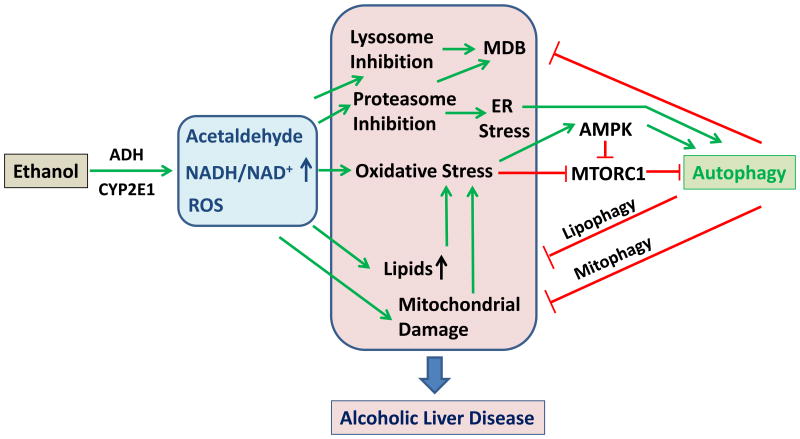
Alcoholic liver disease (ALD) is caused by chronic alcohol abuse and is a serious health concern worldwide. ALD is characterized by steatosis, inflammation, fibrosis, which can progress to cirrhosis. It is generally considered that the pathogenesis of ALD is intimately related to oxidative stress, derived from reactive intermediates including acetaldehyde, increased NADH/NAD+ ratio and ROS generation [1-4] (Figure 1). An increased level of fatty acids and ROS may result in lipid peroxidation and increased production of inflammatory cytokines, which can contribute to liver injury and fibrosis. Autophagy plays important roles in mitigating ethanol-induced hepatocyte apoptosis and liver injury [5].
Figure 1. The activation and function of autophagy in alcoholic liver disease.
Ethanol metabolism through alcohol dehydrogenase (ADH), and/or P450 CYP2E1 results in an increased amount of acetaldehyde, an increased NADH/NAD+ ratio and an increased production of reactive oxygen species (ROS). The oxidative stress is further enhanced with the accumulation of lipids and mitochondrial damage, which can cause additional production of ROS and lipid peroxidation. Chronic use of alcohol can also result in proteasome and lysosome inhibition, leading to ER stress and accumulation of SQSTM1/p62-positive ubiquitinated aggregated proteins, known as Malory-Denk body (MDB). These pathological cellular changes contribute to the development of alcohol fatty liver disease. On the other hand, oxidative stress can activate autophagy by increased AMPK activity and decreased MTORC1 activity. Proteasome inhibition can also enhance autophagy via ER stress, although lysosome inhibition can suppress autophagy. Autophagy may protect the liver against ethanol-induced damage by removing MDB, lipids and damaged mitochondria. Pharmacological enhancement of autophagy could thus be an effective and feasible way to treat alcoholic liver disease.
1. Autophagy Pathways and Regulations
Autophagy is an evolutionarily conserved cellular process essential for development, differentiation, homeostasis and survival [6, 7]. Autophagy (from the Greek, “auto” oneself, “phagy” to eat) refers to cellular degradation that involves the delivery of cytoplasmic cargo (macromolecules or organelles) to the lysosome. There are three types of autophagy, macroautophagy, microautophagy and chaperone-mediated autophagy, which differ in the way by which cargo is delivered to the lysosomes [8]. Macroautophagy is a process, in which cytosolic materials are sequestered by autophagosomes, which transport them to the lysosome for degradation. Macroautophagy can be activated by many signals and is perhaps the most active form of autophagy in terms of the turnover of the cytosolic materials.
The process of microautophagy includes direct engulfment of cytoplasmic cargo at a boundary membrane of the lysosome, which mediate both invagination and vesicle scission into the lumen of lysosomes [9]. Microautophagy is mainly defined in the yeast, but has been observed in mammalian cells [10]. Chaperone mediated autophagy (CMA) involves direct shuttling of specific proteins across the lysosomal membrane for degradation in the lumen [11]. All proteins internalized in lysosomes through CMA contain in their amino acid sequence a pentapeptide motif that is necessary and sufficient for their targeting to lysosomes. These sequences can be recognized by chaperone proteins.
The cargo-carrying vesicular structure formed during autophagy is known as the autophagosome. The origin of the autophagosomal membrane is not quite clear. Various models have been proposed [12]. These include the model that the membrane is synthesized de novo and the model that the membrane is derived from pre-existing cellular membranes such as the endoplasmic reticulum (ER) [13, 14], the Golgi complex [15], the mitochondria [16] and the plasma membrane [17]. Recent studies suggest that the ER is the most plausible candidate for the initial membrane source and/or the platform for autophagosome formation following amino acid starvation [18]. Electron tomography studies have found evidence supporting the connection of initial autophagosomal membranes to the ER membrane [19, 20]. An ER membrane structure, known as the omegosome and identified by the molecule DFCP1, seems to be a site where ER membranes may evolve into autophagosomal membranes [21]. In addition, ER-derived COPII-coated vesicles, which bud from a specialized domain of the ER called the ER exit site (ERES), are found to contribute to autophagosome formation [22]. COP II vesicles are generally transporter vesicles, migrating from ER to the Golgi. Consistently, the ER-Golgi intermediate compartment (ERGIC) has also been identified as the most efficient membrane substrate for LC3 lipidation by recruiting the key early autophagic factor Atg14 [15]. It has to be noted that other membranes could also contribute to autophagosome formation and/or maturation at the early or later phase of the process. This may be particularly meaningful in selective autophagy in which specific subcellular organelles are selectively targeted.
In the past decade, researchers have elucidated key molecular pathways that regulate autophagy. These pathways consist mainly of Atg (Autophagy) proteins [23]. In mammalian cells, initiation of autophagosome biogenesis requires the ULK complex and the autophagy-specific Beclin 1 complex. The ULK complex is composed of ULK1 or ULK2 (the mammalian orthologue of yeast Atg1), FIP200, mAtg13 and Atg101, and is required for the activation of autophagy-specific Beclin 1 complex. The latter is composed of Beclin 1 (the mammalian orthologue of yeast Atg6), Atg14, and class III phosphatidylinositol-3-kinase (PI3KC3) subunits Vps34, and Vps15. Notably, Beclin 1 can also interact with UVRAG, which, together with Vps34 and Vps15, forms a separate complex distinct from the Atg14-Beclin 1 complex [24, 25]. Atg14L-Beclin 1 complex is found mainly in ER, isolation membranes and autophagosome and is responsible for autophagy initiation, whereas UVRAG-Beclin 1 complex is mainly found in the later endosome/lysosome, which is more related to endosomal function [24-26]. The relative abundance of the two complexes could be different in different cells, of which the regulation is little known. Rubicon, another Beclin-1 interacting protein, can negatively regulate the autophagy function of the Beclin-1 complex, perhaps by association only with the Beclin-1-UVRAG complex, thus shifting the balance between the Atg14L-Beclin-1 complex and the UVRAG-Beclin 1 complex [25, 26].
Both ULK complex and Atg14-Beclin1 complex are inhibited by mTORC1, but activated by AMPK in opposing ways [27, 28], thus responding to metabolic or environmental changes. The autophagosomal membrane expansion requires two ubiquitin-like conjugation systems, Atg12 conjugation to Atg5 and LC3 conjugation to phosphatidylethanolamine (PE) [29, 30]. Notably, the Atg5-Atg12-Atg16 complex is required for the efficient conjugation of LC3 to PE, thus acting like an E3 ligase functionally [31]. The conjugation of LC3 is required for the autophagosomal membrane to expand and to complete the formation of the vesicular structure. The Atg5-Atg12-Atg16 complex disengages the membranes after LC3 is conjugated. These molecules are thus associated only with the early autophagosomal membrane and are considered as the early markers. LC3 remains on the autophagosomes and is considered as the general marker of autophagosomes from the early to the later stage [32].
After autophagosomes are formed, they migrate to where lysosomes are located and engaged in the fusion with the latter to form autolysosomes. The cytoskeleton seems to be involved in the movement of autophagosomes. Agents such as nocodazole, which are microtubule poisons, can block fusion of the autophagosome with the lysosome, perhaps by preventing the movement of autophagosomes. Recent findings have identified Syntaxin 17 (Stx17) as the autophagosomal SNARE required for fusion with the endosome/lysosome [33]. Stx17 localizes to the outer membrane of completed autophagosomes, and interacts with SNAP-29 and the endosomal/lysosomal SNARE family molecule, VAMP8. Once fused, the inner membrane of the autophagosome and the cytoplasm-derived materials contained in the autophagosome are degraded by lysosomal/vacuolar enzyme. Monomeric units of the digested macromolecules, such as amino acids, are exported to the cytosol for reuse.
2. Regulation of Autophagy by Metabolic and Stress Signals
Multiple stimuli can promote or inhibit autophagy via different mechanisms. Nutrient deprivation is one of the best known autophagy stimulators. Other autophagy inducers include ER stress, oxidative stress, and DNA damage [34]. The presence of extracellular nutrients (i.e, amino acids, fatty acid, and glucose) and growth factors (e.g., insulin and insulin-like growth factor) can inhibit autophagy. The Class I phosphatidylinositol 3-kinase and mTORC1 are major inhibitors of autophagy along the signaling pathways mediated by nutrients and growth factors [35]. AMPK, being a negative inhibitor of mTORC1, can also promote autophagy. It seems that AMPK is particularly responsive to glucose level, and is thus responsible for autophagy triggered by glucose-deprivation [36]. As discussed above, AMPK and mTORC1 can directly modulate ULK1 and Beclin 1 complex to regulate autophagy induction.
ER stress is another well-known autophagy inducer. The ER is a key compartment in the cell to facilitate folding of newly synthesized proteins. A number of factors can serve as ER stress stimuli, including expression of aggregate-prone proteins, glucose deprivation (resulting in reduced glycosylation and decreased energy for chaperone activity), hypoxia and oxidative stress (causing decreased disulfide bond formation), Ca2+ efflux from the ER and inhibition of the proteasome, all leading to the accumulation of unfolded proteins in the ER [37]. When the folding capacity of ER is exceeded, the unfolded protein response (UPR) is triggered. Mammalian UPR involves three distinct signaling pathways mediated byIRE1, ATF6 and PERK, respectively. UPR causes a general reduction of protein synthesis but activation of the transcription of a selective group of proteins to increase ER folding capacity and degradation of the unfolded or mis-folded proteins. The latter is mainly mediated by the proteasome, and the process is known as the ER-associated degradation (ERAD). Autophagy has also been shown to be important for the degradation of the misfolded proteins, particularly in the condition when the proteasome is suppressed [38-40]. This process is also coined as ER-associated autophagy (ERAA). It seems that mTORC1 is eventually suppressed through the activation of UPR, thus contributing to autophagy activation [41].
3. Activation of autophagy by ethanol
Ethanol can activate hepatic autophagy in vivo and in cultured primary hepatocytes [5, 42, Sid, 2013 #15, 43]. This activation requires ethanol metabolism and can thus requires the activity of alcohol dehydrogenase and cytochrome P450 2E1 (CYP2E1) [5, Wu, 2012 #10, 44, 45]. It seems that the metabolite, acetaldehyde, may be responsible for the autophagy activation [45]. Acetaldehyde is a pro-oxidant. Indeed, anti-oxidants, such as N-acetyl cysteine (NAC) [5, 44] can suppress ethanol-induced autophagy. Certainly, ethanol metabolism can lead to increased oxidative stress via several other mechanisms, including the change in NADH/NAD+ ratio and mitochondrial damage [1-4]. Notably, deletion of cyclophilin D, a major component of mitochondrial permeability transition pore, impaired ethanol-induced autophagy [46], possibly due to a reduction in permeability transition and thus reduced reactive oxygen species (ROS) generation.
Both suppression of mTORC1 [47], and activation of AMPK [43] could contribute to ethanol-induced autophagy under oxidative stress (Figure 1). Consistently, activation of autophagy by rapamycin could reduce ethanol-induced liver injury [5, 44, 48]. Recently, it has been shown that FoxO3a is activated during ethanol treatment, and is responsible for the transcription of several autophagy genes [49]. Ethanol treatment of FoxO3a-deficient mice resulted in enhanced liver injury and steatosis. Resveratrol is well known to inhibit ethanol-induced liver injury and steatosis [50], and it can also activate autophagy via increased deacetylation of FoxO3a [49] and activation of SIRT1 [51]. Protection of ethanol-induced damage by globular adiponectin can also be mediated by its enhancement of autophagy via FoxO3a and AMPK [52].
It is likely that other mechanisms could contribute to autophagy activation as well (Figure 1). Ethanol treatment can lead to proteasome inhibition and ER stress, both of which are known to be linked to autophagy activation [40]. Suppression of proteasome can cause compensatory activation of autophagy via ER stress-mediated UPR, in which the IRE-1 and JNK pathway are involved [39]. Indeed, proteasome activity is inversely correlated with autophagy activation in ethanol-treated cells [42]. Finally, metal elements, such as zinc, can also be critical for autophagy during ethanol exposure and under basal level in hepatoma cell lines [53]. Thus zinc addition in the medium stimulated autophagy. Ethanol treatment can change the expression of zinc transporters and metallothionein, thus activating autophagy.
4. Autophagy protects against ethanol-induced liver injury
Autophagy induced by ethanol serves as a protective mechanism [5, 42, 44, 48]. Suppression of autophagy with pharmacologic agents or small interfering RNAs against Atg7 significantly increased hepatocyte apoptosis and liver injury [5, 42, 48]. How autophagy protective against liver injury is not fully understood, but it could involve selective degradation of damaged mitochondrial and/or lipids [47].
Traditionally autophagy is viewed as a non-selective process under nutrient deprivation condition. The non-selective bulk degradation of cytoplasm and organelles by autophagy can provide the basic building materials to support anabolic metabolism during starvation. However, selective removal of specific organelles by autophagy has now been well recognized [54]. All major organelles could be specifically targeted by autophagy, including mitochondria (mitophagy), ribosomes (ribophagy), endoplasmic reticulum (ER-phagy), peroxisomes (pexophagy), and lipid droplets (lipophagy). The selective functions of autophagy indicate that it is important for maintaining cellular homeostasis by removing superfluous or injured organelles.
Selective autophagy can be important in ameliorating alcoholic liver disease (Figure 1). Autophagy induced by ethanol seems to be selective for damaged mitochondria and accumulated lipid droplets, but not long-lived proteins [5]. Mitochondrial damage is well defined in alcoholic liver disease [55]. Ethanol is mainly metabolized by the liver and liver mitochondria can be a primary target for alcohol toxicity. A major catabolic pathway of ethanol begins with alcohol dehydrogenase (ADH), which generates acetaldehyde. The latter is oxidized predominately by the mitochondrial aldehyde dehydrogenase. In both steps NADH was generated and is oxidized indirectly by mitochondrial electron transport system. The excessive amount of NADH and thus the reducing capacity in the mitochondrial electron transport system is thought to cause an increased leakage of mitochondrial reactive oxygen species (ROS), causing alcohol-induced oxidative stress. This has been shown to alter oxidative phosphorylation, mitochondrial proteome and mitochondrial dynamics. Mitochondrial fragmentation was observed in ethanol-treated hepatocytes [47]. Ethanol treatment also increased the sensitivity of mitochondrial permeability transition [46]. Mitochondrial DNA depletion was observed in livers of ethanol-fed mouse [56, 57]. Chronic ethanol consumption causes enhanced oxidative damage to mtDNA along with increased strand breakage, and other alterations in the structural integrity of mitochondrial DNA [58], which has been thought to cause ethanol-related liver pathology.
Mitophagy may be important to eliminate dysfunctional and potentially deleterious mitochondria. Ethanol-induced autophagy can selectively target to the damaged mitochondria as observed in both acute [47] and chronic [59] ethanol consumption. What is the mechanism of this selectivity has yet to be determined. It is possible that the PINK1-Parkin signaling may be involved. Immunoelectron microscopic studies indicated that expression of PINK1 was increased in mitochondria from ethanol-treated rats [59]. In addition, ethanol-mediated oxidative stress can also cause the translocation of the inducible form of heme oxygenase-1(HO-1) to the mitochondria, which in turn increases the recruitment of LC3 to the mitochondria [60]. The recognition of the damaged mitochondria could be mediated by multiple mechanisms and elucidation of these mechanisms is one of the important future works.
Another major mechanism of autophagy against alcohol liver disease is the elimination of intracellular lipids. Autophagosomes can transport the content of lipid droplets to the lysosome, in which lipids are degraded by the lysosomal acid lipase. This process, known as lipophagy [61], is still far from a complete understanding. Nevertheless, lipophagy occurs in ethanol condition [47]. Colocalization of LC3 with lipid droplet can be demonstrated in vivo and in vitro [5, 48, 62]. Hepatic TG level in alcoholic fatty liver disease models was reduced by activating autophagy, and was elevated when autophagy was inhibited [5, 48].
One of the main features of alcohol fatty liver disease is the excessive accumulation of fatty acids. Free fatty acids can be detrimental to hepatocytes. The esterified lipids are sequestered in lipid droplets (LD) and would be considered non-harmful, although de-esterification can occur, which would increase cellular free fatty acids level. Removal of lipid droplet may favor the equilibrium toward the esterification. Indeed, we found that when autophagy function was modulated by suppressive agents, such as chloroquine, or enhancing agents, such as rapamycin, the hepatic triglycerides level was increased or decreased, respectively, in both acute and chronic ALD models [48]. Notably, similar observations can be made in a HFD-induced non-alcoholic fatty liver disease (NAFLD) model [48], suggesting that a common mechanism of lipophagy is involved. The effect of these agents was specific to autophagy as it was confirmed with the use of specific siRNA to knock down autophagy genes [5].
The combined effects of lipophagy and mitophagy may eliminate both the source of ROS and a potential target of ROS, the lipid, which can amplify the oxidative stress. It is possible that mitophagy might be more important in ALD where mitochondrial injury seems to be more prominent, in comparison with the situation in NAFLD. Another piece of evidence to support this notion is that CYP2E1 plays an important role in the toxicity of ethanol and it has been found that autophagy can suppress several adverse effects of CYP2E1 [44, 63]. Thus autophagy can protect against CYP2E1-induced mitochondrial damage and ROS generation.
It would be interesting to compare the role of autophagy in other types of liver injury. The role of autophagy in removing damaged mitochondria and controlling oxidative stress is also thought to be important for ameliorating ischemia-reperfusion induced liver injury [64]. Specific removal of the insoluble mutant alpha-1 anti-trypsin by the autophagosome is considered to be the mechanism by which autophagy protects against liver injury in alpha1-antitrypsin deficiency [65, 66]. In contrast to its protective role in above cases, autophagy can also be detrimental to the liver. Many types of liver injury are accompanied with fibrosis, which can depend on the activity of hepatic stellate cells (HSC). Autophagy is shown to be important for the activation of HSC in the injury caused by carbon tetrachloride or thioacetamide [67, 68]. Autophagy facilitates the degradation of lipids in HSC, which is required for HSC activation. Under these conditions, suppressing autophagy in HSC via cell-specific Atg7 knockout resulted in attenuated fibrosis. It is thus important to dissect the different roles of autophagy in liver injury in order to better understand how this mechanism may be applied in clinical management.
5. Suppression of autophagy by ethanol
The effect of ethanol on autophagy may depend on the duration of ethanol treatment, the level of ethanol in diet, the way ethanol is administered and possibly other dietary or environmental factors. Acute ethanol treatment promotes autophagy. Chronic alcoholic treatment using the low fat Lieber-DeCarli model also showed an elevation of autophagy when ethanol was given at a lower level (accounting for 29% of the caloric need), but signs of suppression when ethanol was given at a higher level (accounting for 36% of the caloric need) [48]. However, in both cases, suppression of autophagy exacerbated liver injury while enhancement of autophagy improved the condition.
Ethanol can cause many other cellular changes, including proteasome inhibition, lysosome inhibition, endoplasmic reticulum (ER) stress and accumulation of aggregated proteins [69-74], all of which can be associated with autophagy function (Figure 1). Early studies showed the rate of hepatic protein degradation in ethanol-fed animals declined significantly [75], which might be due to declines in both proteasome and autophagy function, contributing to the development of hepatomegaly and the development of Mallory-Denk body (MDB). MDB is a characteristic of alcoholic liver disease and is positive for ubiquitin and p62/SQSTM1, a condition found in protein aggregates, and is often seen in autophagy deficiency. SQSTM1 is able to polymerize via an N-terminal PB1 domain and can interact with ubiquitinated proteins via the C-terminal UBA domain. SQSTM1 can bind directly to LC3 via a specific sequence motif [76]. The protein is itself degraded by autophagy and may serve to link ubiquitinated proteins to the autophagosomal membranes to enable their degradation in the lysosome. The presence of MDB suggests decline in autophagy during chronic ethanol condition. Indeed, by augmenting autophagy using rapamycin, an mTORC1 inhibitor, clearance of MDB can be achieved in a mouse model of MDB pathology [77].
Chronic alcoholic treatment could lead to decreases in both the number and the function of the lysosome, therefore reducing autophagic degradation [78]. On the other hand, the fusion of autophagosomes and lysosome could also be affected. This may be due to a number of factors, including a poor distribution of lysosomes at where autophagosomes locate [45] and change in lysosome membrane due to steatosis as observed in high fat diet induced autophagy inhibition [79].
6. Summary
It is quite evident that autophagy is critical in maintaining normal liver function and plays important roles in a variety of liver pathogenesis (Figure 1). Autophagy is an attractive therapeutic target. The benefits of autophagy in protecting against alcoholic liver disease suggest that it may be possible to pharmacologically elevate or restore autophagy function to improve the liver function. Indeed, applications of clinically available agents, such as rapamycin and carbamazepine (CBZ), in mouse models of ALD have demonstrated the anticipated benefits [48]. However, the complexity in the cellular composition of the liver and the diversity in the response of these cells to pathological or physiological stimuli may need a cell type-specific strategy for modulating autophagy function to achieve the beneficial effect.
References
- 1.Lumeng L, Crabb DW. Alcoholic liver disease. Curr Opin Gastroenterol. 2000;16(3):208–18. doi: 10.1097/00001574-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34(1):9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Day CP, James OF. Hepatic steatosis: innocent bystander or guilty party? Hepatology. 1998;27(6):1463–6. doi: 10.1002/hep.510270601. [DOI] [PubMed] [Google Scholar]
- 4.Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46(6):2032–9. doi: 10.1002/hep.22010. [DOI] [PubMed] [Google Scholar]
- 5.Ding WX, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139(5):1740–52. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33(2):437–49. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36(12):2445–62. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–82. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 10.Farre JC, et al. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21(4):522–30. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–17. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25(4):455–60. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110(6):1923–33. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge L, Schekman R. The ER-Golgi intermediate compartment feeds the phagophore membrane. Autophagy. 2014;10(1):170–2. doi: 10.4161/auto.26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau K, Rubinsztein DC. The plasma membrane as a control center for autophagy. Autophagy. 2012;8(5):861–3. doi: 10.4161/auto.20060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karanasios E, et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013;126(Pt 22):5224–38. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi-Nishino M, et al. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2010;6(2):301–3. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 20.Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- 21.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–76. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan D, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A. 2013;110(48):19432–7. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 24.Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 26.Matsunaga K, Noda T, Yoshimori T. Binding Rubicon to cross the Rubicon. Autophagy. 2009;5(6):876–7. doi: 10.4161/auto.9098. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 31.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 32.Ichimura Y, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279(39):40584–92. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 33.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 34.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alers S, et al. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1-2):290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2011;23(2):135–42. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4(2):141–50. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 41.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14(9):1576–82. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 42.Thomes PG, et al. Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem Biophys Res Commun. 2012;417(1):262–7. doi: 10.1016/j.bbrc.2011.11.097. [DOI] [PubMed] [Google Scholar]
- 43.Sid B, Verrax J, Calderon PB. Role of AMPK activation in oxidative cell damage: Implications for alcohol-induced liver disease. Biochem Pharmacol. 2013;86(2):200–9. doi: 10.1016/j.bcp.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Wu D, et al. Alcohol steatosis and cytotoxicity: the role of cytochrome P4502E1 and autophagy. Free Radic Biol Med. 2012;53(6):1346–57. doi: 10.1016/j.freeradbiomed.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomes PG, et al. Multilevel regulation of autophagosome content by ethanol oxidation in HepG2 cells. Autophagy. 2013;9(1):63–73. doi: 10.4161/auto.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King AL, et al. Involvement of the mitochondrial permeability transition pore in chronic ethanol-mediated liver injury in mice. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G265–77. doi: 10.1152/ajpgi.00278.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding WX, Li M, Yin XM. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7(2):248–9. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CW, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58(5):993–9. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni HM, et al. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am J Pathol. 2013;183(6):1815–25. doi: 10.1016/j.ajpath.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajmo JM, et al. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295(4):G833–42. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung S, et al. Regulation of SIRT1 in cellular functions: role of polyphenols. Arch Biochem Biophys. 2010;501(1):79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nepal S, Park PH. Activation of autophagy by globular adiponectin attenuates ethanol-induced apoptosis in HepG2 cells: involvement of AMPK/FoxO3A axis. Biochim Biophys Acta. 2013;1833(10):2111–25. doi: 10.1016/j.bbamcr.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Liuzzi JP, Yoo C. Role of zinc in the regulation of autophagy during ethanol exposure in human hepatoma cells. Biol Trace Elem Res. 2013;156(1-3):350–6. doi: 10.1007/s12011-013-9816-3. [DOI] [PubMed] [Google Scholar]
- 54.Green DR, Levine B. To Be or Not to Be? How Selective Autophagy and Cell Death Govern Cell Fate. Cell. 2014;157(1):65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serviddio G, et al. Targeting mitochondria: a new promising approach for the treatment of liver diseases. Curr Med Chem. 2010;17(22):2325–37. doi: 10.2174/092986710791698530. [DOI] [PubMed] [Google Scholar]
- 56.Mansouri A, et al. Acute ethanol administration oxidatively damages and depletes mitochondrial dna in mouse liver, brain, heart, and skeletal muscles: protective effects of antioxidants. J Pharmacol Exp Ther. 2001;298(2):737–43. [PubMed] [Google Scholar]
- 57.Mansouri A, et al. An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice. Gastroenterology. 1999;117(1):181–90. doi: 10.1016/s0016-5085(99)70566-4. [DOI] [PubMed] [Google Scholar]
- 58.Cahill A, et al. Chronic ethanol consumption causes alterations in the structural integrity of mitochondrial DNA in aged rats. Hepatology. 1999;30(4):881–8. doi: 10.1002/hep.510300434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eid N, et al. Elevated autophagic sequestration of mitochondria and lipid droplets in steatotic hepatocytes of chronic ethanol-treated rats: an immunohistochemical and electron microscopic study. J Mol Histol. 2013;44(3):311–26. doi: 10.1007/s10735-013-9483-x. [DOI] [PubMed] [Google Scholar]
- 60.Bansal S, Biswas G, Avadhani NG. Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol. 2014;2:273–83. doi: 10.1016/j.redox.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kashima J, et al. Immunohistochemical study of the autophagy marker microtubule-associated protein 1 light chain 3 in normal and steatotic human livers. Hepatol Res. 2013 doi: 10.1111/hepr.12183. [DOI] [PubMed] [Google Scholar]
- 63.Wu D, Cederbaum AI. Inhibition of autophagy promotes CYP2E1-dependent toxicity in HepG2 cells via elevated oxidative stress, mitochondria dysfunction and activation of p38 and JNK MAPK. Redox Biol. 2013;1(1):552–65. doi: 10.1016/j.redox.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evankovich J, et al. Calcium/calmodulin-dependent protein kinase IV limits organ damage in hepatic ischemia-reperfusion injury through induction of autophagy. Am J Physiol Gastrointest Liver Physiol. 2012;303(2):G189–98. doi: 10.1152/ajpgi.00051.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279(5):G961–74. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- 66.Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 67.Thoen LF, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55(6):1353–60. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Hernandez-Gea V, et al. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59(1):98–104. doi: 10.1016/j.jhep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baraona E, et al. Alcoholic hepatomegaly: accumulation of protein in the liver. Science. 1975;190(4216):794–5. doi: 10.1126/science.1198096. [DOI] [PubMed] [Google Scholar]
- 70.Kaplowitz N, Ji C. Unfolding new mechanisms of alcoholic liver disease in the endoplasmic reticulum. J Gastroenterol Hepatol. 2006;21(Suppl 3):S7–9. doi: 10.1111/j.1440-1746.2006.04581.x. [DOI] [PubMed] [Google Scholar]
- 71.Donohue TM, Jr, et al. Role of the proteasome in ethanol-induced liver pathology. Alcohol Clin Exp Res. 2007;31(9):1446–59. doi: 10.1111/j.1530-0277.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 72.Zatloukal K, et al. From Mallory to Mallory-Denk bodies: what, how and why? Experimental cell research. 2007;313(10):2033–49. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 73.Ji C. Dissection of endoplasmic reticulum stress signaling in alcoholic and non-alcoholic liver injury. J Gastroenterol Hepatol. 2008;23(Suppl 1):S16–24. doi: 10.1111/j.1440-1746.2007.05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riley NE, et al. The Mallory body as an aggresome: in vitro studies. Exp Mol Pathol. 2002;72(1):17–23. doi: 10.1006/exmp.2001.2413. [DOI] [PubMed] [Google Scholar]
- 75.Donohue TM, Jr, Zetterman RK, Tuma DJ. Effect of chronic ethanol administration on protein catabolism in rat liver. Alcohol Clin Exp Res. 1989;13(1):49–57. doi: 10.1111/j.1530-0277.1989.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 76.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 77.Harada M, et al. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47(6):2026–35. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 78.Dolganiuc A, et al. Autophagy in alcohol-induced liver diseases. Alcohol Clin Exp Res. 2012;36(8):1301–8. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24(8):3052–65. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]