Summary
Background
How von Willebrand factor (VWF) senses and responds to shear flow remains unclear. In the absence of shear VWF or its fragments can be induced to bind spontaneously to platelet GPIbα.
Objectives
To elucidate the auto-inhibition mechanism of VWF.
Methods
Hydrogen-deuterium exchange (HDX) of two recombinant VWF fragments expressed from baby hamster kidney cells were measured and compared.
Results
The shortA1 protein contains VWF residues 1261–1472 and binds GPIbα with a significantly higher affinity than the longA1 protein that contains VWF residues 1238–1472. Both proteins contain the VWF A1 domain (residues 1272–1458). Many residues in longA1, particularly those in the N- and C-terminal sequences flanking the A1 domain, and in helix α1, loops α1β2 and β3α2, reported markedly reduced HDX than their counterparts in shortA1. The HDX-protected region in longA1 overlaps with the GPIbα-binding interface and is clustered with type 2B von Willebrand disease (VWD) mutations. Additional comparison with the HDX of denatured longA1 and ristocetin-bound longA1 indicates the N- and C-terminal sequences flanking the A1 domain form cooperatively an integrated autoinhibitory module (AIM) that interacts with the HDX-protected region. Binding of ristocetin to the C-terminal part of the AIM desorbs the AIM from A1 and enables longA1 binding to GPIbα.
Conclusion
The discontinuous AIM binds the A1 domain and prevents it from binding to GPIbα, which has significant implications for the pathogenesis of type 2B VWD and the shear-induced activation of VWF activity.
Keywords: Blood platelet, Ristocetin, Tandem Mass Spectrometry, Type 2B von Willebrand Disease, von Willebrand Factor
Plasma protein von Willebrand factor (VWF) plays a critical role in hemostasis and thrombosis [1, 2]. The deficiency or dysfunction of VWF causes von Willebrand disease (VWD), a common hereditary bleeding disorder. In the absence of shear VWF does not bind platelets and is generally in a compact form [3]. Under elevated shear conditions or when immobilized under flow, it becomes extended and exposes its A1 domain, enabling platelet binding [4–6]. Ligation of VWF under flow with glycoprotein (GP)Ibα, the major subunit of the platelet GPIb-IX complex, transmits a signal that eventually leads to platelet aggregation or clearance [6–10]. In contrast to full-length VWF, the isolated VWF A1 domain binds to GPIbα with high affinity in the absence of shear [11]. The difference in binding between VWF and the isolated A1 domain suggests an autoinhibitory mechanism in VWF, but its molecular nature has remained unclear.
Under certain static conditions plasma VWF can bind GPIbα with high affinity. Gain-of-function mutations in VWF that enable its binding to GPIbα in the circulation are well documented in type 2B VWD patients [12, 13]. Clustered in a region of the A1 domain closer to the 1272–1458 disulfide bond than the GPIbα-binding interface, these mutations are thought to stabilize A1 in an “on” conformation to bind GPIbα [14]. Crystal structures of A1 domains bearing type 2B mutations report detectable but small changes, although they are mostly local and do not appear to impact the GPIbα-binding interface [15–17]. The molecular basis underlying the gain-of-function effects of type 2B mutations remains to be defined. In addition, ristocetin induces binding of human VWF to GPIbα [18]. Although residues in or near the A1 domain have been reported to affect its binding to VWF [19], how ristocetin activates VWF remains unclear.
The A1 domain is delimited by the Cys1272-Cys1458 disulfide bond, outside of which few secondary structural elements are observed in reported structures [11, 16, 17, 20, 21]. Crystal structures of the A1/GPIbα complex reveal two discontinuous regions of A1 in contact with GPIbα [11, 16]. The larger interface resides in strand β3, helix α3 and loop α3β4. A separate, smaller interface is comprised of residues from loops α1β2, β3α2 and α3β4. Recombinant A1 proteins of various lengths exhibit different affinities for GPIbα [11, 15, 22–25]. Particularly, the linker sequence between the D3 and A1 domains of VWF (residues 1238–1260) can regulate A1 binding to GPIbα under shear [25–27]. Adding the linker sequence as a peptide to the recombinant A1 protein in trans largely mimics its regulatory effect in cis, suggesting that it interacts with the A1 domain [26, 27].
In this study the hydrogen-deuterium exchange (HDX) profiles of two recombinant A1 fragments with disparate binding affinities for GPIbα were characterized and compared. The results suggest that both N- and C-terminal residues flanking the A1 domain constitute cooperatively an autoinhibitory module (AIM) that masks a region in A1 and impedes its binding to GPIbα. These results have significant implications on the regulation mechanism of VWF activity and on the pathophysiology of type 2B VWD and related bleeding disorders.
Material and Methods
Materials
Monoclonal antibody (MAb) 11A8 was obtained after immunizing mice with recombinant ligand-binding domain of human GPIbα (Green Mountain Antibodies, Burlington, VT). Anti-His-tag MAb was purchased from BioLegend (San Diego, CA) and conjugated with fluorescein isothiocyanate (FITC) as described [10]. Plasma-derived human VWF was from Haematologic Technologies (Essex Junction, VT), and ristocetin from Bio/Data Corporation (Horsham, PA).
Recombinant A1 fragments
DNA fragments encoding human VWF residues 1238–1472 (longA1) and 1261–1472 (shortA1) were amplified from the cDNA of VWF. Oligonucleotides encoding a signal peptide sequence “METDTLLLWVLLLWVPGSTGK” and a decahistidine tag were appended to the 5’- and 3’-ends of both fragments, respectively. The fragments were inserted into the ReNeo plasmid [28] at the Xhol/Notl restriction sites. Each expression vector was transfected into baby hamster kidney-derived (BHK) cells [29]. Stably transfected cells were cultured in DMEM F-12 media supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 10 µg/mL G418. The media were collected and applied to a 5-mL Ni-Sepharose HisTrap column (GE Healthcare, Pittsburgh, PA). After washing with 20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, pH 7.4, the bound protein was eluted with the same buffer containing 500 mM imidazole. The eluent was concentrated to 5 mL using an Amicon Ultracel-10K centrifugal filter, and further purified by gel filtration chromatography using a HiLoad Superdex 200 column (GE Healthcare) equilibrated in phosphate-buffered saline (PBS). Desired fractions were pooled and concentrated using Ultracel-10K. The concentrations exclusive of carbohydrate of both A1 proteins was determined using an extinction coefficient of 0.629 (mg/mL)−1cm−1 at 280 nm. The protein was stored at 4°C, with its purity verified by SDS-PAGE.
Flow cytometry
The informed consent and related protocols were approved by Emory University institutional review boards. Citrated whole blood was obtained from healthy volunteers, from which platelet-rich plasma (PRP) was prepared [30]. Platelet counts were measured using a Sysmex XP-300 analyzer (Lincolnshire, IL). To measure platelet binding, PRP was incubated with 60 nM noted protein for 10 minutes at room temperature. After washing the samples were mixed with 1 µg/mL FITC-labeled anti-His-tag MAb, fixed and analyzed on a BD FACS Canto-II instrument using FlowJo. The signal was quantitated by the median fluorescence intensity (MFI) for the entire cell population (10,000 cells).
ELISA
Each well in a microtiter plate was coated with 100 ng human GPIb-IX complex [31] in 20 mM Tris·HCl, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, pH 7.4 at 4°C overnight and blocked by 2% BSA in the same buffer at room temperature for 2 h. The binding of A1 proteins was detected by mouse anti-His-tag MAb and horse radish peroxidase (HRP)-conjugated goat anti-mouse IgG, and that of human VWF by rabbit anti-VWF polyclonal antibody and HRP-conjugated goat anti-rabbit IgG (Pierce, Rockford, IL). After washing with PBS containing 0.1% Tween-20, bound complexes were detected as described [30]. The binding curve was plotted and fitted using Prism software with a nonlinear OneSite-Total binding equation.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed at 20°C in a Beckman Coulter XLI analytical ultracentrifuge using standard procedures [32]. ShortA1 and longA1 were exchanged into 0.137 M NaCl, 0.01 M phosphate, 2.7 mM KCl, pH 7.37 and 0.01 M Na2HPO4, pH 7.40, respectively, by gel filtration chromatography. Partial specific volumes were calculated from the weight-average amino acid and carbohydrate compositions. SEDNTERP (version 20138013BETA) was used to calculate the partial specific volume of the polypeptide chain. The regions in human VWF corresponding to shortA1 and longA1 contain 2 and 5, predominantly disialyl core 1, O-glycans, respectively [33]. Published values of the partial specific volumes of the component sugar residues of disialyl core 1 O-glycan were used [34]. The calculated partial specific volumes for shortA1 and longA1 were 0.734 and 0.723 mL/g, respectively. ShortA1 and longA1, 0.4 mL each, were loaded at 0.23 mg/mL and 0.22 mg/mL, respectively, into 12 mm path length charcoal-filled Epon double sector cells using the matched buffer in the reference sector. Absorbance scans at 280 nm and interference scans were initiated immediately after reaching a target rotor speed of 50,000 rpm and acquired at 3.8 min intervals for a total of 119 scans. The first 9 scans were discarded to exclude optical artifacts at the meniscus. Absorbance data were collected in continuous mode using a radial spacing of 0.003 cm. Data were analyzed by SEDFIT [35], version 15.01c, using the continuous c(s) distribution model and maximum entropy regularization with a confidence interval of 0.68 [36]. Time-invariant noise (interference and absorbance data) and radial-invariant noise (interference data) also were fitted and subtracted from the graphical data, which were constructed using GUSSI, version 1.2.1 [37]. Nonlinear least-squares fitting was done using both the simplex and Marquardt-Levenberg algorithms with equivalent results.
Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
HDX-MS experiments of shortA1 and longA1 were performed largely as described [38]. Briefly, each protein was prepared in PBS to 0.3 mg/mL. Ristocetin was added at 10 mg/mL. Denatured longA1 was prepared by incubating longA1 in PBS, 4 M GdmCl, 0.5 M Tris(2-carboxyethyl)phosphine (TCEP), pH 7.0, at 95°C for 5 min. Each sample was diluted (v/v) 7 times with 10 mM phosphate, 99.9% D2O, pD 7.0 in an autosampler at 20°C. Denatured longA1 was diluted with the deuterated phosphate buffer also containing 4 M GdmCl. After 10–10,000 s of incubation, the reaction was quenched with an equal volume of precooled quenching buffer (100 mM phosphate, 0.5 M TCEP, 0.8% formic acid, 2% acetonitrile, pH 2.5) at 1°C. Each time point was repeated 4–6 times. Peptic fragments were sequenced and identified through database searching. Mass assignment for each peptide without deuterium exchange was checked manually; any assignment with a mass deviation >0.2 Da was removed. The relative fractional deuterium uptake for each residue amide proton at a given time was averaged and calculated following HDsite equations [39]; and the HDX difference between data sets was calculated by comparing the relative fractional uptake for each residue.
Results
ShortA1 and longA1 are monomeric
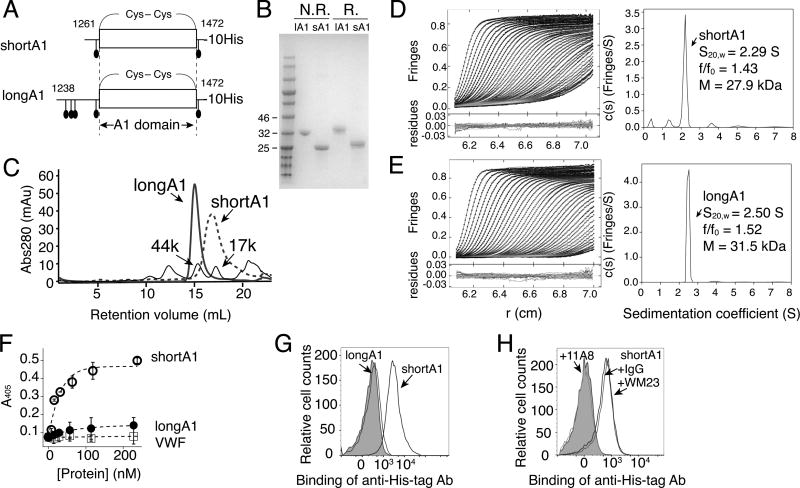
The boundary of the A1 domain has been variably defined [2, 20, 40]. For clarity, the A1 domain in this paper is defined as a structural domain within the Cys1272-Cys1458 disulfide bond [41]. Residues outside of 1272–1458 are not considered a part of A1. ShortA1 and longA1 refer to two recombinant proteins expressed from BHK cells, containing VWF residues 1261–1472 and 1238–1472, respectively (Fig. 1A,B). Both shortA1 and longA1 contain the 1272–1458 disulfide bond as they displayed visibly different mobilities in SDS gel electrophoresis under non-reducing and reducing conditions [25, 42]. Compared to shortA1, longA1 contains additional N-terminal 23 residues and 3 O-glycans therein [43, 44] (Fig. 1A). When stored at 4°C for up to a week, both proteins were primarily monomeric as judged by sedimentation velocity analytical ultracentrifugation and gel filtration chromatography (Fig. 1C–E). The estimated molecular weights of shortA1 and longA1 based on amino acid sequence and assuming 2 and 5 disialyl core 1 O-glycans are 27.44 and 32.64 kDa, respectively. Molecular weight estimates of shortA1 by sedimentation velocity analytical ultracentrifugation using interference and interference optics were 27.9 and 24.0 kDa, respectively, and for longA1 were 32.7 and 31.5 kDa, respectively (Fig. 1D–E, Table S1). These results are in good agreement with the expected values for monomeric shortA1 and longA1. Frictional ratio estimates, a measure of asymmetry, were greater for longA1 compared to shortA1, consistent with additional O-glycans in the former. It is noteworthy that the shortA1 peak in the gel filtration chromatogram was considerably wider than the longA1 peak, suggesting that shortA1 may be more dynamic or interact more with the column matrix than longA1. Consistently, during purification shortA1 exhibited a higher propensity for aggregation and protein loss (data not shown). Thus, the characterization of shortA1 described below was performed within 72 hours of its purification.
Figure 1. Recombinant shortA1 and longA1 proteins are primarily monomeric, and only ShortA1, but not longA1, binds GPIbα.
(A) Illustrations of shortA1 and longA1 proteins. Both proteins contain a C-terminal decahistidine tag. Corresponding VWF residues in each protein are marked by the starting and ending residue numbers. O-glycosylation sites (filled ovals) and the 1272–1458 disulfide bond delimiting the A1 domain are marked. (B) SDS-PAGE of purified longA1 (lA1) and shortA1 (sA1) under non-reducing (N.R.) and reducing (R.) conditions. The gel was stained by Coomassie blue. Nearby molecular weight markers are marked on the left. (C) Superimposed gel filtration chromatograms of shortA1, longA1 and molecular weight standards as indicated by arrows. (D, E) Sedimentation velocity results for shortA1 (D) and longA1 (E) showing fitted interference scans and residuals (left) and sedimentation coefficient distributions (right). Only every 2nd scan and every 3rd data points are shown for clarity. (F) Isotherms of shortA1 (open circle), longA1 (filled circle) and human VWF (open squares) binding to immobilized GPIb-IX measured by ELISA (n=3). (G) Flow cytometry histograms showing binding of shortA1 and lack of binding of longA1 to human platelets. Gray: platelets incubated with only FITC-labeled irrelevant IgG. (H) Overlaid histograms showing that binding of shortA1 to human platelets is inhibited by anti-GPIbα ligand-binding domain MAb 11A8 but not by anti-GPIbα macroglycopeptide region MAb WM23.
Differential binding of shortA1 and longA1 to GPIbα
ShortA1 bound to the immobilized human GPIb-IX complex with an EC50 of about 25 nM, whereas neither longA1 nor human VWF showed significant binding at concentrations up to 250 nM (Fig. 1F). The difference in binding affinities of shortA1 and longA1 is consistent with previous reports [11, 25]. Consistent with ELISA results, significant binding of shortA1, but not longA1, to human platelets was observed (Fig. 1G). The binding was inhibited by MAb 11A8 that recognizes the ligand-binding domain of GPIbα, but not by mouse IgG or MAb WM23 that recognizes the macroglycopeptide region of GPIbα [45] (Fig. 1H). Overall, these results confirmed that shortA1 binds GPIbα with a much higher affinity than longA1.
The HDX-protected region in longA1 overlaps with the GPIbα-binding interface and is clustered with type 2B VWD mutations
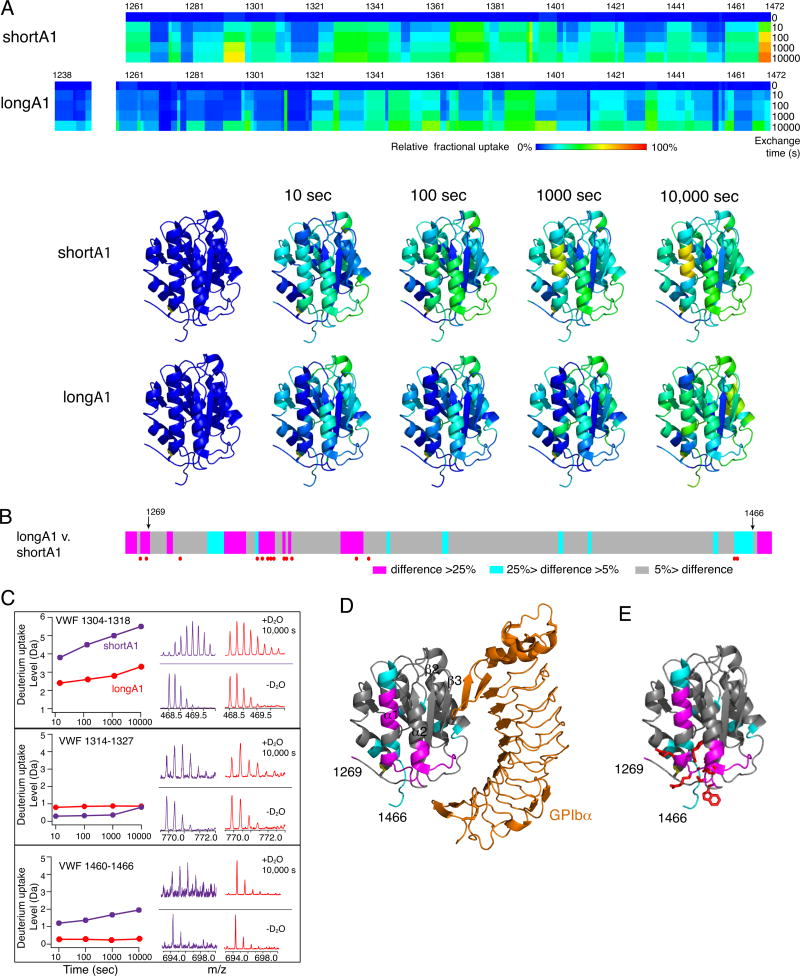
150 and 204 peptic fragments were sequenced and mapped for HDX analysis of shortA1 and longA1, respectively (Fig. S1). No peptides covering residues of 1251–1257 in longA1 were identified, likely because vicinal residues 1255 and 1256 are both O-glycosylated [43]. The extents of deuterium uptake for the identified peptides over time periods ranging from 10 to 10,000 seconds were measured, from which the relative deuterium uptake for each residue was calculated (Fig. 2A). As N- and C-terminal flanking residues are not included or resolved in crystal structures of the A1 domain [16, 20], HDX of the residues outside of 1269–1466 are shown as heat maps overlaid onto the primary sequence but not the A1 structure (Fig. 2A,D).
Figure 2. Difference in HDX between shortA1 and longA1.
Comparison of HDX between shortA1 and longA1. (A) Residual HDX heat maps of shortA1 and longA1 for the noted exchange time. The line for 0 sec denotes the results obtained without exchange. Relative fractional deuterium uptake was calculated for each residue amide from the measured deuterium uptakes of peptic fragments as described in Materials and Methods, and plotted using the rainbow color scale in the figure. Residues 1251–1257 were not detected and are left blank. HDX data of shortA1 and longA1 at the indicated exchange time are mapped to the structure of A1 domain (PDB: 1SQ0; only residues 1269–1466 are shown in the structure). The ribbon diagram of the complex structure is generated using PyMOL. (B) The difference in relative fractional uptake between the same residues of longA1 and shortA1 after 10,000 s of exchange, as defined by the color code in the figure. Residues with reported type 2B VWD mutations [13] are marked by red dots. Residues 1269 and 1466, starting and ending residues of the structure of A1, respectively, are marked by arrows. (C) Representative plots of HDX and mass spectra of three peptic fragments from shortA1 and longA1. Each peptide is identified by the starting and ending residue numbers of VWF. (D) The difference in HDX mapped to a structure of A1 in complex with the ligand-binding domain of GPIbα. Certain secondary structure elements in A1 are labeled. (E) The HDX-protected region in the A1 domain (residues 1269–1466) is clustered with type 2B VWD mutations. Each mutation site is illustrated by its side chain shown in red sticks.
In shortA1, residues in strands β1 and β2 reported the slower exchange rates than those in peripheral helices (Fig. 2A). This is consistent with the expectation that the central hydrophobic core of a folded domain exchanges at the slowest rate, often via global unfolding [46]. However in longA1, in addition to residues in β1 and β2, those in helix α1 and the loop α1β2, which are on the periphery of the A1 domain, also reported slow exchange rates (Fig. 2A). Moreover, several residues in longA1 exchanged at a slower pace than their counterparts in shortA1 (Fig. 2B,C). Importantly, many of these HDX-protected residues in longA1 map to a region in A1 around the disulfide bond, helix α1, loops α1β2 and β3α2, indicating that this region is somewhat shielded from the solvent in longA1, but not in shortA1 (Fig. 2D, S2). This HDX-protected region overlaps with the GPIbα-binding interface, particularly at loops α1β2 and β3α2, which potentially explains the lower binding affinity of longA1 for GPIbα compared to shortA1. It is also remarkable that this HDX-protected region is clustered with type 2B VWD mutations (Fig. 2E, Table S2, Video S1). Only three residues with reported type 2B mutations —Cys1272, Val1316 and Arg1341 — fall outside the HDX-protected sequences. Close inspection of the structure reveals that side chains of Val1316 and Arg1341 are located in the protected region, interacting with helix α1. Cys1272, already located in the protected region, exchanges slowly in shortA1 (Fig. 2A). Overall, comparison of HDX rates of shortA1 and longA1 reveals a shielded region in longA1 that overlaps with the GPIbα-binding interface and is clustered with type 2B VWD mutations.
Autoinhibitory modules cooperatively shield the HDX-protected region in longA1
Because longA1 contains residues 1238–1260 and shortA1 does not (Fig. 1A), this sequence is an obvious candidate for the region that binds to the HDX-protected region in longA1. Indeed, it was reported recently that residues 1238–1260 bind to the A1 domain and modulate its interaction with GPIbα [26, 27, 47]. Consistently, many residues in 1238–1260 of longA1 reported relatively slow HDX (Fig. 2A, S2). However, residues 1261–1268 and 1460–1472 in longA1, which flank the A1 domain (1272–1458) from both sides, also reported slow HDX (Fig. 2A). In contrast, their counterparts in shortA1 reported faster HDX (Fig. 2B).
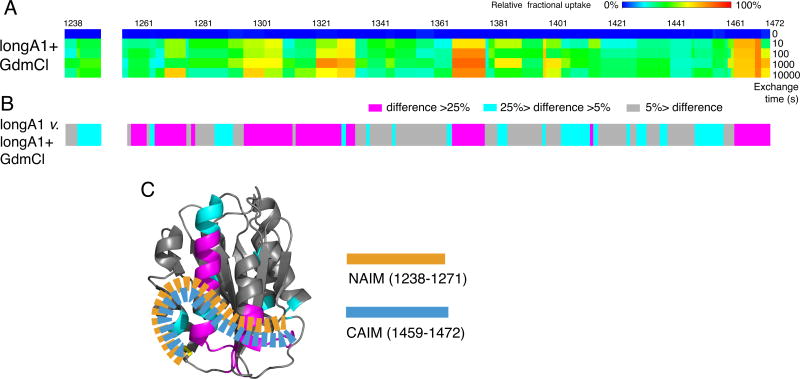
To gauge directly the intrinsic exchange rates of residues 1261–1268 and 1460–1472, longA1 was denatured in 4 M GdmCl at 95°C for 5 min and cooled to room temperature before being mixed with D2O (Fig. S3). LongA1 expressed from bacteria is largely denatured in 4 M urea [27]. Although it is unclear how O-glycans affect the stability of longA1, HDX was significantly increased for all residues of denatured longA1 in 4 M GdmCl compared to longA1 under non-denaturing conditions (Fig. 3A). Particularly, N- and C-terminal residues flanking the A1 domain reported significantly increased HDX compared to their counterparts in folded longA1 (Fig. 2,3), thereby excluding the possibility that the slow exchange of these residues in folded longA1 is due to their intrinsic exchange rates.
Figure 3. AIM consists of two discontinuous segments flanking the A1 domain.
(A) Residual HDX heat maps of longA1 in 4 M GdmCl (longA1+GdmCl) for the noted exchange time, following the same format as described in Figure 3A. (B) The difference bars following the same format as Figure 3B. (C) Illustration of the AIM masking the HDX-protected region in the A1 domain of longA1. AIM is shown as orange and celestial blue dashed ribbons, indicative of well-integrated NAIM and CAIM sequences.
Overall, these results indicate that the presence of residues 1238–1260 in longA1 decreased HDX of residues 1261–1268 and 1460–1472. Although residues 1238–1268 and 1460–1472 are separated in the primary sequence, they are spatially close to each other due to the 1272–1458 disulfide bond. Thus, our results suggest that residues 1238–1268 interact directly with 1460–1472 in longA1, and together they form a structural module that interacts with the A1 domain at the region clustered by the type 2B VWD mutations, thereby providing HDX protection for all the residues involved (Fig. 3C). This module is named autoinhibitory module (AIM). AIM consists of two discontinuous segments: N-terminal residues 1238–1271 and C-terminal residues 1459–1472 that flank the A1 domain, designated here NAIM and CAIM, respectively.
Ristocetin binding desorbs the AIM from the A1 domain
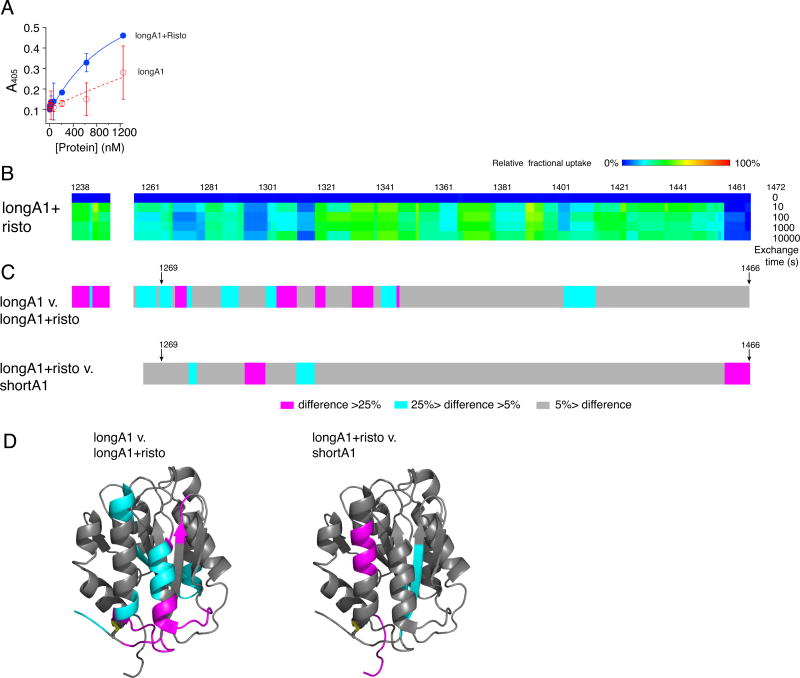
Ristocetin can induce binding of VWF to GPIbα in the absence of shear. Polymorphic variations in NAIM and CAIM, or peptides derived from either sequence, can inhibit or reduce ristocetin-induced activation of VWF [48–51]. To investigate how ristocetin induces VWF binding to GPIbα, the effect of ristocetin on the binding activity and dynamics of longA1 was characterized. Figure 4A shows that addition of 1.5 mg/ml ristocetin to longA1 significantly increased its binding to the immobilized GPIb-IX (Fig. 4A), suggesting structural elements in longA1 are sufficient to recapitulate the activation of full-length VWF by ristocetin. To test whether the conformation of longA1with ristocetin would mimic that of shortA1, HDX of ristocetin-bound longA1 was analyzed (Fig. S3). In addition to residues 1252–1258 that were not identified due to glycosylation as described above, no peptides containing residues 1467–1472 were detected and assigned for unknown reasons (Fig. 4B). However, the adjacent residues (1458–1466) reported little HDX (Fig. 4B), suggesting that ristocetin binds to residues 1458–1472 (i.e. CAIM).
Figure 4. Addition of ristocetin desorbs the AIM from A1 and facilitates longA1 binding to GPIbα.
(A) Binding isotherms of longA1 in the presence (blue) and absence (red) of 1.5 mg/mL ristocetin. (B) Residual HDX heat maps of longA1 mixed with ristocetin (longA1+risto) for the noted exchange time, following the same format as Figure 3A. Residues 1252–1258 and 1467–1472 were not detected and are left blank. (C) The plots of difference in the relative fractional uptake between the same residues of longA1 and longA1+risto, and between longA1+risto and shortA1, following the same format as Figure 3B. (D) The differences are mapped to structures of the A1 domain (PDB: 1SQ0).
Differences in HDX were calculated for longA1 versus ristocetin-bound longA1 (longA1 v. longA1+risto) as well as for ristocetin-bound longA1 versus shortA1 (longA1+risto v. shortA1), and mapped to the A1 structure (Fig. 4C,D). Comparison of the HDX differences indicates that the A1 domain in the ristocetin-bound longA1 mimics that in shortA1 more than longA1. In other words, binding of ristocetin induces exposure of the HDX-protected region in longA1, thus enabling it to bind GPIbα. Moreover, most residues in the NAIM of ristocetin-bound longA1 reported much higher HDX than those in longA1, indicating that these residues were also de-protected by ristocetin binding. Thus, it suggests that ristocetin binding to CAIM disrupts the integrity of the AIM and disrupts its interaction to the A1 domain.
Discussion
In the absence of shear VWF is autoinhibited from binding to GPIbα. However, the location and structure of the autoinhibitory element in VWF has remained unclear as has the mechanism by which the shear force overcomes the autoinhibition. Previous studies of A1-containing fragments of VWF suggest that the A1 domain is sufficient to bind GPIbα with high affinity and that at least one AIM is located close to A1 [11, 15, 17, 23, 24]. In particular, residues 1238–1260 have been suggested to bind A1 and modulate its binding to GPIbα, partly because longA1 reported a GPIbα-binding affinity of 4.5 µM and shortA1 reported an affinity of 30 nM [11, 25, 27]. The difference in binding affinity between longA1 and shortA1 was reproduced in the present study (Fig. 1). More importantly, the difference in HDX between longA1 and shortA1 (Fig. 2–4) provides new critical evidence for the identity of an AIM in VWF and its mode of action. First, compared to residues in shortA1, residues 1238–1260 in longA1 that are absent in shortA1 reduce HDX of residues 1261–1268 and 1460–1472 in longA1 (Fig. 2, 3), suggesting that a crosstalk between NAIM and CAIM enables them to form an integrated and structured AIM that provides HDX protection. The crosstalk is consistent with the increase of HDX of NAIM residues induced by binding of ristocetin to CAIM (Fig. 4), and with the report that peptides of residues 1237–1251 and 1457–1471 inhibit VWF binding to GPIbα much more effectively than either peptide alone [51]. Second, several longA1 residues in the A1 domain showed significantly less HDX than their counterparts in shortA1 (Fig. 2, S1). Since longA1 and shortA1 share similar structure and thermodynamic stability [27], the reduced HDX supports a model in which the AIM interacts with the HDX-protected region in A1. Third, the HDX-protected region overlaps with the GPIbα-binding interface (Fig. 2D). Thus the AIM association with A1 may produce a steric hindrance impeding its binding to GPIbα (Fig. 5). Fourth, ristocetin induces longA1 binding to GPIbα and de-protects HDX-protected regions in longA1 (Fig. 4). This suggests that the AIM interaction with A1 may be modulated, which has significant implications on the pathogenesis of type 2B VWD.
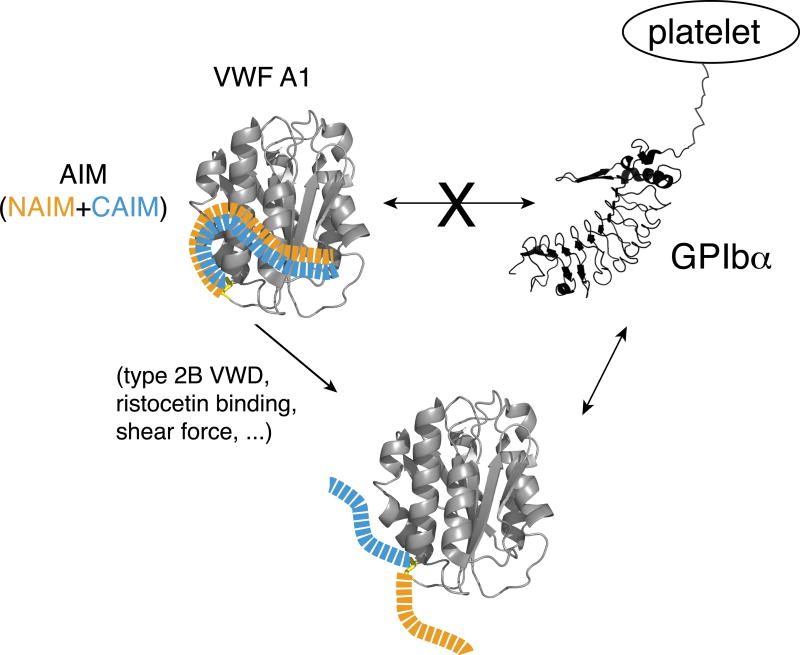
Figure 5. Model of the AIM-masking of A1.
A model of AIM masking of the A1 domain in VWF. In VWF, the N- and C-terminal sequences flanking the A1 domain, designated as NAIM (orange) and CAIM (celestial blue), respectively, cooperatively form the AIM that binds the A1 domain and blocks its association with GPIbα. Various factors, including ristocetin, type 2B VWD mutations and shear force, may induce VWF binding to GPIbα by destabilizing the AIM/A1 association and/or disrupting AIM.
Mutant VWF from patients with type 2B VWD exhibit heightened spontaneous binding to GPIbα, thereby activating platelets and leading to platelet clearance and thrombocytopenia [10, 12]. Thus type 2B mutations are considered as gain-of-function mutations [14]. However, the extent of thrombocytopenia and bleeding diathesis is diverse in patients with type 2B VWD. It is therefore puzzling that different type 2B mutations share a common feature of inducing VWF binding to GPIbα but concurrently differ greatly from one another in their effects on platelets. To date no adequate explanation has been provided. An important finding in the present study is that the HDX-protected region of the A1 domain in longA1 is clustered with most type 2B VWD mutations (Fig. 2E). Some type 2B mutations are also located in NAIM and CAIM (Fig. 2B). Our results suggest that a type 2B mutation induces VWF binding to GPIbα by disrupting or destabilizing the inhibitory association of AIM with A1 without significantly affecting the A1 structure (Fig. 5). A mutation in the HDX-protected region of A1 (e.g. I1309V [15], V1316M [52]) or in the AIM (e.g. P1266L [53], A1461D [54]), or different mutations of the same residue (e.g. R1306W/A/P [13, 52, 55]), could accomplish the same loss-of-inhibitory-function effect. Separately, the interaction of A1 to GPIbα may not require the presence of AIM . Moreover, VWF binding-induced platelet signaling depends more on the force resistance of the A1/GPIbα interaction than the binding affinity [10]. It is conceivable that type 2B mutations may differ from one another in modulating the mechanomolecular parameters of the A1/GPIbα interaction [56, 57]. Thus a combination of shared loss of the autoinhibitory function and individual effects on force-regulated GPIbα binding may be needed to fully explain the common and disparate symptoms of type 2B VWD.
Ristocetin may also induce VWF binding to GPIbα by disrupting the AIM/A1 association (Fig. 4). Comparison of HDX profiles of longA1 in the presence and absence of ristocetin suggests that ristocetin binds directly to CAIM but not NAIM. Binding of ristocetin to CAIM could somehow disrupt the crosstalk between CAIM and NAIM, resulting in the increased HDX of NAIM residues and the disintegration of AIM. This is consistent with a previous report that MAb 6G1 binds to VWF residues 1463–1471 and blocks ristocetin activation of VWF, but its binding alone can activate VWF like ristocetin [58]. It is also consistent with reports that sequence polymorphisms in CAIM could affect ristocetin activation of VWF but not VWF binding to GPIbα [49, 50]. Although ristocetin binding to NAIM, specifically residues Glu1239-Gly1242, was suggested [59], no corroborating HDX evidence was obtained in this study. Consistent with this, murine VWF, which does not bind ristocetin, also contains Glu1239-Gly1242.
Although the study reported here did not involve shear stress, the results suggest a mechanism of shear-induced activation of VWF. It does not escape our attention that the AIM consists of two discontinuous sequences. Thus, it is conceivable that when VWF is under shear stress [41], NAIM and CAIM could be physically pulled apart and dissociated from the A1 domain, thus enabling A1 binding to GPIbα. That the mechanism of AIM dissociation from A1 may be shared by ristocetin- and shear-induced activation of VWF is consistent with the observation that ristocetin-induced binding of VWF to platelets is better than botrocetin-induced binding at mimicking the VWF activity under shear flow [60]. In contrast to ristocetin, botrocetin binds to helices α4 and α5 of A1, distal from the HDX-protected region or the AIM, and induces A1 binding to GPIbα by a different mechanism [61]. Whether the AIM is a mechanosensory module, how much force is required to open AIM, and the structure of the AIM are the subjects of future investigation. Elucidating the structural, energetic, and mechanical underpinning for the AIM/A1 association in the future will undoubtedly elevate our understanding of VWF regulation and activity.
Supplementary Material
Essentials.
The mechanism for the auto-inhibition of von Willebrand factor (VWF) remains unclear.
Hydrogen exchange of two VWF A1 fragments with disparate activities was measured and compared.
Discontinuous residues flanking A1 form a structural module that blocks A1 binding to the platelet.
Our results suggest a potentially unified model of VWF activation.
Acknowledgments
We thank Drs. Evan Sadler and Matthew Auton for helpful discussions. This work was supported in part by Emory WHSC Synergy Award, NIH grants HL082808 (R.L.) and HL112309 (P.L.).
Footnotes
Addendum
W. Deng, Y. Wang, P. Lollar and R. Li designed research; W. Deng, Y. Wang, S. A. Druzak, A. K. Syed and P. Lollar performed research; W. Deng, Y. Wang, S. A. Druzak, A. K. Syed, P. Lollar and R. Li analyzed results; W. Deng, Y. Wang, P. Lollar and R. Li prepared figures; J. F. Healy and P. Lollar provided critical reagents; W. Deng, P. Lollar and R. Li wrote the paper.
Disclosure of Conflict of Interests
The authors state that they have no conflict of interest.
References
- 1.Wagner DD. Cell biology of von Willebrand factor. Annu Rev Cell Biol. 1990;6:217–246. doi: 10.1146/annurev.cb.06.110190.001245. [DOI] [PubMed] [Google Scholar]
- 2.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Slayter H, Loscalzo J, Bockenstedt P, Handin RI. Native conformation of human von Willebrand protein. Analysis by electron microscopy and quasi-elastic light scattering. J. Biol.Chem. 1985;260:8559–8563. [PubMed] [Google Scholar]
- 4.Dong JF, Moake JL, Nolasco L, Bernardo A, Arceneaux W, Shrimpton CN, Schade AJ, McIntire LV, Fujikawa K, Lopez JA. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 5.Siedlecki CA, Lestini BJ, Kottke-Marchant KK, Eppell SJ, Wilson DL, Marchant RE. Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood. 1996;88:2939–2950. [PubMed] [Google Scholar]
- 6.Savage B, Saldivar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 7.Chow TW, Hellums JD, Moake JL, Kroll MH. Shear stress-induced von Willebrand factor binding to platelet glycoprotein Ib initiates calcium influx associated with aggregation. Blood. 1992;80:113–120. [PubMed] [Google Scholar]
- 8.Yuan Y, Kulkarni S, Ulsemer P, Cranmer SL, Yap CL, Nesbitt WS, Harper I, Mistry N, Dopheide SM, Hughan SC, Williamson D, de la Salle C, Salem HH, Lanza F, Jackson SP. The von Willebrand factor-glycoprotein Ib/V/IX interaction induces actin polymerization and cytoskeletal reorganization in rolling platelets and glycoprotein Ib/V/IX-transfected cells. J. Biol.Chem. 1999;274:36241–36251. doi: 10.1074/jbc.274.51.36241. [DOI] [PubMed] [Google Scholar]
- 9.Du X. Signaling and regulation of the platelet glycoprotein Ib-IX-V complex. Curr. Opin. Hematol. 2007;14:262–269. doi: 10.1097/MOH.0b013e3280dce51a. [DOI] [PubMed] [Google Scholar]
- 10.Deng W, Xu Y, Chen W, Paul DS, Syed AK, Dragovich MA, Liang X, Zakas P, Berndt MC, Di Paola J, Ware J, Lanza F, Doering CB, Bergmeier W, Zhang XF, Li R. Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat. Commun. 2016;7:12863. doi: 10.1038/ncomms12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Iba and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 12.Ruggeri ZM, Pareti FI, Mannucci PM, Ciavarella N, Zimmerman TS. Heightened interaction between platelets and factor VIII/von Willebrand factor ina new subtype of von Willebrand’s. N. Engl. J. Med. 1980;302:1047–1051. doi: 10.1056/NEJM198005083021902. [DOI] [PubMed] [Google Scholar]
- 13.Hampshire DJ, Goodeve AC. The international society on thrombosis and haematosis von Willebrand disease database: an update. Semin Thromb Hemost. 2011;37:470–479. doi: 10.1055/s-0031-1281031. [DOI] [PubMed] [Google Scholar]
- 14.Cooney KA, Ginsburg D. Comparative analysis of type 2b von Willebrand disease mutations: implications for the mechanism of von Willebrand factor binding to platelets. Blood. 1996;87:2322–2328. [PubMed] [Google Scholar]
- 15.Miyata S, Goto S, Federici AB, Ware J, Ruggeri ZM. Conformational changes in the A1 domain of von Willebrand factor modulating the interaction with platelet glycoprotein Ibalpha. J. Biol. Chem. 1996;271:9046–9053. doi: 10.1074/jbc.271.15.9046. [DOI] [PubMed] [Google Scholar]
- 16.Dumas JJ, Kumar R, McDonagh T, Sullivan F, Stahl ML, Somers WS, Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Iba complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J. Biol. Chem. 2004;279:23327–23334. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- 17.Blenner MA, Dong X, Springer TA. Structural basis of regulation of von Willebrand factor binding to glycoprotein Ib. J. Biol. Chem. 2014;289:5565–5579. doi: 10.1074/jbc.M113.511220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard MA, Firkin BG. Ristocetin--a new tool in the investigation of platelet aggregation. Thromb Diath Haemorrh. 1971;26:362–369. [PubMed] [Google Scholar]
- 19.Sadler JE. Redeeming ristocetin. Blood. 2010;116:155–156. doi: 10.1182/blood-2010-04-276394. [DOI] [PubMed] [Google Scholar]
- 20.Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib. J. Biol. Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- 21.Tischer A, Campbell JC, Machha VR, Moon-Tasson L, Benson LM, Sankaran B, Kim C, Auton M. Mutational Constraints on Local Unfolding Inhibit the Rheological Adaptation of von Willebrand Factor. J. Biol. Chem. 2016;291:3848–3859. doi: 10.1074/jbc.M115.703850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura S, Fujimura Y, Sugimoto M, Kawasaki T, Ikeda Y, Titani K, Yoshioka A. Structural elements influencing von Willebrand factor (vWF) binding affinity for platelet glycoprotein Ib within a dispase-digested vWF fragment. Blood. 1994;84:1553–1558. [PubMed] [Google Scholar]
- 23.Miura S, Li CQ, Cao Z, Wang H, Wardell MR, Sadler JE. Interaction of von Willebrand factor domain A1 with platelet glycoprotein Ibα-(1–289). Slow intrinsic binding kinetics mediate rapid platelet adhesion. J. Biol. Chem. 2000;275:7539–7546. doi: 10.1074/jbc.275.11.7539. [DOI] [PubMed] [Google Scholar]
- 24.Ulrichts H, Udvardy M, Lenting PJ, Pareyn I, Vandeputte N, Vanhoorelbeke K, Deckmyn H. Shielding of the A1 domain by the D’D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J. Biol. Chem. 2006;281:4699–4707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- 25.Cruz MA, Handin RI, Wise RJ. The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX. The role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain protein. J. Biol. Chem. 1993;268:21238–21245. [PubMed] [Google Scholar]
- 26.Ju L, Dong JF, Cruz MA, Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibalpha. J. Biol. Chem. 2013;288:32289–32301. doi: 10.1074/jbc.M113.504001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tischer A, Cruz MA, Auton M. The linker between the D3 and A1 domains of vWF suppresses A1-GPIbalpha catch bonds by site-specific binding to the A1 domain. Protein Sci. 2013;22:1049–1059. doi: 10.1002/pro.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Healey JF, Lubin IM, Nakai H, Saenko EL, Hoyer LW, Scandella D, Lollar P. Residues 484–508 contain a major determinant of the inhibitory epitope in the A2 domain of human factor VIII. J. Biol. Chem. 1995;270:14505–14509. doi: 10.1074/jbc.270.24.14505. [DOI] [PubMed] [Google Scholar]
- 29.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. High level expression of recombinant porcine coagulation factor VIII. J. Biol. Chem. 2002;277:38345–38349. doi: 10.1074/jbc.M206959200. [DOI] [PubMed] [Google Scholar]
- 30.Liang X, Russell SR, Estelle S, Jones LH, Cho S, Kahn ML, Berndt MC, Bunting ST, Ware J, Li R. Specific inhibition of ectodomain shedding of glycoprotein Ibalpha by targeting its juxtamembrane shedding cleavage site. J. Thromb. Haemost. 2013;11:2155–2162. doi: 10.1111/jth.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan R, Mo X, Paredes AM, Dai K, Lanza F, Cruz MA, Li R. Reconstitution of platelet glycoprotein Ib-IX complex in phospholipid bilayer nanodiscs. Biochemistry. 2011;50:10598–10606. doi: 10.1021/bi201351d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci. 2013;20.12:1–49. doi: 10.1002/0471140864.ps2012s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solecka BA, Weise C, Laffan MA, Kannicht C. Site-specific analysis of von Willebrand factor O-glycosylation. Journal of Thrombosis & Haemostasis. 2016;14:733–746. doi: 10.1111/jth.13260. [DOI] [PubMed] [Google Scholar]
- 34.Perkins SJ, Miller A, Hardingham TE, Muir H. Physical properties of the hyaluronate binding region of proteoglycan from pig laryngeal cartilage.Densitometric and small-angle neutron scattering studies of carbohydrates and carbohydrate-protein macromolecules. Journal of Molecular Biology. 1981;150:69–95. doi: 10.1016/0022-2836(81)90325-9. [DOI] [PubMed] [Google Scholar]
- 35.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brautigam CA. Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Batsuli G, Deng W, Healey JF, Parker ET, Baldwin WH, Cox C, Nguyen B, Kahle J, Konigs C, Li R, Lollar P, Meeks SL. High-affinity, non-inhibitory pathogenic C1 domain antibodies are present in patients with hemophilia A and inhibitors. Blood. 2016 doi: 10.1182/blood-2016-02-701805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan ZY, Walters BT, Mayne L, Englander SW. Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc. Natl. Acad. Sci. USA. 2013;110:16438–16443. doi: 10.1073/pnas.1315532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Celikel R, Varughese KI, Madhusudan, Yoshioka A, Ware J, Ruggeri ZM. Crystal structure of the von Willebrand factor A1 domain in complex with the function blocking NMC-4 Fab. Nat Struct Biol. 1998;5:189–194. doi: 10.1038/nsb0398-189. [DOI] [PubMed] [Google Scholar]
- 41.Springer TA. von Willebrand factor, Jedi knight of the bloodstream. Blood. 2014;124:1412–1425. doi: 10.1182/blood-2014-05-378638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azuma H, Dent JA, Sugimoto M, Ruggeri ZM, Ware J. Independent assembly and secretion of a dimeric adhesive domain of von Willebrand factor containing the glycoprotein Ib-binding site. J. Biol. Chem. 1991;266:12342–12347. [PubMed] [Google Scholar]
- 43.Solecka BA, Weise C, Laffan MA, Kannicht C. Site-specific analysis of von Willebrand factor O-glycosylation. J. Thromb. Haemost. 2016;14:733–746. doi: 10.1111/jth.13260. [DOI] [PubMed] [Google Scholar]
- 44.Canis K, McKinnon TA, Nowak A, Panico M, Morris HR, Laffan M, Dell A. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J. Thromb. Haemost. 2010;8:137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- 45.Berndt MC, Du XP, Booth WJ. Ristocetin-dependent reconstitution of binding of von Willebrand factor to purified human platelet membrane glycoprotein Ib-IX complex. Biochemistry. 1988;27:633–640. doi: 10.1021/bi00402a021. [DOI] [PubMed] [Google Scholar]
- 46.Li R, Woodward C. The hydrogen exchange core and protein folding. Protein Sci. 1999;8:1571–1590. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auton M, Sowa KE, Behymer M, Cruz MA. N-terminal flanking region of A1 domain in von Willebrand factor stabilizes structure of A1A2A3 complex and modulates platelet activation under shear stress. J. Biol. Chem. 2012;287:14579–14585. doi: 10.1074/jbc.M112.348573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmberg L, Dent JA, Schneppenheim R, Budde U, Ware J, Ruggeri ZM. von Willebrand factor mutation enhancing interaction with platelets in patients with normal multimeric structure. J. Clin. Invest. 1993;91:2169–2177. doi: 10.1172/JCI116443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flood VH, Gill JC, Morateck PA, Christopherson PA, Friedman KD, Haberichter SL, Branchford BR, Hoffmann RG, Abshire TC, Di Paola JA, Hoots WK, Leissinger C, Lusher JM, Ragni MV, Shapiro AD, Montgomery RR. Common VWF exon 28 polymorphisms in African Americans affecting the VWF activity assay by ristocetin cofactor. Blood. 2010;116:280–286. doi: 10.1182/blood-2009-10-249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flood VH, Friedman KD, Gill JC, Morateck PA, Wren JS, Scott JP, Montgomery RR. Limitations of the ristocetin cofactor assay in measurement of von Willebrand factor function. J. Thromb. Haemost. 2009;7:1832–1839. doi: 10.1111/j.1538-7836.2009.03594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohri H, Fujimura Y, Shima M, Yoshioka A, Houghten RA, Ruggeri ZM, Zimmerman TS. Structure of the von Willebrand factor domain interacting with glycoprotein Ib. J. Biol. Chem. 1988;263:17901–17904. [PubMed] [Google Scholar]
- 52.Casana P, Martinez F, Espinos C, Haya S, Lorenzo JI, Aznar JA. Search for mutations in a segment of the exon 28 of the human von Willebrand factor gene: new mutations, R1315C and R1341W, associated with type 2M and 2B variants. Am. J. Hematol. 1998;59:57–63. doi: 10.1002/(sici)1096-8652(199809)59:1<57::aid-ajh11>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 53.Zhang ZP, Blomback M, Nyman D, Anvret M. Mutations of von Willebrand factor gene in families with von Willebrand disease in the Aland Islands. Proc. Natl. Acad. Sci. USA. 1993;90:7937–7940. doi: 10.1073/pnas.90.17.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hilbert L, Gaucher C, Mazurier C. Effects of different amino-acid substitutions in the leucine 694-proline 708 segment of recombinant von Willebrand factor. Br. J. Haematol. 1995;91:983–990. doi: 10.1111/j.1365-2141.1995.tb05423.x. [DOI] [PubMed] [Google Scholar]
- 55.Hulstein JJ, de Groot PG, Silence K, Veyradier A, Fijnheer R, Lenting PJ. A novel nanobody that detects the gain-of-function phenotype of von Willebrand factor in ADAMTS13 deficiency and von Willebrand disease type 2B. Blood. 2005;106:3035–3042. doi: 10.1182/blood-2005-03-1153. [DOI] [PubMed] [Google Scholar]
- 56.Tischer A, Madde P, Moon-Tasson L, Auton M. Misfolding of vWF to pathologically disordered conformations impacts the severity of von Willebrand disease. Biophys. J. 2014;107:1185–1195. doi: 10.1016/j.bpj.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J, Hudson NE, Springer TA. Force-induced on-rate switching and modulation by mutations in gain-of-function von Willebrand diseases. Proc. Natl. Acad. Sci. USA. 2015;112:4648–4653. doi: 10.1073/pnas.1501689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Luca M, Facey DA, Favaloro EJ, Hertzberg MS, Whisstock JC, McNally T, Andrews RK, Berndt MC. Structure and function of the von Willebrand factor A1 domain: analysis with monoclonal antibodies reveals distinct binding sites involved in recognition of the platelet membrane glycoprotein Ib-IX-V complex and ristocetin-dependent activation. Blood. 2000;95:164–172. [PubMed] [Google Scholar]
- 59.Scott JP, Montgomery RR, Retzinger GS. Dimeric ristocetin flocculates proteins, binds to platelets, and mediates von Willebrand factor-dependent agglutination of platelets. J. Biol. Chem. 1991;266:8149–8155. [PubMed] [Google Scholar]
- 60.Dong JF, Berndt MC, Schade A, McIntire LV, Andrews RK, Lopez JA. Ristocetin-dependent, but not botrocetin-dependent, binding of von Willebrand factor to the platelet glycoprotein Ib-IX-V complex correlates with shear-dependent interactions. Blood. 2001;97:162–168. doi: 10.1182/blood.v97.1.162. [DOI] [PubMed] [Google Scholar]
- 61.Fukuda K, Doggett T, Laurenzi IJ, Liddington RC, Diacovo TG. The snake venom protein botrocetin acts as a biological brace to promote dysfunctional platelet aggregation. Nature Struct. Mol. Biol. 2005;12:152–159. doi: 10.1038/nsmb892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.