Abstract
Basic region leucine zipper (bZIP) proteins represent a class of transcription factors that bind DNA using a simple, dimeric, α-helical recognition motif. The cAMP response element-binding protein (CREB) is a member of the CREB/ATF subfamily of bZIP proteins. CREB discriminates effectively in vivo and in vitro between the 10 bp cAMP response element (ATGACGTCAT, CRE) and the 9 bp activating protein 1 site (ATGACTCAT, AP-1). Here we describe an alanine scanning mutagenesis study designed to identify those residues within the CREB bZIP element that control CRE/AP-1 specificity. We find that the preference of CREB for the CRE site is controlled in a positive and negative way by acidic and basic residues in the basic, spacer and zipper segments. The CRE/AP-1 specificity of CREB is increased significantly by four glutamic acid residues located at positions 24, 28, 35 and 41; glutamic acid residues at positions 10 and 48 contribute in a more modest way. Specificity is decreased significantly by two basic residues located at positions 21 and 23; basic residues at positions 14, 18, 33 and 34 and V17 contribute in a more modest way. All of the residues that influence specificity significantly are located on the solvent-exposed face of the protein–DNA complex and likely participate in interactions between and among proteins, not between protein and DNA. The finding that the CRE/AP-1 specificity of CREB is dictated by the presence or absence of charged residues has interesting implications for how transcription factors seek and selectively bind sequences within genomic DNA.
INTRODUCTION
Basic region leucine zipper (bZIP) proteins represent a class of transcription factors that bind DNA using a simple, dimeric, α-helical recognition motif (1,2). Each monomer of the folded, dimeric motif contains ∼60 amino acids of contiguous α-helix. Each helix is comprised of three regions that each perform a unique function. Towards the N-terminus is a region rich in basic residues that contact DNA (this segment is often called the ‘basic region’). Near the C-terminus is a region rich in non-polar residues that stabilize the protein dimer (this segment is called the ‘zipper segment’). Connecting the basic and zipper segments is a six residue spacer that sets the register between the two attached functional regions (3–7). High resolution structures of several bZIP–DNA complexes show the zipper segments of two protein monomers assembled into a parallel coiled coil that lies perpendicular to the DNA helix axis and diverges near the DNA to position the two basic segments into adjacent major grooves (3–6,8–10). Although the helical structure of the coiled coil is maintained in the presence and absence of DNA at high protein concentrations, the basic segment exists as a structured α-helix only in the presence of specific DNA (11–14).
The cAMP response element-binding protein (CREB) is a member of the CREB/ATF subfamily of bZIP proteins. CREB is expressed in a variety of cells, including brain, heart, liver, testis, kidney and spleen, where its primary role is regulation of transcription in response to cAMP (15–19). Increased cAMP levels lead to phosphorylation of CREB at Ser133 by cAMP-dependent protein kinase A. Once phosphorylated, CREB interacts with the co-activator CREB-binding protein (CBP); subsequent interactions between CBP and the general transcriptional machinery up-regulate transcription of cAMP-dependent genes (15,20,21). Thus, CREB is a transcriptional end-point of the cAMP signal transduction cascade.
Like other members of the CREB/ATF subfamily, CREB effectively discriminates between two closely related DNA sequences. One, the 10 bp cAMP response element (CRE), is comprised of two ATGAC half-sites arranged as an inverted repeat (ATGACGTCAT). The other, the 9 bp activating protein 1 (AP-1) site, is composed of two ATGA half-sites separated by a single CG base pair (ATGACTCAT) (22–25). Despite the similarity between these two DNA sequences, CREB displays a strong half-site spacing preference (ΔΔG = 2.5 kcal·mol–1) for the CRE site over the AP-1 site (26). Not all bZIP proteins display a half-site spacing preference; for example, GCN4 binds approximately equally to the CRE and AP-1 sites (4,5,27,28).
Crystallographic analysis of the specific DNA complexes formed by CREB and GCN4 homodimers and the Fos–Jun heterodimer identified five conserved residues within the basic region (N–AA–S/CR) that directly contact DNA bases (Fig. 1; 3–5,10,28). Taken together, these bZIP–DNA structures provide a clear image of how this set of conserved basic and hydrophobic residues recognize the sequence ATGA with high affinity. Yet they provide little or no information on precisely which residues are responsible for discriminating between the CRE and AP-1 sites. Previous biochemical work has localized the residues responsible for half-site spacing specificity (CRE/AP-1 specificity) to the basic and spacer segments (23,24,29–33).
Figure 1.
The bZIP element sequences of B70 (CREB271–341), GCN4 (225–281), c-Fos (139–193) and c-Jun (263–317). Residues that contact bases directly are in italic; residues changed to alanine in this study are underlined; leucines within the zipper region are in bold. Residues which when changed to alanine caused enhanced CRE/AP-1 specificity are in blue; residues which when changed to alanine caused diminished CRE/AP-1 specificity are in red.
Here we describe an alanine scanning mutagenesis study designed to identify those residues within the CREB bZIP element that govern CRE/AP-1 specificity. We find that the preference of CREB for the CRE site is controlled in a positive and negative way by discrete acidic and basic residues in the basic, spacer and zipper segments. Specificity is increased significantly by four glutamic acid residues located at positions 24, 28, 35 and 41. Conversion of any one of these residues to alanine diminished CRE/AP-1 specificity by at least ΔΔG = –1.3 kcal·mol–1. Glutamic acid residues at positions 10 and 48 contribute, but less significantly; conversion of either of these residues to alanine diminished CRE/AP-1 specificity by between ΔΔG = –0.6 and –0.8 kcal·mol–1. CRE/AP-1 specificity is decreased significantly by two basic residues located at positions 21 and 23. Conversion of either of these residues to alanine improved CRE/AP-1 selectivity by ΔΔG > 2.3 kcal·mol–1. Basic residues at positions 14, 18, 33 and 34 and V17 also contribute, but less significantly; conversion of any one of these residues to alanine increased CRE/AP-1 specificity by between ΔΔG = –1.0 and –1.3 kcal·mol–1. Most of the residues that change specificity significantly are located on the solvent-exposed face of the protein–DNA complex and likely participate in electrostatic interactions between and among proteins, not between protein and DNA. The finding that the CRE/AP-1 specificity of CREB is controlled by charged residues has interesting implications for how transcription factors seek out and selectively bind to precise sequences within genomic DNA and provide guidelines for the design of exceptionally selective artificial transcription factors.
MATERIALS AND METHODS
Peptides
A peptide containing residues 271–341 of CREB (B70) and alanine derivatives thereof were overexpressed in Escherichia coli strain BL21(DE3)plysS from plasmids prepared from pAED-4B70 by Kunkel mutagenesis (34). All peptides were purified by FPLC using an SP-Sepharose column (Pharmacia) and were ≥98% pure as judged by HPLC. The identity of each peptide was confirmed by amino acid analysis using homoserine and norleucine as internal standards and by ESI mass spectrometry. Aliquots of each peptide were stored at concentrations of between 5 and 50 µM in 5 mM DTT and 10 mM Tris (pH 7.4) at –20°C. Stock peptide concentrations were as follows: B70, 1 µM; Q6A, 220 nM; E9A, 40 nM; E10A, 1 µM; R13A, 400 nM; K14A, 200 nM; R15A, 200 nM; E16A, 10 µM; V17A, 1 µM; R18A, 200 nM; L19A, 180 nM; M20A, 10 µM; K21A, 200 nM; R23A, 180 nM; E24A, 200 nM; R27A, 200 nM; E28A, 200 nM; R31A, 1 µM; K32A, 1 µM; K33A, 5 µM; K34A, 1µM; E35A, 5 µM; V37A, 1 µM; C39A, 40 nM; E41A, 200 nM; N42A, 400 nM; E48A, 40 nM. Several peptides (B70, E9A, R13A, R15A, E16A, L19A, M20A, R23A, E24A, E28A, C39A, N42A and E48A) aggregated upon long-term storage. These peptides were treated with 10 mM DTT, heated in a 55°C water bath for 10 min and frozen at –20°C before use.
DNA
The oligodeoxyribonucleotides CRE24 and AP-123 were synthesized on the 0.2 µmol scale using a Millipore Expedite DNA/RNA Synthesizer Model 8909 (35). CRE24A d(AGTGGAGATGACGTCATCTCGTGC) and its complement CRE24B d(GCACGAGATGACGTCATCTCCACT) contain the CRE target site, whereas AP-123A d(AGTGGAGATGACTCATCTCGTGC) and its complement AP-123B d(GCACGAGATGAGTCATCTCCACT) contain the AP-1 site. Oligodeoxyribonucleotides were labeled on the 5′-end with T4 polynucleotide kinase and [γ-32P]ATP and annealed to the complementary strand by heating the mixture to 95°C for 2 min and slowly cooling to room temperature.
Electrophoretic mobility shift assays
Binding reactions were performed in a buffer containing 1.4 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, 137 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 0.4 mg/ml acetylated BSA and 5% glycerol (pH 7.4) (23,24). Briefly, a set of serially diluted peptides were incubated with <50 pM [γ-32P]-labeled duplex DNA for 30 min at room temperature and then applied to a non-denaturing 8% polyacrylamide gel (80:1 acrylamide:N,N′-methylene bisacrylamide) prepared in 1× TBE. The temperature of the gel and the running buffer (0.5× TBE) were maintained at 25°C using a circulating, temperature controlled water bath. The samples were electrophoresed at 500 V for 30 min. The gels were analyzed using a Storm 840 Phosphorimager (Molecular Dynamics). Amounts of free and bound DNA were analyzed using the program Kaleidagraph (Abelbeck Software). Dissociation constants (Kapp) were determined by fitting the data to the Langmuir equation F = c × {1/(1+ Kapp/[peptide]T2)}. In these equations F = (c.p.m. in protein–DNA complex)/(c.p.m. in protein–DNA complex + c.p.m. in free DNA), [peptide]T is the total peptide concentration and c is an adjustable parameter representing the maximum value of F (c ≤ 1, for many peptides c was defined as 1) (23,36,37). Values reported represent the average ± standard error of at least three independent trials. Error bars on the plots represent the standard error for each data point. ΔG values were calculated from the relationship ΔG = –RTlnKapp–1, where R is the universal gas constant (1.987 × 103 kcal·mol–1·K–1) and T is the temperature in Kelvin.
RESULTS
Quantitative analysis of CRE/AP-1 specificity
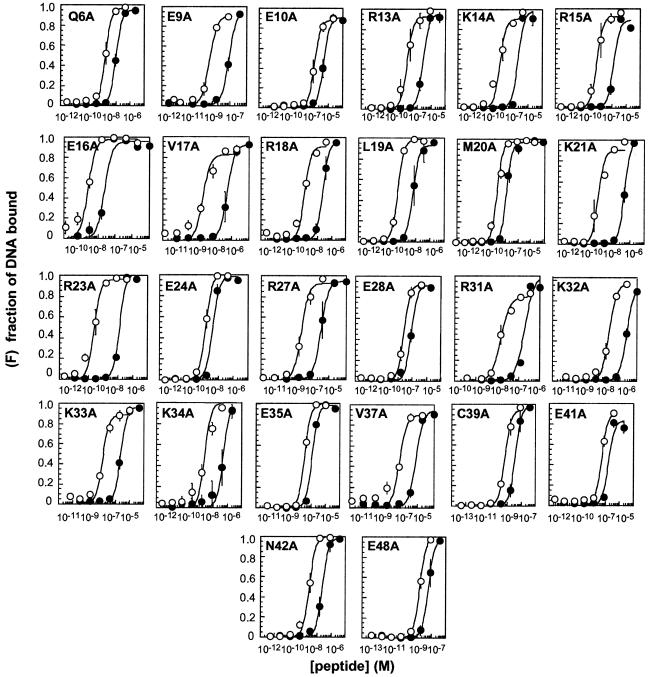
To identify which residues within CREB contribute to CRE/AP-1 selectivity, we prepared 26 CREB variants in which a single residue in the bZIP element (residues 271–341) was replaced by alanine. The positions of alanine substitution were located throughout the basic and spacer segments and in the N-terminal region of the zipper segment, as illustrated schematically in Figure 1. The affinity of each CREB variant for a duplex oligonucleotide containing a CRE (ATGACGTCAT) or AP-1 (ATGACTCAT) target site (CRE24 and AP-123, respectively) was assayed using a quantitative electrophoretic mobility shift assay. The affinity of the variant peptide was then compared to that of a peptide containing the wild-type CREB sequence (B70) (Figs 2 and 3B and Table 1). The CRE/AP-1 specificity of each peptide (ΔΔGspec) was defined by the free energy relationship ΔΔG = –RTln(KappAP-1/KappCRE). Under our experimental conditions B70 displayed a CRE/AP-1 specificity of –2.5 ± 0.2 kcal·mol–1, in good agreement with the value reported previously (26; Fig. 4 and Table 1). Interestingly, 22 of the 26 CREB variants displayed higher DNA affinity than the wild-type CREB sequence (B70).
Figure 2.
Binding isotherms illustrating the CRE/AP-1 specificities of B70 and variants thereof. Plots show the fraction of CRE24 (open symbols) and AP-123 (closed symbols) bound (F) as a function of peptide concentration at 25°C. Curves represent the best fit of the data to the equation C·{1/(1 + Kapp/[peptide]2)}, where Kapp and C are adjustable parameters. The equation used to determine values for Kapp is independent of whether the bZIP peptide forms a dimer before or after the DNA binding event (58). Each point represents the average of at least three independent determinations. Error bars denote the standard error.
Figure 3.
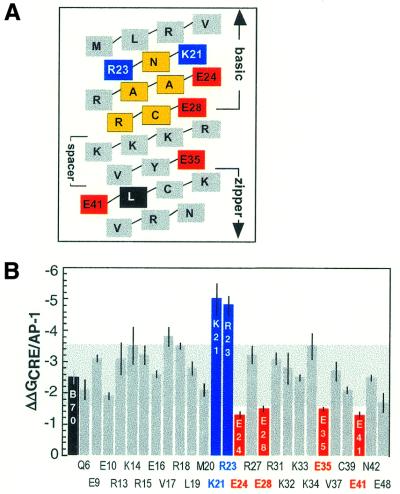
(A) A helical net illustrating the basic spacer and partial zipper segments of B70. Residues shaded yellow are highly conserved among bZIP proteins and contact bases directly in all known bZIP–DNA structures. Residues shaded blue illustrate positions where alanine substitution enhanced CRE/AP-1 specificity; residues shaded red illustrate positions where alanine substitution diminished CRE/AP-1 specificity. The residue shaded black is the first leucine in the zipper region. (B) The specificity values of all CREB variants compared to B70 (colored black). The shaded area behind the bars represents the region within 1 kcal·mol–1 of the specificity of B70 (2.5 kcal·mol–1). Color coding is identical to that in (A).
Table 1. Equilibrium dissociation constants of CREB–CRE and CREB–AP-1 complexes at 25°C determined by electrophoretic mobility shift analysis.
| Peptide |
ΔGCRE24 (kcal·mol–1) |
ΔGAP-123 (kcal·mol–1) |
ΔΔGCRE (ΔGmut – ΔGB70) (kcal·mol–1) |
ΔΔGAP-1 (ΔGmut – ΔGB70) (kcal·mol–1) |
ΔΔGspec (kcal·mol–1) |
| B70 | –21.5 ± 0.1 | –19.0 ± 0.2 | –2.5 ± 0.2 | ||
| Q6A | –24.0 ± 0.2 | –21.9 ± 0.1 | –2.5 | –2.9 | –2.1 ± 0.3 |
| E9A | –25.0 ± 0.2 | –21.9 ± 0.3 | –3.5 | –2.9 | –3.1 ± 0.1 |
| E10A | –22.4 ± 0.2 | –20.5 ± 0.3 | –0.9 | –1.5 | –1.9 ± 0.1 |
| R13A | –23.8 ± 0.5 | –20.7 ± 0.1 | –2.3 | –1.7 | –3.1 ± 0.5 |
| K14A | –24.6 ± 0.4 | –21.1 ± 0.3 | –3.1 | –2.1 | –3.5 ± 0.6 |
| R15A | –24.7 ± 0.4 | –21.5 ± 0.1 | –3.2 | –2.5 | –3.2 ± 0.3 |
| E16A | –23.4 ± 0.1 | –20.8 ± 0.1 | –1.9 | –1.8 | –2.6 ± 0.1 |
| V17A | –23.6 ± 0.1 | –19.8 ± 0.2 | –2.1 | –0.8 | –3.8 ± 0.3 |
| R18A | –24.3 ± 0.3 | –20.8 ± 0.3 | –2.8 | –1.8 | –3.5 ± 0.1 |
| L19A | –25.2 ±0.1 | –22.4 ± 0.4 | –3.7 | –3.4 | –2.8 ± 0.2 |
| M20A | –23.7 ± 0.2 | –21.6 ± 0.2 | –2.2 | –2.6 | –2.1 ± 0.2 |
| K21A | –25.1 ± 0.6 | –20.1 ± 0.2 | –3.6 | –1.1 | –5.0 ± 0.5 |
| R23A | –26.3 ± 0.4 | –21.5 ± 0.1 | –4.8 | –2.5 | –4.8 ± 0.3 |
| E24A | –24.2 ± 0.1 | –22.9 ± 0.1 | –2.7 | –3.9 | –1.3 ± 0.1 |
| R27A | –23.6 ± 0.3 | –20.4 ± 0.1 | –2.1 | –1.4 | –3.2 ± 0.3 |
| E28A | –23.5 ± 0.2 | –22.0 ± 0.2 | –2.0 | –3.0 | –1.5 ± 0.1 |
| R31A | –21.4 ± 0.1 | –18.3 ± 0.2 | +0.1 | +0.7 | –3.1 ± 0.2 |
| K32A | –20.8 ± 0.3 | –18.0 ± 0.2 | +0.7 | +1.0 | –2.8 ± 0.5 |
| K33A | –21.6 ± 0.1 | –18.1 ± 0.1 | –0.1 | +0.9 | –3.5 ± 0.1 |
| K34A | –23.4 ± 0.2 | –19.9 ± 0.6 | –1.9 | –0.9 | –3.5 ± 0.4 |
| E35A | –20.5 ± 0.1 | –19.0 ± 0.1 | +1.0 | 0 | –1.5 ± 0.05 |
| V37A | –22.0 ± 0.3 | –19.3 ± 0.3 | –0.5 | –0.3 | –2.7 ± 0.3 |
| C39A | –25.1 ± 0.1 | –23.0 ± 0.03 | –3.6 | –4.0 | –2.1 ± 0.1 |
| E41A | –22.5 ± 0.1 | –21.2 ± 0.2 | –1.0 | –2.2 | –1.3 ± 0.1 |
| N42A | –23.3 ± 0.2 | –20.8 ± 0.3 | –1.8 | –1.8 | –2.5 ± 0.1 |
| E48A | –24.2 ± 0.3 | –22.5 ± 0.3 | –2.7 | –3.5 | –1.7± 0.3 |
All values represent the average ± SE of at least three independent determinations. Values of ΔG were calculated from the equation ΔG = –RTlnKapp–1, where Kapp is the equilibrium dissociation constant, R = 1.987 × 103 kcal·mol–1·K–1 and T is the temperature in Kelvin.
Figure 4.
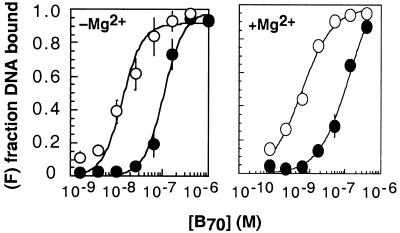
Role of Mg2+ in CRE/AP-1 specificity. Binding isotherms illustrating the CRE24 (open symbols)/AP-123 (closed symbols) specificity of B70 (CREB271–341) in the presence and absence of 10 mM Mg2+. The plot illustrates the fraction of DNA bound (F) as a function of B70 concentration at 25°C. The curves represent the best fit of the data to the equation described in the legend to Figure 2. Each plot represents the average of at least three independent determinations. Error bars represent the standard error.
Variants displaying decreased CRE/AP-1 selectivity
Four CREB variants, E24A, E28A, E35A and E41A, displayed a diminished ability to discriminate between the CRE and AP-1 sites when compared to B70. Each of these variants exhibited a specificity value at least 1.0 kcal·mol–1 lower than that exhibited by B70 itself (Figs 2 and 3B and Table 1). Although many CREB variants bound DNA with affinities higher or lower than B70, in all other cases the change in affinity for CRE24 roughly paralleled the change in affinity for AP-123. It is notable that two other variants containing alanine in place of glutamic acid (E9A and E41A) displayed levels of CRE/AP-1 specificity that were comparable to B70. This finding suggests that the altered specificities displayed by E24A, E28A, E35A and E41A were not due to changes in overall peptide charge but rather to the specific replacement of these four glutamic acid residues by alanine.
The four glutamic acid residues implicated by our experiments as important for specificity are not localized to one small region of the bZIP element, but rather are scattered throughout the basic, spacer and zipper segments (Fig. 3A). Two, at positions 24 and 28, are located in the basic segment, one, at position 35, is located in the spacer segment and one, at position 41, is located in the N-terminal region of the zipper segment (Fig. 5). Interestingly, none of these single site alanine substitutions eliminated CRE/AP-1 specificity entirely. Comparison of the specificity values exhibited by E24A, E28A, E35A and E41A (Table 1) suggested that the four glutamic acids do not function additively, that is, the sum of the specificities (ΔΔG) displayed by each variant exceeded the specificity of B70 (38,39). Loss of any one glutamic acid residue diminished but did not abolish specificity, suggesting that CREB specificity is encoded in a somewhat redundant manner.
Figure 5.
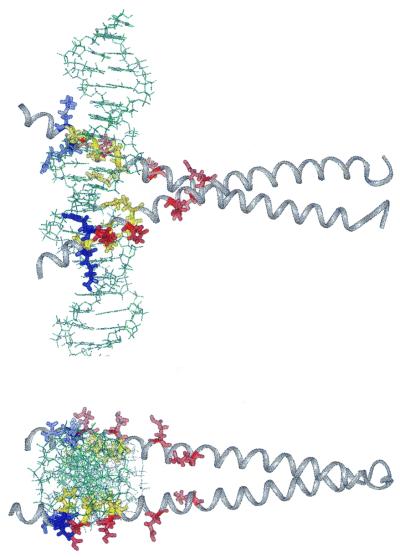
Two perpendicular views of the CREB–CRE complex to illustrate the location of residues that influence CRE/AP-1 specificity. The two basic residues that lower CRE/AP-1 specificity are colored blue, while the four glutamic acid residues that raise CRE/AP-1 specificity are colored red. The conserved quintet is shaded yellow. The residues involved in CRE/AP-1 selectivity all reside on the outside surface of the helix. This observation emphasizes the potential of protein–protein interactions for modulating specificity. Coordinates were generously made available by Professor Brennan in advance (3).
Variants displaying increased CRE/AP-1 selectivity
Two CREB variants, K21A and R23A, displayed an enhanced preference for the CRE site over the AP-1 site when compared to B70. Each exhibited a specificity value that was nearly double that of B70, –4.8 ± 0.3 and –5.0 ± 0.5 kcal·mol–1 for K21A and K23A, respectively (Figs 2 and 3B and Table 1). In contrast to the lack of proximity between those residues whose conversion to alanine lowered CRE/AP-1 specificity, these two basic residues are proximal in sequence as well as structure. In fact, they flank the most highly conserved DNA contact residue within the bZIP motif, N22 (Figs 1, 3A and 5).
The interplay between affinity and specificity
It is useful to consider the changes in CRE and AP-1 affinity that result in the observed changes in selectivity. First consider the four variants displaying diminished selectivity, E24A, E28A, E35A and E41A. In three cases (E24A, E28A and E41A) CRE/AP-1 specificity is reduced in the variant peptide by virtue of enhanced affinity for the AP-1 site (Table 1). For example, the E24A–AP-1 complex is 3.9 kcal·mol–1 more stable than the B70–AP-1 complex, whereas the E24A–CRE complex is only 2.7 kcal·mol–1 more stable than the B70–CRE complex. Similarly, the E28A–AP-1 complex is 3.0 kcal·mol–1 more stable than the B70–AP-1 complex, whereas the E28A–CRE complex is only 2.0 kcal·mol–1 more stable than the B70–CRE complex. Surprisingly, the trend also holds for a residue in the bZIP zipper segment that is located outside the expected range of DNA contact. The E41A–AP-1 complex is 2.2 kcal·mol–1 more stable than the B70–AP-1 complex, whereas the E41A–CRE complex is only 1.0 kcal·mol–1 more stable than the B70–CRE complex. The one exception to this trend is the E35A variant. E35A displays diminished CRE/AP-1 specificity solely by virtue of decreased (1.0 kcal·mol–1) affinity for the CRE site. We conclude that the glutamic acid residues located at positions 24, 28 and 41 contribute to CRE/AP-1 specificity by lowering the stability of CREB–AP-1 complexes; these three residues select against an interaction with the AP-1 site. In contrast, the glutamic acid residue at position 35 selectively increases the stability of CREB–CRE complexes.
Next we consider the two variants that displayed enhanced CRE/AP-1 selectivity, K21A and R23A. In these cases the specificity of the variant is increased by virtue of enhanced affinity for the CRE site (Table 1). For example, the K21A–CRE complex is 3.6 kcal·mol–1 more stable than the B70–CRE complex, whereas the K21A–AP-1 complex is only 1.1 kcal·mol–1 more stable than the B70–AP-1 complex. Similarly, the R23A–CRE complex is 4.8 kcal·mol–1 more stable than the B70–CRE complex, whereas the R23A–AP-1 complex is only 2.5 kcal·mol–1 more stable than the B70–AP-1 complex. For both variants the change in affinity for the CRE site relative to wild-type CREB is twice as large as the difference in affinity for the AP-1 site. We conclude that the basic residues located at positions 21 and 23 in wild-type CREB modulate CRE/AP-1 specificity by lowering the stability of CREB–CRE complexes. The large increase in specificity resulting from these changes provides guidelines for the successful design of other peptides that show exaggerated preference for the CRE site.
DISCUSSION
The molecular mechanism that accounts for the CRE/AP-1 specificities of bZIP proteins has been a focus of research since this DNA recognition motif was first identified in 1987 (1,3,40–42). The phenomenon of CRE/AP-1 specificity attracted attention because of the remarkable similarity between the two DNA sequences in question. The CRE and AP-1 target sites share the same consensus half-site and differ by only a single, central G·C base pair. Coupled with the similarity between the two oligonucleotides is a remarkable similarity among bZIP proteins, whether these proteins discriminate between the two sites (such as CREB/ATF proteins) (22,25) or not (such as GCN4) (4,5,10,28).
Previous proposals for CRE/AP-1 specificity
Various proposals to explain CRE/AP-1 selectivity have been advanced since 1987. One early proposal suggested that the coiled coil in bZIP proteins that display CRE/AP-1 specificity (such as CREB/ATF proteins) might be rigid enough to orient the basic segment helices to specifically recognize one site and not the other. Proteins that do not display CRE/AP-1 specificity (such as GCN4) were envisioned to be more flexible and permit recognition of both sites (41,43,44). This proposal was supported by the observation that certain bZIP peptide models containing unnatural dimerization domains displayed high levels of CRE/AP-1 specificity (36,37). However, extensive experimentation has shown that, irrespective of its role in peptide models, the coiled coil in bZIP proteins is not the primary determinant of CRE/AP-1 specificity (23,24,45).
A second proposal to explain CRE/AP-1 selectivity focused on potential differences in the flexibility of the spacer segment, as opposed to the coiled coil. Indeed, replacement of the six residue spacer segment of GCN4 (which displays low CRE/AP-1 specificity) with that of CRE-BP1 (which displays high CRE/AP-1 specificity) modestly increased specificity (23). Similar results were observed when three residues in the spacer segment of GCN4, L247, R249 and M250, were replaced with the corresponding residues K247, Y249 and V250 from CREB (46). These studies, along with those on bZIP peptide models containing non-natural dimerization domains (described above), indicate that subtle sequence-dependent conformational adjustments in the spacer can alter CRE/AP-1 selectivity in certain cases (36,37,47). However, subsequent ‘segment swapping’ experiments demonstrated clearly that for CREB the primary determinants of selectivity were located within the basic segment, not the spacer segment (23).
A third proposal to explain the origins of CRE/AP-1 specificity focused instead on differential DNA bending (or bendability) within the nucleic acid portion of the bZIP–DNA partnership. This proposal was based on experiments that revealed the existence of an intrinsic bend in the CRE site. This bend was removed, and the DNA straightened, upon binding of the CRE site by the CREB/ATF protein CRE-BP1 (21,26,48–51). No detectable conformational adjustments occurred in the DNA upon binding of GCN4, providing an attractive explanation for how GCN4 binds both sites with equal affinity. However, significant bending of both sites occurred upon binding of the CREB/ATF proteins CRE-BP1 and CREB (26,48,49), suggesting that the differential cost of DNA bending could dictate CRE/AP-1 specificity. Although numerous experiments support differential DNA bending as a mechanism for achieving specificity (52–54), subsequent studies of variant CRE/BP1–GCN4 proteins demonstrated that DNA bending and specificity were unrelated in this case (24).
Co-crystal structure of CREB–CRE adds molecular detail to thermodynamic data
The recent co-crystal structure of CREB bound to the CRE site found in the human somatostatin promoter (CREB–CRE) (3) adds considerable molecular detail to the thermodynamic data described herein. The constructs employed for crystallography are similar, although not identical, to those used here. In particular, the crystallography work employed a fragment of CREB (residues 285–307) that is shorter than B70 (residues 271–341) and a minimal hairpin CRE site (TGACGTCA) (3). Nevertheless, the overall structure of CREB–CRE is remarkably similar to previously characterized bZIP–DNA complexes (4–6,28). First we consider the environment surrounding the three residues that contribute positively to selectivity by selecting against the AP-1 site (E24, E28 and E41) and the one residue (E35) that selects for the CRE site. Next we consider the environment surrounding the two residues that lower CRE/AP-1 selectivity by selecting against the CRE site (K21 and R23).
Residues that select against the AP-1 site: E24, E28 and E41
Each of the three glutamic acid residues that select against the AP-1 site (E24, E28 and E41) directly contact protein or DNA in the CREB–CRE crystal structure (Fig. 5). E24 on one monomer (E295) contributes two hydrogen bonds; one from the side chain to a non-bridging phosphate oxygen between G1 and C–1 and another from the main chain carbonyl to the side chain of R27 (R298). E24 on the second monomer (referred to as E′24) contributes a hydrogen bond from the side chain to the ɛN of R′27. The side chain of E28 (E299) donates a hydrogen bond to the side chain on R31 (R302). Finally, E41 (E312) accepts a hydrogen bond from the phenolic proton of Y′36 (Y307) on the other protein chain. Changing each of these residues to alanine lowers CRE/AP-1 specificity by selectively increasing affinity for the AP-1 site (Table 1).
It is notable that most of these interactions are among and between proteins and not nucleic acid; both E28 and E41 are located on the outside of the protein–DNA complex and point away from the DNA. It is also notable that the Y36F CREB variant forms a less stable bZIP dimer in the absence of DNA than the wild-type protein (3), yet loss of the hydrogen bonding partner for Y36, E′41, leads to a variant (E41A) that binds the CRE site with higher than wild-type affinity. This paradox underscores the difficulty of assigning thermodynamic values to any particular interaction. We suggest that these inter- and intra-protein contacts maintain a conformation of the basic segment that complements the CRE site but clashes with the AP-1 site. Moreover, the importance of intra- and intermolecular protein–protein interactions emphasize how easily the specificity of a DNA-binding protein can be changed by mutations far from the protein–DNA interface.
Residues that select for the CRE site: E35
Of the four residues that contribute to CRE/AP-1 specificity, only one, E35 (E306), does so by selectively increasing affinity for the CRE site. Changing this residue to alanine lowers CRE/AP-1 specificity solely by virtue of a decreased (1.0 kcal·mol–1) affinity for the CRE sequence. In the CREB–CRE structure the side chain of E35 is also located on the solvent-exposed face of the protein–DNA complex and donates a hydrogen bond to the side chain on R31 (R302) (3). This intramolecular contact may also be important in orienting CREB for optimal interaction with the CRE target site. Since mutation of E35 to alanine exhibited no effect on the energetics of the CREB–AP-1 complex, we predict that the side chain of E35 does not contact R31 in the CREB–AP-1 complex. These data emphasize that there may be subtle but energetically relevant differences between the structures of CREB–AP-1 and CREB–CRE.
Residues that select against the CRE site: K21 and R23
The two basic residues that select against the CRE site both contact DNA in the CREB–CRE crystal structure. The side chain of R23 (R294) donates a hydrogen bond to a non-bridging oxygen between G1 and T2 within the consensus DNA site. Interestingly, K21 (K292) donates a hydrogen bond to a non-bridging phosphate between G′–7 and G′–6, outside the consensus 10 bp target site (Fig. 5) (3). Our data suggest that one or both of these contacts are not optimal for increasing affinity for the CRE sites. The fact that specificity can be enhanced by changing one of these residues to alanine provides clear guidelines for the design of non-natural molecules that possess this property.
The role of Mg2+
One of the most interesting details of the CREB–CRE complex as determined by crystallography was the appearance of a hexahydrated Mg2+ ion bound in a cavity between the basic region and DNA. Subsequent experimental work indicated that the presence of 10 mM Mg2+ increased the stability of CREB285–341–CRE by at least 14-fold (3,55). Mg2+ did not enhance binding of CREB to the AP-1 site. Although the experiments reported herein were performed in a buffer that lacked Mg2+ and with a smaller CREB domain (B70 contains residues 271–341 of CREB), we find that CRE/AP-1 specificity is decreased (to ΔΔGspec = –1.8 kcal·mol–1), not increased, by addition of 10 mM Mg2+ (Fig. 4). Although the affinity of B70 for the CRE site was increased 2-fold by addition of 10 mM Mg2+, a value consistent with the 14-fold effect reported earlier, we find that affinity for the AP-1 site was increased more. This result is consistent with a recent report describing no effect of Mg2+ on the affinity of CREB for several non-consensus CRE sequences (55). In conclusion, we find no evidence that Mg2+ increases the CRE/AP-1 specificity of CREB.
CONCLUSION
There is considerable current interest in understanding the molecular mechanisms used by proteins to locate their target sites within a large genome (56–61) and also on how to design non-natural transcription factors able to regulate gene expression in a prescribed manner (62–73). Here we show that the selectivity of a simple natural transcription factor is controlled in both a positive and negative way by electrostatic interactions. Notably, many of these interactions are between and among proteins and not between protein and DNA (at least not directly). A similar situation describes the role of E44 in integration host factor, which specifies T:A/A:T discrimination without directly contacting DNA (74). The finding that the CRE/AP-1 specificity of CREB is controlled by charged residues has interesting implications about how transcription factors seek and selectively bind to precise sequences within genomic DNA. This result emphasizes the ease with which specificity can be altered through the formation of different heterodimers or through combinatorial interactions with cellular or viral accessory factors (56,59,75–78). In addition, these findings provide additional criteria useful for the design of exceptionally selective artificial transcription factors (66,73).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Richard Brennan for providing the coordinates of the CREB–CRE complex. We thank the National Science foundation for predoctoral fellowships to both J.K.M. and L.S.S. This work was supported by the NIH.
References
- 1.McKnight S.L. (1991) Molecular zippers in gene regulation. Sci. Am., 264, 54–64. [DOI] [PubMed] [Google Scholar]
- 2.Kerppola T.K. and Curran,T. (1991) Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell, 66, 317–326. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher M., Goodman,R. and Brennan,R. (2000) The structure of a CREB bZIP:somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced binding. J. Biol. Chem., 275, 35242–35247. [DOI] [PubMed] [Google Scholar]
- 4.König P. and Richmond,T.J. (1993) The X-ray structure of the GCN4-bZIP bound to ATF CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol., 233, 139–154. [DOI] [PubMed] [Google Scholar]
- 5.Keller W., König,P. and Richmond,T.J. (1995) Crystal-structure of a bZIP-DNA complex at 2.2 Å—determinants of DNA specific recognition. J. Mol. Biol., 254, 657–667. [DOI] [PubMed] [Google Scholar]
- 6.Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha-helices—crystal-structure of the protein-DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 7.Pu W.T. and Struhl,K. (1991) The leucine zipper symmetrically positions the adjacent basic regions for specific DNA binding. Proc. Natl Acad. Sci. USA, 88, 6901–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Shea E.K., Klemm,J.D., Kim,P.S. and Alber,T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science, 254, 539–544. [DOI] [PubMed] [Google Scholar]
- 9.O’Neil K.T., Hoess,R.H. and DeGrado,W.F. (1990) Design of DNA-binding peptides based on the leucine zipper motif. Science, 249, 774–778. [DOI] [PubMed] [Google Scholar]
- 10.Glover J.N.M. and Harrison,S.C. (1995) Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature, 373, 257–261. [DOI] [PubMed] [Google Scholar]
- 11.Weiss M.A., Ellenberger,T., Wobbe,C.R., Lee,J.P., Harrison,S.C. and Struhl,K. (1990) Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature, 347, 575–578. [DOI] [PubMed] [Google Scholar]
- 12.Weiss M. (1990) Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: implications for scissor grip recognition. Biochemistry, 29, 8020–8024. [DOI] [PubMed] [Google Scholar]
- 13.O’Neil K.T., Shuman,J.D., Ampe,C. and Degrado,W.F. (1991) DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry, 30, 9030–9034. [DOI] [PubMed] [Google Scholar]
- 14.Saudek V., Pasley,H.S., Gibson,T., Gausepohl,H., Frank,R. and Pastore,A. (1991) Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry, 30, 1310–1317. [DOI] [PubMed] [Google Scholar]
- 15.Shaywitz A.J. and Greenberg,M.E. (1999) CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem., 68, 821–861. [DOI] [PubMed] [Google Scholar]
- 16.Lalli E. and Sassone-Corsi,P. (1994) Signal-transduction and gene-regulation—the nuclear response to cAMP. J. Biol. Chem., 269, 17359–17362. [PubMed] [Google Scholar]
- 17.Lee K.A.W. and Masson,N. (1993) Transcriptional regulation by CREB and its relatives. Biochim. Biophys. Acta, 1174, 221–233. [DOI] [PubMed] [Google Scholar]
- 18.Meyer T.E. and Habener,J.F. (1993) Cyclic adenosine-3′,5′-monophosphate response element-binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr. Rev., 14, 269–290. [DOI] [PubMed] [Google Scholar]
- 19.Hoeffler J.P., Meyer,T.E., Yun,Y., Jameson,J.L. and Habener,J.F. (1988) Cyclic-AMP responsive DNA binding protein: structure determined from a cloned placental DNA. Science, 242, 1430–1433. [DOI] [PubMed] [Google Scholar]
- 20.Fiol C.J., Williams,J.S., Chou,C.H., Wang,Q.M., Roach,P.J. and Andrisani,O.M. (1994) A secondary phosphorylation of CREB(341) at Ser(129) is required for the cAMP-mediated control of gene-expression—a role of glycogen-synthase kinase-3 in the control of gene-expression. J. Biol. Chem., 269, 32187–32193. [PubMed] [Google Scholar]
- 21.deGroot R.P., Delmas,V. and Sassone-Corsi,P. (1994) DNA bending by transcription factors CREM and CREB. Oncogene, 9, 463–468. [PubMed] [Google Scholar]
- 22.Hai T., Liu,F., Coukos,W.J. and Green,M.R. (1989) Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev., 3, 2083–2090. [DOI] [PubMed] [Google Scholar]
- 23.Metallo S.J. and Schepartz,A. (1994) Distribution of labor among CRE-BP1 bZIP segments in the control of DNA affinity and specificity. Chem. Biol., 1, 143–151. [DOI] [PubMed] [Google Scholar]
- 24.Metallo S.J., Paolella,D.N. and Schepartz,A. (1997) The role of a basic amino acid cluster in target site selection and non-specific binding of bZIP peptides to DNA. Nucleic Acids Res., 25, 2967–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellers J.W., Vincent,A.C. and Struhl,K. (1990) Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol. Cell. Biol., 10, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamm M.K. and Schepartz,A. (1995) Studies on the formation of DNA-protein interfaces: DNA specificity and straightening by CREB. Biol. Med. Chem. Lett., 5, 1621–1626. [Google Scholar]
- 27.Alber T. (1993) How GCN4 binds DNA. Curr. Biol., 3, 182–184. [DOI] [PubMed] [Google Scholar]
- 28.Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α-helices: crystal structure of the protein-DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 29.Talanian R.V., McKnight,C.J. and Kim,P.S. (1990) Sequence-specific DNA binding by a short peptide dimer. Science, 249, 769–771. [DOI] [PubMed] [Google Scholar]
- 30.Suckow M., von Wilcken-Bergmann,B. and Müller-Hill,B. (1993) Identification of 3 residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA-binding. EMBO J., 12, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koldin B., Suckow,M., Seydel,A., von Wilcken-Bergmann,B. and Müller-Hill,B. (1995) A comparison of the different DNA-binding specificities of the bZIP proteins C/EBP and GCN4. Nucleic Acids Res., 23, 4162–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suckow M., Schwamborn,K., Kisters-Woike,B., von Wilcken-Bergmann,B. and Müller-Hill,B. (1994) Replacement of invariant bZIP residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res., 22, 4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrisani O.M. and Dixon,J.E. (1991) Involvement of lysine residues 289 and 291 of the cAMP-responsive element-binding protein in the recognition of the cAMP-responsive element. J. Biol. Chem., 266, 21444–21450. [PubMed] [Google Scholar]
- 34.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–377. [DOI] [PubMed] [Google Scholar]
- 35.Beaucage S.L. and Caruthers,M.H. (1981) Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett., 22, 1859–1862. [Google Scholar]
- 36.Cuenoud B. and Schepartz,A. (1993) Altered specificity of DNA-binding proteins with transition metal dimerization domains. Science, 259, 510–513. [DOI] [PubMed] [Google Scholar]
- 37.Cuenoud B. and Schepartz,A. (1993) Design of a metallo-bZIP protein that discriminates between CRE and AP1 target sites: selection against AP1. Proc. Natl Acad. Sci. USA, 90, 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stormo G.D. and Fields,D.S. (1998) Specificity, free energy and information content in protein-DNA interactions. Trends Biochem. Sci., 23, 109–113. [DOI] [PubMed] [Google Scholar]
- 39.Stormo G.D. (1998) Information content and free energy in DNA-protein interactions. J. Theor. Biol., 195, 135–137. [DOI] [PubMed] [Google Scholar]
- 40.Hope I. and Struhl,K. (1987) GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J., 6, 2781–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science, 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- 42.Pathak D. and Sigler,P.B. (1992) Updating structure-function relationships in the bZIP family of transcription factors. Curr. Opin. Struct. Biol., 2, 116–123. [Google Scholar]
- 43.Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1989) The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science, 243, 1681–1688. [DOI] [PubMed] [Google Scholar]
- 44.Shuman J.D., Vinson,C.R. and McKnight,S.L. (1990) Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science, 249, 771–774. [DOI] [PubMed] [Google Scholar]
- 45.Agre P., Johnson,P. and McKnight,S. (1989) Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science, 246, 922–926. [DOI] [PubMed] [Google Scholar]
- 46.Kim J., Tzamarias,D., Ellenberger,T., Harrison,S.C. and Struhl,K. (1993) Adaptability at the protein-DNA interface is an important aspect of sequence recognition by bZIP proteins. Proc. Natl Acad. Sci. USA, 90, 4513–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson P.F. (1993) Identification of C/EBP basic region residues involved in DNA sequence recognition and half-site spacing preferences. Mol. Cell. Biol., 13, 6919–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paolella D.N., Palmer,C.R. and Schepartz,A. (1994) DNA targets for certain bZIP proteins distinguished by an intrinsic bend. Science, 264, 1130–1133. [DOI] [PubMed] [Google Scholar]
- 49.Paolella D.N., Liu,Y., Fabian,M. and Schepartz,A. (1997) Electrostatic mechanism for DNA bending by bZIP proteins. Biochemistry, 36, 10033–10038. [DOI] [PubMed] [Google Scholar]
- 50.Sloan L.S. and Schepartz,A. (1998) Sequence determinants of the intrinsic bend in the cyclic AMP response element. Biochemistry, 37, 7113–7118. [DOI] [PubMed] [Google Scholar]
- 51.Kerppola T.K. and Curran,T. (1993) Selective DNA bending by a variety of bZIP proteins. Mol. Cell. Biol., 13, 5479–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirzabekov A.D. and Rich,A. (1979) Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc. Natl Acad. Sci. USA, 76, 1118–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steitz T.A. (1990) Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q. Rev. Biophys., 23, 205–280. [DOI] [PubMed] [Google Scholar]
- 54.Jones S., van Heyningen,P., Berman,H.M. and Thornton,J.M. (1999) Protein-DNA interactions: a structural analysis. J. Mol. Biol., 287, 877–896. [DOI] [PubMed] [Google Scholar]
- 55.Craig J.C., Schumacher,M.A., Mansoor,S.E., Farrens,D.L., Brennan,R.G. and Goodman,R.H. (2001) Consensus and variant cAMP-regulated enhancers have different CREB-binding properties. J. Biol. Chem., 276, 11719–11728. [DOI] [PubMed] [Google Scholar]
- 56.Kohler J.J. and Schepartz,A. (2001) Kinetic studies of Fos·Jun·DNA complex formation: DNA binding prior to dimerization. Biochemistry, 40, 130–142. [DOI] [PubMed] [Google Scholar]
- 57.Kohler J.J., Metallo,S.J., Schneider,T.L. and Schepartz,A. (1999) DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl Acad. Sci. USA, 96, 11735–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metallo S.J. and Schepartz,A. (1997) Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nature Struct. Biol., 4, 115–117. [DOI] [PubMed] [Google Scholar]
- 59.Schneider T.L. and Schepartz,A. (2001) Hepatitis B virus protein pX enhances the monomer assembly pathway of bZIP·DNA complexes. Biochemistry, 40, 2835–2843. [DOI] [PubMed] [Google Scholar]
- 60.Berger C., Piubelli,L., Haditsch,U. and Bosshard,H.R. (1998) Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett., 425, 14–18. [DOI] [PubMed] [Google Scholar]
- 61.Wu X.L., Spiro,C., Owen,W.G. and McMurray,C.T. (1998) cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J. Biol. Chem., 273, 20820–20827. [DOI] [PubMed] [Google Scholar]
- 62.Choo Y. and Klug,A. (1997) Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol., 7, 117–125. [DOI] [PubMed] [Google Scholar]
- 63.Kim J.-S. and Pabo,C.O. (1998) Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc. Natl Acad. Sci. USA, 95, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pomerantz J.L., Wolfe,S.A. and Pabo,C.O. (1998) Structure-based design of a dimeric zinc finger protein. Biochemistry, 37, 965–970. [DOI] [PubMed] [Google Scholar]
- 65.White S., Szewczyk,J.W., Turner,J.M., Baird,E.E. and Dervan,P.B. (1998) Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- 66.Zondlo N.J. and Schepartz,A. (1999) Highly specific DNA recognition by a designed miniature protein. J. Am. Chem. Soc., 121, 6938–6939. [Google Scholar]
- 67.Beerli R.R., Dreier,B. and Barbas,C.F. (2000) Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl Acad. Sci. USA, 97, 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choo Y. and Isalan,M. (2000) Advances in zinc finger engineering. Curr. Opin. Struct. Biol., 10, 411–416. [DOI] [PubMed] [Google Scholar]
- 69.Wolfe S.A., Ramm,E.I. and Pabo,C.O. (2000) Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers. Struct. Fold. Des., 8, 739–750. [DOI] [PubMed] [Google Scholar]
- 70.Mapp A.K., Ansari,A.Z., Ptashne,M. and Dervan,P.B. (2000) Activation of gene expression by small molecule transcription factors. Proc. Natl Acad. Sci. USA, 97, 3930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segal D.J. and Barbas,C.F. (2000) Design of novel sequence-specific DNA-binding proteins. Curr. Opin. Chem. Biol., 4, 34–39. [DOI] [PubMed] [Google Scholar]
- 72.Pabo C.O., Peisach,E. and Grant,R.A. (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem., 70, 313–340. [DOI] [PubMed] [Google Scholar]
- 73.Chin J.W. and Schepartz,A. (2001) Concerted evolution of structure and function in a miniature protein. J. Am. Chem. Soc., 123, 2929–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Read E.K., Gumport,R.I. and Gardner,J.F. (2000) Specific recognition of DNA by Integration Host Factor. Glutamic acid 44 of the b subunit specifies the discrimination of a T:A from an A:T base pair without directly contacting the DNA. J. Biol. Chem., 275, 33759–33764. [DOI] [PubMed] [Google Scholar]
- 75.Chin J.W., Kohler,J.J., Schneider,T.L. and Schepartz,A. (1999) Gene regulation: protein escorts to the transcription ball. Curr. Biol., 9, R929–R932. [DOI] [PubMed] [Google Scholar]
- 76.Pflum M.K.H., Schneider,T.L., Hall,D. and Schepartz,A. (2001) Hepatitis B virus X protein activates transcription by bypassing CREB phosphorylation, not by stabilizing bZIP-DNA complexes. Biochemistry, 40, 693–703. [DOI] [PubMed] [Google Scholar]
- 77.Baranger A.M., Palmer,C.R., Hamm,M.K., Giebler,H.A., Brauweiler,A., Nyborg,J.K. and Schepartz,A. (1995) Mechanism of DNA-binding enhancement by the human T-cell leukemia-virus transactivator Tax. Nature, 376, 606–608. [DOI] [PubMed] [Google Scholar]
- 78.Palmer C.R., Gegnas,L.D. and Schepartz,A. (1997) Mechanism of DNA binding enhancement by hepatitis B virus protein pX. Biochemistry, 36, 15349–15355. [DOI] [PubMed] [Google Scholar]