SUMMARY
Treponema denticola is an oral spirochete strongly associated with severe periodontal disease. A prominent virulence factor, the major outer sheath protein (Msp), disorients neutrophil chemotaxis by altering the cellular phosphoinositide balance, leading to impairment of downstream chemotactic events including actin rearrangement, Rac1 activation and Akt activation in response to chemoattractant stimulation. The specific regions of Msp responsible for interactions with neutrophils remain unknown. In this study, we investigated the inhibitory effect of truncated Msp regions on neutrophil chemotaxis and associated signaling pathways. Murine neutrophils were treated with recombinant protein truncations followed by assessment of chemotaxis and associated signal pathway activation. Chemotaxis assays indicate sequences within the C-terminal region; particularly the first 130 amino acids, have the strongest inhibitory effect on neutrophil chemotaxis. Neutrophils incubated with the C-terminal region protein also demonstrated the greatest inhibition of Rac1 activation, increased phosphoinositide phosphatase activity, and decreased Akt activation; orchestrating impairment of chemotaxis. Furthermore, incubation with antibodies specific to only the C-terminal region blocked the Msp induced inhibition of chemotaxis and denaturing the protein restored Rac1 activation. Msp from the strain OTK, with numerous amino acid substitutions throughout the polypeptide, including the C-terminal region compared to strain 35405, showed increased ability to impair neutrophil chemotaxis. Collectively, these results indicate the C-terminal region of Msp is the most potent region to modulate neutrophil chemotactic signaling and that specific sequences and structure is likely required. Knowledge of how spirochetes dampen neutrophil response is limited and Msp may represent a novel therapeutic target for periodontal disease.
Keywords: migration, spirochete, immune, lipid, host-pathogen, signaling
INTRODUCTION
Periodontal disease is a microbial-induced chronic inflammatory condition characterized by destruction of both the soft and hard tooth-supporting tissues1. It is estimated that approximately 47% of the US population suffers from some form of periodontal disease2. Periodontal disease involves both dysbiosis of polymicrobial populations in the oral cavity and the host immune response1,3,4. In addition to being a leading cause of poor oral health and tooth loss, there are also significant links between periodontal disease and oral bacteria with systemic conditions including cardiovascular disease5,6, diabetes7, respiratory disease8,9 and cancer10–12.
Neutrophils are key rapid- response cells of the innate immune system that are recruited to sites of infection to eradicate pathogens, including in the oral cavity13. Neutrophils constitute the majority of the immune cells recruited to the gingival tissue and crevice14–16. The importance of neutrophils in maintaining periodontal health is reflected by the fact that more severe periodontal disease is observed in congenital diseases in which neutrophil recruitment and function are compromised15,17. Neutrophils migrate in a directed fashion towards chemoattractants, such as cytokines, complement peptides and bacterial products and chemicals including formyl group peptides (N-formylmethionyl-leucyl-phenylalanine, fMLP)18. Additionally, peripheral blood neutrophils from subjects with chronic periodontal disease have been reported to display impaired neutrophil chemotaxis in in vitro assays19,20.
Key to neutrophil chemotaxis is dynamic elongation, protrusion and retraction of the cell body, necessitating appropriate cytoskeleton remodeling and signaling mechanisms21,22. This requires asymmetrical distribution of molecules within the cell, including accumulation of the phosphoinositide second messenger molecule, phosphatidylinositol (3,4,5)-triphosphate (PIP3). Appropriate localization of PIP3 and downstream Rac1 activation at the leading edge of the neutrophil has been proposed to act as a cellular “compass”, driving efficient neutrophil migration23–25. PIP3 production is catalyzed from phosphatidylinositol 4,5- bisphosphate (PtdIns[(4,5)]P2) by the lipid kinase phosphatidylinositol 3- kinase (PI3K)26, while also being counteracted by the lipid phosphatases; the phosphatase and tensin homolog (PTEN) and the SH2-containing inositol phosphatase1 (SHIP1), which generate PtdIns[(4,5)]P2 and PtdIns[(3,4)]P2, respectively27,28. PI3K, PTEN and SHIP1 play complex interconnected roles in orchestrating and regulating neutrophil directional sensing and migration29–32. PTEN acts a negative regulator of neutrophil functions, including actin polymerization and sensitivity to chemotaxis32,33
The spirochete Treponema denticola is a key pathogen of the polymicrobial dysbiotic biofilm associated with periodontal disease. T. denticola and other oral spirochetes are a minor component in gingival plaque of healthy individuals, however they proliferate robustly in plaque of individuals with periodontal disease34–37. Spirochetes preferentially localize in deep periodontal pockets at the biofilm-tissue interface38–40, in close association with neutrophils41. The major outer membrane sheath protein (Msp) of T. denticola is a prominent membrane protein42 and one of the organism’s most well-characterized virulence factors. Msp is well known to perturb cell functions and cell-signaling pathways in host cells including neutrophils (for recent review see43,44). Msp is able to upset the effective PIP balance through activation of PTEN and inhibition of PI3K activation45,46, leading to hierarchical inhibition of crucial downstream local neutrophil chemotactic events including selective Rac1 activation and actin cytoskeleton rearrangement47–49.
Msp is thought to constitute a channel-forming porin, which can exist as one or more high molecular weight complex forms50–53. There is also a small central variable region which displays variation between some well-studied laboratory strains54,55, as well as noted heterogeneity in clinical samples56. Regions in the N- terminal half of Msp have been reported to mediate binding to extracellular matrix components54 while the C- terminal domain has been reported to possess pore-forming capability50. Localization of Msp within intact spirochetes is controversial as it has been reported that Msp is surface exposed53,57, while others have reported limited surface exposure58. Initial studies examining localization of Msp in intact spirochetes suggested the N- terminal and central V- region are surface exposed54, however a recent study has suggested that Msp has a bipartite architecture, with an N- terminal domain and a C- terminal domain, of which the C- terminal domain is surface exposed50. Given the differing thoughts on Msp location, structure, and function, it is clear there is still much to be understood about the structure and function of Msp.
Despite the wealth of knowledge about the interactions of Msp with host cells, crucial knowledge of the key protein regions involved is lacking. Regions of Msp responsible for extracellular matrix interaction and porin function have been reported50,54, yet protein region(s) important for modulation of neutrophil function are not known. The goal of this study is to identify the active region(s) of Msp responsible for impairment of neutrophil chemotaxis and to characterize the signaling mechanisms involved. Understanding this process will increase our knowledge of how T. denticola contributes to the impairment of neutrophil function. Moreover, Msp presents a novel target for development of new treatments or therapeutics with potential for improving both oral and overall health.
METHODS
Murine neutrophil isolation
Murine neutrophil isolation has been previously described46. All procedures were approved by the University at Buffalo Institutional Animal Care and Use Committee. Briefly, C57BL/6J wild-type mice (male, 6 weeks old) were purchased from Jackson Laboratory (Bar Harbor, Maine). Femurs and tibias were removed and cells were isolated from bone marrow by fractionation into discontinuous Percoll (Sigma) gradients (80%, 65%, 55%). Mature neutrophils were isolated from the 80%/65% interface.
Bacterial strains and culture conditions
Treponema denticola strains used in this study are listed in Table 1. Wild type strains were routinely grown anaerobically at 37 °C in NOS media while the Msp mutant strain MHE (gift of Dr. Chris Fenno) was grown in NOS containing 40 μg/ml erythromycin59. Cultures were examined for purity, typical morphology and enumerated using dark-field microscopy. For bacteria-neutrophil incubation experiments, strains were grown for 3 days anaerobically, washed 2 times with PBS, followed by counting by darkfield microscopy for use in the assay. All Escherichia coli strains were grown in Luria-Bertani (LB) broth with shaking or on LB agar at 37 °C with appropriate antibiotics.
Table 1.
Bacterial strains and plasmid constructs
| Component | Relevant Characteristics | Source |
|---|---|---|
| Bacteria | ||
| Treponema denticola | ||
| 35405 | Wild type strain | R. Ellen84 |
| MHE | Msp mutant of strain 35405 | J.C. Fenno59 |
| OTK | Wild type strain | J.C. Fenno55 |
| Escherichia coli | ||
| M15 | Expression of recombinant Msp and N, V, and C, strain 35405 | Qiagen |
| C41 (DE3) | Expression of recombinant CA and CB Msp, strain 35405 | Lucigen |
| BL21 (DE3) pLysS | Expression of recombinant Msp, strain OTK | A. Sharma |
| Plasmids | ||
| pQE30 | 3461 bp plasmid, adds N-terminal 6xHis tag | H. Jenkinson54 |
| rMsp | 1590 bp 35405-Msp gene cloned into pQE30 | H. Jenkinson54 |
| rN-Msp | 567 bp N-terminal fragment of 35405 Msp cloned into pQE30 | H. Jenkinson54 |
| rV-Msp | 171 bp V-region fragment of 35405 Msp cloned into pQE30 | H. Jenkinson54 |
| rC-Msp | 816 bp C-terminal fragment of 35405 Msp cloned into pQE30 | H. Jenkinson54 |
| pET23b | 3665 bp plasmid, adds N-terminal T7 tag and C-terminal 6xHis tag | W. Hofman |
| p23-CA | 405 bp CA fragment cloned into pET23b | This study |
| p23-CB | 411 bp CB fragment cloned into pET23b | This study |
| p23-OTK | 1690 bp OTK-Msp gene cloned into pET23b | This study |
Cloning of the Msp proteins and Msp protein fragments
Bacterial strains and plasmid constructs used throughout this study are detailed in Table 1. Plasmids previously transformed into the E. coli strain M15 for expression of recombinant Msp, and the N-, V- and C- region proteins have been previously described54 and were generous gifts from Dr. Howard Jenkinson (The University of Bristol). Plasmid pET23b which adds a N-terminal T7 tag (~1 kDa) and a C-terminal six-histidine tag (~1 kDa) to the construct was a gift from Dr. Wilma Hofman (The University at Buffalo). The 1690-bp nucleotide sequence corresponding to the msp gene from strain OTK lacking the leading sequence, was amplified by PCR using genomic DNA as the template with the primers listed in Table 2. Strain OTK was a gift from Dr. Chris Fenno (The University of Michigan). The OTK Msp PCR product was digested with BamHI and XhoI (Thermo) and ligated into plasmid pET23b similarly digested. The C- terminal region of 35405 was further divided in half with the first 405-bp region termed CA and the remaining 411-bp region called CB, also amplified by PCR with primers detailed in Table 2. Both amplicons were digested with BamHI and XhoI (Thermo) followed by ligation into pET23b. These ligation mixtures were transformed into chemically competent E. coli strain DH5α and grown on Luria Broth (LB) plates containing 100 μg/ml ampicillin. The expression plasmids were named p23-OTK, p23-CA and p23-CB, respectively, and were isolated with Qiagen mini prep plasmid isolation kit following the manufacturer’s instructions. Plasmids were next transformed into E. coli strains for protein expression as follows; p23-CA and p23-CB into strain C41 (DE3) and p23-OTK into strain BL-21 pLysS (DE3).
Table 2.
PCR primers used for cloning of recombinant Msp genes.
| Name | Application | Sequence |
|---|---|---|
| OTKfor23 | Recombinant OTK Msp | GATAGGATCCGGTACTCGTGGGCGGA |
| OTKrev23 | Recombinant OTK Msp | GATACTCGAGGTATGTAAGCTTGAGGCT |
| 405CtermAfor23 | Recombinant 35405 CA | GATAGGATCCGGCAGCAAACAAATATGCT |
| 405CtermArev23 | Recombinant 35405 CA | GATACTCGAGTGCTGATTTAAAGGCAAT |
| 405CtermBfor23 | Recombinant 35405 CB | GATAGGATCCGGCTTCAGGAGATACGAAT |
| 405CtermBrev23 | Recombinant 35405 CB | GATACTCGAGGTAGATAACTTTAACACC |
Bold sequences indicate enzyme sites.
Expression and purification of native and recombinant Msp and Msp fragments
Native Msp complex from strain 35405 was isolated as previously described52,60,61. Briefly, T. denticola cultures (2 l) were grown for 3 days in modified NOS medium62. The Msp preparation was highly enriched by sequential deoxycholate and n-octylpolyoxyethylene extraction, ultracentrifugation, autoproteolysis of the extract, concentration by ultrafiltration (Amicon Concentricon Plus 80), extensive washing in 10 mM Tris (pH 8.0) and distilled H2O ultracentrifugation and extensive dialysis.
Recombinant Msp (strain 35405) and the N-, V- and C- regional truncated proteins of Msp from strain 35405 were purified as previously described with minor modifications54 and the same method was used for recombinant Msp (strain OTK) and the CA and CB protein fragments unless otherwise indicated. The E. coli protein expression strains containing the plasmid constructs (Table 1) were incubated with shaking in 100 ml LB medium containing appropriate antibiotics (ampicillin (100 μg/ml) alone for pET23b and with kanamycin (25 μg/ml) for PQE30) from an overnight culture to OD600 ~0.6. Isopropyl-β-D-thiogalactopyranoside (IPTG) (1mM) was then added and the culture was incubated for 4 hours 37°C. Bacteria were harvested by centrifugation (5,000 × g for 10 min). The rMsp, N, V, and C protein pellets were solubilized in 2 ml lysis buffer (8M urea, 0.1M NaH2PO4, 0.01M Tris, pH 7) and mixed gently for 4 hours at room temperature. Cellular debris was removed by centrifugation (12,000 × g for 15 min). Pellets containing rMsp-OTK, CA and CB proteins were resuspended in 5 ml of lysis buffer and mixed by rocking for 1 hour at room temperature. The suspensions were then sonicated with a Branson Sonifier 450 at setting 5, using an 80% pulsed cycle of four 30 sec bursts with 2-min pauses. The sonicated bacterial lysates were centrifuged 10,000 × g for 20 minutes at 4 °C to remove cellular debris. Phenylmethylsulfonyl fluoride (PMSF 100 μM) (Sigma) was added to all the resulting supernatants containing the proteins.
All resulting supernatants were incubated with HisPur Ni-NTA Resin (Thermo) according to manufacturer’s instructions. Briefly, 5 ml of bead suspension was centrifuged at 3,500 × g for 5 minutes and the storage buffer removed. The beads were equilibrated with the lysis buffer prior to incubation with the bacterial lysate suspension for 30 minutes at room temperature with rocking. A column elution was then performed by washing the resin with bound protein once with 2X resin volume of wash buffer (1M NaCl, 10% EtOH, 2% Tween-20, 10 mM imidazole) followed by two additional washes with lysis buffer containing 10 mM imidazole (2X resin volume). Recombinant proteins were eluted three times with the lysis buffer containing 300 mM imidazole (0.5 ml). Pooled eluates containing purified proteins were then dialyzed stepwise into 6M, 4M and 2M urea solutions, then into 1X PBS three times using Slide-A-Lyzer Dialysis Cassettes (Thermo) of 20,000, 10,000, or 3,500 MWCO, as appropriate. The concentration of the pooled eluates was determined using the BCA protein assay (Thermo). Protein purity and expected molecular weight size of all recombinant protein truncations was confirmed with Coomassie staining of an SDS-PAGE gel (Sup Fig 1) and western blotting with a His-tag antibody (Pierce) and anti-Msp antiserum (rMsp: 53 kDa, N-Msp: 23 kDa, C- Msp: 32 kDa, V- Msp: 8 kDa, CA-Msp: 14 kDa, CB-Msp: 15 kDa), while OTK recombinant Msp reactivity was confirmed with a His-tag antibody (62 kDa) as it does not react with this anti-Msp antiserum (data not shown). The endotoxin activity of the recombinant protein preparations was measured using a Limulus Amebocyte Lysate (LAL) assay (Pierce). All protein preparations were essentially free of endotoxin contamination (range of 0.00269 to 0.0079 EU/μg of protein). All experiments using native Msp complex, recombinant Msp protein or Msp truncations were performed using a concentration of 30 ug/ml as this is within the reported effective dose range to affect multiple functions of both neutrophils and fibroblasts45,47,49,60
Chemotaxis assays
Quantitation of chemotaxis using a Zigmond chamber has been previously described46. Briefly, cells were treated with nMsp, rMsp, N, C or V truncated proteins (30 μg/ml for all) for 30 minutes at room temperature prior to being resuspended in Hank’s Balanced Salt Solution (HBSS), pH 7.4, with 1% gelatin and allowed to attach to 1% BSA coated coverslips (22 × 40 mm) at 37 °C for 10 minutes. Coverslips were inverted onto a Zigmond chamber, and HBSS solution was added to the left chamber while 1 μM fMLP was added to the right chamber. Images were taken every 20 seconds for 15 minutes using a Nikon Eclipse microscope E1000 equipped with a Hamatsu camera (model ORCA-ER). Images were analyzed using ImageJ software and chemotaxis analysis (migration and speed) performed using the manual tracking and chemotaxis tools in ImageJ.
Analysis of chemotaxis using transwell assay was performed according to the manufacturer’s instructions (Transwell, Costar 3472 Clear, Corning). Briefly, media with or without fMLP (1 μM) was placed in the bottom of the plates. Neutrophils (0.5 × 106) were incubated with or without Msp proteins (30 μg/ml) for 30 minutes at room temperature. In the case of chemotaxis experiments with whole bacteria, bacteria at an MOI of 500 (2.5 × 108 bacteria) as determined by counting with a darkfield microscope were added to the media in the bottom of the wells in the absence of fMLP. To examine the effect of nMsp on unstimulated neutrophil movement, neutrophils pre-exposed to nMsp were placed in wells in the absence of fMLP. For all assays, the neutrophil suspensions were added to the top of the transwell and incubated at 37 °C for 1 hour. The top of the membrane was gently wiped clean and the entire membrane was fixed in 4% paraformaldehyde overnight at 4 °C. Membranes were then washed and stained with crystal violet. After subsequent washing with dH2O until all excess dye had been removed, cells fixed to the membrane representing the migrated cell population were counted using an inverted microscope. The cells in 5 different areas of each membrane were counted, with duplicate transwells per condition, to yield the total migrated cells. All data were normalized to the control with fMLP alone (positive control). To determine the impact of antibody interaction on neutrophil chemotaxis, native Msp complex was incubated with a 1:50 dilution of α-N -terminal or α-C-terminal antibodies for 1 hour in PBS prior to exposure to neutrophils for 30 minutes and transwell chemotaxis as described above. Antibodies directed towards the N- and C-terminal region of Msp were kind gifts of Dr. Howard Jenkinson (The University of Bristol) and Dr. Justin Radolf (University of Connecticut Health Center)50,54
Rac1 activation assay
To determine the activation state of Rac1 in response to Msp and Msp truncated fragments, G-LISA assays were used following the manufacturer’s instructions (Cytoskeleton Inc.) as previously described45. Murine neutrophils (1 × 106 cells) were pre-treated with Msp proteins (30 μg/ml) for 30 minutes, followed by fMLP stimulation for 1 minute, washed and lysed. Equal amounts of cell lysate samples were used in the G-LISA assays. Rac1 activation was measured using a luminescence plate reader at 100 ms integration (Molecular Devices FlexStation3).
Akt phosphorylation analysis
Murine neutrophils (1×106 cells) were pre-treated with Msp or Msp truncated proteins for 15 minutes followed by stimulation with fMLP (1 μM) for 1 minute then lysed with SDS sample buffer and boiled. For western blot analysis, equal amount of the total protein lysates were separated by SDS-PAGE, followed by transfer to nitrocellulose. Membranes were blocked in 5% milk/TBS/0.1% Tween-20, incubated overnight in primary antibody anti-Akt Serine 473 at a 1:2000 dilution (Cell Signaling Technology) followed by an HRP- conjugated secondary antibody (1:5000). Following development, HRP was inactivated with 0.2% sodium azide and blots were reprobed with non-phosphorylated Akt antibody (1:2000) and β-actin antibodies (1:5000) (Cell Signaling Technology).
Malachite green phosphatase assay
PIP lipid phosphatase activity was determined by measuring the amount of free phosphate released from a synthetic PIP3 substrate using a modified malachite green phosphatase assay as previously described45,46. Neutrophils (1 × 105) were partially permeabilized with 0.1 volume of 2% n-octyl-β-glucopyranoside for 30 seconds, followed by treatment with Msp proteins. As a positive control, partially permeabilized neutrophils were stimulated with 1μM fMLP. Samples were incubated with 1 mM of Phosphatidylinositol 3,4,5-trisphosphate diC8 (Echelon) as a substrate, with 100 μl of malachite green solution (Echelon) for 30 minutes at room temperature. Absorbance at 650 nm was measured using a microplate reader (FlexStation3 plate reader, Molecular Devices Corporation) and the amount of free phosphate released calculated using a prepared phosphate standard curve.
Statistical analysis
Comparisons between two groups were performed using t-tests using PRISM software (version 6). Results are based on at least 3 independent experiments. Statistical significance was defined as P<0.05. Error bars represent the standard error of the mean (SEM).
RESULTS
Loss of Msp increases neutrophil chemotaxis toward T. denticola
A key function of the neutrophil is to migrate toward and destroy invading bacterial pathogens. Using a transwell chemotaxis assay, we compared neutrophil migration toward wild type T. denticola strain 35405 and the Msp mutant strain, MHE59. Cells exposed to fMLP to stimulate chemotaxis were used as a positive control. Significantly more neutrophils migrated toward the MHE strain than the wild type strain (Fig 1). This suggests that the Msp protein plays an important role in limiting neutrophil chemotaxis, through surface exposure on the whole bacterium.
Fig. 1.
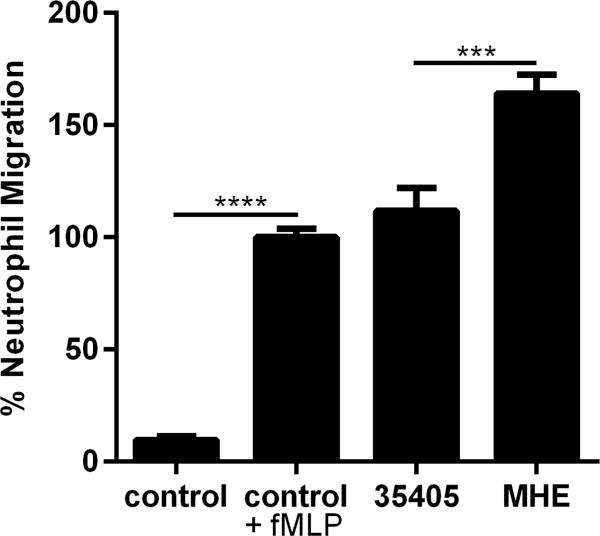
Msp impairs neutrophil chemotaxis in whole bacteria. In a transwell chemotaxis assay, murine neutrophils were exposed to T. denticola 35405 wild type or MHE (Msp mutant) bacteria for 1 hour to allow chemotaxis toward the bacteria through a membrane. Cells that migrated across the membrane were fixed, stained with crystal violet, and counted. Neutrophils exposed to the wild type and MHE bacterial strains were compared to each other, with more neutrophils migrated toward MHE than the wild type strain. Neutrophils alone and unstimulated with fMLP served as a negative control, while neutrophils stimulated with fMLP as the positive control for neutrophil chemotaxis and were compared to each other. Results were normalized to the control +fMLP. Graphs represents mean ± SEM of 3 independent experiments (*** P < 0.001 and **** P < 0.0001 by unpaired t test).
The C- terminal region of Msp strongly inhibits neutrophil chemotaxis
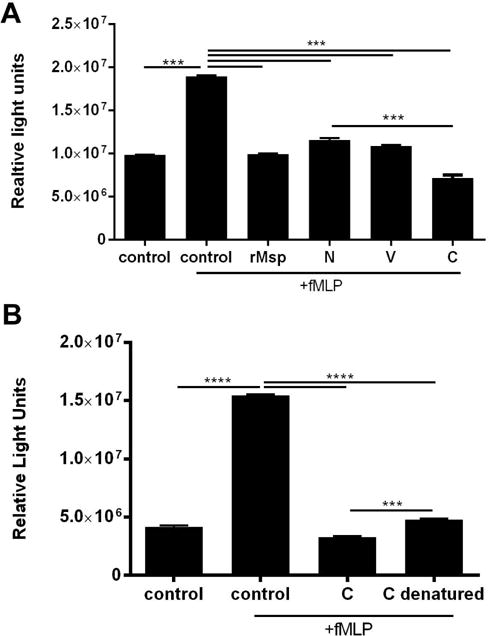
Truncations of the Msp protein, the N-, V- and C- regions (Fig 2), have been studied to identify regions which mediate extracellular substrate binding54. To determine the most active region of Msp that inhibits chemotaxis, we purified these recombinant Msp protein regions and full-length recombinant Msp as described54. Neutrophils were incubated with recombinant Msp proteins or native Msp complex (30 μg/ml) for 30 minutes prior to exposure to fMLP to promote chemotaxis (Fig 3). In a short-term Zigmond chamber chemotaxis assay, neutrophils were exposed to an fMLP gradient for 15 minutes to allow migration. Pre-treatment with the native Msp trimer complex has been shown to impair neutrophil chemotaxis46,48. We show that exposure to recombinant full-length Msp monomer also significantly decreased chemotaxis compared to cells stimulated with fMLP alone and that this protein was as equally effective as the native Msp in this regard (Fig 3A). Cells exposed to the V- and N- regions did not migrate significantly differently than cells stimulated with fMLP alone, but treatment of cells with the C- terminal region resulted in a significant reduction in migration, with cells only moving 4.7 μm compared to 14.4 μm. Our results here indicate that rMsp and the C- terminal protein truncation are similarly effective as the native Msp complex at impeding chemotaxis.
Fig. 2.
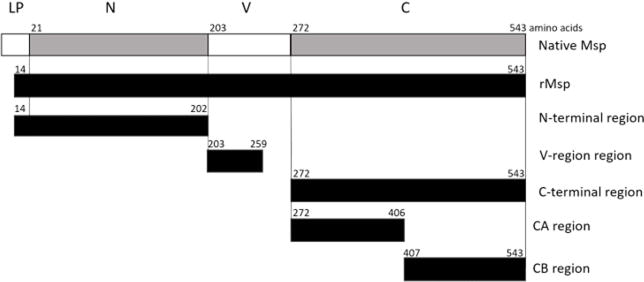
Diagram of the Msp protein from T. denticola strain 35405 and recombinant protein regions. Native Msp: leading peptide (LP) 20 aa, N-terminal region (N) 182 aa, variable region (V) 69 aa, C-terminal region (C) 272 aa. Recombinant Protein Regions: rMsp 530 aa, N-terminal 189 aa, V-region 57 aa, C-terminal 272 aa, CA 135 aa, CB 137 aa. All recombinant proteins also have a 6X histidine tag. Black indicates recombinant truncations. Diagram modified from original version54.
Fig. 3.
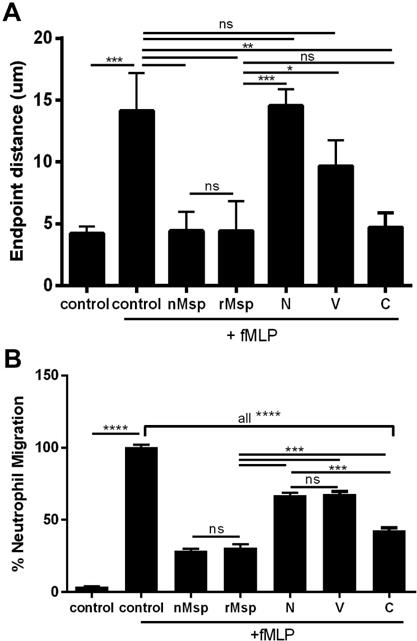
TheC-terminal region of Msp most strongly impairs neutrophil chemotaxis in response to fMLP. A) Murine neutrophils were treated with rMsp or N, V, and C truncations for 30 min followed by exposure to fMLP in a Zigmond chamber for 15 min compared with neutrophils alone or neutrophils +fMLP (controls). The distance traveled by the cells was measured with ImageJ and compared to control +fMLP, with cells exposed to the C protein moving less than the other truncations and comparably to the nMsp and rMsp B) Neutrophils were treated the same as described in A but exposed to fMLP for 1 hour in a transwell chemotaxis assay. Cells that migrated across the membrane were fixed, stained with crystal violet, and counted. Results were normalized and compared to the control +fMLP. Fewer cells treated with nMsp, rMsp, and the C truncation migrated toward fMLP. Graphs represents mean ± SEM of 3 independent experiments (* P < 0.05, ** P < 0.01, *** P < 0.0001, **** P < 0.0001 by unpaired t test).
We next performed transwell chemotaxis assays in which neutrophils were incubated with proteins (30 μg/ml) followed by exposure to fMLP for 1 hour, allowing cells to traverse the membrane (Fig 3B). Exposure to either native Msp complex or rMsp monomer significantly reduced chemotaxis by ~70% compared to control cells, while the N- and V- regions were less able to, only reducing neutrophil chemotaxis by ~33% compared to fMLP stimulation alone. Similar to the results of the Zigmond assay (Fig 3A), treatment with the C- terminal region had the greatest impact of all protein truncations tested, significantly reducing neutrophil chemotaxis by almost 60% (Fig 3B). Due to differences in the molecular weight of the recombinant proteins used in this study, differences in the molar dose must be considered. The majority of our experiments were performed using a concentration of 30 ug/ml for each protein, which equates to differing molarities for each protein. However when transwell chemotaxis assays were performed with all proteins at equal molarities; the overall trend in their ability to impair neutrophil chemotaxis was similar to results obtained using all proteins at 30ug/ml. Overall, the effect of each protein occurs in a molarity dose dependent manner, yet individual effects of each protein region remains the same (data not shown). To confirm involvement of a specific Msp conformational interaction, we performed this assay with proteins denatured by boiling, which restored chemotaxis in response to fMLP stimulation (data not shown). Compared to directed migration towards fMLP characteristic of chemotaxis, there is also a low level of random neutrophil movement observed in migration experiments, therefore we also examined the effect of nMsp exposure on unstimulated neutrophil movement. Fewer cells migrated through the membrane when incubated with nMsp but unstimulated by fMLP compared to untreated cells (Sup Fig 2).
C- terminal specific antibodies block Msp impairment of neutrophils
It has been shown that antibodies directed towards the entire Msp protein are able to prevent Msp-mediated effects53,60,61. We were therefore interested to see if it was possible to block the active region of Msp responsible for impairment of neutrophil chemotaxis, as this would indicate the requirement of specific regions in this interaction. The native Msp complex was incubated with polyclonal antibodies directed towards either the N- terminal or C- terminal region of Msp54 at a 1:50 dilution for 1 hour before exposure to neutrophils and transwell chemotaxis assays as described above (Fig 4). There was no difference in chemotaxis of cells incubated with native Msp alone and those incubated with native Msp pretreated with the antibody specific for the N- terminal region (α-MspN). Both conditions resulted in strong inhibition of chemotaxis, indicating that this antibody does not bind the region of Msp that modulates neutrophil chemotaxis. However, pretreatment of native Msp with a C- terminal antibody (α-MspC), partially blocked inhibition of neutrophil chemotaxis compared to cells stimulated with fMLP alone. This result indicates the α-MspC antibody binds to the region of Msp that interacts with neutrophils to mediate inhibition of neutrophil chemotaxis. Similar results were also obtained with N- terminal and C- terminal antibodies from a second research group (data not shown)50. Strikingly, our results obtained using multiple antibodies provide further evidence supporting the notion that the C- terminal region is the active region of the Msp protein involved in inhibition of neutrophil chemotaxis.
Fig. 4.
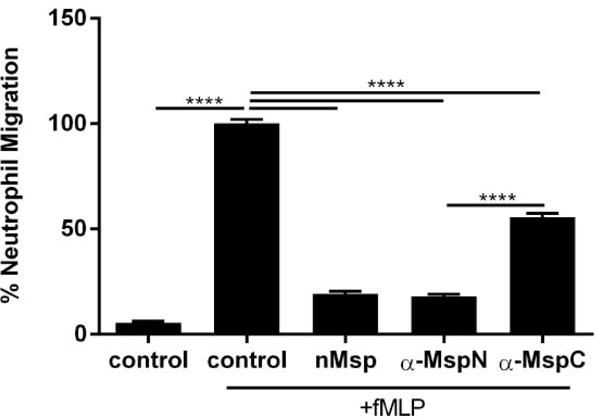
Incubation of Msp with a C-terminal specific antibody blocks the Msp induced inhibition of neutrophil chemotaxis. Treatment of neutrophils with native Msp that was pre-incubated with 1:50 dilutions of antibodies specific to the N or C regions followed by a transwell chemotaxis assay. The numbers of migrated neutrophils incubated with antibody treated nMsp were compared to neutrophils treated with nMsp alone. Untreated neutrophils served as the negative control and untreated neutrophils exposed to fMLP served as the positive control to which the results were normalized and compared. Graphs represents mean ± SEM of 3 independent experiments (**** P < 0.0001 by unpaired t test).
The C- terminal region of Msp alters Rac1 activation
Rac1 is a key signaling component driving neutrophil chemotaxis24. Previously, we have shown that nMsp from T. denticola strain 35405 prevents Rac1 activation in neutrophils downstream of fMLP stimulation45,48. Our data presented within Figures 3 and 4 indicate that the C- terminal region of Msp most strongly impairs neutrophil chemotaxis, therefore we were wanted to determine if this inhibition occurs through a similar signaling mechanism to that of intact Msp. Neutrophils were incubated with the protein truncations for 30 minutes and stimulated with fMLP for 1 minute prior to lysis. Rac1 activation was assessed using a Rac1 GLISA assay (Fig 5). The rMsp monomer protein reduced Rac1 activation in neutrophils almost 2-fold compared to control cells stimulated with fMLP alone (Fig 5A). The effect of treatment with the N- and V- regions was also very similar to the whole rMsp protein. As we anticipated, the C- region was the most effective of all the regions tested at reducing Rac1 activity and was also significantly more effective than the N- region (Fig 5A). To further confirm a conformational requirement for activity of the C- terminal region protein, we denatured the protein by boiling, which partially restored the Rac1 signaling response to fMLP (Fig 5B). Together, our data indicates that similar to intact native Msp complex, the rMsp monomer and the C-terminal region of Msp prevent Rac1 activation downstream of fMLP stimulation and suggests that protein confirmation and amino acid composition are important components for the effectiveness of these proteins.
Fig. 5.
The C- terminal region of Msp inhibits Rac1 activation downstream of fMLP stimulation. Neutrophils alone or stimulated with fMLP were experimental controls for all. A) Neutrophils were treated with Msp proteins or regional truncated proteins (30 μg/ml) for 30 minutes followed by lysis. Rac1 activity in the lysates was measured as relative light units (RLUs) using a G-LISA assay kit. Treatment of neutrophils with the C region yielded the strongest inhibition in Rac1 activity. B) Denaturing the C- terminal protein by boiling for 10 minutes prior to neutrophil treatment caused less inhibition of Rac1 activity in response to fMLP. Graphs represents mean ± SEM of 3 independent experiments (*** P < 0.001 and **** P < 0.0001 by unpaired t test).
The C- terminal region of Msp prevents activation of Akt
The Rac1-Akt pathway is important for proper chemotaxis in neutrophils, with the phosphorylation of Akt as an important downstream component of Rac1 activation63. Akt can also be considered an indicator of PIP3 generation, as PIP3 binds Akt and recruits it to the plasma membrane where Akt is activated by phosphorylation. Msp prevents correct spatial localization of PIP3 and Akt at the plasma membrane together with Akt phosphorylation in response to fMLP46,48 As we have demonstrated that the C- terminal region of Msp prevents Rac1 activation in neutrophils (Fig 5), we wanted to further assess Akt activation. Activation of Akt was assessed by measuring Akt phosphorylation in cell lysates following incubation with intact Msp or Msp regions and stimulation with fMLP by western blot (Fig 6). Stimulation of neutrophils with fMLP alone resulted in increased Akt phosphorylation while treatment of cells with rMsp monomer prior to fMLP stimulation prevented Akt phosphorylation, similar to our previous report for pre-treatment with native Msp complex46. Concurrent with our data reported herein for Rac1 activation (Fig 5), the C- terminal truncation was most effective at preventing Akt phosphorylation while the N-terminal protein was less so (Fig 6). Together these observations indicate that rMsp monomer is highly effective at preventing Akt activation, and the most important residues to influence this action may be located within the C- terminal region.
Fig. 6.
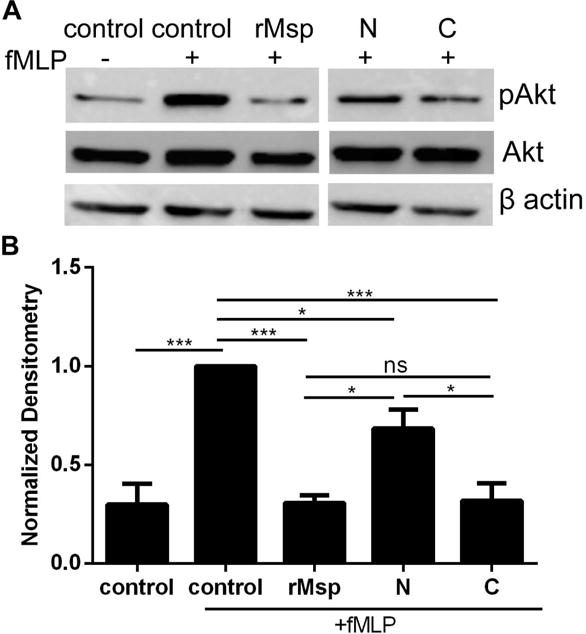
The C-terminal region of Msp impair Akt activation in response to fMLP. Neutrophils were treated with rMsp protein, N and C regions (30 μg/ml) for 30 minutes followed by stimulation with fMLP for 1 minute and lysis by boiling. Neutrophils alone or stimulated with fMLP were experimental controls. Phosphorylation of Akt was assessed as measure of Akt activation. A) The C region is more effective than the N region at inhibiting neutrophil phosphorylation of Akt. Representative western blots of cells lysates were probed with α-pAkt and α-total Akt, with α-β-actin as an additional loading control. B) Densitometry analysis of western blots was performed with ImageJ comparing pAkt to Akt. Results were normalized to Akt with the control + fMLP set to 1. Graphs represents mean ± SEM of 3 independent experiments (* P < 0.05, ** P < 0.01, *** P < 0.001 by unpaired t test).
The C- terminal region of Msp increases lipid phosphatase activity
PTEN is an important regulator of neutrophil chemotaxis, with increased PTEN activity negatively regulating the PI3 kinase pathway responsible for activating chemotaxis64. We were interested to see if the reduction in neutrophil chemotaxis observed after exposure to Msp regional polypeptides involved modulation of PIP3 phosphatase activity, similar to that of native Msp46. To investigate this, neutrophils were treated with Msp proteins followed by a modified malachite green assay to measure the amount of free phosphate released from a synthetic PIP3 substrate as an indirect measure of phosphatase activity. In this experimental setup, we presume we are primarily measuring PTEN activity, as PTEN is most efficient at dephosphorylating PIP3 in vitro65 and using both a PTEN chemical inhibitor and specific PTEN immunoprecipitation assay in combination with malachite green assay, we have previously shown that PTEN activity is increased by Msp in neutrophils46 As expected, stimulation of neutrophils with fMLP decreased free phosphate release from PIP3, indicating reduced phosphatase activity (Fig 7). Exposure of neutrophils to rMsp increased the release of free phosphate from PIP3, similar to our observations for nMsp46. Furthermore, treatment of neutrophils with the N- terminal region did not increase the amount of free phosphate released, while treatment with the C- terminal region resulted in almost 3 fold more free phosphate released compared to the control (Fig 7). Overall, these results indicate that rMsp and the C- terminal region of Msp induce PIP3 phosphatase activity in neutrophils. This provides further evidence for a similar mechanism of action to that of native Msp and is consistent with our results herein for neutrophil chemotaxis.
Fig. 7.
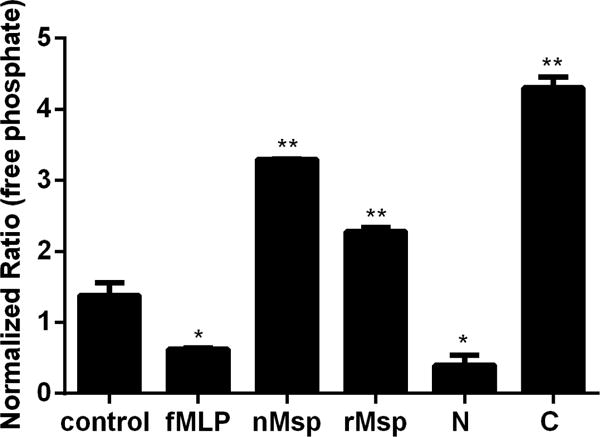
The C-terminal region of Msp increases PIP3 lipid phosphatase activity. Neutrophils were pretreated with nMsp, rMsp, N, and C or truncated Msp followed by assessment of phosphate release from a synthetic PIP3 substrate using a malachite green assay. Neutrophils alone and neutrophils stimulate with fMLP were experimental controls. Results were compared to the control alone, with the C region causing the most phosphate release. Graph represents the mean ± SEM of 1 of 3 independent experiments all showing the same results (* P<0.05, ** P<0.01 unpaired t test).
The first half of the C- terminal region displays neutrophil inhibitory action
Thus far, treatment with the C- terminal region has had the strongest impact of the three regions tested on both reducing neutrophil chemotaxis and limiting the signal pathway regulating this function. We were interested to see if we could identify a more specific region of the C- terminal region of Msp that alters neutrophil response. To preliminary narrow down a target region, we further cloned and expressed two truncations of this C- terminal region, the first half termed CA encompassing the first 135 amino acids (14.3kDa) and the second half termed CB encompassing the later 137 amino acids (15.1 kDa) (Fig 1). Treatment with both of these truncations significantly reduced neutrophil chemotaxis, with the CA region being the most effective of the two (Fig 8A). Both regions also limited Rac1 activation (Fig 8B) and Akt phosphorylation (Fig 8C and D), and while the data fits the trend of CA being the more inhibitory fragment, there was not a significant difference between the two regions in these assays. These results support our previous data and hypothesis that the C- terminal region contains regions that are highly effective at inhibiting neutrophil function but additional experiments are required to determine the most effective region conclusively, including the analysis of overlapping fragments and additional experiments to analyze neutrophil response to smaller truncations of the Msp protein.
Fig. 8.
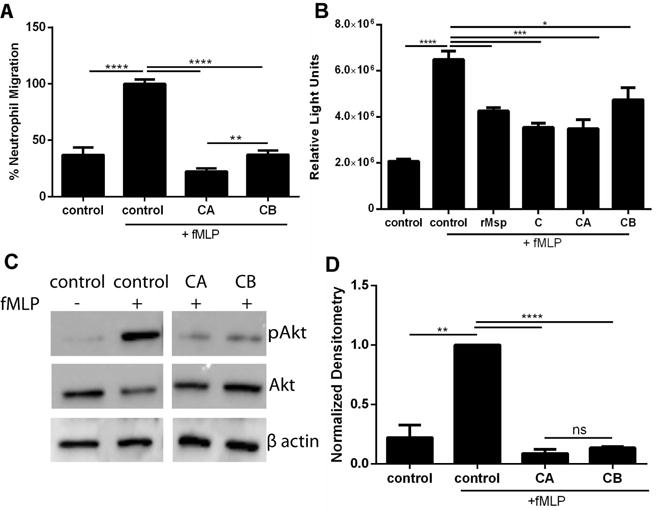
The CA region inhibits neutrophil function. A) Treatment of neutrophils with CA and CB protein truncations followed by transwell chemotaxis assay as described in Fig 3. Results were normalized and compared to the control +fMLP. Treatment with CA results in the strongest chemotaxis inhibition. B) Neutrophils were treated with CA and CB protein truncations (30 μg/ml) for 30 minutes followed by lysis. Rac1 activity in the lysates was measured as relative light units (RLUs) using a G-LISA assay kit. Treatment of neutrophils with both of these region yielded strong inhibition in Rac1 activity. C) Treatment of neutrophils with the CA and CB regions inhibit Akt phosphorylation. Representative western blots of cells lysates were probed with α-pAkt and α-total Akt, with α-β-actin as an additional loading control as described in Fig 6. D) Densitometry analysis of western blots was performed with ImageJ comparing pAkt to Akt. Results were normalized to Akt with the control + fMLP set to 1. Graphs represents mean ± SEM of 3 independent experiments (* P < 0.05 ** P < 0.01 *** P < 0.001 **** P < 0.0001 by unpaired t test).
Sequence variation in Msp alters the impairment of neutrophil chemotaxis
It is known that there is sequence variability between strains of T. denticola, including that of Msp55,56. We were interested to see if this sequence variation in the Msp protein would be enough to have an altered functional effect on chemotaxis. To investigate this, we cloned and expressed the Msp protein from T. denticola strain OTK, which differs significantly in the sequence throughout the protein, including in the C- terminal region55. The OTK rMsp protein reduced chemotaxis significantly more than both 35405 rMsp monomer and native Msp complex (Fig 9A) and also resulted in a stronger inhibition of Akt phosphorylation (Fig 9B and C). Together, these results indicate that rMsp from strain OTK has a stronger effect on neutrophil function. As OTK is known to have significant sequence differences throughout the protein including the C- terminal region compared to strain 35405, this result could be expected and further supports the notion of protein sequence variation and topology affecting pathogenic interaction with neutrophils.
Fig. 9.
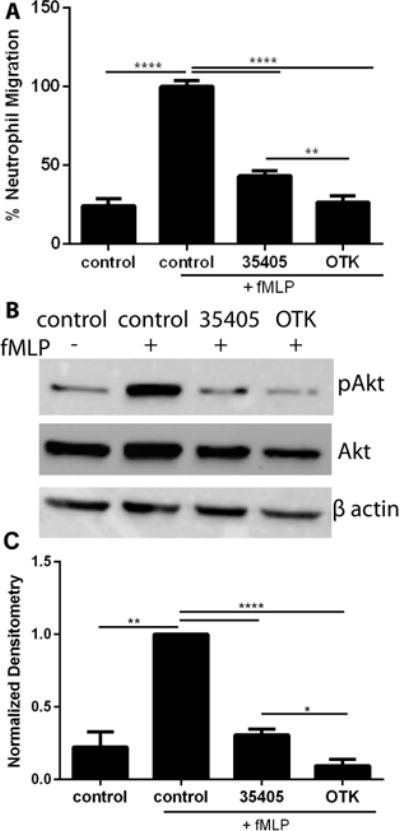
Strain variation alters the inhibitory effect of Msp on neutrophil function. A) Treatment of neutrophils with rMsp proteins from strain 35405 and OTK, with neutrophils alone or stimulated with fMLP as controls, followed by a transwell chemotaxis assay as described in Fig 3. B) Treatment of neutrophils with rMsp from 35405 and OTK followed by lysis and assessment of pAkt levels as described in Fig 6. Representative western blots of cells lysates were probed with α-pAkt and α-total Akt, with α-β-actin as an additional loading control. C) Densitometry analysis was performed with ImageJ comparing pAkt to Akt. Results were normalized and compared to Akt with the control + fMLP set to 1. Graphs represents mean ± SEM of 3 independent experiments (** P < 0.01 and **** P < 0.0001 by unpaired t test).
DISCUSSION
Interactions of spirochetes with both resident cells and infiltrating immune cells during periodontal disease are well known to occur in the oral cavity. Surface proteins are prominent virulence factors of many pathogenic bacteria and play key roles in interactions with both host cells and other organisms. T. denticola is known to modulate neutrophil function, likely dampening the response of one of the key innate immune cells in the periodontal environment. T. denticola has been reported to be resistant to some antimicrobial peptides, induce less release of molecules from neutrophil granules, induce a differential cytokine response compared to other oral bacteria and possibly evade or delay neutrophil phagocytosis66–70. Furthermore, Msp is able to impair neutrophil chemotaxis through perturbation of actin assembly, calcium flux and selective Rac1 inhibition47–49. Moreover, we have reported that Msp impairs the local balance of PIP3 in the cell, through activation of PTEN together with concomitant inhibition of PI3K activation46. Upset of this crucial PIP3 balance required for the establishment of neutrophil directionality leads to downstream impairment of neutrophil chemotaxis. Here we provide evidence for identification of active regions of Msp involved in inhibition of neutrophil chemotaxis.
We speculate that Msp in intact bacteria may interact directly with host cells to mediate its effects or may be shed from the spirochete in some fashion. While direct secretion of Msp by T. denticola has not been reported to date, Msp can be released as a component of bacterial outer membrane vesicles58,71 which may represent a novel way for Msp to interact with host cells both locally in the gingival tissue and periodontal pocket as well as at distant sites. Msp may mediate its effect on intracellular components through modulation of host cell signaling pathways by external engagement upon contact with the plasma membrane. Msp appears to remain extracellular or in close association with the plasma membrane61 with host cell responses observed rapidly within minutes of exposure48,49,61. While specific interacting partners or receptors for Msp on host cells remain elusive, it has been reported that Msp interacts with a 65-kDa surface protein in HeLa cells72.
Native Msp in T. denticola has long been thought to exist as a membrane spanning β-barrel protein, which forms a trimeric complex associated with the protease dentilisin, in the bacterial membrane51,73,74, but the exact topology and conformation of Msp is controversial. More recently, it has been reported that Msp can exist as both distinct membrane associated and periplasmic trimer complex forms and these forms may have distinct physical properties50. Recombinant Msp proteins have been reported to display porin activity and binding to a range of extracellular matrix components such as laminin, fibronectin, collagen type I and fibrinogen50,53,54,74. Here we demonstrate that recombinant Msp in the monomer form is also able to impair neutrophil migration (Fig 3) through upset of the crucial PIP3 balance leading to prevention of Rac1 signaling (Fig 5) and downstream Akt activation (Fig 6), similarly to the native Msp trimer complex, expanding the functional knowledge and importance of the Msp protein.
Despite a wealth of studies investigating the impact of Msp on host cells, knowledge of protein regions or domains involved in specific biological interactions is limited. Regions in the N- terminal half of Msp have been reported to mediate binding to extracellular matrix components54 while the C- terminal domain has been reported to possess pore-forming capability50. Localization of Msp within intact spirochetes is controversial as it has been reported that Msp is surface exposed53,57, while others have reported limited surface exposure58. Initial studies examining localization of Msp in intact spirochetes using specific Msp regional antibody labeling approaches suggested that only antibodies directed towards the N- terminal and central V- region (~ first 260 amino acids) reacted with surface exposed epitopes54 however more recently, a study using antibodies towards the C- and N- terminal domains of Msp has demonstrated that only the C- terminal domain (amino acids 332 – 543) is surface exposed50. These discrepancies may be due to differences in antibody specificity, antibody and protein accessibility and the techniques used to examine localization. Our data herein comparing neutrophil chemotaxis following exposure to wild type or Msp mutant strains (Fig 1) indicates that Msp in the native state with presumed surface exposure in intact organisms plays a significant role in modulating neutrophil chemotaxis. Likewise, we have previously reported that a strain lacking Msp was less able to alter the host actin cytoskeleton of fibroblasts45 and epithelial cell migration is increased following exposure to a Msp mutant strain75.
Furthermore, our data presented herein indicates that the C- terminal region of Msp, is the most inhibitory region preventing neutrophil chemotaxis downstream of fMLP stimulation (Fig 3). We have shown that polyclonal antisera from two different research groups directed towards similar yet different recombinant C- terminal protein regions are able to block neutrophil chemotaxis inhibitory regions on Msp, while N- terminal antibodies demonstrate minimal effect (Fig 4). As both our truncated C- terminal Msp regions were able to decrease neutrophil chemotaxis, Rac1 activation and Akt phosphorylation (Fig 8), it is possible a region spanning these two regions will contain the most important peptide region. Additional studies using smaller overlapping truncations will be required to definitively identify the most important region of Msp to alter neutrophil function. Overall, our data also supports the notion that the C- terminal region of Msp may be surface exposed.
Msp requires specific interactions with host cells to mediate its action, but these actions can be blocked by Msp specific antibodies as we demonstrated when the C- terminal antibody partially blocked the activity of Msp, allowing for chemotaxis to occur downstream of fMLP stimulation (Fig 4). Likewise, Msp-induced calcium transients and inhibition of collagen binding in fibroblasts can be reversed by pre-treatment with antiserum to the native Msp complex60,61. Inhibition studies have also revealed that exposure to either a native Msp or recombinant Msp antisera prevents adherence of T. denticola to periodontal ligament epithelial cells and Msp regional antibodies can inhibit adhesion to immobilized host extracellular matrix proteins53,54. Overall, our data indicates that the C- terminal domain of Msp is crucial for interactions with neutrophils in vitro and could be expected to be surface exposed. Surface exposed regions of bacterial proteins play key roles in interactions with host cells and represent potential therapeutic targets. For example, development of monoclonal antibodies recognizing active Msp regions may provide a useful therapeutic tool. Alternately, development of complementary peptides to active exposed regions could hold promise, as this approach has been used to develop selective inhibitory peptides to other bacterial porins76
The msp gene displays interstrain variability55 and forms three main phylogenetic lineages defined by differing DNA sequence and antigenic properties. Commonly studied wild type strains 35405, 33520 and OTK represent members of the three distinct Msp groups. While the msp coding regions of strains 35405 and 33520 share high homology with 94.6% identity, the msp of strain OTK shows much lower homology to 35405 with only 50.6% identity overall. With regards to specific areas of variation between 35405 and OTK msp, the 5′ end contains the highest level of homology at 80% while the 3′ ends have only 64% identity55. Each of these Msp groups is also serologically distinct, as antibodies raised against Msp from the strain 35405 react weakly with strain 33520 and do not react with the strain OTK55, possibly through variation in the central region of the amino acid sequence. Molecular methods have also been used to examine msp gene sequences from a variety of isolated strains as well as directly from subgingivial plaque samples where it was reported that these sequences display high similarity within the three established Msp groups56,77,78. Interestingly, serum antibodies from human subjects with moderate to severe gingivitis are also able to discriminate different T. denticola Msp protein groups79.
Due to their differing nucleotide sequence, T. denticola Msp proteins vary in size from approximately 53 kDa to 64 kDa55,80. Msp from the strain OTK has significantly different nucleotide and amino acid sequence compared to strain 35405, including the C- terminal region, and displays a larger molecular weight size of 62 kDa55. To our knowledge, there is little reported information regarding the pathogenic potential of the strain OTK, or Msp of this strain, interacting with host cells. Neither are there any functional comparisons of Msp proteins from different strains, Msp groups or serotypes. While little functional comparison between OTK, 35405 and other strains has been reported, it has been shown that strain OTK displays less dentilisin protease activity81. Here we demonstrate that OTK Msp appears to be able to impair neutrophil migration through impairment of PIP3 levels more effectively than 35405 Msp, measured indirectly by downstream Akt activation (Fig 9). We postulate that this could be the result of direct sequence differences in potential crucial active regions within the C- terminal region of Msp, which we predict plays an active role in neutrophil interaction. Alternatively, the difference in amino acids could alter the topology and structure of the protein. These sequence variations may reflect functional differences in binding to host proteins or other protein interactions. Analysis of the predicted secondary structure of Msp OTK is similar to 35405 with multiple β-sheets forming a β barrel structure55, however amino acid variation may result in extension of surface exposed loops or structures (data not shown), which we speculate may reflect differing interactions with host cell molecules. Variability in Msp amino acid sequences could reflect evolution of periodontal disease and pathogenic potential through immune-driven antigenic variation, which is a common pathogenic feature of many spirochetes. Amino acid variation detected in a small number of clinical specimens led to antigenic modification of predicted surface exposed portions of Msp, primarily due to substitution of amino acids with diverse functionality such as polar vs non-polar or vice versa56. Interestingly, OTK Msp has significantly more polar amino acid residues compared to other Msps55, therefore may be reflective of the unique properties of this protein and its role in immune evasion.
While the periodontal clinical parameters of the subject that strain OTK was originally isolated from is not known, clonal abundance of the OTK msp genotype is significantly associated with periodontal disease78, therefore this strain may have unique pathogenic potential. Further analysis and comparison of the functionality of different Msp proteins and pathogenic variability between different strains will aid in further understanding the important contribution of this virulence factor. While topology and physiochemical properties of Msp from 35045 have been reported50, knowledge of these properties for Msp proteins from other strains is not known. More detailed structural, biophysical and functional studies of both intact Msp and regional peptides of diverse Msp proteins, including from the strain OTK, are ongoing.
Similar to native Msp, which impairs the orchestration of neutrophil chemotaxis by upset of the cellular PIP3 balance through activation of PTEN leading to inhibition of Rac1 activation46, the C- terminal region of Msp appears to use a similar mode of action. While all the regional truncated proteins were able to impair neutrophil chemotaxis in response to fMLP to some degree in both types of chemotaxis assays tested and prevent Rac1 activation, the C- terminal region was significantly more effective (Fig 5). A contribution for the N- and V- regional sequences in these processes cannot be completely ruled out by our studies, but the fact that only the C- terminal protein was able to increase PIP3 phosphatase activity (Fig 7) and that Msp-mediated impairment of neutrophil chemotaxis can be reversed by exposure to a C-terminal antibody indicates that this region contains crucial sequences involved in neutrophil interaction.
While denaturation of the C-terminal protein by boiling significantly increased neutrophil Rac1 activation compared to the non-heated C-terminal protein, this treatment was only partially able to restore Rac1 activation (~ 33%) when compared to control fMLP stimulation alone (Figure 5). While heat denaturation of native Msp complex has been reported to almost completely restore host cell interactions such as chemotaxis in neutrophils48,49, collagen binding and calcium signaling responses in fibroblasts60,61 and adherence to host cells74; heat denaturation of the native Msp complex is only able to partially restore Rac1 activation in neutrophils48 to similar levels as the recombinant C-terminal region. Overall, this indicates that heat-sensitive conformational specificity is important in the specific interaction of Msp with neutrophils, however the ability of the heat denatured C-terminal protein to retain inhibitory action towards neutrophils also suggests that primary amino acid sequence specificity may also be crucial in these interactions. Specific amino acid sequences within a region of a Klebsiella pneumoniae porin are involved in interaction with complement proteins, while specific amino acids of a Haemophilus influenzae porin are key to activating MAPK pathways and cytokine release82. Furthermore, spirochete outer membrane proteins including Msp may have unique β-barrel structure and composition, with differing heat modifiability properties compared to classical outer membrane proteins50, therefore we can speculate that less heat-sensitive linear motifs or unstructured protein regions83 may contribute to specific Msp-neutrophil interactions.
While we have previously shown using specific PTEN inhibitor and immunoprecipitation assays that Msp specifically activates PTEN to disrupt the local PIP3 concentration to impair neutrophil chemotaxis46, in this report we cannot rule out the contribution of other lipid phosphatases to the PIP3 phosphatase activity changes we observe. For example, the 5-phosphatase SHIP-1 can also dephosphorylate PIP3 and is known to coordinate with PTEN to control PIP3 signaling and chemotaxis, particularly during adhesion31. Future work will examine other phospholipid metabolism pathways in neutrophils which are manipulated by T. denticola.
In summary, this study has led to the novel observations that 1) sequences within the C- terminus of Msp contain important regions responsible for inhibiting chemotaxis, 2) this interaction can be blocked only with antibodies specific to the C- terminal region, 3) C- terminal Msp impairs PIP3 levels and Rac1 activation similar to the native Msp complex and 4) msp sequence variation among strains may contribute to differences in pathogenic properties. Further experiments are still required to identify the specific essential region of Msp that alters neutrophil functionality, including assessment of overlapping protein regions and analysis of smaller protein regions. Knowledge of how T. denticola, an understudied oral pathogen, uses specific virulence factors to manipulate neutrophil function to evade the immune response is a crucial first step in development of potential therapeutics to improve oral health.
Supplementary Material
Sup. Fig. 1 Purified His-tagged recombinant proteins. Proteins were separated by 10% SDS-PAGE and stained with Coommassie Blue to show the purity of the purified proteins. From left to right, rMsp (53 kDa), N (23 kDa), V (8 kDa), C (32 kDa), CA (14 kDa), CB (15 kDa) from strain 35405 and rMsp from strain OTK (62 kDa). Molecular weight sizes in kDa are noted on the left.
Sup. Fig. 2 Native Msp inhibits stimulated and resting neutrophil movement. Neutrophils were treated with nMsp for 30 minutes then placed in chemotaxis chamber with and without fMLP for 1 hour. Neutrophils untreated with nMsp were stimulated or unstimulated with fMLP were experimental controls. Cells migrated through the membrane were stained and counted as described in Fig 1. The number of migrated neutrophils untreated (control) and stimulated with fMLP for 1 hour were compared to neutrophils treated with nMsp for 30 minutes and to cells treated with both nMsp and fMLP. Cells treated with nMsp alone or nMsp stimulated with fMLP show significant inhibition of chemotaxis compared to both the untreated cells (control) or those treated with fMLP. Graph represents the mean ± SEM of 2 independent experiments (** P<0.01, ****P<0.0001 by unpaired t test).
Acknowledgments
This work was supported by operating funds (MBV) from NIH NIDCR (R03DE024769) and The University at Buffalo School of Dental Medicine Funds and training support from NIH (MMJ) (5T32DE023526-03).
Footnotes
The authors have no conflict to report.
References
- 1.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8(7):481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 2.Eke PI, Dye BA, Wei L, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86(5):611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21(3):172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann Periodontol. 2003;8(1):38–53. doi: 10.1902/annals.2003.8.1.38. [DOI] [PubMed] [Google Scholar]
- 6.Leishman SJ, Do HL, Ford PJ. Cardiovascular disease and the role of oral bacteria. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v3402i3400.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preshaw PM, Alba AL, Herrera D, et al. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55(1):21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann Periodontol. 2003;8(1):54–69. doi: 10.1902/annals.2003.8.1.54. [DOI] [PubMed] [Google Scholar]
- 9.Saini R, Saini S, Sharma S. Periodontitis: A risk factor to respiratory diseases. Lung India. 2010;27(3):189–189. doi: 10.4103/0970-2113.68313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narikiyo M, Tanabe C, Yamada Y, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95(7):569–574. doi: 10.1111/j.1349-7006.2004.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng X-T, Xia L-Y, Zhang Y-G, Li S, Leng W-D, Kwong JSW. Periodontal disease and incident lung cancer risk: a meta-analysis of cohort studies. J Periodontol. 2016;87(10):1158–1164. doi: 10.1902/jop.2016.150597. [DOI] [PubMed] [Google Scholar]
- 12.Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10(3):e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6(3):173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 14.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- 15.Ryder MI. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol 2000. 2010;53:124–137. doi: 10.1111/j.1600-0757.2009.00327.x. [DOI] [PubMed] [Google Scholar]
- 16.Landzberg M, Doering H, Aboodi GM, Tenenbaum HC, Glogauer M. Quantifying oral inflammatory load: oral neutrophil counts in periodontal health and disease. J Periodontal Res. 2015;50(3):330–336. doi: 10.1111/jre.12211. [DOI] [PubMed] [Google Scholar]
- 17.Hajishengallis E, Hajishengallis G. Neutrophil homeostasis and periodontal health in children and adults. J Dent Res. 2014;93(3):231–237. doi: 10.1177/0022034513507956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuzzi PA, Lokuta MA, Huttenlocher A. Analysis of neutrophil chemotaxis. In: Coutts AS, editor. Adhesion Protein Protocols. Totowa, NJ: Humana Press; 2007. pp. 23–35. [DOI] [PubMed] [Google Scholar]
- 19.Kumar RS, Prakash S. Impaired neutrophil and monocyte chemotaxis in chronic and aggressive periodontitis and effects of periodontal therapy. Indian J Dent Res. 2012;23(1):69–74. doi: 10.4103/0970-9290.99042. [DOI] [PubMed] [Google Scholar]
- 20.Roberts HM, Ling MR, Insall R, et al. Impaired neutrophil directional chemotactic accuracy in chronic periodontitis patients. J Clin Periodontol. 2015;42(1):1–11. doi: 10.1111/jcpe.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenteany G, Glogauer M. Cytoskeletal remodeling in leukocyte function. Curr Opin Hematol. 2004;11(1):15–24. doi: 10.1097/00062752-200401000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Wang F. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol. 2009;1(4):a002980. doi: 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuiper JW, Sun C, Magalhaes MA, Glogauer M. Rac regulates PtdInsP(3) signaling and the chemotactic compass through a redox-mediated feedback loop. Blood. 2011;118(23):6164–6171. doi: 10.1182/blood-2010-09-310383. [DOI] [PubMed] [Google Scholar]
- 24.Sun CX, Downey GP, Zhu F, Koh AL, Thang H, Glogauer M. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 2004;104(12):3758–3765. doi: 10.1182/blood-2004-03-0781. [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4(7):513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 26.Hawkins PT, Stephens LR, Suire S, Wilson M. PI3K signaling in neutrophils. Curr Top Microbiol Immunol. 2010;346:183–202. doi: 10.1007/82_2010_40. [DOI] [PubMed] [Google Scholar]
- 27.Damen JE, Liu L, Rosten P, et al. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc Natl Acad Sci U S A. 1996;93(4):1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 29.Hannigan M, Zhan L, Li Z, Ai Y, Wu D, Huang CK. Neutrophils lacking phosphoinositide 3-kinase gamma show loss of directionality during N-formyl-Met-Leu-Phe-induced chemotaxis. Proc Natl Acad Sci U S A. 2002;99(6):3603–3608. doi: 10.1073/pnas.052010699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heit B, Robbins SM, Downey CM, et al. PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat Immunol. 2008;9(7):743–752. doi: 10.1038/ni.1623. [DOI] [PubMed] [Google Scholar]
- 31.Mondal S, Subramanian KK, Sakai J, Bajrami B, Luo HR. Phosphoinositide lipid phosphatase SHIP1 and PTEN coordinate to regulate cell migration and adhesion. Mol Biol Cell. 2012;23(7):1219–1230. doi: 10.1091/mbc.E11-10-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian KK, Jia Y, Zhu D, et al. Tumor suppressor PTEN is a physiologic suppressor of chemoattractant-mediated neutrophil functions. Blood. 2007;109(9):4028–4037. doi: 10.1182/blood-2006-10-055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Prasad A, Jia Y, et al. Pretreatment with phosphatase and tensin homolog deleted on chromosome 10 (PTEN) inhibitor SF1670 augments the efficacy of granulocyte transfusion in a clinically relevant mouse model. Blood. 2011;117(24):6702–6713. doi: 10.1182/blood-2010-09-309864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183(12):3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 36.Dashper SG, Seers CA, Tan KH, Reynolds EC. Virulence factors of the oral spirochete Treponema denticola. J Dent Res. 2011;90(6):691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki N, Yoneda M, Hirofuji T. Mixed red-complex bacterial infection in periodontitis. Int J Dent. 2013;2013:587279. doi: 10.1155/2013/587279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol 2000. 2005;38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 39.Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge X, Rodriguez R, Trinh M, Gunsolley J, Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One. 2013;8(6):e65520. doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47(1):1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Veith PD, Dashper SG, O’Brien-Simpson NM, et al. Major proteins and antigens of Treponema denticola. Biochim Biophys Acta. 2009;1794(10):1421–1432. doi: 10.1016/j.bbapap.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Fenno JC. Treponema denticola interactions with host proteins. J Oral Microbiol. 2012;4 doi: 10.3402/jom.v4i0.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visser MB, Ellen RP. New insights into the emerging role of oral spirochaetes in periodontal disease. Clin Microbiol Infect. 2011;17(4):502–512. doi: 10.1111/j.1469-0691.2011.03460.x. [DOI] [PubMed] [Google Scholar]
- 45.Visser MB, Koh A, Glogauer M, Ellen RP. Treponema denticola major outer sheath protein induces actin assembly at free barbed ends by a PIP2-dependent uncapping mechanism in fibroblasts. PLoS One. 2011;6(8):e23736. doi: 10.1371/journal.pone.0023736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visser MB, Sun CX, Koh A, Ellen RP, Glogauer M. Treponema denticola major outer sheath protein impairs the cellular phosphoinositide balance that regulates neutrophil chemotaxis. PLoS One. 2013;8(6):e66209. doi: 10.1371/journal.pone.0066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amin M, Ho AC, Lin JY, Batista da Silva AP, Glogauer M, Ellen RP. Induction of de novo subcortical actin filament assembly by Treponema denticola major outer sheath protein. Infect Immun. 2004;72(6):3650–3654. doi: 10.1128/IAI.72.6.3650-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magalhaes MA, Sun CX, Glogauer M, Ellen RP. The major outer sheath protein of Treponema denticola selectively inhibits Rac1 activation in murine neutrophils. Cell Microbiol. 2008;10(2):344–354. doi: 10.1111/j.1462-5822.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- 49.Puthengady Thomas B, Sun CX, Bajenova E, Ellen RP, Glogauer M. Modulation of human neutrophil functions in vitro by Treponema denticola major outer sheath protein. Infect Immun. 2006;74(3):1954–1957. doi: 10.1128/IAI.74.3.1954-1957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anand A, Luthra A, Edmond ME, Ledoyt M, Caimano MJ, Radolf JD. The major outer sheath protein (Msp) of Treponema denticola has a bipartite domain architecture and exists as periplasmic and outer membrane-spanning conformers. J Bacteriol. 2013;195(9):2060–2071. doi: 10.1128/JB.00078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haapasalo M, Muller KH, Uitto VJ, Leung WK, McBride BC. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60(5):2058–2065. doi: 10.1128/iai.60.5.2058-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egli C, Leung WK, Muller KH, Hancock RE, McBride BC. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect Immun. 1993;61(5):1694–1699. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenno JC, Muller KH, McBride BC. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178(9):2489–2497. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73(5):2891–2898. doi: 10.1128/IAI.73.5.2891-2898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fenno JC, Wong GW, Hannam PM, Muller KH, Leung WK, McBride BC. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol. 1997;179(4):1082–1089. doi: 10.1128/jb.179.4.1082-1089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaibani P, Pellegrino MT, Rossini G, et al. The central region of the msp gene of Treponema denticola has sequence heterogeneity among clinical samples, obtained from patients with periodontitis. BMC Infect Dis. 2010;10:345–353. doi: 10.1186/1471-2334-10-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Godovikova V, Goetting-Minesky MP, Fenno JC. Composition and localization of Treponema denticola outer membrane complexes. Infect Immun. 2011;79(12):4868–4875. doi: 10.1128/IAI.05701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caimano MJ, Bourell KW, Bannister TD, Cox DL, Radolf JD. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect Immun. 1999;67(8):4072–4083. doi: 10.1128/iai.67.8.4072-4083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fenno JC, Wong GW, Hannam PM, McBride BC. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett. 1998;163(2):209–215. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 60.Batista da Silva AP, Lee W, Bajenova E, McCulloch CA, Ellen RP. The major outer sheath protein of Treponema denticola inhibits the binding step of collagen phagocytosis in fibroblasts. Cell Microbiol. 2004;6(5):485–498. doi: 10.1111/j.1462-5822.2004.00377.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, Ko KS, Kapus A, McCulloch CA, Ellen RP. A spirochete surface protein uncouples store-operated calcium channels in fibroblasts: a novel cytotoxic mechanism. J Biol Chem. 2001;276(25):23056–23064. doi: 10.1074/jbc.M011735200. [DOI] [PubMed] [Google Scholar]
- 62.Dawson JR, Ellen RP. Tip-oriented adherence of Treponema denticola to fibronectin. Infect Immun. 1990;58(12):3924–3928. doi: 10.1128/iai.58.12.3924-3928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, Tang H, Hay N, Xu J, Ye RD. Akt isoforms differentially regulate neutrophil functions. Blood. 2010;115(21):4237–4246. doi: 10.1182/blood-2009-11-255323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo HR, Mondal S. Molecular control of PtdIns(3,4,5)P3 signaling in neutrophils. EMBO Rep. 2015;16(2):149–163. doi: 10.15252/embr.201439466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spinelli L, Leslie NR. Assaying PTEN catalysis in vitro. Methods. 2015:77–78. 51–57. doi: 10.1016/j.ymeth.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Brissette CA, Simonson LG, Lukehart SA. Resistance to human beta-defensins is common among oral treponemes. Oral Microbiol Immunol. 2004;19(6):403–407. doi: 10.1111/j.1399-302x.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- 67.Shin J, Ji S, Choi Y. Ability of oral bacteria to induce tissue-destructive molecules from human neutrophils. Oral Dis. 2008;14(4):327–334. doi: 10.1111/j.1601-0825.2007.01382.x. [DOI] [PubMed] [Google Scholar]
- 68.Ding Y, Haapasalo M, Kerosuo E, Lounatmaa K, Kotiranta A, Sorsa T. Release and activation of human neutrophil matrix metallo- and serine proteinases during phagocytosis of Fusobacterium nucleatum, Porphyromonas gingivalis and Treponema denticola. J Clin Periodontol. 1997;24(4):237–248. doi: 10.1111/j.1600-051x.1997.tb01837.x. [DOI] [PubMed] [Google Scholar]
- 69.Hurlen B, Olsen I, Lingaas E, Midtvedt T. Neutrophil phagocytosis of Treponema denticola as indicated by extracellular release of lactoferrin. Acta Pathol Microbiol Immunol Scand B. 1984;92(3):171–173. doi: 10.1111/j.1699-0463.1984.tb02814.x. [DOI] [PubMed] [Google Scholar]
- 70.Olsen I, Lingaas E, Hurlen B, Midtvedt T. Scanning and transmission electron microscopy of the phagocytosis of Treponema denticola and Escherichia coli by human neutrophils in vitro. Scand J Dent Res. 1984;92(4):282–293. doi: 10.1111/j.1600-0722.1984.tb00893.x. [DOI] [PubMed] [Google Scholar]
- 71.Rosen G, Naor R, Rahamim E, Yishai R, Sela MN. Proteases of Treponema denticola outer sheath and extracellular vesicles. Infect Immun. 1995;63(10):3973–3979. doi: 10.1128/iai.63.10.3973-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathers DA, Leung WK, Fenno JC, Hong Y, McBride BC. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun. 1996;64(8):2904–2910. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abiko Y, Nagano K, Yoshida Y, Yoshimura F. Characterization of Treponema denticola mutants defective in the major antigenic proteins, Msp and TmpC. PLoS One. 2014;9(11):e113565. doi: 10.1371/journal.pone.0113565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto VJ, McBride BC. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66(5):1869–1877. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Inagaki S, Kimizuka R, Kokubu E, Saito A, Ishihara K. Treponema denticola invasion into human gingival epithelial cells. Microb Pathog. 2016;94:104–111. doi: 10.1016/j.micpath.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 76.Cantisani M, Vitiello M, Falanga A, Finamore E, Galdiero M, Galdiero S. Peptides complementary to the active loop of porin P2 from Haemophilus influenzae modulate its activity. Int J Nanomedicine. 2012;7:2361–2371. doi: 10.2147/IJN.S30467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fenno JC, Lee SY, Bayer CH, Ning Y. Treponema msp’s from subgingival plaque and cultivated strains. J Dent Res. 2001;80:S53. [Google Scholar]
- 78.Watt RM, Pun SY, You M. Treponeme major surface (msp) genotypes associated with periodontitis. IADR General Session. 2013 [Google Scholar]
- 79.Capone R, Wang HT, Ning Y, Sweier DG, Lopatin DE, Fenno JC. Human serum antibodies recognize Treponema denticola Msp and PrtP protease complex proteins. Oral Microbiol Immunol. 2008;23(2):165–169. doi: 10.1111/j.1399-302X.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- 80.Weinberg A, Holt SC. Chemical and biological activities of a 64-kilodalton outer sheath protein from Treponema denticola strains. J Bacteriol. 1991;173(21):6935–6947. doi: 10.1128/jb.173.21.6935-6947.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goetting-Minesky MP, Godovikova V, Li JJ, et al. Conservation and revised annotation of the Treponema denticola prcB-prcA-prtP locus encoding the dentilisin (CTLP) protease complex. Mol Oral Microbiol. 2013;28(3):181–191. doi: 10.1111/omi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galdiero S, Falanga A, Cantisani M, et al. Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr Protein Pept Sci. 2012;13(8):843–854. doi: 10.2174/138920312804871120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44(5):1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng SL, Chan EC. The routine isolation, growth, and maintenance of the intermediate-size anaerobic oral spirochetes from periodontal pockets. J Periodontal Res. 1983;18(4):362–368. doi: 10.1111/j.1600-0765.1983.tb00371.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup. Fig. 1 Purified His-tagged recombinant proteins. Proteins were separated by 10% SDS-PAGE and stained with Coommassie Blue to show the purity of the purified proteins. From left to right, rMsp (53 kDa), N (23 kDa), V (8 kDa), C (32 kDa), CA (14 kDa), CB (15 kDa) from strain 35405 and rMsp from strain OTK (62 kDa). Molecular weight sizes in kDa are noted on the left.
Sup. Fig. 2 Native Msp inhibits stimulated and resting neutrophil movement. Neutrophils were treated with nMsp for 30 minutes then placed in chemotaxis chamber with and without fMLP for 1 hour. Neutrophils untreated with nMsp were stimulated or unstimulated with fMLP were experimental controls. Cells migrated through the membrane were stained and counted as described in Fig 1. The number of migrated neutrophils untreated (control) and stimulated with fMLP for 1 hour were compared to neutrophils treated with nMsp for 30 minutes and to cells treated with both nMsp and fMLP. Cells treated with nMsp alone or nMsp stimulated with fMLP show significant inhibition of chemotaxis compared to both the untreated cells (control) or those treated with fMLP. Graph represents the mean ± SEM of 2 independent experiments (** P<0.01, ****P<0.0001 by unpaired t test).


