Abstract
In addition to the cap-dependent mechanism, eukaryotic initiation of translation can occur by a cap-independent mechanism which directs ribosomes to defined start codons enabled by internal ribosome entry site (IRES) elements. IRES elements from poliovirus and encephalomyocarditis virus are often used to construct bi- or oligocistronic expression vectors to co-express various genes from one mRNA. We found that while cap-dependent translation initiation from bicistronic mRNAs remains comparable to monocistronic expression, internal initiation mediated by these viral IRESs is often very inefficient. Expression of bicistronic expression vectors containing the hepatitis B virus core antigen (HBcAg) together with various cytokines in the second cistron of bicistronic mRNAs gave rise to very low levels of the tested cytokines. On the other hand, the HBcAg was well expressed when positioned in the second cistron. This suggests that the arrangement of cistrons in a bicistronic setting is crucial for IRES-dependent translation of the second cistron. A systematic examination of expression of reporter cistrons from bicistronic mRNAs with respect to position was carried out. Using the dual luciferase assay system we show that the composition of reading frames on a bicistronic mRNA and the order in which they are arranged define the strength of IRES-dependent translation. Although the cellular environment and the nature of the IRES element influence translation strength the dominant determinant is the nature and the arrangement of cistrons on the mRNA.
INTRODUCTION
Internal initiation of translation mediated by internal ribosome entry site (IRES) elements (1) in artificial expression constructs has enabled the development of expression vectors in which two or more unrelated reading frames are expressed from a single transcription unit (2–5). The major advantage of this technique, which occurs naturally in prokaryotes but has not been detected in higher eukaryotes, lies in strict co-expression of transgenes in vitro, in cell culture and transgenic animals and plants (6–10). Furthermore, it avoids the side-effects of using multiple promoters in vectors with limited size retroviral vectors (11–14).
IRES elements have been isolated from picornaviruses (15,16), other animal viruses (e.g. hepatitis C virus, hepatitis A virus and retroviruses) and mammalian and Drosophila RNAs (17–26). They differ in their primary sequence. Some IRES elements show similarities in their secondary structures (27,28). In several picornaviruses the internal entry process seems to require the cellular polypyrimidine tract-binding protein (PTB) (29), but PTB binding does not necessarily correlate with its activity (30). It seems that the activity of IRES elements is controlled by different cellular factors (31,32). Since the known IRES elements show differential activity and their activity depends on cell type (33), the composition of cellular factors binding to IRES elements is responsible for translation efficiency.
The strength of translation initiated from different IRES elements differs significantly (7,33,34). The IRES elements from poliovirus and encephalomyocarditis virus (EMCV) are most commonly used for construction of bicistronic expression constructs and vectors (3,35). Making use of IRES elements, tri- and polycistronic expression cassettes can be constructed. Some examples of tricistronic expression have been published (see for example 36–38).
It is generally believed that the strength of internal translational initiation is defined by the nature of the IRES element and the cellular context. However, construction of many bi- and oligocistronic expression constructs has shown us that other parameters resulting from construction of the vectors strongly influence IRES efficiency. This would indicate that protein expression from artificial oligocistronic expression units cannot be predicted. Here we describe some of these observations. In experiments with more defined conditions using the dual luciferase system we show that the constellation of two reading frames in a bicistronic expression setting can alter IRES-dependent translation from different IRES elements by orders of magnitude. This indicates that the nature of the reading frames assembled on one transcription unit dictates the strength of IRES-dependent translation. Further experiments suggest that the nature of the first cistron defines the strength of downstream IRES-dependent translation.
MATERIALS AND METHODS
Plasmid constructs
Constructions were carried out by standard molecular cloning techniques (39). All of the bicistronic plasmids of the pCI series are based on the pCI backbone (Promega). All bicistronic plasmids of the pBS or pBC series are derivatives of pSBC-1 and -2 (3).
Two new vectors based on pSBC were constructed. pMS, a monocistronic expression plasmid contains the SV40 promoter and a downstream multiple cloning site (MCS), followed by a unique NotI site. The second monocistronic plasmid, pMSP, is identical to pMS except that the poliovirus-derived IRES sequence is inserted between the promoter and MCS. The fragment spanning from the IRES to the MCS is flanked by two EagI sites. In the monocistronic plasmid pMC the SV40 promoter in pMS was exchanged for the human cytomegalovirus (CMV) immediate-early enhancer/promoter region derived from pCI. After introducing the gene(s) of interest (e.g. Fluc, Fluz, Rluc or cat) into the MCS of the monocistronic vectors, bicistronic expression plasmids were created by inserting the IRES plus the gene of interest as EagI fragments from pMSP into NotI-restricted monocistronic plasmids derived from pMS or pMC. This cloning step results in a single NotI site 3′ to the second cistron in the bicistronic constructs. This allows generation of tri- and oligocistronic vectors by adding further EagI fragments from monocistronic pMSP plasmids (35).
Monocistronic expression vectors of the pMSP type carrying the EMCV-derived IRES sequence in combination with the gene of interest were constructed using overlap extension PCR in order to fuse the start codon in the correct position.
All hepatitis B virus core antigen (HBcAg) and cytokine expressing vectors are based on the pCI plasmid backbone. The poliovirus IRES sequence containing the MCS site from pVBC-3 (40) was cloned into pCI using EcoRI and NotI, resulting in plasmid pCI P, which contains EcoRI and SalI sites upstream and a NotI site downstream of the IRES sequence.
The HBcAg sequence (40,41) was blunt-end cloned into pCI P using Klenow-treated SalI to create the monocistronic pCI HBc vector containing the poliovirus IRES downstream of the HBcAg. The cytokine genes IFNγ and GM-CSF (42,43) were introduced downstream of the IRES using the NotI site of pCI HBc, creating bicistronic pCI Cyt P HBc vectors encoding HBcAg and either cytokine.
To construct monocistronic plasmids expressing IFNγ or GM-CSF, the gene of interest was introduced into pCI P using EcoRI and SalI. The HBcAg encoding sequence was cloned downstream of the IRES into the monocistronic NotI site of cytokine-expressing vectors, resulting in bicistronic pCI Cyt P HBc vectors.
The IL-12 subunit p35 and p40 genes were cloned into vector pCI P upstream of the IRES sequence to create monocistronic constructs pCI p35 and pCI p40, respectively. Bicistronic vectors pCI-p35-P p40 and pCI p40 P p35 were constructed by inserting either the p40 or p35 gene downstream of the IRES into pCI p35 and pCI p40, respectively.
Series of bicistronic constructs with reporter genes encoding firefly luciferase (Fluc), a synthetic variant of firefly luciferase (Fluz) (Promega), Renilla luciferase (Rluc) or chloramphenicolacetyl transferase (cat) were created. They contain either the poliovirus IRES element (pBSFlucPRluc, pBSRlucPFluc, pBSFluzPRluc, pBCFlucPRluc, pBCRlucPFluc, pBScatPFluc, pBSFlucPcat, pBSFluzPcat, pBScatPRluc and pBSRlucPcat), the EMCV IRES element (pBSFlucERluc, pBSRlucEFluc, pBCFlucERluc and pBCRlucEFluc) or the human NRF IRES (pBSRlucNFluc). The coding sequences of these reporters were derived from de Wet et al. (44) for Fluc, from Promega for Rluc and Fluz and from Gorman et al. (45) for cat. All PCR created parts of the expression plasmids were verified by DNA sequencing.
Cell culture and transient transfections
Mouse C243 (46), mouse LMTK (ATCC no. CCL 1.3), hamster BHK-21 (ATCC no. CCL-10) and chicken LMH cells (a generous gift of Dr H.-J. Schlicht, Ulm, Germany) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS), antibiotics and 2 mM glutamine. Murine C2C12 cells (ATCC no. CRL-9096) were cultured in RPMI 1640 (Gibco BRL) supplemented with 5% FCS, antibiotics and glutamine. DNA was transiently transfected using the calcium phosphate co-precipitation method. For reporter gene expression cells from the exponential growth phase were seeded (7.5 × 104 cells) into six-well plates the day before transfection. The medium was changed 4 h prior to transfection and renewed 18 h post-transfection. For transient transfection 1 µg reporter plasmid together with 4 µg high molecular weight DNA from LMTK– cells per well were used. For transient transfection of the DNA immunisation vectors LMH, BHK-21 and C2C12 cells (2 × 106) were treated with 10 µg plasmid DNA (encoding murine IFNγ, murine GM-CSF and the murine IL-12 p35 and p40 subunits). Transfection efficiency was standardised by co-transfection with one of the monocistronic plasmids expressing either Fluc, Rluc or cat. Two days post-transfection cells or supernatant were harvested and analysed.
All transfections were carried out between three and six times. Although the absolute amounts of expression varied, the standard deviations of the ratios cistron 2/1 were <20%. All data shown derive from representative experiments.
Analysis of gene products
The supernatants were analysed for release of cytokines by conventional double sandwich ELISA as described (42,43). For detection of HBcAg the cells were lysed in PBS containing 1% Triton X-100. Immunoprecipitation was performed using a rabbit anti-C antiserum (αHC1) and protein A–Sepharose. Immune complexes were processed for SDS–PAGE and transferred onto nitrocellulose. Detection of HBcAg was performed using αHX1 and alkaline phosphatase-conjugated anti-rabbit IgG and subsequent staining with BCIP and NBT.
Luciferase activity of firefly and Renilla luciferase was measured with the Dual Luciferase System Assay (Promega) according to the manufacturer’s instructions. cat analysis was performed by cat-ELISA (Boehringer Mannheim) as described in the manufacturer’s instructions. Mean values from double or triple determinations were taken. Extracts from transfected cells were prepared by passive lysis. The amount of specific first cistron reporter protein expression was normalised to the protein content, which was measured with BCA protein assay reagent (Pierce). Experiments were only considered if transfection efficiency between the different experiments was comparable. Relative expression of two different genes in a bicistronic construct was determined by dividing the value for gene activity of the gene in the second cistron by the value obtained for the gene in the first cistron.
RESULTS
Translation of cytokines in the second cistron is very inefficient
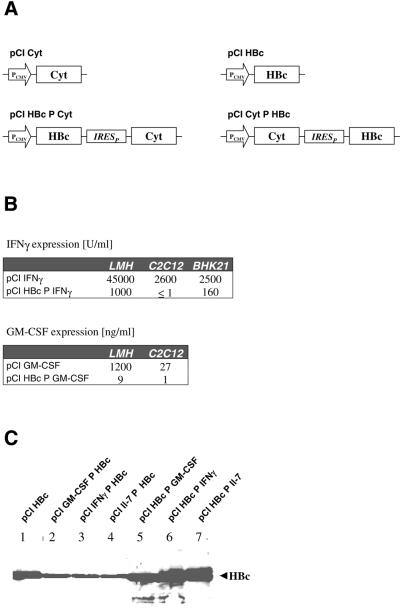
Figure 1A shows the transcription units of expression plasmids encoding the HBcAg and diverse cytokines in monocistronic and bicistronic configurations. Expression of all genes, HBcAg as well as the cytokines positioned in the first cistron and thereby translated in a cap-dependent manner, was clearly detectable. Importantly, no significant differences could be found between mono- and bicistronic configurations (Fig. 1B and C and data not shown). This indicates that comparable steady-state amounts of mRNA from mono- and bicistronic expression plasmids were produced. HBcAg was also well expressed when positioned in the second cistron of the bicistronic expression plasmids (Fig. 1C). The protein amount was ∼3- to 10-fold lower when compared to monocistronic expression or if expressed as the first cistron in a bicistronic set-up. This reduction was expected, since earlier data showed that poliovirus IRES-dependent translation was less efficient than cap-dependent translation (47).
Figure 1.
Expression of cytokines and HBcAg (HBc) from mono- and bicistronic mRNAs. (A) Schematic diagram of mono- and bicistronic vector constructs (pCI based) used to measure protein expression after transient transfection into different cell lines. PCMV in the open arrow marks the CMV promoter. Cyt indicates the cytokine gene for either IFNγ, GM-CSF or Il-7. IRESP represents the poliovirus-derived IRES element. (B) Determination of IFNγ and GM-CSF in the supernatant of the indicated cells by ELISA. The indicated plasmids were transfected into the cell lines LMH, C2C12 and BHK21 and cytokine concentrations were determined after a transient expression period of 48 h. (C) Detection of HBcAg (HBc) in the lysates of LMH cells by western blot analysis. HBcAg was expressed either monocistronically (lane 1), as a first (lanes 5–7) or as a second (lanes 2–4) cistron.
In contrast, expression of the cytokines IFNγ and GM-CSF from second cistrons was very low (Fig. 1B). Compared to expression from the first cistron this amounts to a >100-fold difference. The strength of HBcAg expression from these bicistronic mRNAs (Fig. 1C) indicates inefficient IRES-dependent translation of the cytokines tested.
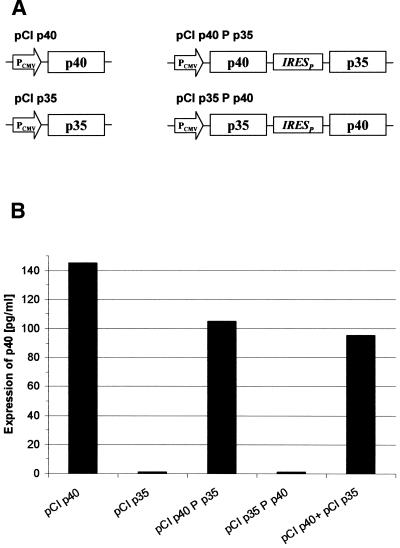
Another example of unexpected low translation from a second cistron was seen when the two peptide chains (p40 and p35) of IL-12 were expressed in a bicistronic manner. p40 was well expressed from monocistronic mRNA or as the first cistron of a bicistronic mRNA. However, it could hardly be detected when it was positioned in the second cistron (Fig. 2).
Figure 2.
Expression of IL-12 subunit p40 from mono- and bicistronic expression plasmids. (A) Schematic presentation of mono- and bicistronic vector constructs used to express the two subunits p35 and p40 of IL-12 in the first or second cistron. The genes encoding p35 and p40 were separated by the poliovirus-derived IRES element (IRESP) and were expressed under control of the CMV promoter (PCMV). (B) Detection of the p40 subunit of IL-12 by ELISA. Plasmids as described in (A) were transfected into LMH cells and the gene product of the p40 subunit was determined in the supernatant after a transient expression period of 48 h. One representative experiment from a series of independent experiments with identical results is shown.
The above-described key observation, namely inefficiency of IRES-dependent translation of certain cytokines, was found in cell lines from different species.
These observations led us to conclude that IRES-dependent translation is not predictable and obviously depends on the composition of the mRNA. Which RNA elements and other factors, like expression strength, contribute to this phenomenon and if they are of a general nature cannot be deduced from these results. We thus tried to verify these findings with other genes and find principles which allow the prediction of IRES-dependent translation efficiency.
Position-specific expression of two luciferase genes
Reporter systems for which translation was found to be unpredictable from IRES-containing mRNAs are the luciferases from firefly (Fluc) and Renilla (Rluc). In our study we have employed both luciferases as reporters for determination of translational efficiency of two reading frames from one mRNA. The composition of different protein coding and non-coding elements in a new context should not only influence transcription, but also mRNA processing, transport, stability, destination and translation (48,49). However, our results showed no significant influence on transcription (data not shown). mRNA stability seems to be dictated by sequence elements which are responsible for rapid degradation. In experiments with the luciferase genes and IRES elements we could detect little influence on the steady-state level (3-fold maximum) of the mRNAs produced by the constructs (data not shown). Our experiments further confirmed that translation of the first (cap-dependent) cistron paralleled the steady-state level of the respective mRNA but was not significantly influenced by the protein coding sequence on the mRNA. For this reason translation of the cap-dependent reading frame could be used as an internal standard for determination of the strength of IRES-dependent translation of the downstream reading frame(s). The ratio between the luciferase activities from the second and the first cistron defines the strength of IRES-dependent translation. This system, with its high accuracy and low error rate of activity determination, significantly increased the reproducibility of the results from transfection assays.
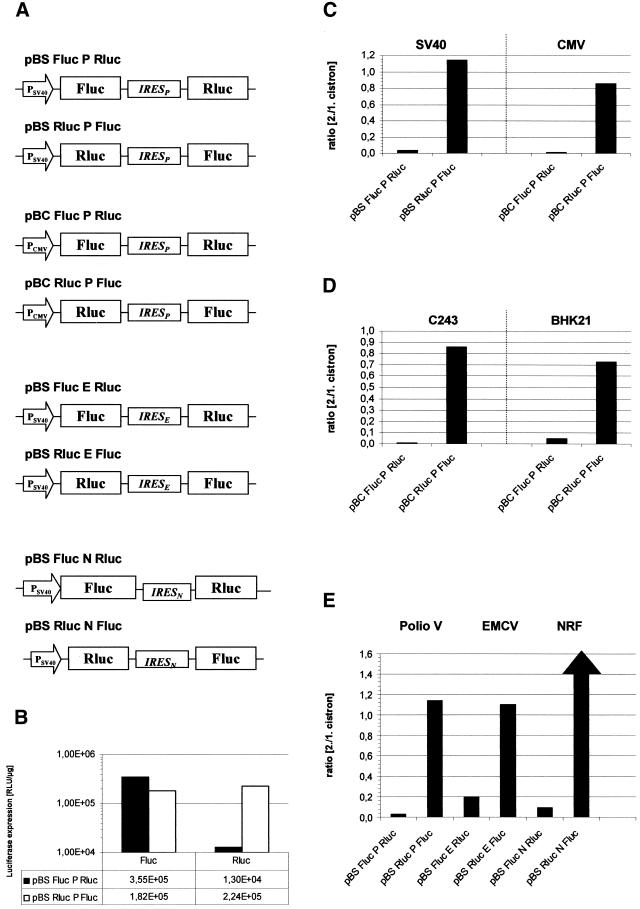
Figure 3 shows expression of the luciferases from bicistronic mRNAs with various compositions of the same sequence elements in C243 cells. The luciferase values for the first cistrons were very similar irrespective of the gene (Fluc or Rluc) and the existence of a downstream IRES-directed second cistron. This is compatible with the assumption that the stabilities of the newly assembled mRNAs are comparable and that cap-dependent translation is not influenced by the downstream elements. This allowed us to use the quotient ratio of cistron 2/1 expression. Expression of the second cistron, however, was dependent on the arrangement of the luciferases on the mRNA. While IRES-dependent translation of Fluc was very strong, that of Rluc appeared ∼30-fold less efficient (Fig. 3B). Thus, expression of the two luciferases reflects the unexpected observations of bicistronic expression of HBcAg and cytokines.
Figure 3.
Determination of different parameters affecting expression level of reporter genes in bicistronic constructs. (A) Schematic diagram of bicistronic vector constructs used to measure protein expression of firefly and Renilla luciferase after transient transfection of C243 cells. PSV40 and PCMV in the open arrow mark the SV40 and CMV promoters, respectively. The genes encoding firefly luciferase (Fluc) and Renilla luciferase (Rluc) are separated by IRES elements from poliovirus (IRESP), EMCV (IRESE) or huNRF (IRESN). The pSBC vector backbone was used. (B) The plasmids pBSFlucPRluc and pBSRlucPFluc were transiently transfected into C243 cells. After 48 h the activities of firefly and Renilla luciferase were determined as described in Materials and Methods. One representative from at least six independent experiments with comparable results is shown. (C) The same bicistronic transcription units as in (B), driven by either the SV40 promoter (pBS) or the CMV promoter (pBC), were transfected into C243 cells. Activities from Fluc and Rluc were determined as described in (B) and the ratio (cistron 2/1) was determined by dividing the luciferase activities from the second cistron by those derived from the first cistron. The absolute luciferase values for the pBS plasmids are indicated in (B); the values for the first cistron from pBC plasmids were 5 × 106 U/µg for Fluc and 4.5 × 106 U/µg for Rluc. (D) The indicated plasmids were transfected into C243 and BHK 21 cells. The samples were assayed and calculated as described in (C). First cistron derived values were 6 × 105 U/µg for Fluc, 7.3 × 105 U/µg for Rluc in C243 cells and 8 × 105 U/µg for Fluc, 9.4 × 105 U/µg for Rluc in BHK21 cells. (E) Three sets of bicistronic plasmids containing IRES elements derived from poliovirus, EMCV or human NRF were assayed in C234 cells. The results were plotted as in (C) and (D). Due to the high strength of the NRF IRES element in the Rluc-IRES-Fluc arrangement the calculated ratio of 30 is not depicted to scale. The absolute luciferase values for all first cistron values were between 5.4 × 105 and 7 × 105 U/µg for Fluc and between 3 × 105 and 6.8 × 105 U/µg for Rluc.
We examined whether this phenomenon could be due to the abundance and composition of cellular factors which influence IRES-mediated translation. This was done by expression of the mRNAs from different promoters and in different cellular backgrounds (Fig. 3C and D, respectively). Indeed, the abundance of steady-state mRNA as judged from expression of the first cistron luciferases differed >10-fold (data not shown). However, the ratio cistron 2/1 was nearly identical. This indicates that specific cellular conditions are not responsible for the differences in IRES-dependent translation. In contrast, it seemed as though the phenomenon is of a general nature.
Influence of the specificity of the IRES element
All bicistronic constructs described above are based on the poliovirus-derived IRES element. We asked whether the nature of the IRES element would influence the efficiency of second cistron translation. Two IRES elements of different origins, types and strengths were used to replace the poliovirus IRES element in the bicistronic expression plasmids. The EMCV-derived IRES element is known to be of similar strength compared to the poliovirus element, although it is classified as a type II IRES (33). The cellular IRES element from the human NRF gene is a type I IRES, but shows superior activity compared to the poliovirus IRES (34). Figure 3E shows the relative expression values from the relevant constructs. Again, IRES-mediated translation of Fluc was strong or very strong in the case of the NRF IRES. However, the inverse orientation of the luciferase cistrons (Fluc-IRES-Rluc) did not allow strong IRES-mediated translation of Rluc. Rluc expression from the second cistron was significantly lower than Fluc expression from the first cistron. The relative efficiency differed amongst the IRES elements. In this conformation IRES-dependent translation is extremely low. Thus, translation from the EMCV IRES showed the best performance. Nevertheless, the phenomenon of mRNA composition-dependent IRES activity was seen with all three IRES elements, indicative of a general mechanism.
Influences of position and sequence of reading frames
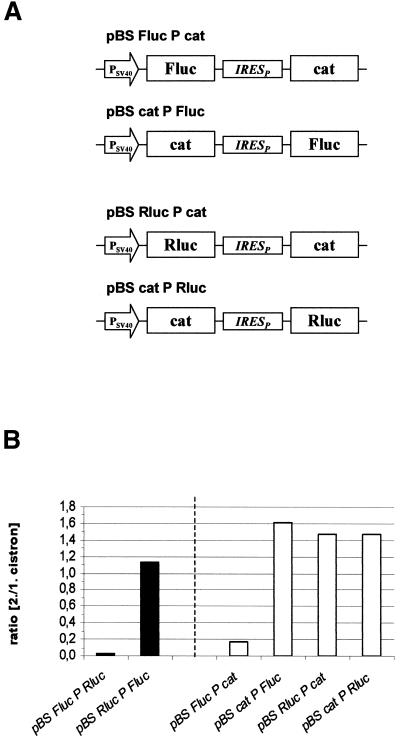
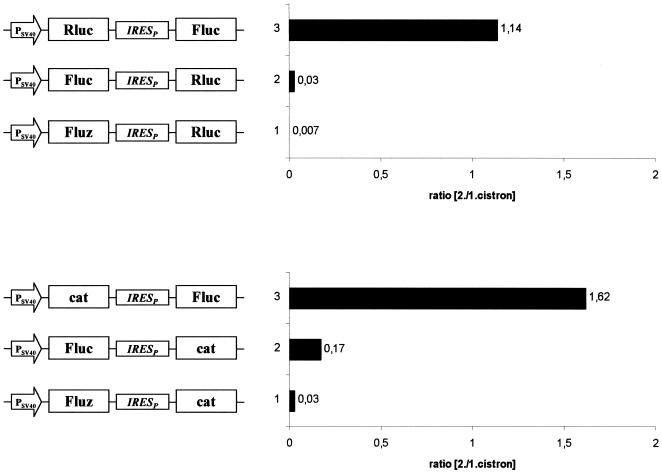
Based on the assumption that one of the luciferase cistrons in the bicistronic construct (Fluc-Rluc configuration) exerts a position-dependent negative effect on IRES-mediated translation, bicistronic expression plasmids in which the reporter gene cat replaced either of the luciferases were constructed (Fig. 4A). Constructs composed of cat and Rluc showed efficient IRES-mediated translation in both combinations, indicating that Rluc does not inhibit its own translation when positioned as a second cistron (Fig. 4B). The results further show that cat has no negative effect on IRES-mediated translation in either position. Constructs harboring cat and Fluc exerted strong translation of the downstream Fluc but very low expression of cat as the second cistron (Fig. 4B). These data suggest that the positioning of Fluc in the first cistron position exerts a negative effect on IRES-driven cistrons (cat and Rluc).
Figure 4.
Effect of arrangement of luciferase reporter genes on cap-dependent and IRES-dependent translation. (A) Schematic representation of expression constructs used to measure reporter gene expression after transient transfection of C243 cells. (B) Expression of firefly luciferase, Renilla luciferase and cat from the indicated plasmids were determined. The data in the left half were determined as described in Figure 3C–E. cat values from comparable monocistronic constructs were converted to luciferase analogous values in order to obtain comparable relative expression values. The absolute values for the first cistrons (luciferases) were 3.8 × 105 U/µg for pBS Fluc PRLuc, 2.4 × 105 U/µg for pBS RLuc PFluc, 4.5 × 105 U/µg for pBS Fluc Pcat and 3.6 × 105 U/µg for pBS LucPcat.
In order to determine whether the translation product of Fluc or the translation process as such was responsible for this effect, different experiments were carried out (not shown). First, monocistronic Fluc was co-expressed with pBScatPRluc. The results show no hindrance of Rluc expression, proving that the presence of cellular Fluc RNA or protein does not inhibit IRES-mediated translation of Rluc. Secondly, the poliovirus-derived protease 2A was expressed together with different bicistronic constructs (pBScatPRluc and pBSFlucPRluc). Protease 2A inhibited cap-dependent translation ∼3-fold. However, the level of IRES-mediated translation was not significantly altered in these experiments. The ratio cistron 2/1 from pBSFlucPRluc and pBSFlucPcat was increased ∼3-fold in the presence of the protease. These data again give no indication of an influence of the cap-dependent Fluc translation process on downstream IRES-mediated translation.
The presented data point towards an influence of sequence or structure of Fluc when positioned upstream of the IRES. Thus, sequence alterations in Fluc in pBSFlucPRluc should lead to a different efficiency of IRES-mediated translation. We used a variant Fluc cDNA, Fluz, in which 76.1% of the coding nucleotide sequence is altered. Fluz replaced Fluc as the first cistron in two bicistronic constructs (Fig. 5). Fluz protein translation showed little alteration in comparison to Fluc and did not change the yield of firefly luciferase activity in C243 cells (data not shown). However, IRES-mediated translation of both Rluc and cat was more strongly reduced (Fig. 5). This indicates that the nucleotide exchanges introduced led to an even stronger negative effect on downstream IRES-mediated translation.
Figure 5.
Effect of arrangement of wild-type and mutant firefly luciferase reporter genes on cap-dependent and IRES-dependent translation. A schematic representation of expression constructs used to measure reporter gene expression after transient transfection of C243 cells is shown on the left. Relative expression of wild-type firefly luciferase, Renilla luciferase, mutant firefly luciferase (Fluz) and cat from the indicated plasmids was determined. cat values from comparable monocistronic constructs were converted to firefly luciferase analogous values in order to obtain comparable relative expression values. The luciferase values (first cistron) for all six constructs were between 2.4 × 105 and 7 × 105 U/µg.
DISCUSSION
Bicistronic expression is frequently used in transgene expression in mammalian cell culture or in transgenic animals where co-expression of heterologous genes is required (35,50). One of the classical applications is the establishment of stable cell lines producing recombinant proteins by co-expression of a selectable marker together with the protein of interest (51). It is further used to achieve overexpression of recombinant proteins in baby hamster kidney (BHK) and Chinese hamster ovary (CHO) cells. Other applications include co-expression of genes which are needed for specific applications, like immunisation to co-express an antigen and a co-stimulatory protein. Although bi- or oligocistronic expression has not yet been convincingly demonstrated as a naturally occurring process in mammalian cells, it is a powerful tool. It is further important to note that many examples of successful bicistronic gene expression have demonstrated IRES-dependent translation of artificial mRNAs. This applies not only to mammalian genes but also to reporter genes from different species. However, failures of bicistronic applications have been observed, although these were often not published. Our motivation was an inability to sufficiently co-express HBcAg antigen and cytokines in DNA immunisation experiments.
With the data shown in this work we demonstrate that synthetic mRNA assembly can exert strong negative effects on IRES-mediated translation. Our data indicate that certain coding sequences can exert a negative effect on IRES-mediated translation, e.g. those encoding the HBcAg and two firefly luciferases. Cap-dependent translation was little influenced by the assembly of the cistrons and IRES elements on the mRNA. This does not mean that cap-dependent translation is completely independent of the mRNA structure. There are numerous reports on the influence of mRNA sequence, in particular the 3′-UTR. For this reason we have kept the same 3′-UTR in the expression plasmids.
Interestingly, IRES elements of different nature and sequence are affected by this negative influence, although to different degrees. This indicates that the inhibitory effect is mediated by a common component(s) of the reinitiation machinery, but one that is not part of the cap-dependent translation machinery.
The EMCV IRES element is the least affected of the three elements tested when the unfavourable Fluc was positioned in the first cistron. This might explain the high frequency of successful use of this IRES element. Interestingly, this IRES element is the only one for which efficient translation in vitro has been reported (52).
Neither the translation process per se nor the resulting translation products seem to be responsible for the inhibitory activity on IRES-mediated translation. The current data direct us more towards the sequence of the inhibitory cistrons. A comparison of the sequences of HBcAg and firefly luciferase does not show any homology. This does not necessarily exclude a sequence-mediated effect, but effects of secondary structure might be more convincing.
The most intriguing question concerns the fact that the inhibitory activity is only exerted from the first cistron position. Since in our experiments we could not achieve complete inhibition of the first cistron by poliovirus 2A protease we cannot fully exclude an effect of the cap-dependent translation process. However, complete resistance of IRES-mediated translation to partial cap-dependent translation inhibition leads us to believe that it is the presence of a particular sequence or structure which mediates the inhibitory effect. How this is transmitted to the downstream IRES-initiated translation process is not understood.
In summary, our data show that certain cistrons can inhibit the activity of IRES elements of different sequence and category in translational reinitiation. We are still far from a mechanistic understanding of the phenomena described. The above interpretation of the results may not necessarily be valid for other constructs. Nevertheless, with this work we have developed a basis for a more systematic construction of new oligocistronic expression plasmids. The frequent use of IRES elements and the lack of alternatives deserves further examination, with the goal of predictable construction of bi- and oligocistronic vectors.
References
- 1.Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- 2.Davies M.V. and Kaufman,R.J. (1992) Internal translation initiation in the design of improved expression vectors. Curr. Opin. Biotechnol., 3, 512–517. [DOI] [PubMed] [Google Scholar]
- 3.Dirks W., Wirth,M. and Hauser,H. (1993) Bicistronic transcription units for gene expression in mammalian cells. Gene, 128, 247–249. [DOI] [PubMed] [Google Scholar]
- 4.Mountford P.S. and Smith,A.G. (1995) Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet., 11, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Salas E. (1999) Internal ribosome entry site biology and its use in expression vectors. Curr. Opin. Biotechnol., 10, 458–464. [DOI] [PubMed] [Google Scholar]
- 6.Dirks W. and Hauser,H. (1994) Equimolar expression of two protein chains in mammalian cells. In Spier,R., Griffiths,B. and Berthold,W. (eds), Animal Cell Technology: Products of Today, Prospects for Tomorrow. Butterworths, Oxford, UK, pp. 610–616.
- 7.Borman A.M., Bailly,J.L., Girard,M. and Kean,K.M. (1995) Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res., 23, 3656–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchhoff S., Köster,M., Wirth,M., Schaper,F., Gossen,M., Bujard,H. and Hauser,H. (1995) Identification of mammalian cell clones exhibiting highly regulated expression from inducible promoters. Trends Genet., 11, 219–220. [DOI] [PubMed] [Google Scholar]
- 9.Wild J., Gruner,B., Metzger,K., Kuhrober,A., Pudollek,H.P., Hauser,H., Schirmbeck,R. and Reimann,J. (1998) Polyvalent vaccination against hepatitis B surface and core antigen using a dicistronic expression plasmid. Vaccine, 16, 353–360. [DOI] [PubMed] [Google Scholar]
- 10.Fussenegger M., Bailey,J., Hauser,H. and Mueller,P.P. (1999) Genetic optimization of recombinant protein production by mammalian cells. Trends Biotechnol., 17, 43–50. [DOI] [PubMed] [Google Scholar]
- 11.Adam M.A., Ramesh,N., Miller,A.D. and Osborne,W.R. (1991) Internal initiation of translation in retroviral vectors carrying picarnovirus 5′ nontranslated regions. J. Virol., 65, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan R.A., Couture,L., Elroy-Stein,O., Ragheb,J., Moss,B. and Anderson,W.F. (1992) Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Res., 20, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramesh N., Kim,S.T., Wei,M.Q., Khalighi,M. and Osborne,W.R. (1996) High-titer bicistronic retroviral vectors employing foot-and-mouth disease virus internal ribosome entry site. Nucleic Acids Res., 24, 2697–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhoeyen E., Hauser,H. and Wirth,D. (2001) Conversion to expression stability by Flp-mediated cassette replacement. Hum. Gene Ther., 12, 933–944. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier J. and Sonenberg,N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 16.Gan W. and Rhoads,R.E. (1996) Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J. Biol. Chem., 271, 623–626. [DOI] [PubMed] [Google Scholar]
- 17.Vagner S., Gensac,M.C., Maret,A., Bayard,F., Amalric,F., Prats,H. and Prats,A.C. (1995) Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol., 15, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds J.E., Kaminski,A., Kettinen,H.J., Grace,K., Clarke,B.E., Rowlands,D.J. and Jackson,R.J. (1995) Unique features of internal initiation of translation of hepatitis C virus translation. EMBO J., 14, 6010–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein J., Sella,O., Le,S.-Y. and Elroy-Stein,O. (1997) PDGF2/c-sis mRNA leader contains a differentiation linked internal ribosomal entry site (D-IRES). J. Biol. Chem., 272, 9356–9362. [DOI] [PubMed] [Google Scholar]
- 20.Ye X.P., Fong,P., Iizuka,N., Choate,D. and Cavener,D.R. (1997) Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol., 17, 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiri G., Nahari,D., Finkelstein,Y., Le,S.Y., Elroy-Stein,O. and Levi,B.-Z. (1998) Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene, 17, 227–236. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.G., Armstrong,R.C., Berndt,J.A., Kim,N.W. and Hudson,L.D. (1998) A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol. Cell. Neurosci., 12, 119–140. [DOI] [PubMed] [Google Scholar]
- 23.Negulescu D., Leong,L.E.-C., Chandy,K.G., Semler,B.L. and Gutman,G.A. (1998) Translation initiation of cardiac voltage-gated potassium channel by internal ribosome entry. J. Biol. Chem., 273, 20109–20113. [DOI] [PubMed] [Google Scholar]
- 24.Stein I., Itin,A., Einat,P., Skaliter,R., Grossman,Z. and Keshet,E. (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol., 18, 3112–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chappell S.A., Edelman,G.M. and Mauro,V. (2000) A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA, 97, 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henis-Korenblit S., Strumpf,N.L., Goldstaub,D. and Kimchi,A. (2000) A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol., 20, 496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson R.J. and Kaminski,A. (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- 28.Le S.-Y. and Maizel,J.V. (1997) A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res., 25, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belsham G.J. and Sonenberg,N. (1996) RNA–protein interactions in regulation of picarnovirus RNA translation. Microbiol. Rev., 60, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huez I., Creancier,L., Audigier,S., Gensac,M.C., Prats,A.C. and Prats,H. (1998) Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol., 18, 6178–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova T.V., Hellen,C.U.T. and Shatsky,I.N. (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosome entry. Mol. Cell. Biol., 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts L.O., Seamons,R.A. and Belsham,G.J. (1998) Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA, 4, 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borman A.M., Le Mercier,P., Girard,M. and Kean,K.M. (1997) Comparison of picornaviral-IRES driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res., 25, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oumard A., Hennecke,M., Hauser,H. and Nourbakhsh,M. (2000) Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol. Cell. Biol., 20, 2755–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller P., Oumard,A., Wirth,D., Kröger,A. and Hauser,H. (2001) Polyvalent vectors for coexpression of multiple genes. In Schleef,M. (ed.), Plasmids for Therapy and Vaccination. Wiley-VCH, Weinheim, Germany, pp. 119–137.
- 36.Zitvogel L., Tahara,H., Cai,Q., Storkus,W.J., Muller,G., Wolf,S.F., Gately,M., Robbins,P.D. and Lotze,M.T. (1994) Construction and characterization of retroviral vectors expressing biologically active human interleukin-12. Hum. Gene Ther., 5, 1493–1506. [DOI] [PubMed] [Google Scholar]
- 37.Fussenegger M., Mazur,X. and Bailey,J.E. (1998) pTRIDENT, a novel vector family for tricistronic gene expression in mammalian cells. Biotechnol. Bioeng., 57, 1–10. [DOI] [PubMed] [Google Scholar]
- 38.Mielke C., Tümmler,M., Schübeler,D., von Hoegen,I. and Hauser,H. (2000) Stabilized, long-term expression of heteromeric proteins from tricistronic mRNA. Gene, 254, 1–8. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook R. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schirmbeck R., von Kampen,J., Metzger,K., Wild,J., Grüner,B., Schleef,M., Kröger,A., Hauser,H. and Reimann,J. (1999) DNA-based vaccination with polycistronic expression plasmids. Methods Mol. Med., 29, 313–322. [DOI] [PubMed] [Google Scholar]
- 41.Schirmbeck R., Wild,J. and Reimann,J. (1998) Similar as well as distinct MHC class I-binding peptides are generated by exogenous and endogenous processing of hepatitis B virus surface antigen. Eur. J. Immunol., 28, 4149–4161. [DOI] [PubMed] [Google Scholar]
- 42.Ladel C.H., Flesch,I.E., Arnoldi,J. and Kaufmann,S.H. (1994) Studies with MHC-deficient knock-out mice reveal impact of both MHC I and MHC II-dependent T cell responses in Listeria monocytogenes infection. J. Immunol ., 153, 3116–3122. [PubMed] [Google Scholar]
- 43.Thoma S., Bonhagen,K., Vestweber,D., Hamann,A. and Reimann,J. (1998) Expression of selectin-binding epitopes and cytokines by CD4+ T cells repopulating scid mice with colitis. Eur. J. Immunol., 28, 1785–1795. [DOI] [PubMed] [Google Scholar]
- 44.de Wet J.R., Wood,K.V., DeLuca,M., Helsinki,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorman C.M., Moffat,L.F. and Howard,B.H. (1982) Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol., 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oie H., Gazdar,A.F., Buckler,C.E. and Baron,S. (1977) High interferon producing line of transformed murine cells J. Gen. Virol., 1, 107–109. [DOI] [PubMed] [Google Scholar]
- 47.Dirks W., Mielke,C., Karreman,S., Haase,B., Wirth,M., Lindenmaier,W. and Hauser,H. (1994) Applications of expression vectors containing bicistronic transcription units in mammalian cells. In Fusenig,N.E. and Graf,H. (eds), Cell Culture in Pharmaceutical Research. Springer Verlag, Heidelberg, Germany, pp. 239–266.
- 48.Ross J. (1995) mRNA stability in mammalian cells. Microbiol. Rev., 59, 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day D.A. and Tuite,M.F. (1998) Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. Endocrinology, 157, 361–371. [DOI] [PubMed] [Google Scholar]
- 50.Attal J., Theron,M.C. and Houdebine,L.M. (1999) The optimal use of IRES (internal ribosome entry site) in expression vectors. Genet. Anal., 15, 161–165. [DOI] [PubMed] [Google Scholar]
- 51.Rees S., Coote,J., Stables,J., Goodson,S., Harris,S. and Lee,M.G. (1996) Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotics-resistant cells to express recombinant protein. Biotechniques, 20, 48–56. [DOI] [PubMed] [Google Scholar]
- 52.Jang S.K., Krausslich,H.G., Nicklin,M.J., Duke,G.M., Palmenberg,A.C. and Wimmer,E. (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol., 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]