Abstract
Fgf-3 is expressed in a complex pattern during mouse development. Previously, an essential regulatory element PS4A was identified in the promoter region, and shown to bind at least three factors. To identify the transcription factor(s), we used a yeast one-hybrid screen and obtained a novel Sox6 cDNA (SOX6D). When introduced into cells it strongly repressed activity from both an Fgf-3 reporter gene as well as an artificial promoter containing three PS4A elements. In situ hybridisation analysis showed that Sox6 and Fgf-3 are co-expressed in the otic vesicle of E9.5 mouse embryos in a mutually exclusive pattern, consistent with a repression of Fgf-3 transcription by SOX6. To characterise additional factor(s) involved in Fgf-3 gene repression, a yeast two-hybrid screen was used with the N-terminal portion of SOX6D. Mouse CtBP2 cDNA clones were isolated and shown to bind SOX6 in yeast and mammalian cells. Furthermore, mutational analysis of SOX6 showed that binding to CtBP2, and its responsiveness to this co-repressor, were dependent on a short amino acid sequence motif PLNLSS. Co-expression studies in NIH3T3 cells showed that SOX6 and CtBP2 co-operate to repress activity from the Fgf-3 promoter through the enhancer element PS4A. These results show that SOX6 can recruit CtBP2 to repress transcription from the Fgf-3 promoter.
INTRODUCTION
Fibroblast growth factors (Fgfs) comprise a large family of related proteins that display a broad spectrum of biological activities in vitro. In vivo, several Fgfs have been implicated as intercellular signalling molecules in embryogenesis, homeostasis and tissue repair (reviewed in 1–5). Fgf-3 is expressed at several locations in the mouse embryo suggesting multiple roles during development. Moreover, targeted germ line disruption of Fgf-3 results in abnormalities of the inner ear and fusion of the caudal vertebrae (6–10).
Treatment of F9 embryonal carcinoma cells with retinoic acid and dibutyryl cyclic AMP (Bt2cAMP) preferentially induces markers of parietal endoderm, including expression of Fgf-3 (11–13). Using F9 cells as a model, we previously identified an essential complex regulatory element (PS4A) within the 5′ proximal region of the Fgf-3 promoter (14,15). At least three factors bind co-operatively to PS4A. One factor identified as GATA-4 mediates induction of transcription by retinoic acid and Bt2cAMP. However, other proteins necessary for co-operative binding with GATA-4 have yet to be identified (16).
SOX proteins constitute a family of more than 25 transcription factors related by homology within their HMG-box DNA-binding domain to the mammalian testis determining factor SRY (17,18; reviewed in 19–21). SOX proteins may bind directly to their target promoters (22–26), or affect transcription indirectly by interacting with other components of the transcription machinery (27–29). The SOX family of transcription factors are expressed in adult tissues as well as embryonic locations where they are often implicated in cell fate decisions (19,20). For example, SOX1, 2, 3, 11 and SOXD have been implicated in neural development (30–32), and SOX1, 2, 3 and 9 in lens development (23,27,33). Recently, an unexpected interaction between SOX proteins and β-catenin has suggested that they may also modulate Wnt signalling (34).
In addition to the inherent activating or repressing activities of transcription factors, promoter activity may also be modulated by the association of co-activators or co-repressors (reviewed in 35). A recently defined class of co-repressor identified by their interactions with the adenovirus E1A protein are the CtBP proteins (36). A Drosophila homologue of human CtBP1 (dCtBP) was shown to interact with the transcriptional repressors Hairy, Knirps and Snail (37,38). In addition, CtBP homologues were shown to associate and act as co-repressors with basic Krüppel-like factor, a zinc finger-homeodomain protein δEF1, and the vertebrate homologues of Polycomb proteins XPc and HPC2 (39–41).
In this study we show that PS4A contains a SOX binding site. Using a yeast one-hybrid screen a splice variant of Sox6 (SOX6D) was isolated and shown by transfection experiments to act as a repressor of Fgf-3 transcription. Whole mount in situ hybridisation analysis on early embryos identified the otic vesicle as a potential site of Fgf-3/SOX6 interaction in vivo. In a subsequent yeast two-hybrid screen using part of the SOX6 protein, we isolated mouse CtBP2 as a co-repressor that interacts with SOX6. Point mutation analysis showed that SOX6 interacts with CtBP2 through the sequence motif PLNLSS. Moreover, reporter assays confirmed that transcription of Fgf-3 is negatively regulated through an interaction of SOX6 with CtBP2.
MATERIALS AND METHODS
Electrophoretic mobility shift assay (EMSA)
Experimental procedures and conditions were as previously described except that the non-specific competitor was poly(dG-dC):poly(dC-dG) (Amersham Pharmacia Biotech) (16). In vitro transcribed and translated SOX6 proteins were synthesised from pCI-neo/Sox6 DNA (described below) using TnT Coupled Reticulocyte Lysate System (Promega). SOX6 protein fused to glutathione S-transferase (GST) was generated from a cDNA fragment cloned into pGEX-5X-1 (Pharmacia Biotech). Bacterial extracts expressing the fusion protein (pGEX/Sox6) were purified using a MicroSpin GST Purification Module (Pharmacia Biotech). The truncated SOX6 protein pGEX/Sox6DB was similarly prepared. It comprised 132 amino acids (523–654) of SOX6D, encompassing the 80 residues of the HMG-box. Anti-SOX6 sera were kindly provided by Dr Shinya Yamashita (Nippon Suisan Kaisha Ltd, Tokyo, Japan) (42) and Dr Véronique Lefebvre (The University of Texas, TX) (28). Both antisera gave similar results.
Yeast one-hybrid and two-hybrid screening
Yeast one-hybrid screening was carried out according to MATCHMAKER One-Hybrid System (Clontech). pHISi-4Ay and pLacZi-4Ay bait plasmids were constructed by inserting a synthetic DNA oligomer corresponding to the central element of the PS4A enhancer contained in the Fgf-3 promoter (16). cDNAs were synthesised from poly(A)+ RNA prepared from differentiated F9 cells using TimeSaver cDNA Synthesis Kit (Pharmacia Biotech), and used as a dF9 cDNA library after fusion to GAL4-AD sequence in pGAD424. Procedures for isolation of total RNA and selection of poly(A)+ RNA were described previously (16). Yeast two-hybrid screening was carried out according to the MATCHMAKER Two-Hybrid System (Clontech). The bait plasmid was constructed by fusion of Sox6 cDNA corresponding to 523 amino acids at the N-terminus to GAL4-DBD sequence in pGBT9. After screening the dF9 cDNA library described above, the CtBP2 cDNA clone obtained was found to lack 30 nt from the 5′ initiation codon. The cDNA was extended beyond the initiation codon by 5′ RACE System (Gibco BRL). For two-hybrid interactions in yeast cells, Sox6 and CtBP2 cDNAs encoding full-length proteins were fused to GAL4-DBD in pGBT9 and GAL4-AD in pGAD424, respectively. Point or deletion mutations were introduced into the Sox6 insert using QuickChange Site-Directed Mutagenesis Kit (Stratagene).
Transfection assay
The luciferase reporter plasmids ptkLuc, p(4Ax3)tkLuc and pFgf-3/Luc used in transfection experiments have been described previously (16). Expression plasmids, pCI-neo/Sox6 and pCI-neo/CtBP2 were constructed by inserting the coding region of the Sox6 and CtBP2 cDNAs, respectively, into the multiple-cloning site of pCI-neo (Promega). Procedures for cell culture, transfection with plasmid DNA by the calcium phosphate–DNA precipitation method, preparation of cell extracts and luciferase assays have been described previously (14–16).
Immunoprecipitation and immunoblotting
The plasmid for expression of SOX6 tagged with six histidine residues at the C-terminus, pCI-neo/Sox6HT was constructed by insertion of the Sox6 coding region into pET-21b(+) (Novagen) and then re-insertion of the coding region with the tag into pCI-neo. The mutations introduced for the two-hybrid interaction experiments described above were transferred to pCI-neo/Sox6HT by replacing the ApaI–EcoNI fragment of the wild-type cDNA with the equivalent fragment for each of the mutated plasmids. The plasmid for expression of CtBP2 tagged with FLAG at the N-terminus, pCMV/FLAG-CtBP2, was constructed by insertion of the CtBP2 coding region into pCMV-Tag 2 (Stratagene). The plasmid DNAs were introduced into Cos7 cells by an electroporation method. Briefly, cells were trypsinised and resuspended in K-PBS (30.8 mM NaCl, 120.7 mM KCl, 8.1 mM Na2HPO4, 1.46 mM KH2PO4 and 5 mM MgCl2). After adding 20 µg DNA for each 10 cm dish of cultured cells the suspension was electrically pulsed at 250 V, 960 µF using Bio-Rad Gene Pulser and cultured for a further 2 days. Cells were lysed in a lysis buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 µg/ml leupeptin, 1 mM PMSF, 1.0% Triton X-100). His-tagged SOX6 and FLAG-tagged CtBP2 proteins in the cell lysates were immunoprecipitated with Protein G–Sepharose (Pharmacia Biotech) coated with anti-His serum (Santa Cruz) and anti-FLAG serum (Eastman Kodak Company), respectively. The immunoprecipitates were separated by SDS–PAGE and analysed by immunoblotting.
Northern blot hybridisation
Procedures for isolation of total RNA, selection of poly(A)+ RNA and northern blot hybridisation were described previously (16). An AccI fragment (1.3 kb, 363–1711) of Sox6 cDNA and an EcoRI fragment (1.1 kb, 1–1079) of CtBP2 cDNA were used for hybridisation as probes. The hEF1a cDNA clone was obtained from Dr Sakamaki (Kyoto University, Kyoto, Japan), and a 2.3 kb BamHI fragment was used for hybridisation.
In situ hybridisation
Whole mount in situ hybridisation of mouse embryos was done according to a method described previously (43). The BamHI–EcoRI fragment (0.6 kb) in the third exon of Fgf-3 and the XbaI–SmaI fragment (0.26 kb) of Sox6 were cloned into LITMUS 28 (New England Biolabs). RNA probes were synthesised from the linearised plasmid DNAs with T7 RNA polymerase (Boehringer Mannheim) using digoxigenin-11-UTP (Boehringer Mannheim). Hybridised probe was detected using BM Purple AP substrate (Boehringer Mannheim) after treatment with Anti-Digoxigenin-AP, Fab fragment (Boehringer Mannheim).
RESULTS
Identification of SOX6 as a binding protein to PS4A of the Fgf-3 promoter
To identify regulatory factors that bind to the central region of the PS4A enhancer, a yeast one-hybrid screen was used with a bait containing just the central binding site from PS4A, with a cDNA library prepared from F9 cells. Sixteen primary colonies were isolated from 1.5 × 107 transformants on His– plates, and two of these were also positive in a secondary screen using a LacZ reporter (see Materials and Methods). Sequencing revealed an insert encoding Sox6 cDNA.
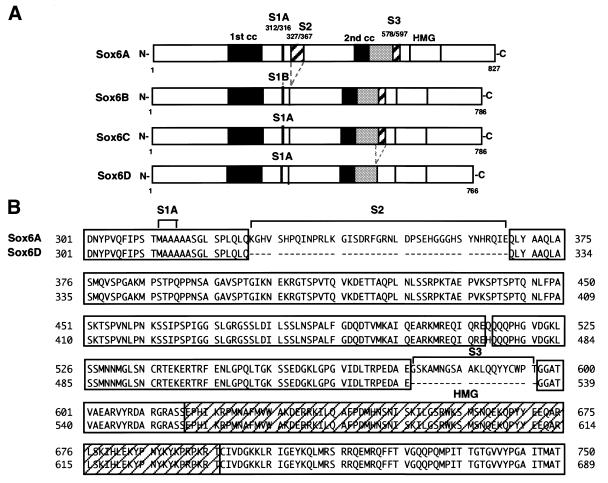
A schematic representation of the previously described SOX6 isoforms and the variant that we have designated SOX6D is shown in Figure 1A (44). SOX6D most closely resembles SOX6C, but has 20 amino acids missing (S3) between the second potential coiled-coil domain and the HMG-box (Fig. 1B). RT–PCR was used to establish the presence of Sox6D mRNA in F9 cells. Primers flanking the variable S2 region (Fig. 1A) produced two products corresponding to the size predicted for Sox6A (578 bp) and Sox6B/C/D (455 bp), respectively. Similarly, primers flanking the variable S3 region produced two products, a signature band of 159 bp for Sox6D and another of 219 bp indicative of Sox6A/B/C (data not shown). Hence, signature fragments for Sox6D and Sox6A were detected, confirming their expression in F9 cells. However, at present there is no evidence to suggest specific functional differences amongst the SOX6 isoforms.
Figure 1.
Sequence comparison of SOX6 proteins. (A) Schematic comparison of SOX6 isoforms, SOX6A, SOX6B and SOX6C (28) together with the variant SOX6D. S1, S2 and S3 designate segments that differ between the isoforms. Two putative coiled-coil domains (1st cc and 2nd cc) and the HMG-box DNA binding domain common to all isoforms are indicated. (B) Comparison of the central region of SOX6A and SOX6D showing the divergent amino acid sequences. Identical residues are shaded.
Binding of SOX6 proteins to the central element of PS4A
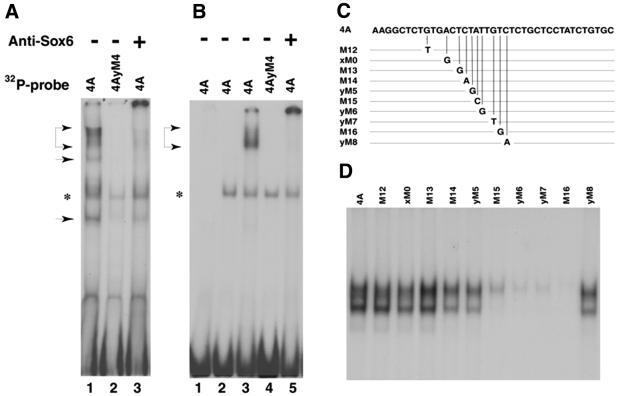
To confirm SOX6 binding to PS4A, we initially used in vitro transcribed and translated SOX6 protein in an EMSA (Fig. 2A). Several complexes with different mobilities were found associated with a PS4A probe (Fig. 2A, lane 1). The complexes were absent (Fig. 2A, lane 2) when a mutated probe (4AyM4) was used. This mutated probe has two base substitutions and was previously shown to abolish factor binding to PS4A, as well as to cause a complete loss of Fgf-3 promoter activity in cell transfection assays (16). Inclusion of SOX6 antiserum in the EMSA shifted the SOX6 specific bands to the top of the gel confirming its presence in the complexes (Fig. 2A, lane 3). These results demonstrate the binding of SOX6 protein to the central element of PS4A. A non-specific band (Fig. 2, asterisk) in the EMSA was identified by its appearance with rabbit reticulocyte lysate alone (Fig. 2B, lane 2).
Figure 2.
Binding and recognition sequence of SOX6 proteins in PS4A. (A) EMSA using in vitro transcribed and translated SOX6 proteins and 32P-labeled probes. Lanes 1 and 3, 4A; lane 2, mutated probe 4AyM4. The + indicates addition of SOX6 antiserum after complex formation. (B) EMSA using recombinant SOX6 protein alone (lane 1) or mixed with rabbit reticulocyte lysate (lanes 3–5). Rabbit reticulocyte lysate alone (lane 2). 32P-radiolabelled probes are indicated above the lanes. Positions of complexes and a non-specific band are indicated by arrowheads and an asterisk, respectively. The two smaller complexes [lane 1 in (A)] probably arise from initiation of translation at internal AUG codons. (C) Schematic depiction showing the sequences of wild-type and point mutated PS4A probes used to establish the SOX6 binding site. (D) EMSA using 32P-labelled PS4A probes shown in (C), mixed with purified GST-Sox6DB protein.
Full-length recombinant SOX6 protein binds PS4A poorly in an EMSA, unless accompanied by rabbit reticulocyte lysate (Fig. 2), indicating that efficient SOX6 binding is co-operative and requires other factors. This finding is consistent with our previous point mutation analysis showing co-operative binding to PS4A (16). Moreover, others have shown that the coiled-coil domains of full-length SOX6 permit the formation of homo- or heterodimers with other SOX proteins. These bind to pairs of HMG-box motifs, and can interfere with SOX protein binding to a single HMG-box motif, as found in PS4A (28,42). In confirmation of previous observations, recombinant full-length SOX6 was found to bind strongly to a HMG-box dimer motif (data not shown). Specificity of SOX6 binding was further demonstrated by using a truncated recombinant SOX6 protein (GST-SOX6DB) that lacks the coiled-coil domain implicated in dimer formation. Purified GST-SOX6DB was used in EMSA with a series of oligonucleotide probes, each containing a single base substitution across the central region of PS4A (Fig. 2C). Mutations 4AM15, 4AyM6, 4AyM7 and 4AM16 severely diminished binding while 4AM14 and 4AyM5 significantly reduced it, from which (CT)ATTGT could be deduced as the core recognition sequence for SOX6 (Fig. 2D). This is similar to the previously determined recognition sequence for the central site in PS4A. The complementary strand of the identified core binding sequence is close to the consensus sequence (A/T)(A/T)CAA(A/T)G reported for several other SOX proteins (20).
Repression of the Fgf-3 promoter activity by SOX6
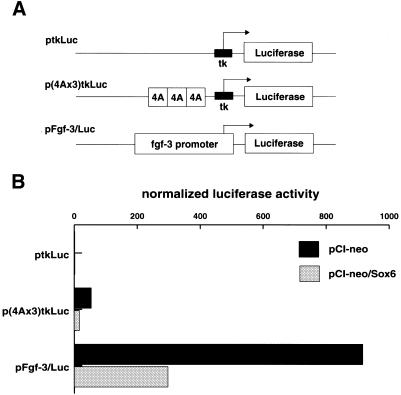
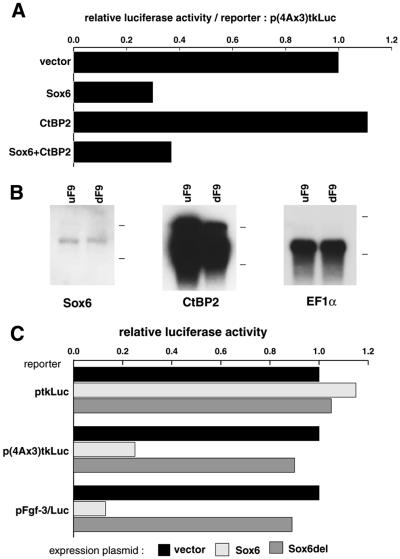
To determine whether SOX6 expression could affect transcription from the Fgf-3 promoter, differentiated F9 cells were co-transfected with one of two luciferase reporter plasmids (Fig. 3A), together in the presence or absence of a SOX6 expression plasmid (Fig. 3B). The reporter constructs contained either three copies of the PS4A enhancer fused to a minimal tk promoter [p(4Ax3)tkLuc], or a 1.7 kb fragment encompassing the Fgf-3 proximal promoter region (pFgf-3/Luc) (14). Co-transfection of a SOX6 expression plasmid had little effect on transcription from the control vector ptkLuc. However, transcription from the reporters p(4Ax3)tkLuc and pFgf-3/Luc was substantially reduced when co-expressed with SOX6, showing that SOX6 is able to repress transcription from an Fgf-3 promoter. Similar results were also obtained in co-transfection experiments using undifferentiated F9, NIH3T3 and Cos7 cells, although the effect was less marked since these cell lines only express weakly from the Fgf-3 reporter (data not shown). Although SOX6 is known to be an enhancer of transcription, these observations suggest that it can also function in gene repression, perhaps by recruiting a co-repressor (35). As the SOX binding site in PS4A is essential for Fgf-3 promoter activity, it is not possible using base mutations of the site to demonstrate a relief of the repression by SOX6.
Figure 3.
Effects of SOX6 expression on the Fgf-3 proximal promoter and an artificial promoter containing PS4A sites. (A) Schematic representation of the control and two reporter plasmids (16) used to test the effect of SOX6 on the Fgf-3 promoter. (B) Differentiated F9 cells co-transfected with a reporter plasmid (3 µg) as indicated, and either control vector (6 µg) pCI-neo or SOX6 expression vector (6 µg) pCI-neo/Sox6. A β-galactosidase reporter (1 µg) pRL-CMV DNA was used as an internal control to normalize transfection efficiency.
Analysis of F9 cells using degenerate RT–PCR showed the presence of Sox6, but also several other Sox transcripts (Sox2, Sox7, Sox13, Sox17) consistent with the pluripotential nature of F9 cells, and the role of SOX proteins in lineage determination (data not shown). Therefore, to provide evidence for a potential interaction between these proteins in vivo, we examined sites of Fgf-3 and Sox6 expression.
Co-expression of Sox-6 and Fgf-3 in the embryonic inner ear
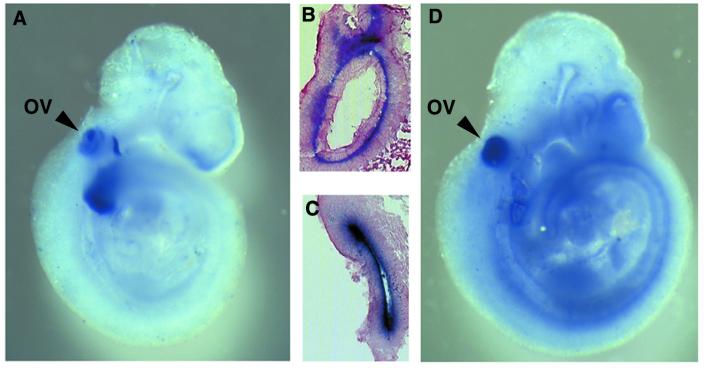
Previous studies have reported that Fgf-3 and Sox6 are both expressed in the otic vesicle during early ear development (7,9,45,46). Using embryos from embryonic day 9.5 (E9.5), we confirmed the otic vesicle as a site of expression for both these genes (Fig. 4). To determine which cells were expressing Fgf-3 and Sox6 RNA, the stained embryos were sectioned for histological examination. Fgf-3 RNA was found to be localised to the presumptive sensory epithelia, which forms the inner ear, as well as to the delaminating neuronal precursors that migrate to form the cochleovestibular ganglion (Fig. 4A and B). The otic vesicle at E9.5 was the only site of discernible Sox6 expression (Fig. 4D) (46). In contrast to the sites of Fgf-3 expression, Sox6 was associated with the inner epithelial layer of the otic vesicle. Hence, Sox6 and Fgf-3 were expressed in a mutually exclusive pattern, consistent with the idea that SOX6 could act to repress Fgf-3 transcription within a sub-region of the otic vesicle epithelium.
Figure 4.
Whole mount in situ hybdridisation showing Fgf-3 and Sox6 expression in mouse embryos. Mouse embryos (E9.5) were hybridised with either antisense Fgf-3 (A and B) or antisense Sox6 (C and D) probes labelled with digoxigenin and detected with anti-digoxigenin alkaline phosphatase conjugated antiserum as described in Materials and Methods. The major common expression site is in the otic vesicle (OV). Sections through the otic vesticles seen in (A) and (D) are shown in (B) and (C), respectively, and demonstrate the cellular location of the labelled cells.
Identification of CtBP2 as a SOX6-binding protein
Previously, SOX6 was shown to act as an activator, rather than a repressor of transcription, suggesting that its regulatory activity may be dependent on its interaction with either a co-activator or co-repressor (28). Therefore, we used a yeast two-hybrid screen to search for co-repressor proteins that could bind SOX6. As bait, a polypeptide of 523 amino acids from the N-terminal part of SOX6D was fused to a GAL4 DNA binding domain that served to replace the HMG-box DNA binding region. This vector was used to screen the same differentiated F9 cDNA-GAL library described above. Two positive clones were obtained after screening 4.3 × 106 transformants. Both clones were found to encode mouse CtBP2, a recently described co-repressor protein (39). The two clones were slightly shorter than the published sequence, and were extended using 5′ RACE to obtain a full-length coding sequence.
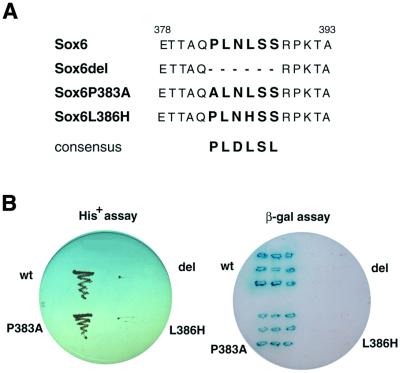
The amino acid sequence PLDLSL has been identified in some transcription factors to be a consensus motif for binding CtBP2 (37–41). A similar sequence, PLNLSS, is present in SOX6D (amino acids 383–388). Therefore, mutations were introduced into this motif to assess their effect on SOX6 binding to CtBP2 in a yeast two-hybrid system (Fig. 5). Deletion of the complete motif PLNLSS abolished the interaction. This was shown by the lack of growth on His– plates and the absence of β-galactosidase induction. Substitution of proline by alanine (P383A) had little effect, whereas substitution of the leucine by histidine (L386H) abolished the interaction. These results indicate that the PLNLSS sequence in SOX6 is required for interaction with CtBP2 in yeast cells.
Figure 5.
Interaction of SOX6 with CtBP2 in yeast depends on the PLNLSS sequence motif in SOX6. (A) Amino acid sequences of the putative recognition site for CtBP2 in wild-type SOX6 and mutated SOX6. (B) HF7c yeast cells were co-transformed with plasmids expressing wild-type or a mutant GAL4DBD-Sox6 and GAL4AD-CtBP2. For each transformation single colonies were tested for growth on His– plates (left) and for β-galactosidase activity (right), respectively.
To confirm that the binding between SOX6 and CtBP2 also occurs in mammalian cells, these proteins were co-expressed in Cos7 cells. Protein extracts derived from these cells were subjected to immunoprecipitation followed by immunoblotting (Fig. 6). FLAG-tagged CtBP2 proteins were immunoprecipitated with an anti-FLAG serum and then the His-tagged SOX6 was revealed by immunoblotting with an anti-HIS serum (Fig. 6A, lane 1). Reciprocally, SOX6 proteins were immunoprecipitated with an anti-HIS serum and CtBP2 revealed by immunoblotting with the anti-FLAG serum (Fig. 6B, lane 1). These experiments demonstrate that SOX6 and CtBP2 can form a complex in Cos7 cells. As anticipated, neither the SOX6 PLNLSS-deletion mutant nor SOX6 with the L386H substitution formed significant complexes with CtBP2 (Fig. 6A and B, lanes 2 and 4, respectively). Moreover, in this analysis the P383A mutant also failed to form a stable complex with CtBP2 (Fig. 6A and B, lanes 3). These combined results demonstrate that in mammalian cells, the PLNLSS sequence is necessary for complex formation between SOX6 and CtBP2.
Figure 6.
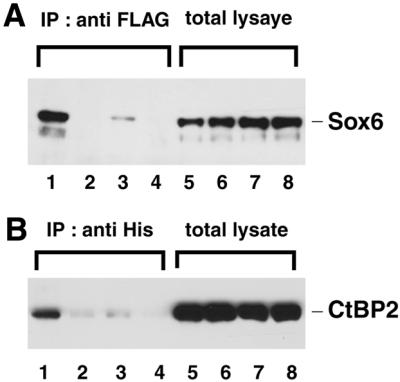
Interaction of SOX6 with CtBP2 in Cos7 cells depends on the PLNLSS sequence in SOX6. Histidine-tagged SOX6 and FLAG-tagged CtBP2 were co-expressed in Cos7 cells. Lanes 1 and 5, SOX6 wild-type; lanes 2 and 6, SOX6del; lanes 3 and 7, SOX6P383A; lanes 4 and 8, SOX6L386H (see Fig. 5 for mutated amino acid sequences). (A) Total cell lysates (lanes 5–8) or cell lysates immunoprecipitated with anti-FLAG serum (lanes 1–4) were analysed by immunoblotting with ant-His serum. (B) As described in (A) except cell lysates were immunoprecipitated with anti-His serum and immunoblotted with anti-FLAG serum.
Repression by SOX6 by recruitment of CtBP2
As SOX6 is able to repress transcription from the native Fgf-3 promoter or an artificial promoter containing three PS4A elements (Fig. 3), we examined the effect of co-expressing CtBP2 on SOX6 activity. In differentiated F9 cells, the introduction of CtBP2 had little additional effect on Fgf-3 promoter activity above that of SOX6 alone (Fig. 7A). However, subsequent northern blot analysis indicated that endogenous CtBP2 is already expressed at very high levels compared with Sox6 in these cells (Fig. 7B). Hence, the normal level of CtBP2 is most likely in excess of that required for maximal repression by SOX6. Functional evidence for the interaction between SOX6 and CtPB2 in the repression of the Fgf-3 promoter was evident from the behaviour of the Sox6del mutant lacking the PLNLSS motif in reporter assays (Fig. 7C). The mutated SOX6 protein that was unable to bind CtBP2 was also unable to repress transcription. An EMSA using Sox6del showed that the loss of repression was not due to loss of binding activity for PS4A (data not shown).
Figure 7.
Effect of over-expressing CtBP2 on the repression of the Fgf-3 promoter by SOX6 in differentiated F9 cells. (A) Differentiated F9 cells were co-transfected with the p(4Ax3)tkLuc reporter together with effector plasmids for the expression of SOX6, CtBP2 or SOX6 and CtBP2. Relative luciferase activities were normalised to the value obtained with the vector alone. (B) Endogenous expression of Sox6 and CtBP2 mRNAs in F9 cells. Poly(A)+ RNA (1 µg) from undifferentiated F9 (uF9) and differentiated F9 (dF9) cells was analysed by northern blotting using 32P-labelled Sox6, CtBP2 and EF1α as a control for RNA loading. Positions of 28S and 18S ribosomal RNAs are indicated to the right of each panel. (C) SOX6 deletion mutant lacking PLNLSS does not repress transcription from the Fgf-3 promoter. Differentiated F9 cells were co-transfected with one of the reporter plasmids and an effector plasmid for expression of SOX6 or SOX6del. Relative luciferase activities are shown. Amounts of DNAs for transfection are the same as those in Figure 3.
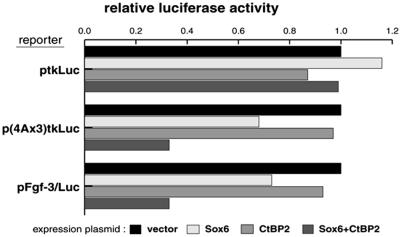
The high endogenous level of CtBP2 in F9 cells makes them unsuitable to demonstrate a co-operative effect between co-expressed Sox6 and CtBP2. Therefore we used NIH3T3 cells which were found to express very low levels of endogenous CtBP2. In this cell line the co-operative effect of co-expressing SOX6 and CtBP2 could be clearly demonstrated (Fig. 8). Thus the reporter assays using both dF9 and NIH3T3 cells clearly demonstrate that SOX6 must recruit CtBP2 to repress the activity of the Fgf-3 promoter through the PS4A enhancer.
Figure 8.
Effects of CtBP2 on the repression by SOX6 in NIH3T3 cells. NIH3T3 cells were co-transfected with each of the reporter plasmids together with expression plasmids for the expression of SOX6 and/or CtBP2 as indicated. Relative luciferase activities are normalized to the vector alone. Amounts of DNAs for transfection are the same as those used in Figure 3.
DISCUSSION
In a previous study we identified PS4A as an essential regulatory element proximally located in the Fgf-3 promoter (16). Here we show that SOX6 can bind to the central region of this essential regulatory element at the sequence motif ACAAT, a site that is similar to the published consensus for other SOX protein binding sites (20). Binding of SOX6 to the Fgf-3 promoter, or a minimal artificial promoter containing three PS4A regulatory elements, results in a strong repression of transcription. Using a yeast one-hybrid screen, SOX6 was identified as an expressed protein in F9 cells. However, subsequent screening of F9 cells using RT–PCR revealed several other SOX proteins which should also bind to the SOX consensus site. Therefore, to obtain evidence that SOX6 might act as a repressor of Fgf-3 transcription in vivo, we used in situ hybridisation analysis on early mouse embryos to look for their sites of expression. The otic vesicle was the one site of Sox6 expression found in E9.5 embryos, and this was also a site for Fgf-3 expression. The expression pattern of Sox6 and Fgf-3 is complementary and mutually exclusive, consistent with SOX6 acting as a repressor of the Fgf-3 promoter. Interestingly SOX13, another group D SOX protein, is also expressed in the inner ear of mouse embryos at E13.5 but was not detected at E9.5 (47; A.Murakami, unpublished results). The SOX13 sequence also encodes potential CtBP2 binding sites and will repress the Fgf-3 promoter if over-expressed in F9 cells (A.Murakami, unpublished results). Hence it is possible that SOX13 may play a role in regulating Fgf-3 expression, but at a later stage of development. If Sox6 null mice become available then this proposal might be readily tested. In this context, mice deficient for Fgf-3 have been found to have an inner ear abnormality (10), suggesting that likewise, a similar abnormality might occur in mice deficient for Sox6.
The collagen type II promoter (Col2a1) which is expressed in chondrocytes, is a recently identified target gene for SOX6 (28). But in this context, SOX6 functions as an activator of transcription rather than a repressor. It was shown to co-operate with SOX5 and SOX9 in the activation of Col2a1 through binding at four HMG-like sites in the minimal enhancer region (28). Based on the sequence of the HMG-box DNA-binding domain, SOX genes have been divided into several groups. For example SOX5, SOX6 and SOX13 belong to group D while SOX9 belongs to group E (19). Hetero- and homodimerizaton between group D SOX proteins appears to be mediated by the coiled-coil domain located at their N-termini (28). In contrast to Col2a1 that can accommodate SOX dimers, the Fgf-3 promoter contains only a single consensus HMG site corresponding to the central PS4A element. From this, and an earlier study, this site shows co-operative protein binding (16). This must therefore involve SOX6 and a non-SOX protein.
Although the SOX protein family has been principally associated with the activation of transcription (22–26), there are now examples of their being involved in the repression of transcription. For instance, Oct-4 mediated activation of the OPN gene can be repressed by SOX2 (48). In contrast to OPN promoter, the δ1-crystallin promoter is activated by SOX2 as well as SOX1, and repressed by SOX14 and SOX21 (49). Thus, depending on the promoter context, SOX2, like SOX6, can act as both a repressor and activator of transcription.
To explain the repression of Fgf-3 transcription by SOX6, we sought other proteins that could interact with this transcription factor, and identified mouse CtBP2. Binding was shown to require the amino acid motif PLNLSS which is similar to CtBP2 binding regions identified in other transcription factors (4,41). Furthermore, co-expression studies showed that CtBP2 co-operates with SOX6 to repress transcription of Fgf-3, and this repression was dependent upon the integrity of the PLNLSS motif in SOX6. CtBP was first identified as a cellular protein (human CtBP) that bound the C-terminal region of the adenovirus E1A protein (36) and attenuated its ability to activate transcription (50). More recently, two mouse homologues, CtBP1 and CtBP2, were found to associate with, and repress transcriptional activation through the basic Krüppel-like factor (39). Both CtBP1 and CtBP2 enhance transcriptional repression by δEF1 and are widely expressed in developing embryos with different tissue preferences (41).
Thus, depending on the context, SOX6 can act as an activator and repressor of transcription. This suggests a general mechanism where SOX family members recruit either co-activators or co-repressors to modulate transcription. Furthermore, a given promoter, such as δ1-crystallin may be transcriptionally activated by one set of SOX proteins and yet repressed by other members of the same protein family. We have evidence that a similar situation pertains to the Fgf-3 promoter. Preliminary data suggests that in F9 cells, SOX7 mediates activation of Fgf-3 transcription (A.Murakami, unpublished results), while from this study, in the otic vesicle SOX6 is a good candidate for its repression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Véronique Lefebvre and Shinya Yamashita for generously providing antiserum against SOX6, and Drs Malcolm Parker and Vera Fantl for critical reading of the manuscript. This work was supported by a grant from the Human Frontier Science Program to A.M. and C.D.
References
- 1.Ornitz D.M. (2000) FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays, 22, 108–112. [DOI] [PubMed] [Google Scholar]
- 2.McKeehan W.L., Wang,F. and Kan,M. (1998) The heparan-sulfate fibroblast growth-factor family – diversity of structure and function. Prog. Nucleic Acids Res. Mol. Biol., 59, 135–176. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi T.P. and Rossant,J. (1995) Fibroblast growth-factors in mammalian development. Curr. Opin. Genet. Dev., 5, 485–491. [DOI] [PubMed] [Google Scholar]
- 4.Martin G.R. (1998) The roles of FGFs in the early development of vertebrate limbs. Genes Dev., 12, 1571–1586. [DOI] [PubMed] [Google Scholar]
- 5.Goldfarb M. (1996) Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev., 7, 311–325. [DOI] [PubMed] [Google Scholar]
- 6.Mahmood R., Kiefer,P., Guthrie,S., Dickson,C. and Mason,I. (1995) Multiple roles for FGF-3 during cranial neural development in the chicken. Development, 121, 1399–1410. [DOI] [PubMed] [Google Scholar]
- 7.McKay I.J., Lewis,J. and Lumsden,A. (1996) The role of FGF-3 in early inner ear development: An analysis in normal and kreisler mutant mice. Dev. Biol., 174, 370–378. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson D.G., Peters,G., Dickson,C. and McMahon,A.P. (1988) Expression of FGF-related proto-oncogene int-2 during gastrulation and neurulation in the mouse. EMBO J., 7, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson D.G., Bhatt,S. and McMahon,A.P. (1989) Expression pattern of FGF-related proto-oncogene int-2 suggests multiple roles in fetal development. Development, 105, 131–136. [DOI] [PubMed] [Google Scholar]
- 10.Mansour S.L., Goddard,J.M. and Capecchi,M.R. (1993) Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development, 117, 13–28. [DOI] [PubMed] [Google Scholar]
- 11.Smith R., Peters,G. and Dickson,C. (1988) Multiple RNAs expressed from the int-2 gene in mouse embryonal carcinoma cell lines encode a protein with homology to fibroblast growth factors. EMBO J., 7, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobovits A., Shackleford,G., Varmus,H. and Martin,G. (1986) Two proto-oncogenes implicated in mammary carcinogenesis, int-1 and int-2, are independently regulated during mouse development. Proc. Natl Acad. Sci. USA, 83, 7806–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strickland S. and Mandhavi,V. (1978) The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell, 15, 393–403. [DOI] [PubMed] [Google Scholar]
- 14.Murakami A., Grinberg,D., Thurlow,J. and Dickson,C. (1993) Identification of positive and negative regulatory elements involved in the retinoic acid/cAMP induction of Fgf-3 transcription in F9 cells. Nucleic Acids Res., 21, 5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinberg D., Thurlow,J., Watson,R., Smith,R., Peters,G. and Dickson,C. (1991) Transcriptional regulation of the int-2 gene in embryonal carcinoma cells. Cell Growth Differ., 2, 137–143. [PubMed] [Google Scholar]
- 16.Murakami A., Thurlow,J. and Dickson,C. (1999) Retinoic acid-regulated expression of fibroblast growth factor 3 requires the interaction between a novel transcription factor and GATA-4. J. Biol. Chem., 274, 17242–17248. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair A.H., Berta,P., Palmer,M.S., Hawkins,J.R., Griffiths,B.L., Smith,M.J., Foster,J.W., Frischauf,A.-M., Lovell-Badge,R. and Goodfellow,P.N. (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346, 240–244. [DOI] [PubMed] [Google Scholar]
- 18.Gubbay J., Collignon,J., Koopman,P., Capel,B., Economou,A., Munsterberg,A., Vivian,N., Goodfellow,P. and Lovell-Badge,R. (1990) A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature, 346, 245–250. [DOI] [PubMed] [Google Scholar]
- 19.Pevny L.H. and Lovell-Badge,R. (1997) Sox genes find their feet. Curr. Opin. Genet. Dev., 7, 338–344. [DOI] [PubMed] [Google Scholar]
- 20.Wegner M. (1999) From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res., 27, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowles J., Schepers,G. and Koopman,P. (2000) Phylogeny of the SOX family of developmental transcription factors based on sequence and structure indicators. Dev. Biol., 227, 239–255. [DOI] [PubMed] [Google Scholar]
- 22.Hosking B.M., Muscat,G.E.O., Koopman,P.A., Dowhan,D.H. and Dunn,T.L. (1995) Trans-activation and DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res., 23, 2626–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamachi Y., Cheah,K.S.E. and Kondoh,H. (1999) Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol. Cell. Biol., 19, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanai Y., Kanai-Azuma,M., Noce,T., Saido,T.C., Shiroishi,T., Hayashi,Y. and Yazaki,K. (1996) Identification of two Sox17 messenger RNA isoforms, with and without the high mobility box region, and their differential expression in mouse spermatogenesis. J. Cell Biol., 133, 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudbeck P., Schmitz,M.L., Baeuerle,P.A. and Scherer,G. (1996) Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nature Genet., 13, 230–232. [DOI] [PubMed] [Google Scholar]
- 26.van de Wetering M., Oosterwegel,M., Norren,K. v. and Clevers,H. (1993) Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J., 12, 3847–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamachi Y., Sockanathan,S., Liu,Q., Breitman,M., Lovell-Badge,R. and Kondoh,H. (1995) Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J., 14, 3510–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre V., Li,P. and Crombrugghe,B.D. (1998) A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J., 17, 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan H., Corbi,N., Bascilico,C. and Dailey,L. (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev., 9, 2635–2645. [DOI] [PubMed] [Google Scholar]
- 30.Mizuseki K., Kishi,M., Shiota,K., Nakanishi,S. and Sasai,Y. (1998) SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron, 21, 77–85. [DOI] [PubMed] [Google Scholar]
- 31.Pevny L.H., Sockanathan,S., Placzek,M. and Lovell-Badge,R. (1998) A role for SOX1 in neural determination. Development, 125, 1967–1978. [DOI] [PubMed] [Google Scholar]
- 32.Uwanogho D., Rex,M., Catwright,E.J., Pearl,G., Healy,C., Scotting,P.J. and Sharpe,P.T. (1995) Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neural development. Mech. Dev., 49, 23–36. [DOI] [PubMed] [Google Scholar]
- 33.Nishiguchi S., Wood,H., Kondoh,H., Lovell-Badge,R. and Episkopou,V. (1998) Sox1 directly regulates the γ-crystallin genes and is essential for lens development in mice. Genes Dev., 12, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zorn A.M., Barish,G.D., Williams,B.O., Lavender,P., Klymkowsky,M.W. and Varmus,H.E. (1999) Regulation of Wnt signaling by Sox proteins: XSox17α/β and XSox3 physically interact with β-catenin. Mol. Cell, 4, 487–498. [DOI] [PubMed] [Google Scholar]
- 35.Torchia J., Glass,C. and Rosenfeld,M.G. (1999) Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol., 10, 373–383. [DOI] [PubMed] [Google Scholar]
- 36.Schaeper U., Boyd,J.M., Verma,S., Uhlmann,E., Subramanian,T. and Chinnadurai,G. (1995) Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl Acad. Sci. USA, 92, 10467–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryogenic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nibu Y., Zhang,H. and Levine,M. (1998) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science, 280, 101–103. [DOI] [PubMed] [Google Scholar]
- 39.Turner J. and Crossley,M. (1998) Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J., 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sewalt R.G.A.B., Gunster,M.J., van der Vlag,J., Satijn,D.P.E. and Otte,A.P. (1999) C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate polycomb proteins. Mol. Cell. Biol., 19, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furusawa T., Moribe,H., Kondoh,H. and Higashi,Y. (1999) Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol. Cell. Biol., 19, 8581–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamatsu N., Kanda,H., Tsuchita,I., Yamada,S., Ito,M., Kabeno,S., Shiba,T. and Yamashita,S. (1995) A gene that is related to Sry and is expressed in the testes encodes a leucine zipper-containing protein. Mol. Cell. Biol., 15, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson D.G. and Nieto,M.A. (1993) Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. In Wassarman,P.M. and DePamphilis,M.L. (eds), Guide to Techniques in Mouse Development. Academic Press, Inc., San Diego, CA, Vol. 225, pp. 361–373. [DOI] [PubMed]
- 44.Cohen-Barak O., Hagiwara,N., Arlt,M., Horton,J. and Brilliant,M. (2001) Cloning, characterization and chromosome mapping of the human SOX6 gene. Gene, 265, 157–164. [DOI] [PubMed] [Google Scholar]
- 45.Pirvola U., Spencer-Dene,B., Liang,X.Q., Kettunen,I., Thesleff,I., Fritzsch,B., Dickson,C. and Ylikoski,J. (2000) FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J. Neurosci., 20, 6125–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connor F., Wright,E., Denny,P., Koopman,P. and Ashworth,A. (1995) The Sry-related HMG-containing gene Sox6 is expressed in the adult testis and developing nervous system of the mouse. Nucleic Acids Res., 23, 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roose J., Korver,W., Oving,E., Wilson,A., Wagenaar,G., Markman,M., Lamers,W. and Clevers,H. (1998) High expression of the HMG box factor sox-13 in arterial walls during embryonic development. Nucleic Acids Res., 26, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Botquin V., Hess,H., Fuhrmann,G., Anastassiadis,C., Gross,M.K., Vriend,G. and Scholer,H.R. (1998) New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox2. Genes Dev., 12, 2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchikawa M., Kamachi,Y. and Kondoh,H. (1999) Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev., 84, 103–120. [DOI] [PubMed] [Google Scholar]
- 50.Sollerbrant K., Chinnadurai,G. and Svensson,C. (1996) The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res., 24, 2578–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]