Abstract
Recapitulation of developmental signals represents a promising strategy for treating intervertebral disc degeneration. During development, embryonic notochord-derived cells (NDCs) are the direct progenitors of cells that populate the adult nucleus pulposus (NP) and are an important source of secreted signaling molecules. The objective of this study was to define global gene expression profiles of NDCs at key stages of embryonic disc formation. NDCs were isolated from Shh-cre;ROSA:YFP mice at embryonic day 12.5 and postnatal day 0, representing opposite ends of the notochord to NP transformation. Differences in global mRNA abundance across this developmental window were established using RNA-Seq. Protein expression of selected molecules was confirmed using immunohistochemistry. Principal component analysis revealed clustering of gene expression at each developmental stage with more than 5000 genes significantly differentially expressed between E12.5 and P0. There was significantly lower mRNA abundance of sonic hedgehog pathway elements at P0 vs E12.5, while abundance of elements of the transforming growth factor-beta and insulin-like growth factors pathways, and extracellular matrix components including collagen 6 and aggrecan, were significantly higher at P0. This study represents the first transcriptome-wide analysis of embryonic NDCs. Results suggest signaling and biosynthesis of NDCs change dramatically as a function of developmental stage.
Introduction
The intervertebral discs are the partially movable joints that consecutively connect the vertebrae of the spine. Each intervertebral disc is comprised of three substructures: centrally, the proteoglycan-rich nucleus pulposus (NP); peripherally, the fibrocartilaginous annulus fibrosus; and superiorly and inferiorly, two endplates of hyaline cartilage1–4. These three substructures act synergistically to absorb and transfer compressive forces and facilitate complex motion of each intervertebral joint. Embryonic formation of the intervertebral discs centers on the notochord, a highly specialized, mesoderm-derived, transient, midline structure that is present in all chordates during early development5. The notochord initially serves as a primitive structural axial skeleton in the developing embryo and as the main signaling center for providing patterning information for surrounding tissues during early development through the regulation of secreted molecular factors6–11. In higher vertebrates, this singular structure eventually transforms into distinct NPs within the intervertebral discs during late stages of embryonic development12–14. In mice, this transformation occurs across an 8 day window commencing at embryonic day 12.515. The molecular mechanisms that regulate transformation of the notochord into the NPs remain poorly understood.
Fate mapping studies in mice have demonstrated that all cells in the adult NP are derived from the notochord16, 17. In humans, the phenotype of NP cells changes markedly during growth and aging18–20 and by skeletal maturity, these cells have lost the majority of their notochordal characteristics and assume physical and molecular characteristics that more closely resemble those of articular cartilage chondrocytes21. However, molecular profiling studies have demonstrated that these adult NP cells do continue to maintain high expression of some notochordal markers22.
Degeneration of the intervertebral discs, which is ubiquitous amongst the aging adult population, is strongly implicated as a cause of low back pain, a condition that will affect around 85% of all people at some point in their lifetime and costs over $100 billion annually in the United States23–26. Disc degeneration is a cascade of cellular, structural, and biomechanical changes that is closely linked with aging23, 27. The earliest manifestations of disc degeneration typically occur in the NP, where reduced proteoglycan content compromises mechanical function leading to progressive structural deterioration of the entire intervertebral joint. Current treatments for low back pain include physical therapy, steroid injections, or where surgery is warranted, spinal fusion or artificial total disc replacement28. These treatments, however, seek to manage symptoms without maintaining or restoring native disc structure or biomechanical function. As such, a key focus of current research efforts is to develop new biological treatment strategies that can both address symptoms and regenerate native disc tissue.
An attractive strategy with high promise for long term reconstitution of healthy disc tissue is cell-based regeneration using therapeutic cell types such as mesenchymal stem cells (MSCs) or induced pluripotent stem cells (IPSCs)29, 30. One of the major impediments to successful stem cell-driven disc regeneration is the requirement for delivered cells to regenerate multiple tissues, each with distinct architecture and composition, and each comprised of cells with disparate developmental lineages.
Given the established function of the notochord as a source of secreted signaling molecules that regulate embryonic disc formation, there is intense interest in identifying notochordal cell-secreted factors and applying them to develop improved therapeutic strategies for disc regeneration31. Previous studies suggest that such factors may potentiate anabolic disease-modifying effects32–36. As the notochord is a transient structure and disappears in early development, harvesting notochordal cells for direct therapeutic use for disc regeneration is not feasible; however, improved understanding of embryonic NP formation may enable recapitulation of important developmental signals that are necessary for the formation of the NP and its proteoglycan-rich extracellular matrix (ECM).
The overall objective of this study was to undertake a global analysis of the molecular regulation of embryonic disc formation by mapping the transcriptome of notochord-derived cells (NDCs) at developmental stages representing opposite ends of the notochord to NP transformation. In our analysis, we focused on signaling pathways with established roles regulating skeletal patterning and growth, cell differentiation, and extracellular matrix deposition. Additionally, we examined changing expression levels of key ECM molecules and putative NP cell-specific markers as previously defined in the literature37.
Materials and Methods
Animals and Tissue Collection
For these studies, we used the Shh-cre;ROSA:YFP mouse model, previously established and shown to express YFP in SHH-expressing notochord cells and their progeny at any developmental stage16 (Fig. 1). Animals were raised under NIH guidelines for the care and use of animals in research, and all experimental studies were carried out with approval from the Institutional Animal Care and Use Committee of the University of Florida. Animals were euthanized at two developmental stages, embryonic day 12.5 (E12.5) and postnatal day 0 (P0), representing opposite ends of the notochord to NP transformation15. Euthanasia of pregnant mothers was achieved via cervical dislocation followed by tracheotomy, after which the E12.5 embryos were collected and placed into nuclease-free PBS. P0 pups were euthanized by decapitation and placed into nuclease-free PBS. Biological replicates (n = 4 at both time points) each consisted of pooled embryos or pups (~6) from a single litter.
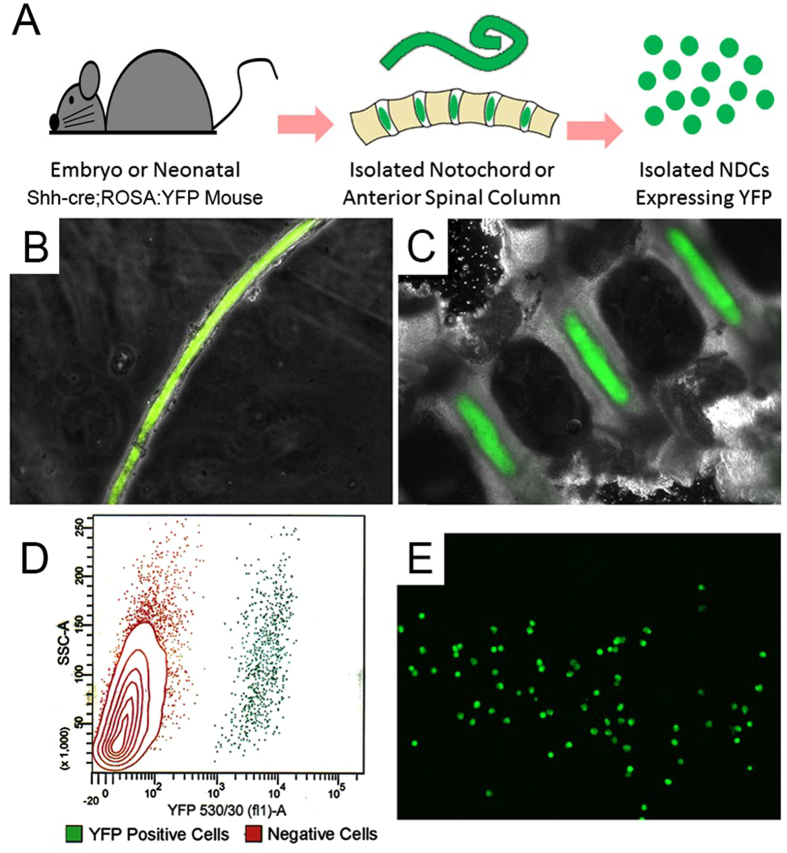
Figure 1.
Isolation of YFP-positive NDCs from Shh-cre;ROSA:YFP mice. (A) Cell isolation schematic. (B) YFP-positive E12.5 notochord. (C) YFP-positive nucleus pulposi in a P0 spine. (D) FACS plot for P0 NDC sort. (E) Isolated YFP-positive NDCs.
Following euthanasia, for E12.5 embryos, whole, intact notochords (Fig. 1B) were harvested under a dissecting microscope, any extraneous adjoining tissue carefully removed, immediately placed into TRIzol (Ambion; Austin, TX) and frozen. For P0 pups, anterior vertebral columns (Fig. 1C) were harvested, and each disc space in the lumbar and thoracic regions was carefully bisected axially under a dissecting microscope on ice to expose the NPs (Schematic, Fig. 1A). Gentle mechanical agitation in PBS buffer with 25 mM HEPES and 0.1% BSA to minimize cell stickiness was then used to dissociate NP cells from the surrounding tissue, and the resulting cell mixture was strained through a 50 µm mesh filter. Enzymatic cell isolation was specifically avoided as it has been shown that exposure to enzymes such as collagenase can confound global mRNA expression patterns38. The collected cells underwent fluorescence assisted cell sorting (FACS) in the PBS buffer described above and YFP-positive NDCs (~10,000–30,000 cells per litter) were collected directly into TRIzol LS (Ambion; Austin, TX) and frozen (Fig. 1D,E).
RNA Extraction, Library Preparation, and Whole-transcriptome Sequencing
Whole-transcriptome sequencing (RNA-Seq) is a powerful tool that can accurately quantify global mRNA expression in many samples in parallel, which allows for direct comparisons of expression levels of many genes-of-interest at once. Another advantage of RNA-Seq is the ability to measure transcript levels over a large range of expression with limited background noise and signal saturation, which allows for detection of rare and lowly expressed transcripts.
RNA was extracted using serial TRIzol-chloroform extractions, then in-column treated with RNase-free DNase (Qiagen; Valencia, CA) on Direct-zol RNA MiniPrep columns (Zymo Research; Irvine, CA) and eluted following the manufacturer’s protocols. Separate aliquots of extracted RNA samples were stored at −80 °C for either RNA quality analysis or RNA-Seq library preparation in order to reduce the number of freeze-thaw cycles.
The quality of each RNA sample was verified using an Agilent BioAnalyzer and RNA 6000 Pico Kit (Santa Clara, CA). Only high quality total RNA (RNA integrity number >7) was used to prepare RNA-Seq libraries. Libraries were prepared using the TotalScript Kit (Illumina; San Diego, CA), which has been optimized for use in synthesizing RNA-Seq libraries from low amounts of total starting RNA (<5 ng). Single-end, 100-base pair sequencing was performed using the Illumina HiSeq 2500 platform at the Next Generation Sequencing Core of the University of Pennsylvania. Results were aligned to the mouse genome build GRCm38.p3 (NCBI) and genomic features annotated from the M4 build from Gencode (www.gencodegenes.org).
RNA-Seq Analysis
Fastq files containing raw sequence and quality scores were mapped to the mouse genome (GRCm38) using the STAR aligner (https://www.ncbi.nlm.nih.gov/pubmed/23104886). Genomically mapped reads were counted against reference genes as annotated in version M4 from Gencode (version M4, https://www.ncbi.nlm.nih.gov/pubmed/26187010) using htseq-count (https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu638). DESeq239 was used to generate regularized log2-transformed expression values for all genes in all samples, which were visualized to assess inter-sample variability using Principal Components Analysis (PCA) as implemented in Partek Genomics Suite (v6.6, Partek, Inc., St. Louis, MO). DESeq2 was used to calculate statistics for differential expression between the E12.5 and P0 groups. Genes were considered significant if they showed a false-discovery-rate adjusted p-value of <0.05. We carried out pathway analysis (Ingenuity Pathway Analysis; Qiagen, Valencia, CA), through which we applied a structured approach to our analysis of specific pathways and molecules, focusing on signaling pathways with well-defined roles in tissue patterning and growth including cell migration, proliferation and differentiation, families of extracellular matrix molecules that are important for tissue assembly, structure and mechanical function, and previously identified markers of the NP cell phenotype.
Histology and Immunohistochemistry
Immunohistochemical detection of protein expression was undertaken for selected molecules identified through RNA-Seq. Representative samples of whole embryos (E12.5) or anterior spinal columns (P0) from five litters were placed into 4% paraformaldehyde overnight. Following fixation, P0 spines were gently decalcified in 0.5 M EDTA at pH 8.0 for 2 days. E12.5 whole embryos and P0 spines were then processed into paraffin and sectioned at 8 µm. For each antibody, E12.5 and P0 samples were stained in parallel to standardize the timing of each step throughout the protocol. Antigen retrieval was carried out on rehydrated sections using a heat-mediated technique in a 95 °C buffer bath of 10 mM sodium citrate with 0.05% Tween 20, pH 6.0 for 20 minutes. Sections were permeabilized in TBS +0.025% Triton X-100 (TBS-T) and blocked using Background Buster (Accurate Chemical & Scientific Corporation; Westbury, NY) for 30 minutes at room temperature. Sections were then incubated with primary antibodies against sonic hedgehog (SHH), transforming growth factor beta 1 (TGF-β1), insulin-like growth factor-1 (IGF-1), aggrecan (ACAN), and collagens 1, 2, and 6 (COL1, COL2, COL6) (primary antibody information is provided in Supplemental Table S1) diluted in Background Buster overnight at 4 °C. Slides were rinsed twice, 5 minutes each in TBS-T, then incubated in 0.3% H2O2 in TBS for 15 minutes to suppress endogenous peroxidase activity. Slides were rinsed three times, 2 minutes each with TBS. Antibody staining was visualized using the SuperPicture Polymer Detection kit, DAB, rabbit (ThermoFisher Scientific; Waltham, MA) using the manufacturer’s protocol. Slides were rinsed well in diH2O, and then hematoxylin QS (Vector Laboratories; Burlingame, CA) was applied for 30 seconds. Slides were then rinsed with tap H2O, dehydrated and cleared, and coverslipped with Permount Mounting Medium (ThermoFisher Scientific; Waltham, MA). Slides were imaged under bright field light microscopy (Eclipse 90i; Nikon; Tokyo, Japan). As negative controls, sections without primary antibodies were treated in parallel with the same protocol, except with only Background Buster during the overnight primary antibody incubation (representative images, Supplemental Fig. S1). Additionally, to assess overall tissue composition and morphology, sections were double-stained with either Alcian blue and picrosirius red (ABPR) for glycosaminoglycans (GAG) and collagen, respectively, or hematoxylin and eosin (H&E) to demonstrate cellularity and imaged under bright field light microscopy.
Data Availability Statement
The datasets generated and analyzed during this study are available in the NCBI Gene Expression Omnibus (GEO) repository, accession number: GSE100934.
Results
Morphological Appearance of the Spinal Column at E12.5 and P0
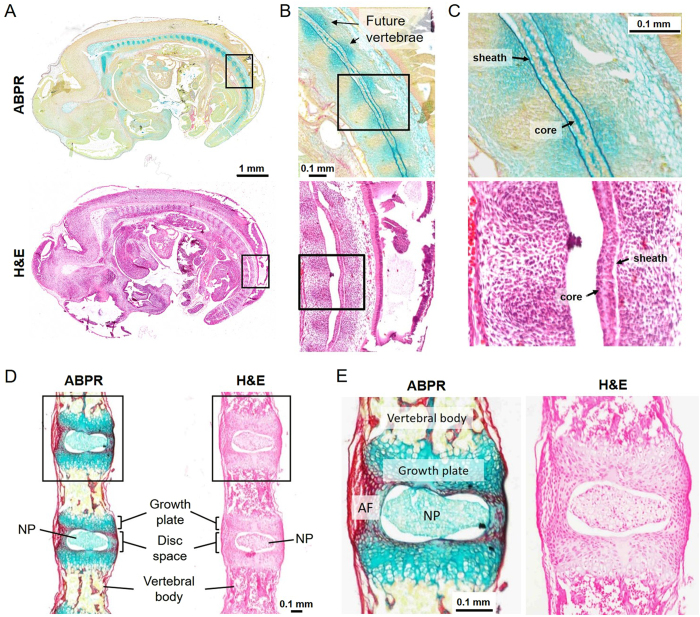
Histological examination confirmed the expected morphological characteristics of the vertebral column at each developmental stage (Fig. 2). At E12.5, the notochord was observed as a single discrete structure spanning the length of the axial skeleton and was encased in a GAG-rich sheath (Fig. 2A–C). Mesenchymal condensations, representing future vertebral bodies, stained positively for GAG (Fig. 2B,C). The central region of the notochord contained a core that stained intensely for GAGs and appeared relatively acellular (Fig. 2C). At P0, NPs were fully formed within the intervertebral discs (Fig. 2D). The population of cells within the NPs at P0 appeared morphologically homogeneous (Fig. 2E, H&E staining). The lamellar architecture of the annulus fibrosus of the discs was established, and in the vertebrae, primary ossification was complete with secondary ossification yet to commence.
Figure 2.
Overall morphological characteristics of E12.5 embryos and P0 spines. (A) ABPR and H&E stained E12.5 embryo sections. (B) Black box insets of Fig. 2A. Mesenchymal condensations indicating locations of future vertebrae are labeled. (C) Black box insets of Fig. 2B. GAG-rich, relatively acellular outer sheath and inner core of notochord are labeled. (D) ABPR and H&E stained P0 spine sections showing key anatomical features. (E) Black box insets of Fig. 2D.
RNA-Seq and Pathway Analysis
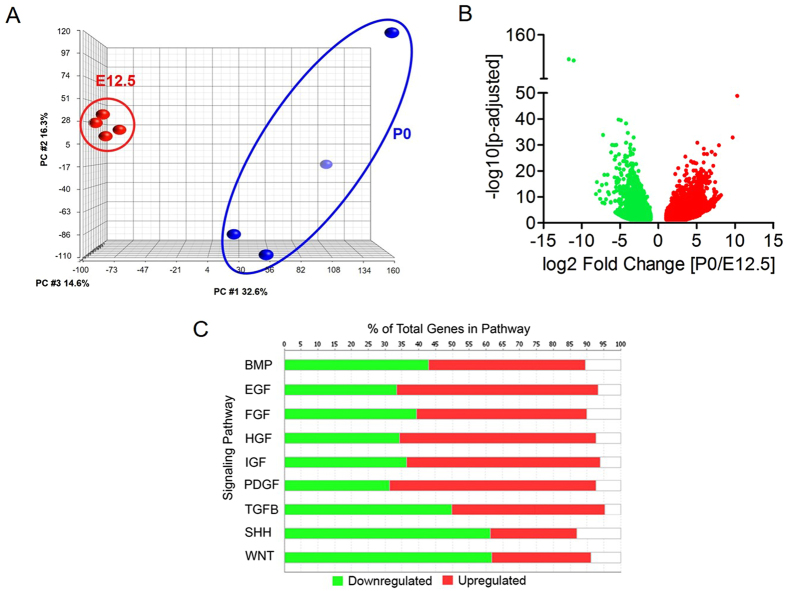
Principal component analysis (PCA) revealed clustering of samples at E12.5 and P0 indicating clear effects of developmental stage on global mRNA abundance (Fig. 3A). There were 5015 genes significantly differentially expressed with fold-changes greater than 2 between the two developmental stages: 2022 genes were upregulated and 2993 genes were downregulated at P0 compared to E12.5 (Fig. 3B). Pathway analysis revealed signaling pathways previously established as important regulators of embryonic tissue morphogenesis as exhibiting significant differential expression at P0 compared to E12.5. These included the sonic hedgehog (Shh), transforming growth factor-beta (TGF-β), and insulin-binding growth factor (IGF) signaling pathways, amongst others (Fig. 3C).
Figure 3.
Global RNA-Seq analysis. (A) Principle component analysis of RNA-Seq data. (B) Volcano plot showing log2-fold change of mRNA abundance as a ratio of P0 expression to E12.5 expression. (C) Pathway analysis of RNA expression data showing percent of total genes significantly differentially expressed at P0 compared to E12.5 for key developmental pathways. N = 4.
Validation of Secreted Signaling Molecules, Pathway Elements, and Targets
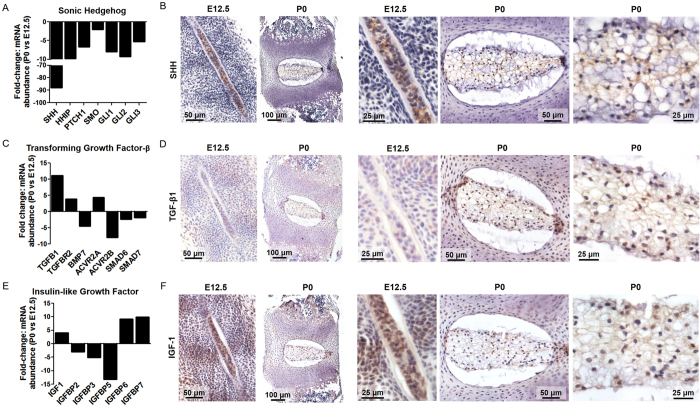
There was significantly lower mRNA abundance of Shh pathway activators including ligand (Shh), receptors (Patched1 (Ptch1); Smoothened (Smo)), and transcription factors (Gli1, 2, 3) (Table 1, Fig. 4A) at P0 when compared to E12.5. Shh mRNA showed the largest overall change with 87.8-fold lower abundance at P0 compared to E12.5. Immunohistochemistry was performed to confirm the downregulation of SHH protein expression at P0 compared to E12.5. In line with RNA-Seq data, there was intense SHH-positive staining in the notochord at E12.5 and light diffuse SHH staining in the NP at P0 (Fig. 4B).
Table 1.
Fold-change in mRNA expression of Shh pathway genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression P0 vs E12.5 | p value |
|---|---|---|---|
| Gli1 | GLI family zinc finger 1 | −7.99 | 1.05E-18 |
| Gli2 | GLI family zinc finger 2 | −9.23 | 1.00E-12 |
| Gli3 | GLI family zinc finger 3 | −5.27 | 5.35E-07 |
| GliS1 | GLIS family zinc finger 1 | −7.43 | 1.09E-07 |
| GliS2 | GLIS family zinc finger 2 | −3.13 | 2.31E-04 |
| Hhip | Hedgehog interacting protein | −9.86 | 6.54E-15 |
| Ptch1 | Patched 1 | −6.72 | 3.34E-17 |
| Ptch2 | Patched 2 | −5.05 | 1.36E-04 |
| Shh | Sonic Hedgehog | −87.82 | 8.84E-12 |
| Smo | Smoothened | −2.03 | 5.07E-03 |
| Sufu | SUFU negative regulator of hedgehog signaling | −2.05 | 1.89E-03 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Figure 4.
Fold-change in mRNA expression of P0 vs E12.5 NDCs for selected developmental pathways and immunohistochemical analysis of component growth factors at E12.5 and P0. (A) Fold-change in mRNA expression of Shh signaling pathway elements showing significant downregulation at P0. (B) SHH protein staining showing intense positive staining of the notochord at E12.5 and diffuse staining of the NP at P0. (C) Fold-change in mRNA expression of TGF-β pathway elements showing significant upregulation of both Tgfb1 and Tgfbr2 at P0. (D) TGF-β1 protein staining showing few positively stained notochord cells at E12.5, and widespread positive staining of NP cells and diffuse NP ECM staining at P0. (E) Fold-change in mRNA expression of IGF pathway elements showing significant upregulation of Igf1 at P0. (F) IGF-1 protein staining showing positive staining of the notochord at E12.5, and widespread positive staining of NP cells and diffuse NP ECM staining at P0. N = 4; all fold changes p < 0.05, P0 compared to E12.5, analyzed by DESeq2 and adjusted for false discovery rate. Refer to Fig. 2 for annotation of structural regions.
Around 95% of genes associated with the TGF-β pathway were differentially expressed at P0 compared to E12.5 (Fig. 3C), including 11.2 and 3.9-fold higher mRNA abundance of the secreted ligand Tgfb 1 and its receptor Tgfbr2, respectively (Table 2, Fig. 4C). Immunohistochemistry revealed widespread TGF-β1 positive cells in the mesenchymal progenitor population surrounding the notochord at E12.5 and TGF-β1 staining of relatively few cells within the notochord itself (Fig. 4D). At P0, there was widespread staining of cells within the NP and diffuse staining within the ECM (Fig. 4D). Around 93% of IGF pathway elements exhibited differential expression at P0 compared to E12.5 (Fig. 3C), including a 4.12-fold mRNA increase in the secreted ligand Igf1 as well as 9.11-fold and 9.92-fold increases in IGF binding proteins 6 and 7, respectively (Table 3, Fig. 4E). Immunohistochemistry for IGF-1 protein expression revealed positive staining at E12.5 within the notochord as well as in the surrounding mesenchymal progenitor population (Fig. 4F). Similarly to TGF-β1, there was widespread IGF-1 staining of cells in the NP and diffuse IGF-1 staining of the ECM at P0 (Fig. 4F).
Table 2.
Fold-change in mRNA expression of TGF-β pathway genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression | p value |
|---|---|---|---|
| P0 vs E12.5 | |||
| Acvr2a | activin A receptor type IIA | 4.39 | 1.71E-04 |
| Acvr2b | activin A receptor type IIB | −7.92 | 3.35E-16 |
| Bmp1 | Bone morphogenetic protein 1 | −2.04 | 1.28E-02 |
| Bmp2k | BMP2-inducible protein kinase | 2.66 | 7.52E-03 |
| Bmp3 | Bone morphogenetic protein 3 | −5.93 | 2.35E-08 |
| Bmp4 | Bone morphogenetic protein 4 | −3.13 | 1.98E-05 |
| Bmp7 | Bone morphogenetic protein 7 | −4.45 | 3.10E-08 |
| Bmpr1b | Bone morphogenetic protein receptor type-1B | −4.05 | 9.93E-05 |
| Fst | Follistatin | −3.79 | 9.09E-07 |
| Grb2 | growth factor receptor bound protein 2 | 2.64 | 1.32E-04 |
| Grem1 | Gremlin 1 | 3.77 | 1.11E-03 |
| Nfkb1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 3.49 | 2.63E-07 |
| Nfkb2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | 3.28 | 8.34E-04 |
| Nog | noggin | −28.36 | 3.52E-19 |
| Smad5 | SMAD family member 5 | −2.24 | 4.29E-06 |
| Smad6 | SMAD family member 6 | −2.39 | 7.15E-03 |
| Smad7 | SMAD family member 7 | −1.89 | 3.37E-02 |
| Sostdc1 | sclerostin domain containing 1 | 9.09 | 1.83E-10 |
| Tfe3 | transcription factor binding to IGHM enhancer 3 | 2.04 | 5.43E-03 |
| Tgfb1 | Transforming growth factor beta 1 | 11.18 | 2.38E-11 |
| Tgfbr2 | Transforming growth factor beta receptor 2 | 3.92 | 2.86E-08 |
| Znf423 | zinc finger protein 423 | −9.70 | 3.89E-23 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Table 3.
Fold-change in mRNA expression of IGF pathway genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression P0 vs E12.5 | p value |
|---|---|---|---|
| Elk1 | ELK1, member of ETS oncogene family | −1.86 | 1.61E-02 |
| Fos | FBJ murine osteosarcoma viral oncogene homolog | 23.33 | 4.50E-12 |
| Foxo1 | forkhead box O1 | 1.82 | 3.55E-02 |
| Grb10 | growth factor receptor bound protein 10 | −3.99 | 6.37E-07 |
| Grb2 | growth factor receptor bound protein 2 | 2.64 | 1.32E-04 |
| Igf1 | insulin-like growth factor 1 | 4.12 | 2.64E-03 |
| Igf2 | insulin-like growth factor 2 | −10.88 | 1.45E-15 |
| Igfbp2 | insulin-like growth factor binding protein 2 | −3.04 | 1.09E-02 |
| Igfbp3 | insulin-like growth factor binding protein 3 | −5.06 | 7.78E-06 |
| Igfbp5 | insulin-like growth factor binding protein 5 | −13.16 | 3.92E-23 |
| Igfbp6 | insulin-like growth factor binding protein 6 | 9.11 | 2.15E-04 |
| Igfbp7 | insulin-like growth factor binding protein 7 | 9.92 | 1.05E-09 |
| Igf2bp1 | insulin-like growth factor 2 mRNA binding protein 1 | −9.22 | 5.53E-29 |
| Igf2bp2 | insulin-like growth factor 2 mRNA binding protein 2 | −12.52 | 7.07E-31 |
| Igf2bp3 | insulin-like growth factor 2 mRNA binding protein 3 | −1.87 | 2.46E-03 |
| Igf2os | insulin-like growth factor 2, opposite strand | −10.24 | 2.11E-09 |
| Igf2r | insulin-like growth factor 2 receptor | −3.18 | 3.43E-04 |
| Irs1 | insulin receptor substrate 1 | −3.22 | 1.04E-03 |
| Jak1 | Janus kinase 1 | 2.77 | 3.88E-05 |
| Jak2 | Janus kinase 2 | 1.98 | 3.85E-03 |
| Stat3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 2.89 | 6.16E-06 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Over 90% of Wnt signaling pathway elements exhibited significant differential mRNA expression at P0 compared to E12.5 (Fig. 3C), including pathway ligands Wnt 1, 3, 6, 11, 5a, 7a, 7b and 9b (all lower abundance at P0) and Wnt16 (higher abundance at P0), modulators such as Sfrps (secreted frizzled-related proteins), and downstream target genes, Axin2, Cd44, and Myc (Supplemental Table S2). Additional differential gene expression of molecules in other key signaling pathways (epidermal growth factor - Egf, fibroblast growth factor - Fgf, platelet-derived growth factor - Pdgf) identified through pathway analysis, as well as gene expression data on commonly expressed kinases and phosphatases, are presented in Supplemental Tables S3–S6.
Extracellular Matrix
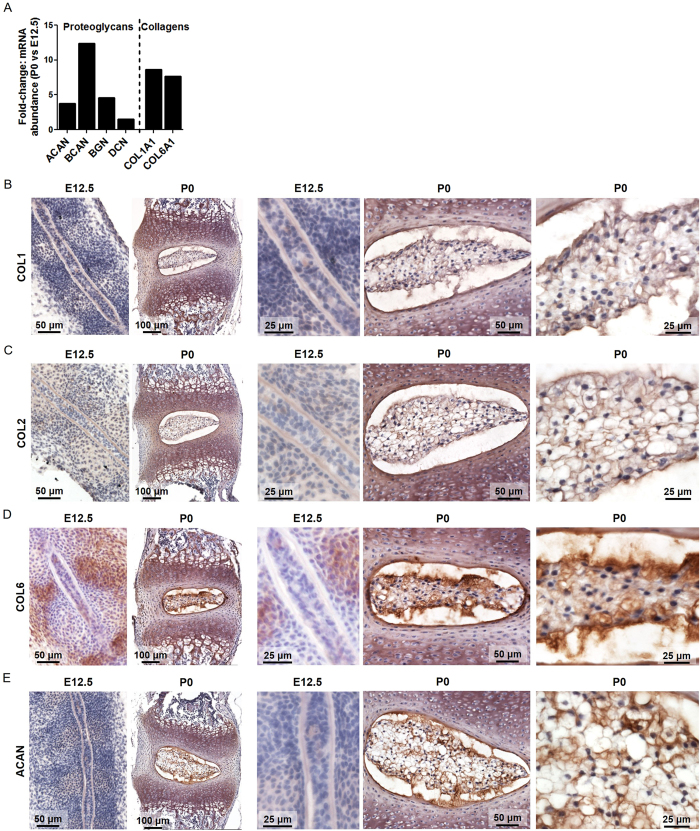
There was significantly higher mRNA abundance of ECM structural molecules, including proteoglycans (aggrecan (Acan); brevican (Bcan); biglycan (Bgn); decorin (Dcn)) and collagens (Col1a1, Col6a1), at P0 compared to E12.5 (Tables 4 and 5, Fig. 5A). Protein expression of COL1, COL2, COL6, and ACAN at each developmental stage was examined using immunohistochemistry (Fig. 5B–E). Overall, COL1, COL6, and ACAN showed diffuse staining in non-cellular regions (core and sheath) of the notochord at E12.5. At P0, COL6, and ACAN exhibited diffuse positive protein staining throughout the NP ECM, with intense staining in the outer boundary of the NP (Fig. 5D,E). Diffuse positive COL1 staining was present throughout the NP at P0 (Fig. 5B). There was no significant difference in the mRNA abundance of Col2a1 between E12.5 and P0, and IHC revealed diffuse protein expression of COL2 in the notochord and NP at each respective developmental stage (Fig. 5C).
Table 4.
Fold-change in mRNA expression of proteoglycan genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression | p value |
|---|---|---|---|
| P0 vs E12.5 | |||
| Acan | Aggrecan | 3.78 | 4.14E-03 |
| Agrn | Agrin | −7.00 | 1.56E-14 |
| Bcan | Brevican | 12.39 | 2.25E-04 |
| Bgn | Biglycan | 4.56 | 4.11E-08 |
| Epyc | Epiphycan | −7.46 | 3.39E-04 |
| Fmod | Fibromodulin | 4.91 | 5.29E-05 |
| Gpc2 | Glypican-2 | −8.12 | 1.07E-16 |
| Gpc3 | Glypican-3 | −4.66 | 2.95E-06 |
| Ncan | Neurocan | −2.59 | 1.35E-02 |
| Omd | Osteomodulin | 5.28 | 2.45E-02 |
| Prelp | Prolargin | 2.66 | 1.92E-02 |
| Sdc1 | Syndecan-1 | −4.63 | 1.02E-10 |
| Sdc3 | Syndecan-3 | 2.70 | 5.15E-03 |
| Sdc4 | Syndecan-4 | 3.80 | 2.80E-05 |
| Vcan | Versican | −20.00 | 1.37E-26 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Table 5.
Fold-change in mRNA expression of collagen genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression P0 vs E12.5 | p value |
|---|---|---|---|
| Col1a1 | Collagen I alpha 1 | 8.65 | 1.20E-07 |
| Col1a2 | Collagen I alpha 2 | 5.54 | 7.67E-09 |
| Col3a1 | Collagen III alpha 1 | −3.87 | 4.94E-05 |
| Col4a1 | Collagen IV alpha 1 | −6.88 | 3.44E-11 |
| Col4a2 | Collagen IV alpha 2 | −5.70 | 1.97E-09 |
| Col4a5 | Collagen IV alpha 5 | −6.18 | 1.63E-16 |
| Col4a6 | Collagen IV alpha 6 | −5.10 | 1.48E-03 |
| Col9a1 | Collagen IX alpha 1 | −7.31 | 4.85E-09 |
| Col9a2 | Collagen IX alpha 2 | −20.46 | 6.12E-26 |
| Col9a3 | Collagen IX alpha 3 | −4.34 | 1.98E-05 |
| Col5a3 | Collagen V alpha 3 | 4.29 | 1.07E-03 |
| Col6a1 | Collagen VI alpha 1 | 7.68 | 8.97E-08 |
| Col6a2 | Collagen VI alpha 2 | 5.80 | 2.15E-05 |
| Col10a1 | Collagen X alpha 1 | 8.88 | 2.41E-12 |
| Col11a1 | Collagen XI alpha 1 | −3.33 | 1.67E-03 |
| Col12a1 | Collagen XII alpha 1 | −2.79 | 3.45E-04 |
| Col14a1 | Collagen XIV alpha 1 | −4.59 | 6.75E-11 |
| Col19a1 | Collagen XIX alpha 1 | −4.94 | 1.79E-05 |
| Col17a1 | Collagen XVII alpha 1 | −3.42 | 3.93E-03 |
| Col18a1 | Collagen XVIII alpha 1 | −5.20 | 8.01E-07 |
| Col20a1 | Collagen XX alpha 1 | −4.20 | 1.12E-04 |
| Col22a1 | Collagen XXII alpha 1 | 4.18 | 2.15E-05 |
| Col23a1 | Collagen XXIII alpha 1 | −4.69 | 8.56E-06 |
| Col25a1 | Collagen XXV alpha 1 | −4.18 | 3.20E-05 |
| Col26a1 | Collagen XXVI alpha 1 | −9.80 | 1.02E-13 |
| Col27a1 | Collagen XXVII alpha 1 | −6.74 | 5.99E-09 |
| Col28a1 | Collagen XXVIII alpha 1 | 5.28 | 1.64E-02 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Figure 5.
Fold-change in mRNA expression of P0 vs E12.5 NDCs and immunohistochemical analysis of key ECM molecules at E12.5 and P0. (A) Fold-change in mRNA expression of proteoglycans and collagens showing significant upregulation at P0. Protein staining for (B) COL1, (C) COL2, (D) COL6, and (E) ACAN, showing weak staining in non-cellular regions (core and sheath) of the E12.5 notochord and intense positive staining of the NP at P0. N = 4, all fold changes p < 0.05, P0 compared to E12.5, analyzed by DESeq2 and adjusted for false discovery rate. Refer to Fig. 2 for annotation of structural regions.
With respect to surrounding tissue structures, at E12.5, condensations in the regions of the future discs exhibited positive staining for COL1 and COL6 protein (Fig. 5B,D). At P0, there was positive staining for COL2 protein in the inner AF, cartilaginous epiphyses, growth plates, and primary spongiosa (Fig. 5C). COL1 protein staining was observed in the AF and ossified regions of the vertebral bodies (Fig. 5B). COL6 and ACAN staining were present in the growth plates (Fig. 5D,E).
Nucleus Pulposus Cell-Specific Markers
Finally, we examined differential mRNA expression of molecules considered to be specific markers of the NP cell phenotype37. Several such markers exhibited stable mRNA expression across the E12.5 to P0 developmental window, including brachyury (T), keratins 8 and 18 (Krt8, Krt18), and hypoxia-induced factor (Hif1a) (Table 6). Other NP markers exhibited significant changes in mRNA expression from E12.5 to P0, including cluster of differentiation 24 (Cd24, 2.48-fold increase), keratin 19 (Krt19, 2.9-fold increase), carbonic anhydrase 3 (Car3, 8.3-fold increase), carbonic anhydrase 12 (Car12, 4.6-fold decrease), and vimentin (Vim, 5.7-fold increase) (Table 6).
Table 6.
Fold-change in mRNA expression of putative NP marker genes at P0 compared to E12.5.
| Gene | Name | Fold Change in Expression P0 vs E12.5 | p value |
|---|---|---|---|
| Car12 | Carbonic anhydrase 12 | −4.46 | 0.006 |
| Car3 | Carbonic anhydrase 3 | 7.41 | 3.79E-07 |
| Cd24 | Cluster of differentiation 24 | 2.48 | 0.003 |
| Krt19 | Cytokeratin 19 | 3.24 | 0.003 |
| Sostdc1 | Sclerostin domain-containing protein 1 | 9.09 | 1.83E-10 |
| Vim | Vimentin | 5.22 | 2.91E-08 |
N = 4, Differential expression analyzed by DESeq2 and p values adjusted for false discovery rate.
Discussion
Recapitulation of developmental signaling responsible for embryonic NP formation represents a promising strategy for regenerating the intervertebral discs. During embryonic development, the cells of the notochord function both as a source of secreted signaling molecules to regulate the patterning of the discs and surrounding tissues and as the direct progenitors of NP cells. Notochordal NP cells from juvenile discs have been demonstrated to possess anabolic disease-modifying effects40–45, and thus, the application of notochord-derived factors in disc regeneration strategies has received considerable interest. In this study, we advance the molecular-level understanding of notochord cell function and disc development by using RNA-Seq to define the global changes in mRNA expression of NDCs at key stages of embryonic disc formation. It should be emphasized that the data analysis presented here, together with the immunohistochemical validation of a small number of molecules is a first step, and future work will delve further into specific pathways and gene families responsible for formation of the intervertebral discs. We will build on these results by extending RNA-Seq studies to additional pre- and postnatal developmental stages.
In the last decade, there have been important advances in our understanding of intervertebral disc development. Fate mapping studies in mice have provided conclusive evidence of the developmental origin of NP cells16, 17 and support the premise that these cells are the direct progenitors of adult NP cells. While these fate-mapping studies do not conclusively prove the notochordal origin of all NP cells in higher species, including humans, transcriptome analysis has demonstrated that adult human and bovine NP cells do highly express some notochordal cell-associated molecules46, 47. With respect to molecular regulation of embryonic disc formation, Shh and TGF-β signaling have both been established as indispensable for normal disc development48–50. SHH is a long range morphogen, expressed at various stages of embryonic development by cells of the primitive node, notochord, the neural tube floor plate, and posterior limb bud, which regulates spatial patterning, cell survival, and proliferation in both the axial skeleton and central nervous system6, 14. Shh signaling is critical for early embryonic patterning of the discs50, 51 and disruption of Shh signaling in mouse embryos through removal of Smo from the notochord and floor plate results in reduced NP size and absence of AF lamellar structure50. SHH is also required for formation of the sheath surrounding the notochord, and in its absence, notochord cells migrate aberrantly throughout surrounding tissues50. It has also been shown that SHH expressed solely by notochordal cells is sufficient for disc formation51. Interestingly, if SHH is removed subsequent to sheath formation the discs develop normally, suggesting that SHH is of foremost importance in the early stages of embryonic disc development50. Our finding in the current study that Shh signaling is significantly diminished at P0 compared to E12.5, evidenced by lower mRNA abundance of the ligand, receptor, and downstream transcriptional regulators and confirmed via immunohistochemistry (Fig. 4B), is consistent with this early embryonic role. A previous study showed that there continues to be Shh mRNA expressed in the mouse NP at E15.5, which is mid-way through the notochord to NP transformation52. While our results suggest a significantly diminished role for Shh signaling in the NP following embryonic development, there is evidence for a continuing postnatal role for this pathway53, 54, although the exact nature of this role and its importance relative to other pathways for regenerative therapeutics remains to be elucidated.
TGF-β signaling is critical for embryonic formation of both the NP and AF48, 55, 56. In mice where Tgfbr2 is specifically removed from type II collagen expressing cells, the NPs that form are abnormally small suggesting that TGF-β signaling is required to drive NP growth55. TGF-β has been localized to human notochord cells and was shown to increase in expression from 12 to 14 weeks gestation57. The presence of all TGF-β isoforms and type I and II receptors has been demonstrated in the adult mouse NP, with expression decreasing progressively with aging7. In the current study, we found an 11-fold increase in Tgfb1 mRNA abundance at P0 relative to E12.5 with protein expression confirmed using immunohistochemistry at both ages, and a corresponding 4-fold increase in Tgfbr2 mRNA abundance. These results support the previously described central role for TGF-β signaling in the late embryonic and early postnatal differentiation and growth of the disc38, 58.
While the Shh and TGF-β pathways are perhaps the best characterized with respect to disc development, our results also demonstrated differential expression of secreted signaling molecules in other pathways. IGFs are important regulators of proliferation and differentiation during development across multiple skeletal cell types, including chondrocytes and osteoblasts59, 60. In NDCs, Igf1 and Igf2 exhibited opposite expression patterns, exhibiting higher and lower mRNA abundance at P0 compared to E12.5, respectively (Table 3). In general, these findings are consistent with existing literature for other tissues where Igf1 has been shown to increase in expression during postnatal growth and into adulthood while Igf2 is highest during embryonic and fetal development11. Igf1r-deficient mice exhibit signs of accelerated disc degeneration, suggesting Igf1 is required to maintain the healthy adult disc10. In culture studies, IGF-1 has been shown to exert anabolic and anti-apoptotic effects on disc cells18, 20, 29, 30, 32, 61, 62 and has also been shown to exert anabolic effects on disc cells in vivo 31.
The ECM components of the NP are important in supporting tissue structure and biomechanical function as well as for the binding and distribution of the many secreted growth factors that mediate tissue morphogenesis, homeostasis, repair, and remodeling. The ECM composition of the adult NP has been widely studied. The two components that are principally important for mechanical function (specifically, resisting compressive forces) are COL2 and ACAN. Both COL2 and ACAN proteins have been shown previously to be expressed in the NP during embryonic development12, 63. COL2 is prominent in the human fetal NP from ~12 weeks gestation with ACAN also present as both diffuse and focal extracellular deposits12. COL2 is critical for the notochord to NP transformation64: in Col2-deficient mice the notochord persists as a rod-like structure until birth and intervertebral discs do not form. Here, we show that Col2a1 exhibits stable mRNA abundance as the notochord transforms into the NP, while Acan abundance is higher at P0 compared to E12.5. Protein for both ACAN and COL2 was detected immunohistochemically at both developmental stages. Interestingly, we found increased mRNA abundance of Col1, not typically associated with the healthy NP matrix, at P0 relative to E12.5. In the adult disc, Col1 is predominantly confined to the outer annulus fibrosus1 except during aging and degeneration when the NP becomes more fibrous63. Consistent with our results, however, Col1 mRNA and protein have been demonstrated previously in the developing notochord and NP in humans and mice12, 54, 57, 65. We also found significantly elevated Col6 mRNA expression at P0 (Table 4, Fig. 5A). COL6 protein, shown to be present in the human fetal disc from 12 weeks gestation and to increase with postnatal development and into adulthood63, exhibits predominantly pericellular localization8 and may play an important role in cell mechanotransduction similar to articular cartilage9. We also found significantly increased mRNA abundance of the small leucine rich repeat proteoglycans (SLRPs) Bgn, Fmod, and Prelp at P0. These molecules have established roles in the regulation of ECM assembly through binding to collagen fibrils and growth factors such as TGF-β and IGF-166–68.
Finally, we examined the abundance of putative NP phenotypic markers shown previously to be expressed by immature and adult NP cells38, 41, 42, 61, 63. The search for NP markers has attracted fervent interest over the past decade due to their potential utility in validating NP-lineage specific differentiation of stem cells and to facilitate NP-specific analyses of gene function in transgenic animal models37, 46, 47, 67, 69. Here, we identify several such markers that exhibited stable or increased expression across the E12.5 to P0 developmental window that may be the most faithful indicators of notochordal and early postnatal NP cell phenotypes.
A limitation of this study was that we analyzed NDCs assuming a homogeneous population. Previous work demonstrated heterogeneity within the mouse NP cell population and the associated ECM, which manifests during later stages of development and into adulthood as a central notochordal-rich region surrounded by a halo of GAG-rich matrix-containing cells that are more chondrocyte-like. While our histological observations at P0 (Fig. 2D,E) superficially suggest a homogeneous cell population, it may well be the case that at the molecular level, phenotypically distinct subpopulations of cells are already beginning to emerge at this early postnatal developmental stage. Future work will seek to establish the extent to which altered gene expression at P0, and at later stages of postnatal development, reflects progressive heterogeneity in NP cellularity as a consequence of differentiation. While whole-transcriptome profiling studies such as this one provide comprehensive and high-fidelity gene-level information across a specified developmental window, we can only speculate as to the specific function and relative importance of many of those genes in disc development. To answer these questions, we anticipate that the results presented here will motivate future tissue-specific knockdown or overexpression studies in transgenic animal models.
In addition to improving fundamental understanding of molecular regulation of embryonic disc formation, our long term goal is to use these results to develop improved biologic strategies for regenerating NP tissue. While direct application of notochordal cells in cell-based therapeutics is limited due to their prenatal source, it may be possible to reprogram more readily available therapeutic cell types such as mesenchymal or induced pluripotent stem cells to mimic the secretory profile of notochordal cells, enhancing their potential for NP-specific tissue reconstitution. A promising strategy may be to drive stem cells towards a mature biosynthetic NP cell phenotype through sequential exposure to specific growth factors. A similar paradigm has been used previously to derive cartilage cells from pluripotent stem cell-derived progenitors69, and the results of this study provide a preliminary roadmap towards achieving this for disc regeneration.
Conclusions
We have conducted the first transcriptome-wide analysis of notochord-derived cells during embryonic intervertebral disc formation. Overall, our results suggest that the secretory profile of NDCs changes from one optimized to direct tissue patterning through secretion of morphogens such as SHH at E12.5, to one focused on regulating cell differentiation, proliferation, and tissue growth through increased secretion of growth factors such as TGF-β1 and IGF-1 at P0 combined with increased synthesis of ECM molecules required to support the structure and mechanical function of the NP.
Electronic supplementary material
Acknowledgements
This work was funded by grants from the University of Pennsylvania Institute on Aging, the Department of Veteran’s Affairs (I01RX001321), and the National Institutes of Health (F32AR071298, R21AR070959, and R21AR062690). Additional support was received from the Penn Center for Musculoskeletal Disorders (P30AR069619) and the Sharpe Foundation. The authors acknowledge technical support provided by staff members at the Next Generation Sequencing Core and the Flow Cytometry Facility at the University of Pennsylvania.
Author Contributions
S.H.P. designed and performed the experiments and drafted the manuscript. K.K.M. maintained the mouse line and dissected notochords/spines from animals. J.W.T. carried out statistical analysis of RNA-Seq data. N.R.M. and B.D.H. provided conceptual input on experimental design. L.J.S. designed the experiments and drafted the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10692-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Urban, J. In The Lumbar Spine Vol. 1 (eds Wiesel, S. W. et al.) 271–280 (Saunders, 1996).
- 2.Humzah MD, Soames RW. Human intervertebral disc: structure and function. Anat Rec. 1988;220:337–356. doi: 10.1002/ar.1092200402. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy JJ, Hiltner A, Baer E. Hierarchical structure of the intervertebral disc. Conn. Tissue Res. 1989;23:75–88. doi: 10.3109/03008208909103905. [DOI] [PubMed] [Google Scholar]
- 4.Rajasekaran S, Babu JN, Arun R, Armstrong BR, Shetty AP. ISSLS prize winner: A study of diffusion in human lumbar discs: a serial magnetic resonance imaging study documenting the influence of the endplate on diffusion in normal and degenerate discs. Spine. 2004;29:2654–2667. doi: 10.1097/01.brs.0000148014.15210.64. [DOI] [PubMed] [Google Scholar]
- 5.Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- 6.Christ B, Huang R, Scaal M. Formation and differentiation of the avian sclerotome. Anat Embryol. 2004;208:333–350. doi: 10.1007/s00429-004-0408-z. [DOI] [PubMed] [Google Scholar]
- 7.Danos MC, Yost HJ. Linkage of cardiac left-right asymmetry and dorsal-anterior development in Xenopus. Development. 1995;121:1467–1474. doi: 10.1242/dev.121.5.1467. [DOI] [PubMed] [Google Scholar]
- 8.Fouquet B, Weinstein BM, Serluca FC, Fishman MC. Vessel patterning in the embryo of the zebrafish: guidance by notochord. Developmental Biology. 1997;183:37–48. doi: 10.1006/dbio.1996.8495. [DOI] [PubMed] [Google Scholar]
- 9.Pourquie O, Coltey M, Teillet M, Ordahl C, Douarin N. Control of dorsoventral patterning of somitic derivatives by notochord and floor plate. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5242–5246. doi: 10.1073/pnas.90.11.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada T, Pfaff SL, Edlund T, Jessell TM. Control of cell pattern in the neural tube: motor neuron induction by diffusible factors from notochord and floor plate. Cell. 1993;73:673–686. doi: 10.1016/0092-8674(93)90248-O. [DOI] [PubMed] [Google Scholar]
- 11.Yamada T, Placzek M, Tanaka H, Dodd J, Jessell TM. Control of cell pattern in the developing nervous system: polarizing activity of the floor plate and notochord. Cell. 1991;64:635–647. doi: 10.1016/0092-8674(91)90247-V. [DOI] [PubMed] [Google Scholar]
- 12.Linsenmayer TF, Gibney E, Schmid TM. Segmental appearance of type X collagen in the developing avian notochord. Developmental Biology. 1986;113:467–473. doi: 10.1016/0012-1606(86)90182-X. [DOI] [PubMed] [Google Scholar]
- 13.Smits, P., Lefebvre, V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development130 (2003). [DOI] [PubMed]
- 14.Swiderski RE, Solursh M. Localization of type II collagen, long form alpha 1(IX) collagen, and short form alpha 1(IX) collagen transcripts in the developing chick notochord and axial skeleton. Dev Dyn. 1992;194:118–127. doi: 10.1002/aja.1001940205. [DOI] [PubMed] [Google Scholar]
- 15.Smith LJ, Nerurkar NL, Choi KS, Harfe BD, Elliott DM. Degeneration and regeneration of the intervertebral disc: lessons from development. Dis Model Mech. 2011;4:31–41. doi: 10.1242/dmm.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953–3958. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73–82. doi: 10.1242/dmm.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearce RH, Grimmer BJ, Adams ME. Degeneration and the chemical composition of the human lumbar intervertebral disc. J Orthop Res. 1987;5:198–205. doi: 10.1002/jor.1100050206. [DOI] [PubMed] [Google Scholar]
- 19.Roughley PJ, Melching LI, Heathfield TF, Pearce RH, Mort JS. The structure and degradation of aggrecan in human intervertebral disc. Eur Spine J. 2006;15(Suppl 3):S326–332. doi: 10.1007/s00586-006-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JA, Schmatz C, Schultz AB. Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine (Phila Pa 1976) 1988;13:173–178. doi: 10.1097/00007632-198802000-00008. [DOI] [PubMed] [Google Scholar]
- 21.Sive JI, et al. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Molecular Pathology. 2002;55:91–97. doi: 10.1136/mp.55.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter CJ, Matyas JR, Duncan NA. Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: a species comparison. J Anat. 2004;205:357–362. doi: 10.1111/j.0021-8782.2004.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology. 2009;48:5–10. doi: 10.1093/rheumatology/ken396. [DOI] [PubMed] [Google Scholar]
- 24.Andersson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 25.Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88(Suppl 2):21–24. doi: 10.2106/JBJS.E.01273. [DOI] [PubMed] [Google Scholar]
- 26.Bogduk N. The lumbar disc and low back pain. Neurosurg Clin N Am. 1991;2:791–806. [PubMed] [Google Scholar]
- 27.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–130. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirza SK, Deyo RA. Systematic review of randomized trials comparing lumbar fusion surgery to nonoperative care for treatment of chronic back pain. Spine. 2007;32:816–823. doi: 10.1097/01.brs.0000259225.37454.38. [DOI] [PubMed] [Google Scholar]
- 29.Leung VY, Chan D, Cheung KM. Regeneration of intervertebral disc by mesenchymal stem cells: potentials, limitations, and future direction. Eur Spine J. 2006;15:S406–S413. doi: 10.1007/s00586-006-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai D. Future perspectives of cell-based therapy for intervertebral disc disease. Eur Spine J. 2008;17:452–458. doi: 10.1007/s00586-008-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda K, Oegema TR, Jr., An HS. Growth factors and treatment of intervertebral disc degeneration. Spine. 2004;29:2757–2769. doi: 10.1097/01.brs.0000146048.14946.af. [DOI] [PubMed] [Google Scholar]
- 32.Masuda K, An HS. Prevention of disc degeneration with growth factors. Eur Spine J. 2006;15:422–432. doi: 10.1007/s00586-006-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda K, et al. Osteogenic protein-1 injection into a degenerated disc induces the restoration of disc height and structural changes in the rabbit anular puncture model. Spine. 2006;31:742–754. doi: 10.1097/01.brs.0000206358.66412.7b. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JP, Oegema TJ, Bradford DS. Stimulation of mature canine intervertebral disc by growth factors. Spine. 1991;16:253–260. doi: 10.1097/00007632-199103000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Gruber HE, Hoelscher GL, Ingram JA, Bethea S, Hanley EN. IGF-1 rescues human intervertebral annulus cells from in vitro stress-induced premature senescence. Growth Factors. 2008;26:220–225. doi: 10.1080/08977190802273814. [DOI] [PubMed] [Google Scholar]
- 36.Gruber HE, Norton HJ, Hanley EN., Jr. Anti-apoptotic effects of IGF-1 and PDGF on human intervertebral disc cells in vitro. Spine (Phila Pa 1976) 2000;25:2153–2157. doi: 10.1097/00007632-200009010-00002. [DOI] [PubMed] [Google Scholar]
- 37.Risbud MV, et al. Defining the Phenotype of Young Healthy Nucleus Pulposus Cells: Recommendataions of the Spine Research Interest Group at the 2014 Annual ORS Meeting. J Orthop Res. 2015;33:283–293. doi: 10.1002/jor.22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matta A, Karim MW, Isenman DE, Erwin WM. Molecular Therapy for Degenerative Disc Disease: Clues from Secretome Analysis of the Notochordal Cell-Rich Nucleus Pulposus. Scientific Reports. 2017;7 doi: 10.1038/srep45623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biology. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129–137. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 41.Korecki CL, Taboas JM, Tuan RS, Iatridis JC. Notochordal cell conditioned medium stimulates mesenchymal stem cell differentiation toward a young nucleus pulposus phenotype. Stem cell research & therapy. 2010;1 doi: 10.1186/scrt18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purmessur D, et al. Notochordal conditioned media from tissue increases proteoglycan accumulation and promotes a healthy nucleus pulposus phenotype in human mesenchymal stem cells. Arthritis Res Ther. 2011;13 doi: 10.1186/ar3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erwin WM, Ashman K, O’Donnel P, Inman RD. Nucleus pulposus notochord cells secrete connective tissue growth factor and up-regulate proteoglycan expression by intervertebral disc chondrocytes. Arthritis and rheumatism. 2006;54:3859–3867. doi: 10.1002/art.22258. [DOI] [PubMed] [Google Scholar]
- 44.Erwin WM, Islam D, Inman RD, Fehlings MG, Tsui FW. Notochordal cells protect nucleus pulposus cells from degradation and apoptosis: implications for the mechanisms of intervertebral disc degeneration. Arthritis research & therapy. 2011;13 doi: 10.1186/ar3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gantenbein B, et al. Activation of intervertebral disc cells by co-culture with notochordal cells, conditioned medium and hypoxia. BMC Musculoskelet Disord. 2014;15 doi: 10.1186/1471-2474-15-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010;62:3695–3705. doi: 10.1002/art.27710. [DOI] [PubMed] [Google Scholar]
- 47.Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12 doi: 10.1186/ar2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baffi MO, Moran MA, Serra R. Tgfbr2 regulates the maintenance of boundaries in the axial skeleton. Developmental Biology. 2006;296:363–374. doi: 10.1016/j.ydbio.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohn P, Cox M, Chen D, Serra R. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010;10 doi: 10.1186/1471-213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi KS, Harfe BD. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9484–9489. doi: 10.1073/pnas.1007566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi KS, Lee C, Harfe BD. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev. 2012;129:255–262. doi: 10.1016/j.mod.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiPaola CP, Farmer JC, Manova K, Niswander LA. Molecular signaling in intervertebral disk development. J Orthop Res. 2005;23:1112–1119. doi: 10.1016/j.orthres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Postnatal growth, differentiation, and aging of the mouse intervertebral disc. Spine (Phila Pa 1976) 2009;34:447–455. doi: 10.1097/BRS.0b013e3181990c64. [DOI] [PubMed] [Google Scholar]
- 54.Winkler, T., Mahoney, E. J., Sinner, D., Wylie, C. C. & Dahia, C. L. Wnt signaling activates Shh signaling in early postnatal intervertebral discs, and re-activates Shh signaling in old discs in the mouse. Plos One9 (2014). [DOI] [PMC free article] [PubMed]
- 55.Baffi MO, et al. Conditional deletion of the TGF-beta type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Developmental Biology. 2004;276:124–142. doi: 10.1016/j.ydbio.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 56.Kaczmarska M, et al. The influence of very small doses of alpha radiation on the stability of erythrocytes. Microsc Res Tech. 2017;80:131–143. doi: 10.1002/jemt.22803. [DOI] [PubMed] [Google Scholar]
- 57.Gotz W, Osmers R, Herken R. Localisation of extracellular matrix components in the embryonic human notochord and axial mesenchyme. J Anat. 1995;186(Pt 1):111–121. [PMC free article] [PubMed] [Google Scholar]
- 58.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Intercellular signaling pathways active during and after growth and differentiation of the lumbar vertebral growth plate. Spine (Phila Pa 1976) 2011;36:1071–1080. doi: 10.1097/BRS.0b013e3181f7a3ca. [DOI] [PubMed] [Google Scholar]
- 59.Trippel SB, et al. Effect of somatomedin-C/insulin-like growth factor I and growth hormone on cultured growth plate and articular chondrocytes. Pediat Res. 1989;25:76–82. doi: 10.1203/00006450-198901000-00017. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, et al. Osteoblast-specific Knockout of the Insulin-like Growth Factor (IGF) Receptor Gene Reveals an Essential Role of IGF Signaling in Bone Matrix Mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 61.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis. Developmental Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Pratsinis H, Kletsas D. PDGF, bFGF and IGF-I stimulate the proliferation of intervertebral disc cells in vitro via the activation of the ERK and Akt signaling pathways. Eur Spine J. 2007;16:1858–1866. doi: 10.1007/s00586-007-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nerlich AG, Boos N, Wiest I, Aebi M. Immunolocalization of major interstitial collagen types in human lumbar intervertebral discs of various ages. Virchows Arch. 1998;432:67–76. doi: 10.1007/s004280050136. [DOI] [PubMed] [Google Scholar]
- 64.Aszodi A, Chan D, Hunziker E, Bateman JF, Fassler R. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. J Cell Biol. 1998;143:1399–1412. doi: 10.1083/jcb.143.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sowa KM, Last A, Korecki P. Grid-enhanced X-ray coded aperture microscopy with polycapillary optics. Scientific Reports. 2017;7 doi: 10.1038/srep44944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bi Y, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 67.Schaefer L, et al. Decorin-mediated regulation of fibrillin-1 in the kidney involves the insulin-like growth factor-I receptor and Mammalian target of rapamycin. Am J Pathol. 2007;170:301–315. doi: 10.2353/ajpath.2007.060497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schaefer L, et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. Journal of Clinical Investigation. 2005;115:2223–2233. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craft AM, et al. Generation of articular chondrocytes from human pluripotent stem cells. Nat biotechnol. 2015;33:638–645. doi: 10.1038/nbt.3210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during this study are available in the NCBI Gene Expression Omnibus (GEO) repository, accession number: GSE100934.