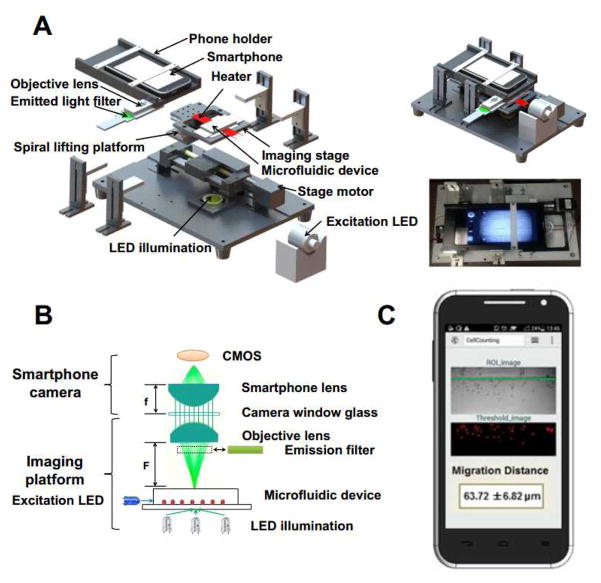
The smartphone-based imaging platform is illustrated in Fig. 2A–B. A commercial brand smartphone (H30-T100, HUAWEI, China) was used for the Mkit prototype. This phone operates with the Android system. It has an 8 MPixel CMOS module at the maximal resolution of 2560×1920 pixels and its built-in lens has a focal length of f = 4 mm. In general, the Mkit can work with different types of camera phones and operating systems.
Fig. 2. Illustration of the smartphone imaging platform.
(A) Expanded view of the imaging platform. The system is 260 (L) × 180 (W) × 100 (H) mm; The illustration and a picture of the real assembled imaging platform are shown on the right; (B) Optical configuration of the imaging platform; (C) Smartphone app interface showing the cell migration distance analysis. The green line denotes the edge of the gradient channel next to the docking structure. The final result is presented as the average migration distance of all cells ± standard error of the mean (s.e.m.) in μm.
In addition to the smartphone, the imaging platform consists of the following components and configuration:
A U-shape smartphone holder and an objective lens (focal length F ≈ 6.5 mm) installed in a plastic holder and connected with the U-shape holder's bottom facet. The add-on objective lens was aligned with the smartphone camera to enable cell imaging in the microfluidic device.
An imaging stage and a spiral lifting platform were used for vertical focus adjustment. In addition, a stage motor (SLT-BB-0412-ELE0150RG, IGUS, Germany) was used for driving the horizontal stage movement. In this study, the optics of the imaging platform matches the dimension of the microfluidic device, allowing the two test units to be imaged within a single field-of-view (FOV). The motorized stage further permits programmable multi-position image acquisition when more test units cannot fit to a single FOV. The system can be made cheaper by using a manual stage, which is sufficient for end-point image acquisition but not multi-position time-lapse imaging.
A low-power white LED (3 W, 3.2–3.3 V, COB, LED, China) was used for illumination.
A blue excitation LED (470 nm, 700 mA, XRE, CREE, USA) and an emission filter (520 nm, JSL520-25, Zhuoli Hanguang Beijing, China) aligned with the objective lens holder were configured for fluorescence imaging. The current configuration was for imaging FITC-Dextran as a measure of gradient generation. The fluorescence imaging unit can be modified to image other fluorophores.
A transparent heater and a controller (Minco, MN) were mounted on the imaging stage to control the temperature of the microfluidic device at 37 °C for cell migration experiments. The glass bottom of the microfluidic device was placed on top of the heater. The heating element was embedded in the transparent plastic sheet. The temperature was controlled by an ON/OFF controller, which was pre-calibrated using a digital thermometer.
2.1.3. Image acquisition and analysis apps
Two customized Android apps were developed for capturing single or time-lapse cell migration images and for image processing and data analysis.
The image acquisition app controls the acquisition of cell migration images at the user-defined frame rate and period. For example, neutrophil migration images were captured at 4 frames/min for 15 min. The exposure time and electronic gain were automatically adjusted. Other image acquisition parameters including brightness, contrast and image compression rate can be user-defined. The captured images were stored in the smartphone's memory card in JPEG format.
The image processing and data analysis app identifies cells in the images and calculates cell migration distance along the gradient direction as a measure of chemotaxis (Fig. 2C). It has the option to import the images directly from the smartphone camera or from the memory card for analysis. In the current experiments, the images were first collected by the acquisition app and then imported to the analysis app. The target region of interest (ROI) for analysis is user-defined. The cell recognition algorithm is based on threshold controlled segmentation, which generally gives over 90% of accuracy. In addition to the simple cell migration distance analysis by the app, the time-lapse images can be exported to a third party software such as ImageJ (NIH, version 1.44) for further cell tracking analysis. Quantitative cell migration parameters such as the commonly used chemotactic index (CI: cell displacement in the gradient direction divided by the total migration distance) and migration speed (total cell migration distance divided by experiment time) were calculated from the tracking data following the established methods (Lin and Butcher 2006).
2.1.4. Microfluidic chip
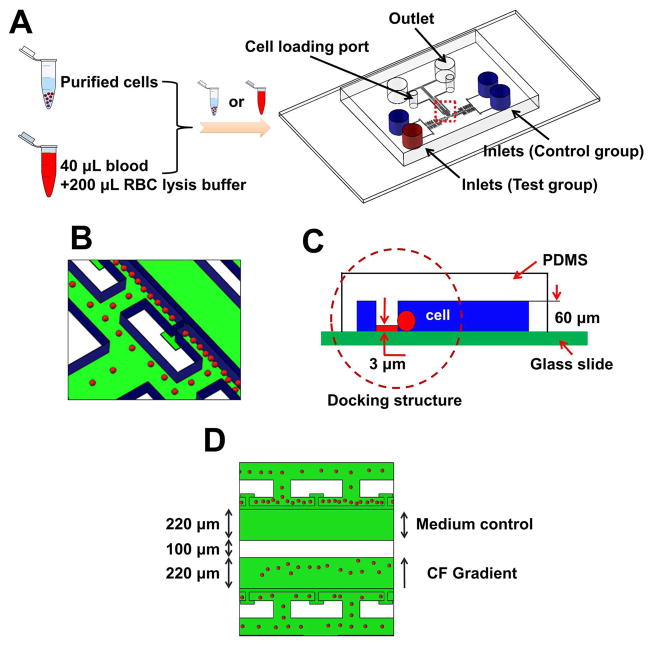
The polydimethylsiloxane (PDMS)-based microfluidic device was fabricated using standard photolithographic and soft lithographic techniques (Dertinger et al. 2001). Different from the previous design that only allows a single migration experiment of one condition (Wu et al. 2016), the current design has two parallel independently controlled test units on a single chip (Fig. 3). Briefly, the device was designed using AutoCAD software and printed on a transparency mask using a high resolution printer with 24,000 dpi resolution. The multi-height device mold was fabricated using SU-8 photoresist on a Si wafer by photolithography following the previously established protocol (Taylor et al. 2005). The first layer (~3 μm thick) was used to form the cell-docking structure. The second layer (~60 μm thick) was aligned on the top of the first layer to form the flow channels, source inlets, cell loading ports and outlets. The SU-8 mold was stamped with PDMS (Sylgard 184, Dow Corning) to fabricate the negative replica by soft-lithography. The PDMS replica was peeled off from the mold. The inlets and outlets were punched out of PDMS using sharpened punchers. The PDMS replica was then bonded to a glass slide using a plasma cleaner (Harrick Plasma) to complete the microfluidic device fabrication. The microfluidic channel was coated with fibronectin (BD Biosciences) for 1 h followed by 0.4% bovine serum albumin (BSA) blocking for another 1 h all at room temperature. The fibronectin coating provides a substrate for cell migration.
Fig. 3. Illustration of the microfluidic device for cell migration test.
(A) Microfluidic device illustration to test purified cells or directly from whole blood with RBC lysis. (B–C) Enlarged view of the microfluidic channel with cell docking. The red spheres indicate the cells, which were initially aligned by the thin barrier channel because the cell size is larger than the height of the barrier channel. Upon chemoattractant stimulation, cells will change their morphology to crawl through the barrier channel into the gradient channel. (D) Illustration of a cell migration experiment for parallel chemotaxis test and medium control. CF indicates chemotactic factor. Blue color indicates the channel walls and green color reflects the glass surface.
2.2 Cell preparation
Human blood samples were obtained by venipuncture from healthy donors under an ethics protocol approved by the Joint-Faculty Research Ethics Board at the University of Manitoba. Informed written consent form was obtained from all participants by the recruiting staff at the Victoria General Hospital in Winnipeg. Neutrophils were negatively selected using a magnetic neutrophil isolation kit directly from the whole blood (EasySep Direct, STEMCELL Technologies). Isolated neutrophils were suspended in RPMI-1640 medium before use for experiment within 8 h. Alternatively, 40 uL whole blood sample was mixed with 1×RBC lysis buffer in 1:5 ratio in an Eppendorf tube and incubated at room temperature for 5 min to enrich the white blood cell population for cell migration experiment. Human metastatic breast cancer cell line, MDA-MB-231 cells, were obtained from ATCC and cultured in DME/F12 with 10% FBS in an incubator at 37 °C with 5% CO2 and passaged regularly. Cancer cells were detached with 0.25% trypsin for 2 min and re-suspended in DME/F12 with 1% FBS before migration experiment.
2.3 Cell migration experiment
The cell migration experiment using the Mkit is illustrated in Fig. 1B, Fig. 3 and Supplemental Movie 1. Cells were loaded to the two test units of the microfluidic device from the cell loading ports and allowed to align beside the cell docking structure. For whole blood, 30 uL RBC lysed blood sample was loaded to the microfluidic device followed by washing with the migration medium. Then, equal volume of chemoattractant solution (e.g. fMLP in RPMI-1640 with 0.4% BSA for neutrophil experiment; EGF in DME/F12 with 1% FBS for cancer cell experiment) and medium (e.g. RPMI-1640 with 0.4% BSA for neutrophil experiment; DME/F12 with 1% FBS for cancer cell experiment) were added to the source wells of the test unit, respectively. FITC-Dextran was added to the chemoattractant solution for gradient measurement. For the control unit, only the migration medium was loaded into both source wells. Similar to the previously reported strategy (Ge et al. 2015), this device allows rapid flow-based chemical gradient generation based on the pressure difference between the inlets and outlet without requiring external pumps, which simplifies the assay operation. The microfluidic device was placed on the imaging stage of the smartphone system. Gradient generation was checked by measuring the intensity profile of FITC-Dextran in the microfluidic channel (Fig. 4). Then neutrophil migration was imaged for 15 min and analyzed using the smartphone apps (Fig. 2C) or by cell tracking analysis. The temperature of the microfluidic device was controlled at 37 °C using the stage heater. Alternatively, if cell tracking analysis was not required, cells in the device were fixed in 2% PFA at the end of the migration assay, and only the final cell image was acquired and analyzed using the smartphone system. For cancer cells, the migration assay was incubated in a conventional incubator at 37 °C with 5% CO2 for 9 h, followed by cell fixing and imaging analysis using the smartphone system. The cell migration assay was repeated independently three times for each condition. Forty cells were analyzed for each experiment. The on-site test and COPD sample test were repeated two times. The Student's t-test was used to compare the cell migration parameters between different conditions. The difference was considered statistically significant at p < 0.05.
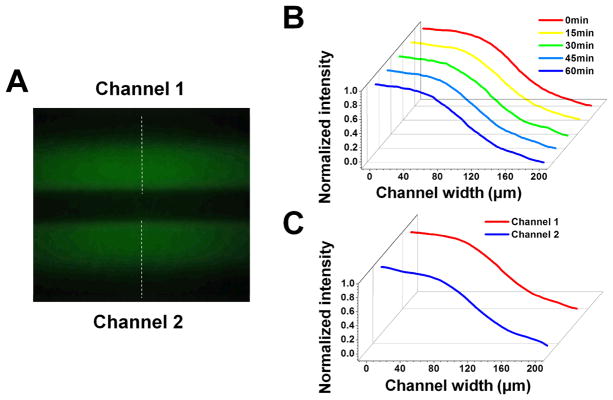
Fig. 4. Gradient measurements by the smartphone imaging system.
(A) Gradient of FITC-Dextran in the two gradient channels captured by the smartphone system; (B) Gradient profile at different time points in one gradient channel; (C) Gradient profiles in two gradient channels.
3. Results
3.1 Imaging performance using the Mkit
We tested the imaging performance of the smartphone system using the 1951 USAF resolution target. The smartphone system can resolve Element 4 in Group 7 of the target. The smallest lines that can be resolved are indicated by the red arrow (Fig. S1A) and the corresponding peak intensity separation (Fig. S1B). The corresponding resolution is ~2.76 μm, which is sufficient for cell imaging applications and is consistent with the smartphone imaging resolution reported by previous work (Arpa et al. 2012). Indeed, we demonstrated that the smartphone microscope can capture clear bright field images of both human blood neutrophils and breast cancer cells in the microfluidic channels (Fig. 5A). The optical magnification was selected so the maximal FOV of the current system is more than 0.5 mm2, which allows imaging two gradient channels in a single FOV. Increasing or decreasing the optical magnification will allow either enlarged visualization of less cells or larger FOV, but will not affect the general chemotaxis measurement. The fluorescence images were taken using the smartphone fluorescent microscopy module. We demonstrated that the intensity profile of FITC-Dextran in the microfluidic channel can be quantitatively measured using the smartphone system (Fig. 4A). The gradient profile is stable over time in each channel for up to one hour (Fig. 4B) and identical gradients can be configured in both channels (Fig. 4C). Here we measured the gradient at 1.5 mm below the junction of the inlet channels. The stable gradient is sufficient for neutrophil migration experiment. For cancer cell migration experiment, chemoattractant solution and migration medium were added to the inlets and the waste solution was removed from the outlet every 1 h to maintain the gradient over a longer period. Taken together, the smartphone-based imaging system is capable of imaging biological cells at the single cell resolution and can image fluorescent intensity profile for gradient measurement. These imaging capabilities are required for performing smartphone-based cell migration and chemotaxis experiments.
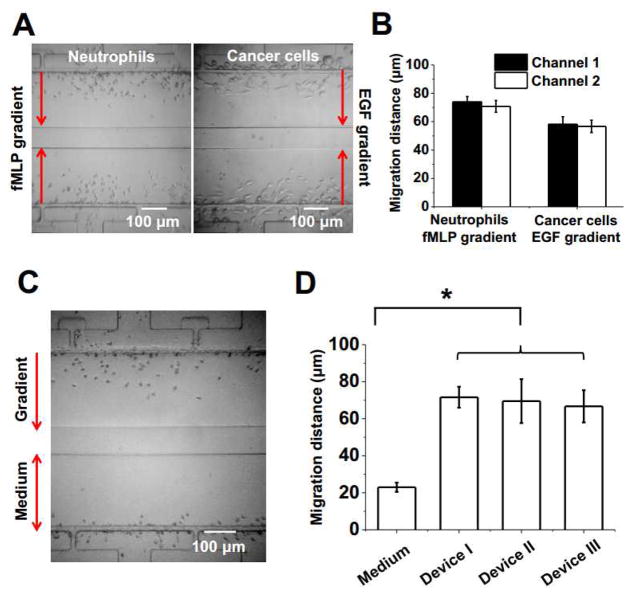
Fig. 5. Chemotaxis test of neutrophils and cancer cells using the Mkit.
(A) Final distribution of purified neutrophils and cancer cells in the gradient channels in response to a 100 nM fMLP gradient or a 30 nM EGF gradient respectively. The experiment was done in duplicate on each device for purified neutrophils or cancer cells. (B) Cell migration distance analysis of chemotaxis experiments using purified neutrophil and cancer cells. (C) Final distribution of neutrophils from whole blood to a 100 nM fMLP gradient or in the medium control; (D) Cell migration distance analysis of neutrophil from whole blood in 3 different devices. The bars show the average migration distance of all cells to the fMLP gradient for each device and the error bars are the standard error of the mean (s.e.m.). The migration distance of the medium control shown in the graph is the average of all cells from one device and it is similar in all three devices.
3.2 Cell migration assay using the Mkit
3.2.1 Chemotaxis test of purified neutrophils and cancer cells
First, we validated the developed Mkit by testing chemotaxis of purified human blood neutrophils and human breast cancer cells to the gradient of their respective known chemoattractant (i.e. 100 nM fMLP gradient for neutrophils; 30 nM EGF gradient for cancer cells). Each cell migration experiment was done in duplicate by testing the same condition in the two test channels of the same microfluidic device. The results show that cells clearly migrated toward the gradient and the results in the two channels are consistent (Fig. 5A–B). In this set of experiments, the average migration distance of neutrophils is 70.8 μm and 73.8 μm in the two channels respectively; the average migration distance of human breast cancer cells is 56.7 μm and 58.1 μm in the two channels respectively. The difference of migration distance between the two channels is not statistically significant (by the Student's t-test), demonstrating good reproducibility. Medium control experiment was performed separately, which measured much lower migration distance comparing to the gradient experiment (Fig. 6A for purified neutrophils; the average migration distance of cancer cells in the medium control is ~20 μm).
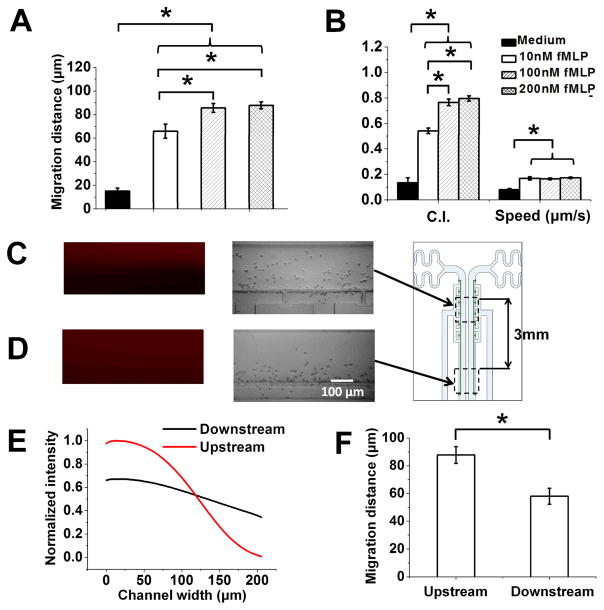
Fig. 6. Chemotaxis test of neutrophils to fMLP gradients of different doses and profiles using the Mkit.
(A) Cell migration distance of neutrophils to fMLP gradients of different doses; (B) Cell tracking analysis by calculating CI and cell speed to different fMLP gradient doses; (C–D) Rhodamine 6 G gradient images and the corresponding final cell distribution images at the upstream and downstream of the gradient channel; (E) Plot of gradient profiles at the upstream and downstream (normalized to the upstream gradient) of the gradient channel; (F) Cell migration distance analysis of neutrophil in upstream and downstream to a 100 nM fMLP gradient. The bars show the average value of the parameter of all cells for each condition and the error bars are the standard error of the mean (s.e.m.). The data shown are from a set of representative experiments (one experiment for each condition)
3.2.2 Test neutrophil chemotaxis directly from whole blood
Next, we demonstrated that neutrophil chemotaxis can be rapidly tested from a drop of whole blood. Previously, we developed an all-on-chip chemotaxis assay by integrating on-chip magnetic neutrophil selection and chemotaxis test in a single microfluidic device (Wu et al. 2016). This method is fully compatible with the smartphone system. Here we further showed that neutrophil chemotaxis can be directly tested from whole blood with RBC lysis and on-chip washing using the Mkit. The whole assay from-sample-to-result can be done in 30 min. Such a method eliminated the requirement of magnetic cell separation module. On-chip DAPI staining confirmed that all the migrated cells into the gradient channel are polymorphonuclear (PMN) cells (Fig. S2). For the cell migration experiments, we configured chemotaxis test to a 100 nM fMLP gradient and medium control in parallel in each microfluidic device (Fig. 5C), and repeated the experiment multiple times (n=3). The results showed significantly higher migration distance to the fMLP gradient comparing to the medium control and the results are consistent in repeating experiments with good reproducibility (Fig. 5D).
3.2.3 Effect of chemoattractant dose and gradient profile on neutrophil chemotaxis
To further demonstrate the use of Mkit for chemotaxis experiment, we tested purified neutrophil migration to fMLP gradients over a range of doses (i.e. 10 nM, 100 nM, 200 nM) along with parallel medium control. The results showed the chemoattractant dose-dependent chemotaxis. All three fMLP gradients induced significantly higher cell migration distance comparing to the medium control, and the migration distance to the 100 nM fMLP gradient and the 200 nM gradient is significantly higher than the 10 nM fMLP gradient (Fig. 6A). Single cell tracking analysis was further performed and the results are consistent with the migration distance analysis (Fig. 6B). CI for all three fMLP gradients is significantly higher than the medium control, and CI for the 100 nM fMLP gradient and the 200 nM gradient is significantly higher than the 10 nM fMLP gradient. The migration speed for all three fMLP gradients is significantly higher than the medium control, and the speed is comparable for different fMLP gradients. Such dosedependent neutrophil chemotaxis to fMLP gradient was not clear in our previous study using a different microfluidic device without the cell docking feature (Wu et al. 2015). We believe this is because cells are randomly seeded in the gradient channel without the docking structure so their starting positions in the gradient are different, which reduced the difference of averaged cell migration parameters (e.g. CI) between different gradients. With the docking structure, cells have the same starting position relative to the gradient so the gradient dose-dependent chemotaxis is shown more clearly by the cell migration parameters (e.g. cell migration distance or CI).
In addition to the chemoattractant dose, the quantitative gradient profile can also influence chemotaxis. Our microfluidic device can generate steeper nonlinear gradient at the upstream of the channel and smoother linear gradient at the relatively downstream of the channel due to laminar flow mixing of the chemoattractant solution and the medium along the channel (Fig. 6C–E), allowing us to compare the gradient profile dependent chemotaxis in a single device. It is worth pointing out that the molecular weight of FITC-Dextran (10 kD) used in our experiments is significantly higher than fMLP (< 0.5 kD). To more accurately confirm the fMLP gradient profile, we in this experiment further used Rhodamine 6 G with similar molecular weight of fMLP as the fluorescence indicator to measure the gradients at the upstream (1.5 mm below the junction of the inlet channels) and downstream (4.5 mm below the junction of the inlet channels) of the gradient channel. The upstream region and the downstream region are separated by 3 mm in our experiment. As expected, the results (again using purified human blood neutrophils) showed the migration distance is significantly higher at the upstream gradient comparing to the downstream gradient, suggesting the influence of gradient steepness and nonlinearity on chemotaxis (Fig. 6F). Such a gradient profile effect was shown by a previous study that breast cancer cells chemotax to nonlinear but not linear EGF gradient (Wang et al. 2004). Our previous study also found the similar effect for human blood T cell that T cells can migrate toward a nonlinear chemokine gradients (Lin and Butcher 2006) but not a linear chemokine gradient (unpublished) using microfluidic devices.
3.2.4. On-site and clinical tests using Mkit
To demonstrate the effective on-site use of the Mkit, we performed chemotaxis test of neutrophils from healthy donors directly at a local hospital (Victoria General Hospital in Winnipeg) without specialized cell culture and imaging facilities. The results showed neutrophils chemotaxis to a 100 nM fMLP gradient using the Mkit (Fig. S3A). To demonstrate the effective use of the Mkit for potential clinical diagnostic application, we tested human blood neutrophils from healthy donor to a gradient of sputum supernatant from patient with chronic obstructive pulmonary disease (COPD) using the Mkit. The COPD sputum samples were obtained at the Seven Oaks General Hospital in Winnipeg under an ethics protocol approved by the University of Manitoba. The sputum was processed to obtain the supernatant following the previously established method (Wu et al. 2015). We have previously showed neutrophil chemotaxis to a COPD sputum gradient using a microfluidic device and conventional imaging system for potential clinical diagnostic application (Wu et al. 2015). Consistently, our current results using the Mkit showed neutrophil chemotax to the COPD sputum gradient (Fig. S3B).
4. Discussion
Although the current prototype system of Mkit has shown promise for research and clinical applications, its performance can be strengthened. Particularly, the microfluidic device and data analysis app can be further developed to improve their functionalities for different applications. The current cell migration test takes less than 30 min so it is well within the battery time of the phone for on-site test. Further incorporation of CO2 and humidity controls will allow direct time-lapse cell migration imaging with the smartphone system over a longer period of time without requiring a separate incubator for more advanced cell tracking analysis. For such a test, the phone may need to be plugged into a power outlet or a portable battery charger.
Aside from targeting cell migration assay as a specific application, the smartphone-based imaging platform can be generally useful for a broad range of cell-based applications in and out of a research laboratory. The imaging quality of the smartphone-based imaging platform can be further improved by configuring more sophisticated optical system to work with the smartphone camera. Different visible wavelengths of fluorescent signal can be measured using the smartphone-based imaging platform by configuring the suitable light sources and filter sets. Furthermore, we envision that 3D printing technology can be employed for fabricating integrated smartphone-based cell imaging system and to provide the necessary biological containment to allow its safe use out of a research lab (Knowlton et al. 2015; Skandarajah et al. 2014; Yafia et al. 2015).
Cell functional assays represent a higher level of validation for scientific hypothesis and development in biological and biomedical research, and can also be used for clinical disease assessment (Albini and Benelli 2007; Hong and Zu 2013; Kepp et al. 2011; Liang et al. 2007; Martins et al. 2012). In addition to chemotaxis assay as described in this study, we envision that the MS2 technology can be developed to enable a range of cell functional assays such as cell viability assay, apoptosis assays, cell proliferation assay, cell differentiation assay, cell invasion assay, wound healing assays, cell killing assay, live cell signaling assays, and cell-based organ-on-chip imaging and monitoring. Fast growth of microfluidic technology has made its way to connect with all main areas of cell biology research (Duncombe et al. 2015). It has been increasingly enabling sophisticated experiments in miniaturized devices, and many microfluidics-based cell functional assays have been developed (Duncombe et al. 2015). However, a bottleneck that hinders the adoption of microfluidic cell functional assays in life science research and medical applications remains due to knowledge gap and the facility and skill requirement for effective use of microfluidic assays. In this context, extending MS2 technology to cell functional assays provides a solution to bridge and integrate the two fields. With suitable biological containment and safety procedures, the MS2-based cell functional assays can be performed in different scientific educational settings to target both academic audience and the general public. This will effectively promote science knowledge and healthcare awareness by taking advantage of the high public acceptance of smartphone.
5. Conclusion
In conclusion, we successfully developed a MS2-based cell migration assay, the Mkit, as an example of MS2 technology for cell functional assays. The imaging performance and some representative applications of the Mkit were successfully demonstrated. With the optical add-on components, the smartphone-based imaging platform can achieve ~3 μm resolution, which is adequate for imaging many cell types with comparable quality of using a conventional microscope at low magnification. The smartphone-based imaging platform is also capable of fluorescence microscopy, which is consistent with a previously reported MS2 system (Zhu et al. 2011). The Mkit was successfully validated for testing chemotaxis of purified human blood neutrophils or directly from a drop of blood and breast cancer cell line. The Mkit also offers a promising solution for on-site test at the hospital and testing patient samples. Collectively, this Mkit has the potential to be broadly adopted for various cell migration related research and clinical applications. Further development of different MS2-based cell functional assays will provide useful tools for scientific research, medical diagnosis and education.
Supplementary Material
Acknowledgments
This work is supported by Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC)(RGPIN-2014-04789) and the Canadian Institutes of Health Research (CIHR)(CPG-134755), and financial assistance from CMC Microsystems (Project Reference ID:3699). F.L. thanks the Winnipeg Rh Institute Foundation and the University of Manitoba for a Rh Award; J.D.W. thanks Mitacs for a postdoc fellowship. H.P-S thanks NSERC for a postdoc fellowship. We thank the Nano-Systems Fabrication Laboratory at the University of Manitoba for their technical support. We thank the Clinical Institute of Applied Research and Education at the Victoria General Hospital in Winnipeg for managing human blood samples and Seven Oaks General Hospital in Winnipeg for managing COPD patient samples. We thank Dongxue Xie and Xiaoou Ren for technical research assistance.
References
- Agrawal N, Toner M, Irimia D. Lab Chip. 2008;8(12):2054–2061. doi: 10.1039/b813588f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, Benelli R. Nat Protoc. 2007;2(3):504–511. doi: 10.1038/nprot.2006.466. [DOI] [PubMed] [Google Scholar]
- Arpa A, Wetzstein G, Lanman D, Raskar R. 2012 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops; IEEE; 2012. pp. 23–28. [Google Scholar]
- Butler KL, Ambravaneswaran V, Agrawal N, Bilodeau M, Toner M, Tompkins RG, Fagan S, Irimia D. PloS One. 2010;5(7):e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HN, Shu Y, Xiong B, Chen Y, Chen Y, Tian Q, Michael SA, Shen B, Wu H. ACS Sensors. 2015;1(3):227–234. [Google Scholar]
- Chen A, Wang R, Bever CR, Xing S, Hammock BD, Pan T. Biomicrofluidics. 2014a;8(6):064101. doi: 10.1063/1.4901348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Chen WY, Yen YC, Wang CW, Chang HT, Chen CF. Anal Chem. 2014b;86(14):6843–6849. doi: 10.1021/ac5008688. [DOI] [PubMed] [Google Scholar]
- Chun HJ, Park YM, Han YD, Jang YH, Yoon HC. BioChip J. 2014;8(3):218–226. [Google Scholar]
- Condeelis J, Segall JE. Nat Rev Cancer. 2003;3(12):921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio MV, Bakalar M, Bennuru S, Reber C, Skandarajah A, Nilsson L, Switz N, Kamgno J, Pion S, Boussinesq M. Sci Transl Med. 2015;7(286):286re284–286re284. doi: 10.1126/scitranslmed.aaa3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S, Rosowski EE, Huttenlocher A. Nat Rev Immunol. 2016;16(6):378–391. doi: 10.1038/nri.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger SK, Chiu DT, Jeon NL, Whitesides GM. Anal Chem. 2001;73(6):1240–1246. [Google Scholar]
- Duncombe TA, Tentori AM, Herr AE. Nat Rev Mol Cell Bio. 2015;16(9):554–567. doi: 10.1038/nrm4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson D, O'Dell D, Jiang L, Oncescu V, Gumus A, Lee S, Mancuso M, Mehta S. Lab Chip. 2014;14(17):3159–3164. doi: 10.1039/c4lc00142g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Nat Rev Cancer. 2003;3(5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Fronczek CF, San Park T, Harshman DK, Nicolini AM, Yoon JY. Rsc Advances. 2014;4(22):11103–11110. [Google Scholar]
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Cell. 2002;109(5):611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Ge Y, An Q, Gao Y, Chen Y, Li D. Microsyst Technol. 2015;21(8):1797–1804. [Google Scholar]
- Hamza B, Irimia D. Lab Chip. 2015;15(12):2625–2633. doi: 10.1039/c5lc00245a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Zu Y. Theranostics. 2013;3(6):377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cui X, Gong Y, Xu X, Gao B, Wen T, Lu TJ, Xu F. Biotechnol adv. 2016;34(3):305–320. doi: 10.1016/j.biotechadv.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Huang CW, Cheng JY, Yen MH, Young TH. Biosens Bioelectron. 2009;24(12):3510–3516. doi: 10.1016/j.bios.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Hutchison JR, Erikson RL, Sheen AM, Ozanich RM, Kelly RT. Analyst. 2015;140(18):6269–6276. doi: 10.1039/c5an01304f. [DOI] [PubMed] [Google Scholar]
- Isikman SO, Greenbaum A, Lee M, Bishara W, Mudanyali O, Su TW, Ozcan A. Anal Cell Pathol. 2012;35(4):229–247. doi: 10.3233/ACP-2012-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HH, Lee SH, Kim JM, Kim HE, Kim YG, Yoo JY, Chang WS, Lee CS. Biosens Bioelectron. 2010;26(2):351–356. doi: 10.1016/j.bios.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Jeong HH, Lee SH, Lee CS. Biosens Bioelectron. 2013;47:278–284. doi: 10.1016/j.bios.2013.03.031. [DOI] [PubMed] [Google Scholar]
- Jiang L, Mancuso M, Lu Z, Akar G, Cesarman E, Erickson D. Sci Rep. 2014;4:4137. doi: 10.1038/srep04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Wong D, Li J, Luo Z, Guo Y, Liu B, Wu Q, Ho CM, Fei P. Sci Rep. 2015;5:18483. doi: 10.1038/srep18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CN, Hoang AN, Martel JM, Dimisko L, Mikkola A, Inoue Y, Kuriyama N, Yamada M, Hamza B, Kaneki M. J Leukocyte Biol. 2016;100(1):241–247. doi: 10.1189/jlb.5TA0715-310RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CN, Moore M, Dimisko L, Alexander A, Ibrahim A, Hassell BA, Warren HS, Tompkins RG, Fagan SP, Irimia D. PloS One. 2014;9(12):e114509. doi: 10.1371/journal.pone.0114509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Nat Rev Drug Discov. 2011;10(3):221–237. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, Khademhosseini A. Lab Chip. 2011;11(10):1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Koo KI, Bae H, Dokmeci MR, Hamilton GA, Bahinski A, Kim SM, Ingber DE, Khademhosseini A. Lab Chip. 2012;12(20):3976–3982. doi: 10.1039/c2lc40345e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton S, Sencan I, Aytar Y, Khoory J, Heeney M, Ghiran I, Tasoglu S. Sci Rep. 2015;5:15022. doi: 10.1038/srep15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, Kubes P. Nat Rev Immunol. 2013;13(3):159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Laird DJ, von Andrian UH, Wagers AJ. Cell. 2008;132(4):612–630. doi: 10.1016/j.cell.2008.01.041. [DOI] [PubMed] [Google Scholar]
- Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J. Sci transl Med. 2015;7(273):273re271–273re271. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu T, Zou H, Chen X, Sun D, Yang M. Biosens Bioelectron. 2016 doi: 10.1016/j.bios.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL. Nat Protoc. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Liao SC, Peng J, Mauk MG, Awasthi S, Song J, Friedman H, Bau HH, Liu C. Sensor Actuat B: Chem. 2016;229:232–238. doi: 10.1016/j.snb.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj PB, Huang MC, Truong N, Ho CM. Lab Chip. 2013;13(15):2950–2955. doi: 10.1039/c3lc50306b. [DOI] [PubMed] [Google Scholar]
- Lin F, Butcher EC. Lab Chip. 2006;6(11):1462–1469. doi: 10.1039/b607071j. [DOI] [PubMed] [Google Scholar]
- Liu X, Lin TY, Lillehoj PB. Ann Biomed Eng. 2014;42(11):2205–2217. doi: 10.1007/s10439-014-1055-z. [DOI] [PubMed] [Google Scholar]
- Lopez-Ruiz N, Curto VF, Erenas MM, Benito-Lopez F, Diamond D, Palma AJ, Capitan-Vallvey LF. Anal Chem. 2014;86(19):9554–9562. doi: 10.1021/ac5019205. [DOI] [PubMed] [Google Scholar]
- Luster AD, Alon R, von Andrian UH. Nat Immunol. 2005;6(12):1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- Lynch AE, Triajianto J, Routledge E. PloS One. 2014;9(8):e103547. doi: 10.1371/journal.pone.0103547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SA, Trabuco JR, Monteiro GA, Chu V, Conde JP, Prazeres DMF. Trend Biotechnol. 2012;30(11):566–574. doi: 10.1016/j.tibtech.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Lab Chip. 2012;12(15):2678–2686. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinonen-Martin AJ, Veltman DM, Kalna G, Insall RH. PloS One. 2010;5(12):e15309. doi: 10.1371/journal.pone.0015309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preechaburana P, Gonzalez MC, Suska A, Filippini D. Angew Chem Internat Edit. 2012;51(46):11585–11588. doi: 10.1002/anie.201206804. [DOI] [PubMed] [Google Scholar]
- Pushkarsky I, Liu Y, Weaver W, Su TW, Mudanyali O, Ozcan A, Di Carlo D. Sci Rep. 2014;4:4717. doi: 10.1038/srep04717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann EK, Berthier E, Young EW, Shelef MA, Wernimont SA, Huttenlocher A, Beebe DJ. Blood. 2012;120(14):e45–e53. doi: 10.1182/blood-2012-03-416453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014a;507(7491):181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- Sackmann EKH, Berthier E, Schwantes EA, Fichtinger PS, Evans MD, Dziadzio LL, Huttenlocher A, Mathur SK, Beebe DJ. 2014b;111(16):5813–5818. doi: 10.1073/pnas.1324043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Honda K, Asaka C, Ueki S, Ishikawa K. Allergol Int. 2016;65(3):280–285. doi: 10.1016/j.alit.2016.01.001. [DOI] [PubMed] [Google Scholar]
- San Park T, Li W, McCracken KE, Yoon JY. Lab Chip. 2013;13(24):4832–4840. doi: 10.1039/c3lc50976a. [DOI] [PubMed] [Google Scholar]
- Skandarajah A, Reber CD, Switz NA, Fletcher DA. PloS One. 2014;9(5):e96906. doi: 10.1371/journal.pone.0096906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthews D, Wachsmann-Hogiu S. PLoS One. 2011;6(3):e17150. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Tourlousse DM, Seyrig G, Stedtfeld TM, Kronlein M, Price S, Ahmad F, Gulari E, Tiedje JM, Hashsham SA. Lab Chip. 2012;12(8):1454–1462. doi: 10.1039/c2lc21226a. [DOI] [PubMed] [Google Scholar]
- Stemple CC, Angus SV, San Park T, Yoon JY. J Lab Autom. 2014;19(1):35–41. doi: 10.1177/2211068213498241. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat Methods. 2005;2(8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas P, Terriac E, Lennon-Duménil AM, Piel M. J Vis Exp. 2014;(84):51099. doi: 10.3791/51099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzik MP, Vollmar V, Lachnit T, Dietz H, Haug S, Bachmann H, Fath M, Aschenbrenner D, Mofrad SA, Friedrich O. Biosens Bioelectron. 2015;64(15):639–649. doi: 10.1016/j.bios.2014.09.061. [DOI] [PubMed] [Google Scholar]
- Wang H, Li Yj, Wei Jf, Xu, Wang Yh, Zheng Gx. Anal Bioanal Chem. 2014;406(12):2799–2807. doi: 10.1007/s00216-014-7715-x. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhao X, Khimji I, Akbas R, Qiu W, Edwards D, Cramer DW, Ye B, Demirci U. Lab Chip. 2011;11(20):3411–3418. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Saadi W, Lin F, Nguyen CMC, Jeon NL. Exp Cell Res. 2004;300(1):180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Wang X, Gartia MR, Jiang J, Chang TW, Qian J, Liu Y, Liu X, Liu GL. Sensor Actuat B: Che. 2015;209:677–685. [Google Scholar]
- Wei Q, Qi H, Luo W, Tseng D, Ki SJ, Wan Z, Göröcs Zn, Bentolila LA, Wu TT, Sun R. ACS nano. 2013;7(10):9147–9155. doi: 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hillier C, Komenda P, de Faria RL, Levin D, Zhang M, Lin F. PloS One. 2015;10(5):e0126523. doi: 10.1371/journal.pone.0126523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hillier C, Komenda P, de Faria RL, Santos S, Levin D, Zhang M, Lin F. Technology. 2016;4(02):104–109. [Google Scholar]
- Wu J, Ouyang L, Wadhawan N, Li J, Zhang M, Liao S, Levin D, Lin F. Biomed Microdevices. 2014;16(4):521–528. doi: 10.1007/s10544-014-9854-4. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu X, Lin F. Lab Chip. 2013;13(13):2484–2499. doi: 10.1039/c3lc50415h. [DOI] [PubMed] [Google Scholar]
- Yafia M, Ahmadi A, Hoorfar M, Najjaran H. Micromachines. 2015;6(9):1289–1305. [Google Scholar]
- Yang K, Peretz-Soroka H, Liu Y, Lin F. Lab Chip. 2016;16(6):943–958. doi: 10.1039/c5lc01524c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SJ, Choi K, Cuc BT, Hong NN, Bao DT, Ngoc NM, Le MQ, Hang NLK, Thach NC, Mallik SK. Theranostics. 2016;6(2):231. doi: 10.7150/thno.14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You DJ, San Park T, Yoon JY. Biosens Bioelectron. 2013;40(1):180–185. doi: 10.1016/j.bios.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu Q. Biosens Bioelectron. 2016;75(15):273–284. doi: 10.1016/j.bios.2015.08.037. [DOI] [PubMed] [Google Scholar]
- Zhang YS, Ribas J, Nadhman A, Aleman J, Selimovi3 Š, Lesher-Perez SC, Wang T, Manoharan V, Shin SR, Damilano A. Lab Chip. 2015;15(18):3661–3669. doi: 10.1039/c5lc00666j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Mavandadi S, Coskun AF, Yaglidere O, Ozcan A. Anal Chem. 2011;83(17):6641–6647. doi: 10.1021/ac201587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sencan I, Wong J, Dimitrov S, Tseng D, Nagashima K, Ozcan A. Lab Chip. 2013;13(7):1282–1288. doi: 10.1039/c3lc41408f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sikora U, Ozcan A. Analyst. 2012;137(11):2541–2544. doi: 10.1039/c2an35071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.