Supplemental Digital Content is available in the text
Keywords: asthma, meta-analysis, polymorphism, TLR2, TLR4
Abstract
Background:
Several epidemiological studies have focused on the association between polymorphisms in toll-like receptors (TLRs) and asthma. However, the results remained inconclusive.
Methods:
We systematically reviewed the database of PubMed, EMBASE, Web of Science, CNKI, and Google scholar for all related articles on TLR polymorphisms and asthma. We used the software STATA 12.0 to conduct the meta-analysis. The heterogeneity and publication bias were examined, respectively.
Results:
Eighteen studies consisting of 3538 asthma cases and 4090 controls were selected into the meta-analysis. The pooled odds ratios (ORs) show that rs3804099 was associated with asthma in dominant model (OR = 1.51, 95% CI = 1.17–1.96, P = .002), and rs4986791 was associated with asthma in additive model (OR = 0.81, 95% CI = 0.64–1.02, P = .07) and dominant model (OR = 0.76, 95% CI = 0.60–0.97, P = .025).
Conclusion:
The combined results show that rs3804099 in TLR2 and rs4986791 in TLR4 were significantly associated with asthma risk. Polymorphisms in TLRs play important roles in asthma.
1. Introduction
Asthma is a chronic inflammatory disorder of the airways, which is characterized by reversible airflow obstruction and airway hyper-responsiveness.[1] It has been one of the most common disease in the world.[2] Although the exact mechanisms in asthma have not been completely elucidated,[3] it is generally accepted that both genetic and environmental factors play important roles in this disease.[4] Currently, more than 200 genetic variations have been reported to be associated with asthma.[5]
Toll-like receptors (TLRs) are a class of proteins that belong to the family of transmembrane receptors. They are counted among the key molecules in pathogen recognition and activation of innate immunity.[6] The TLRs include 11 members in humans (TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, and TLR11).[6] TLR2 and TLR4 are the most important members of them. Several variants in them have been proved associated with asthma.[7–11] However, the results remained conflicting. Among the reported studies, most of them found that these polymorphisms were associated with asthma, while others reported contrary opinions. Meantime, most of these studies were conducted with small numbers of subjects. Only 1 meta-analysis was conducted to examine the relationship between TLR gene polymorphisms and asthma 2 years ago.[12] During the past years, several studies about TLR gene polymorphisms and asthma have been conducted.[13,14] So, we conducted the update meta-analysis expecting to give a more determinately conclusion.
2. Methods
2.1. Search strategy
We systematically searched the database of Pubmed, EMBASE, Web of Science (ISI), China National Knowledge Infrastructure (CNKI), and Google scholar for all related articles on TLR polymorphisms and asthma. We used the following keywords: “TLR2” or “TLR4,” “polymorphism,” and “asthma.” Both English and Chinese language articles were included in the analysis. The included studies were published from January 1, 2000 to September 30, 2016. In addition, the references and citations of the originally retrieved articles, which were not captured by our database searches, were identified through manual searching.
The study was approved by the institutional ethics committees of Tongji Hospital.
2.2. Inclusion criteria
Although several single nucleotide polymorphisms (SNPs) in TLR2 and TLR4 have been previously studied, only those most widely studied variants (reported in more than 3 studies) were analyzed in this analysis. Finally, 2 SNPs (rs5743708 and rs3804099) in TLR2 and 2 SNPs (rs4986790 and rs4986791) in TLR4 were included in the analysis. The title and abstract of the articles identified through electronic literature search were scanned. If the article could not be verified, the full text was further examined. The articles that met the following inclusion criteria were included in the analysis: case–control study; aims at the association between TLR2 (rsrs5743708, rs3804099) or TLR4 (rs4986790, rs4986791) and asthma; and sufficient data to perform the meta-analysis. The case-only studies, reviews, case reports and studies without sufficient information were excluded. If the study subjects were reported in more than 1 publication, we selected the study with the largest sample size.
2.3. Data extraction
Two reviewers (JLZ and HHS) extracted the data separately. Characters extracted from the studies including: journal, first author, year of publication, ethnicity, genotyping methods, asthma diagnosis, source of controls, average age at baseline, male percentage, distribution of genotypes for each polymorphism among cases and controls, and the odds ratio (OR) with their 95% confidence intervals (CIs). Four polymorphisms were extracted respectively from selected studies. The results of items were compared and all of them reached consistencies in the end.
2.4. Statistical analysis
We examined the Hardy–Weinberg equilibrium (HWE) for each polymorphism by using the Chi-square test. The association between polymorphisms and asthma was estimated by means of OR and corresponding 95% CIs comparing cases to controls. Additive model (M vs N), dominant model (MM vs MN + NN), and recessive model (MM + MN vs NN) were used to estimate statistical significance between TLR polymorphisms and asthma risk, respectively. The heterogeneity represents the total percentage of variation across the selected studies. We used the Chi-square-based Q statistic and I2 statistic to evaluate the heterogeneity. The percentages of I2 around 25%, 50%, and 75% indicate low, medium, and high heterogeneity, respectively.[15]P < .10 was representative of significant heterogeneity.[16] When there was no significant heterogeneity (P > .10 and I2 less than 50%), the fixed-effects model was used to calculate the pooled ORs; otherwise, the random-effects model was used.[17] In addition, we performed sensitivity analysis to investigate the influence of individual study on the overall meta-analysis. Finally, we evaluated publication bias of selected analysis by funnel plot and Egger tests. We performed the analyses by using the STATA 12.0 software (StataCorp, College Station, TX).[18] A P < .05 was considered statistically significant and all statistical testing were 2-sided.
3. Results
3.1. Search results
On the initial search, 342 papers were identified by the keywords mentioned in the search strategy. A total of 258 papers were excluded for the reason that they were not aiming at the association of TLR2 or TLR4 and asthma. When we reviewed abstracts of the remained papers, 64 articles were excluded. For the reason that these studies were case reports, gave insufficient data for subsequent analysis, or subjects of these studies had other diseases. Then we reviewed the full texts and excluded 2 articles. The first excluded from our meta-analysis was reported by Liang et al.[19] Neither the Asp299Gly nor Thr399Ile polymorphism was found by DNA sequencing in the study. So, we excluded this study in the meta-analysis. Another study conducted by Saçkesen et al was case-only study and was excluded as well.[20] Finally, 18 literatures were selected into the meta-analysis.[10,13,14,21–35]Figure 1 outlines our study selection process. The characteristics of studies included in the analysis were listed in Table 1. The raw data we got from the 18 studies consist of 3538 asthma cases and 4090 controls. The general characteristics of each study, genotype frequencies, and HWE examination results were presented in Table 2.
Figure 1.

Flow chart for the process of selecting eligible publications.
Table 1.
Study characteristics in the analysis of TLR polymorphisms and asthma.
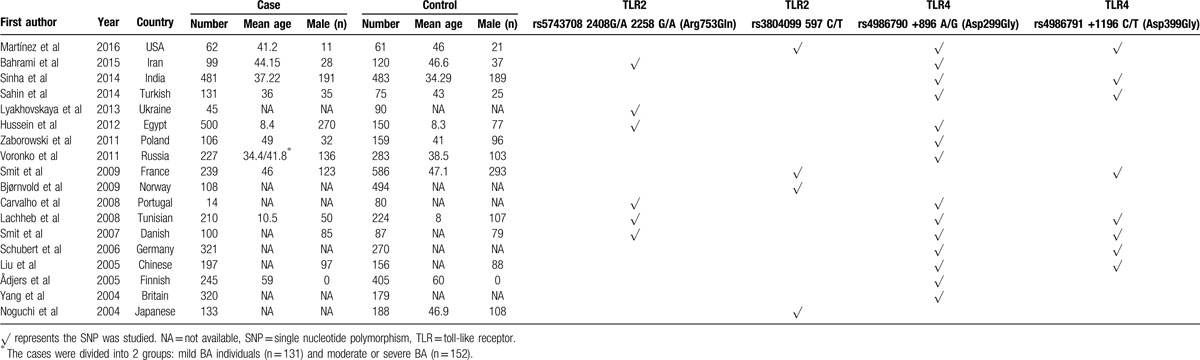
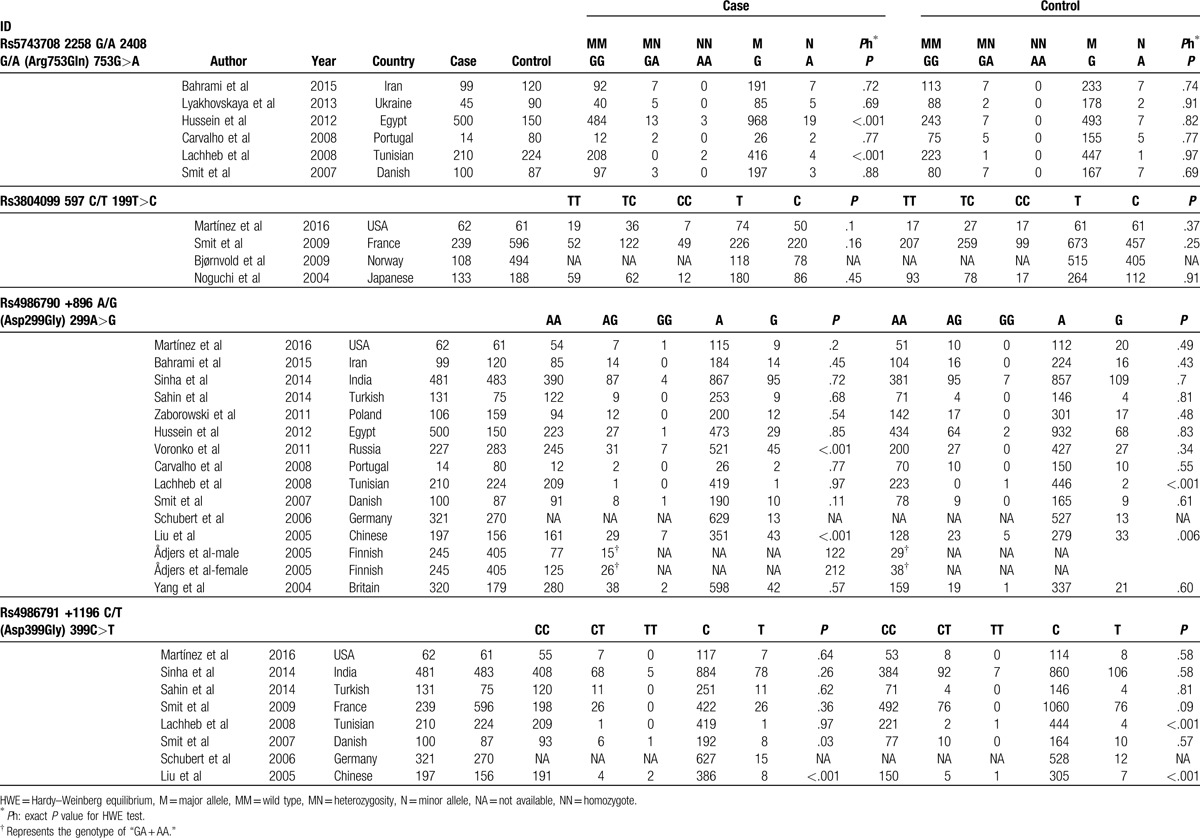
Table 2.
Genotypes of the polymorphisms in selected studies.
3.2. Association of the polymorphisms rs5743708 and rs3804099 in TLR2 with asthma
The polymorphism rs5743708 is also known as 2408G/A, which causes a nonsynonymous amino acid change (Arg753Gln) in this protein. Six studies analyzed the polymorphism rs5743708 and were selected in our study. They consisted of 968 asthma cases and 751 healthy controls. When we pooled all eligible publications into the meta-analysis, no significant association was found between rs5743708 and asthma in additive model (OR = 1.40, 95% CI = 0.85–2.32, P = .186), as shown in Fig. 2. Among the 6 studies, the genotypes of 2 studies conducted by Hussein et al[23] and Lachheb et al[28] were not in HWE. Therefore, we excluded these 2 studies and the pooled OR was consistent in direction (OR = 1.23, 95% CI = 0.67–2.26, P = .496). Under recessive model and dominant model, no significant association was found between rs5743708 and asthma as well, as shown in Table 3.
Figure 2.
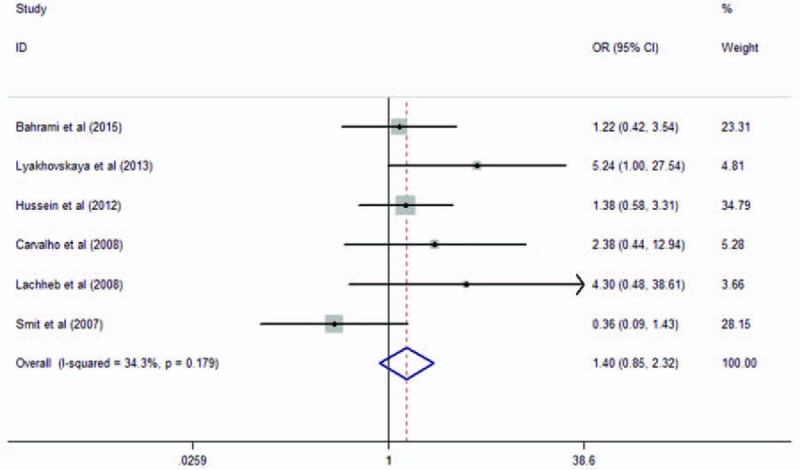
Meta-analysis of the association between TLR2 rs5743708 and asthma in additive model. TLR = toll-like receptor.
Table 3.
Pooled measures on the relation of TLR polymorphisms and asthma.
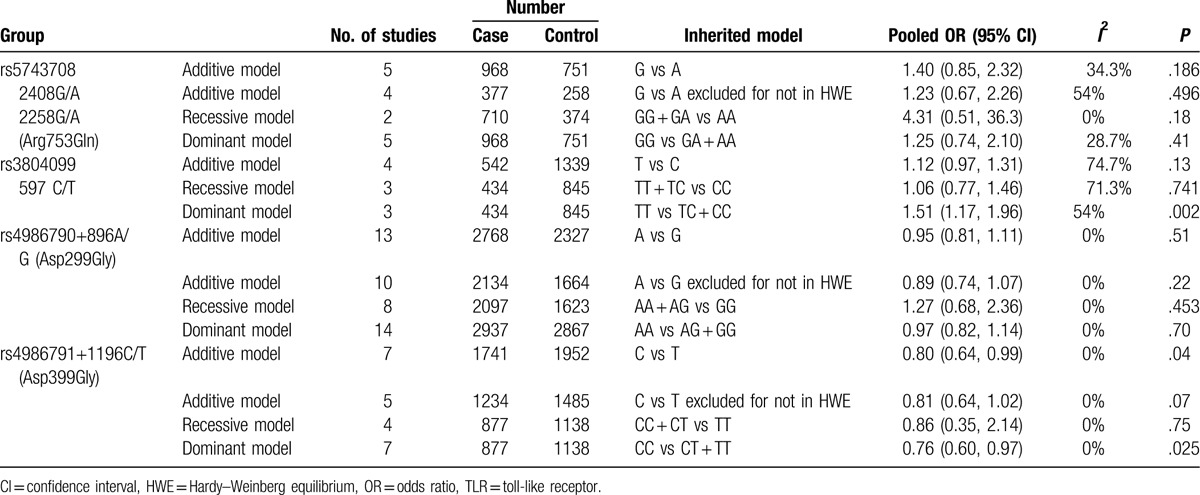
Four studies analyzed the polymorphism rs3804099. The study conducted by Bjørnvold et al[26] only reported the frequency of each alleles of rs3804099, but did not report the frequency of each genotype. When we pooled all eligible publications into the meta-analysis, significant association was found between rs3804099 and asthma in dominant model (OR = 1.51, 95% CI = 1.17–1.96, P = .002), as shown in Fig. 3. No significant association was found in additive model (OR = 1.12, 95% CI = 0.97–1.31, P = .13) and recessive model (OR = 1.06, 95% CI = 0.77–1.46, P = .74).
Figure 3.
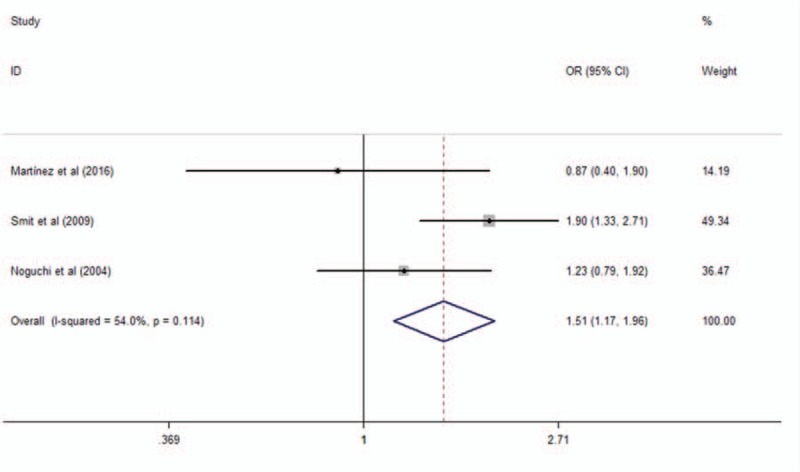
Meta-analysis of the association between TLR2 rs3804099 in TLR2 and asthma in dominant model. TLR = toll-like receptor.
3.3. Association of the TLR4 rs4986790 and rs4986791 polymorphisms with asthma
A total of 14 studies encompassing 3011 cases and 2728 controls investigated the association of the polymorphism rs4986790 with asthma risk. The study conducted by Ådjers et al[32] contains 92 male cases and 151 male controls, as well as 151 female cases and 250 female controls. Therefore, we extracted these data separately. When we pooled all data extracted from the eligible publications, no significant association was found between rs5743708 and asthma (additive model: OR = 0.95, 95% CI = 0.81–1.11, P = .51, as shown in Fig. 4; dominant model: OR = 0.89, 95% CI = 0.74–1.07, P = .22; and recessive model: OR = 1.27, 95% CI = 0.68–2.36, P = .453), as shown in Table 3. Low heterogeneity was observed among these studies (I2 = 0%).
Figure 4.
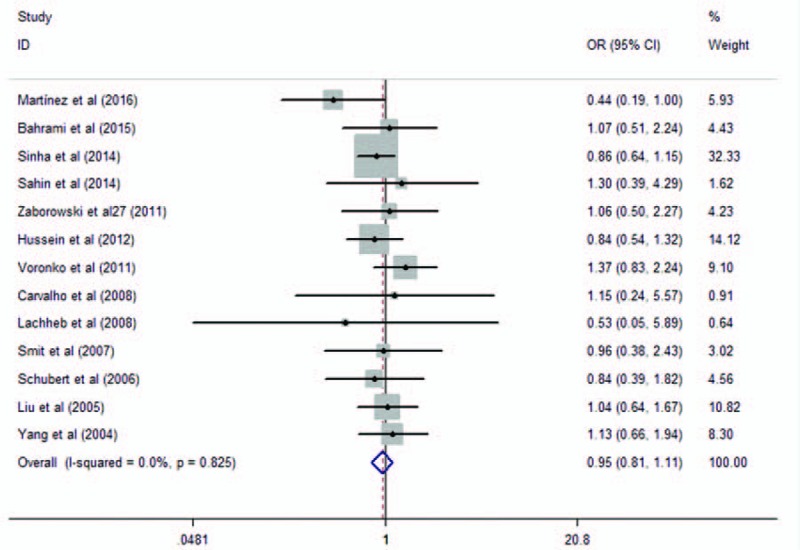
Meta-analysis of the association between TLR4 rs4986790 in TLR4 and asthma in additive model. TLR = toll-like receptor.
Eight studies analyzed the association of the variant rs4986791 with asthma risk. They contained 1741 asthma cases and 1952 healthy controls. When we pooled all selected studies together, significant association was found between rs4986791 and asthma in additive model (OR = 0.80, 95% CI = 0.64–0.99, P = .04). The genotypes of the study conducted by Liu et al[31] were not in HWE. When we excluded the study not in HWE, a barely significant association was found between rs4986791 and asthma in additive model (OR = 0.81, 95% CI = 0.64–1.02, P = .07), as shown in Fig. 5. Significant association was found between dominant model (OR = 0.76, 95% CI = 0.60–0.97, P = .025, as shown in Fig. 6), while not in recessive model (OR = 0.86, 95% CI = 0.35–2.14, P = .75).
Figure 5.
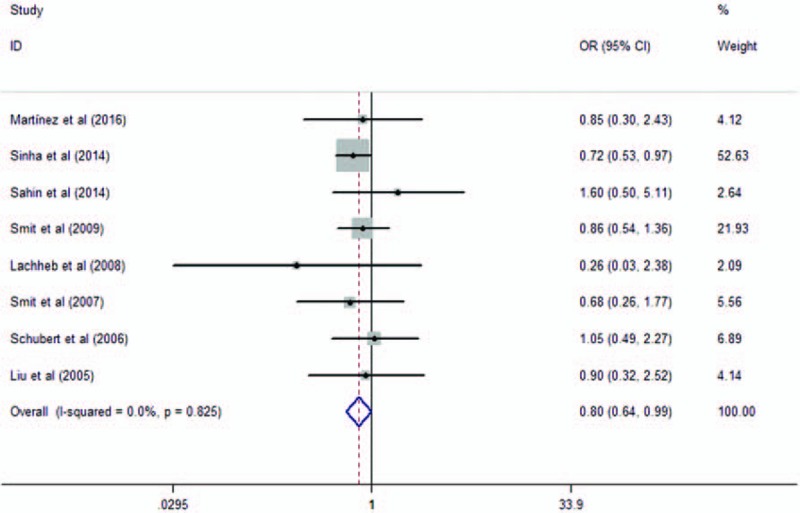
Meta-analysis of the association between TLR4 rs4986791 in TLR4 and asthma in additive model. TLR = toll-like receptor.
Figure 6.
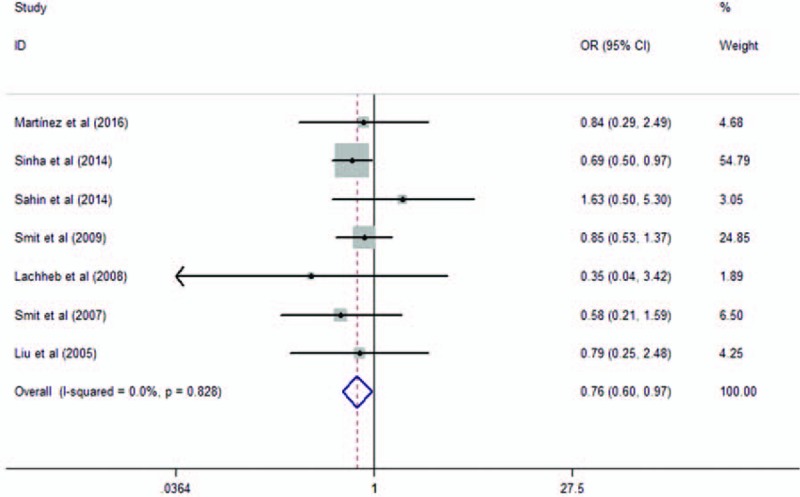
Meta-analysis of the association between TLR4 rs4986791 in TLR4 and asthma in dominant model. TLR = toll-like receptor.
3.4. Sensitivity analysis and publication bias
In the sensitivity test, we evaluated the sensitivity by exclusion of 1 study at a time, and the results indicated that the pooled ORs were stable, which confirmed the stability and reliability of the pooled results. The Begg funnel plot and Egger test were used to assess the publication bias of the selected studies. The results did not suggest any publication bias in our meta-analysis (as shown in Supplemental Figures). Therefore, publications bias did not have significant influence on the pooled results.
4. Discussion
In the meta-analysis, 18 studies consisting of 3538 asthma cases and 4090 healthy controls were selected to evaluate the association between polymorphisms in TLRs and asthma. The combined results show that rs3804099 in TLR2 and rs4986791 in TLR4 were significantly associated with asthma risk.
Up to now, more than 200 variants have been reported to be associated with asthma risk, but only a few of them have been replicated.[36] Lack of reproducibility has become a big challenge in genetic association studies. Noguchi et al[33] firstly reported that the variant rs3804099 in TLRs was marginally associated with asthma risk. Since then, many subsequent replication studies have evaluated the association between variants in TLRs and asthma risk. However, the results were inconclusive and further assessment was needed. In current study, we conducted an extensive, up-to-date, and unbiased meta-analysis combining all together 18 studies consisting of 3538 asthma cases and 4090 healthy controls to evaluate the association between 4 polymorphisms (rs5743708 and rs3804099 in TLR2; rs4986790 and rs4986791 in TLR4) and asthma. Our study demonstrated that variants in TLR2 and TLR4 might influence the risk of asthma.
Genome-wide association study (GWAS) has been a useful methods in discovering new genetic variants associated with complex disease.[37] However, most complex diseases, such as asthma, are usually caused by accumulation of several genetic determinants and environmental factors.[5] Each of these variants gets a small effect, and therefore it is difficult to find out new susceptibility locus in small sample size analysis. Many variants look promising in the original GWAS while do not meet the levels of significance. Meanwhile, the results of independent cohorts were not conclusive, and meta-analysis gives more conclusive results. For these variants did not reach statistical significance in single analysis, we get the chance to detect novel variants by combining results from single analysis to meta-analysis.
In current meta-analysis, we extracted data from a combined sample of 7628 individuals and the results show that rs3804099 in TLR2 and rs4986791 in TLR4 contribute to the increased risk of asthma. During the past years, several studies have analyzed variants in TLR2 and TLR4 with asthma, while the results were discordance due to small sample size and some other reasons. By combining results from single analysis, we got more conclusive results. Furthermore, our results further confirmed the hypothesis that common genetic variants with modest or small effects in single study might contribute to the risk of complex disease and most of them were not identified. Our results highlight the power of meta-analysis in genetic association studies. However, further studies with larger sample size and prospective study are needed to confirm these findings, and biological study is needed to investigate the mechanisms underling these variants and asthma.
5. Conclusion
This meta-analysis provides the evidence that variations rs3804099 in TLR2 and rs4986791 in TLR4 were associated with asthma risk. However, further prospective studies with larger sample size are needed to clarify the association between other variation in TLRs and asthma.
Supplementary Material
Acknowledgments
The authors thank all the participants in this study.
Footnotes
Abbreviations: CI = confidence interval, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, SNP = single nucleotide polymorphism, TLR = toll-like receptor.
Authorship: Conceived and designed the study: XL, MX, and JX. Analyzed the data: JZ, HS, and XC. Contributed reagents/materials/analysis tools: XL. Wrote the paper: JZ, YH, XF, and SZ. All authors read and approved the final manuscript.
Funding/support: This study was supported by the National Natural Science Foundation of China (No. 81670048).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].National Asthma Education and Prevention Program. Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol 2007;120(5 Suppl):S94–138. [DOI] [PubMed] [Google Scholar]
- [2].Ambrosino N, Paggiaro P. The management of asthma and chronic obstructive pulmonary disease: current status and future perspectives. Expert Rev Respir Med 2012;6:117–27. [DOI] [PubMed] [Google Scholar]
- [3].Ribatti D, Puxeddu I, Crivellato E, et al. Angiogenesis in asthma. Clin Exp Allergy 2009;39:1815–21. [DOI] [PubMed] [Google Scholar]
- [4].Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 2006;7:95–100. [DOI] [PubMed] [Google Scholar]
- [5].Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 2011;378:1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol 2003;21:335–76. [DOI] [PubMed] [Google Scholar]
- [7].Raby B, Klimecki W, Laprise C, et al. Polymorphisms in toll-like receptor 4 are not associated with asthma or atopy-related phenotypes. Am J Respir Crit Care Med 2002;166:1449–56. [DOI] [PubMed] [Google Scholar]
- [8].Eder W, Klimecki W, Yu L, et al. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol 2004;113:482–8. [DOI] [PubMed] [Google Scholar]
- [9].Fagerås Böttcher M, Hmani-Aifa M, Lindström A, et al. A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J Allergy Clin Immunol 2004;114:561–7. [DOI] [PubMed] [Google Scholar]
- [10].Yang I, Barton S, Rorke S, et al. Toll-like receptor 4 polymorphism and severity of atopy in asthmatics. Genes Immun 2004;5:41–5. [DOI] [PubMed] [Google Scholar]
- [11].Kormann M, Depner M, Hartl D, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol 2008;122:86. [DOI] [PubMed] [Google Scholar]
- [12].Tizaoui K, Kaabachi W, Hamzaoui K, et al. Association of single nucleotide polymorphisms in toll-like receptor genes with asthma risk: a systematic review and meta-analysis. Allergy Asthma Immunol Res 2015;7:130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ortiz-Martinez MG, Frias-Belen O, Nazario-Jimenez S, et al. A case-control study of innate immunity pathway gene polymorphisms in Puerto Ricans reveals association of toll-like receptor 2 +596 variant with asthma. BMC Pulm Med 2016;16:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bahrami H, Daneshmandi S, Heidarnazhad H, et al. Lack of association between toll like receptor-2 and toll like receptor-4 gene polymorphisms and other feature in Iranian asthmatics patients. Iran J Allergy Asthma Immunol 2015;14:48–54. [PubMed] [Google Scholar]
- [15].Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 2006;11:193–206. [DOI] [PubMed] [Google Scholar]
- [16].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [18].Bagos PG. Meta-analysis in Stata using gllamm. Res Synth Methods 2015;6:310–32. [DOI] [PubMed] [Google Scholar]
- [19].Liang XH, Cheung W, Heng CK, et al. Absence of the toll-like receptor 4 gene polymorphisms Asp299Gly and Thr399Ile in Singaporean Chinese. Ther Clin Risk Manag 2005;1:243–6. [PMC free article] [PubMed] [Google Scholar]
- [20].Saçkesen C, Karaaslan C, Keskin O, et al. The effect of polymorphisms at the CD14 promoter and the TLR4 gene on asthma phenotypes in Turkish children with asthma. Allergy 2005;60:1485–92. [DOI] [PubMed] [Google Scholar]
- [21].Sahin F, Yildiz P, Kuskucu A, et al. The effect of CD14 and TLR4 gene polimorphisms on asthma phenotypes in adult Turkish asthma patients: a genetic study. BMC Pulm Med 2014;14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lyakhovskaya N, Shlykova O, Bobrova N, et al. The content of mediators of allergic inflammation in the blood serum of patients with atopic asthma, depending on the polymorphism 2258G/A gene TLR2. АСТМА ТА АЛЕРГІЯ 2013;UDK: [577.21:616.5-002]-053.3/.5. [Google Scholar]
- [23].Hussein Y, Awad H, Shalaby S, et al. Toll-like receptor 2 and Toll-like receptor 4 polymorphisms and susceptibility to asthma and allergic rhinitis: a case-control analysis. Cell Immunol 2012;274:34–8. [DOI] [PubMed] [Google Scholar]
- [24].Voron’ko OE, Dmitrieva-Zdorova EV, Latysheva EA, et al. [CARD15 and TLR4 genes polymorphisms in atopic bronchial asthma]. Mol Biol (Mosk) 2011;45:831–9. [PubMed] [Google Scholar]
- [25].Smit L, Siroux V, Bouzigon E, et al. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med 2009;179:363–8. [DOI] [PubMed] [Google Scholar]
- [26].Bjørnvold M, Munthe-Kaas M, Egeland T, et al. A TLR2 polymorphism is associated with type 1 diabetes and allergic asthma. Genes Immun 2009;10:181–7. [DOI] [PubMed] [Google Scholar]
- [27].Carvalho A, Pasqualotto A, Pitzurra L, et al. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis 2008;197:618–21. [DOI] [PubMed] [Google Scholar]
- [28].Lachheb J, Dhifallah I, Chelbi H, et al. Toll-like receptors and CD14 genes polymorphisms and susceptibility to asthma in Tunisian children. Tissue Antigens 2008;71:417–25. [DOI] [PubMed] [Google Scholar]
- [29].Smit L, Bongers S, Ruven H, et al. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case-control study. Clin Exp Allergy 2007;37:1602–8. [DOI] [PubMed] [Google Scholar]
- [30].Schubert K, von Bonnsdorf H, Burke M, et al. A comprehensive candidate gene study on bronchial asthma and juvenile idiopathic arthritis. Dis Markers 2006;22:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu RM, Wu JM, Cui T, Li Y. Asp299Gly and Thr399Ile polymorphisms in the tol-l like receptor 4 and the susceptivity of allergic asthma. Chin J Microbiol Immunol 2005;25:94–7. [in Chinese]. [Google Scholar]
- [32].Adjers K, Karjalainen J, Pessi T, et al. Epistatic effect of TLR4 and IL4 genes on the risk of asthma in females. Int Arch Allergy Immunol 2005;138:251–6. [DOI] [PubMed] [Google Scholar]
- [33].Noguchi E, Nishimura F, Fukai H, et al. An association study of asthma and total serum immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy 2004;34:177–83. [DOI] [PubMed] [Google Scholar]
- [34].Zaborowski T, Wojas-Krawczyk K, Krawczyk P, et al. The effect of CD14 and TLR4 gene polymorphisms on the occurrence of atopic and non-atopic asthma. Adv Clin Exp Med 2011;413–21. [Google Scholar]
- [35].Sinha S, Singh J, Jindal SK, et al. Role of TLR4 C>1196T (Thr399Ile) and TLR4 A>896G (Asp299Gly) polymorphisms in a North Indian population with asthma: a case-control study. Int J Immunogenet 2014;41:463–71. [DOI] [PubMed] [Google Scholar]
- [36].Martinez FD, Vercelli D. Asthma Lancet 2013;382:1360–72. [DOI] [PubMed] [Google Scholar]
- [37].Lasky-Su J, Himes BE, Raby BA, et al. HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy 2012;42:1724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


