Abstract
Crustose coralline algae (CCA) are major benthic calcifiers that play crucial roles in coral reef ecosystems. Two diseases affecting CCA have recently been investigated: coralline white band syndrome (CWBS) and coralline white patch disease (CWPD). These diseases can trigger major losses in CCA cover on tropical coral reefs, but their causative agents remain unknown. Here, we provide data from the first investigation of the bacterial communities associated with healthy and diseased CCA tissues. We show that Neogoniolithon mamillare diseased tissues had distinct microbial communities compared to healthy tissues and demonstrate that CWBS and CWPD were associated with different pathobiomes, indicating that they had different disease causations. CWBS tissues were composed of opportunistic bacteria, and the origin of the disease was undetermined. In contrast, a vibrio related to Vibrio tubiashii characterized the CWPD pathobiome, suggesting that it could be a putative disease agent and supporting the case of a temperature dependent disease associated with global warming.
Keywords: disease, bacterial communities, crustose coralline algae, bacterial pathogen, causative agent, coral reefs
Introduction
In recent decades, there has been an increasing number of reports of diseases in marine organisms, such as corals, mollusks, turtles, and mammals (Harvell et al., 1999, 2004). Infectious diseases have been acknowledged as a major factor of change in marine ecosystems. They can lead to a loss of keystone species and alter critical ecosystem processes (Harvell et al., 2002). However, their causal agents often remain unknown, thus slowing the development of management and remediation tools (Harvell et al., 2004). Together with scleractinian corals, crustose coralline algae (CCA) are major framework builders and carbonate producers on tropical reefs (Rasser and Riegl, 2002). Two disease symptoms have been recently investigated: the coralline white band syndrome (CWBS) and the coralline white patch disease (CWPD) (Quéré et al., 2015b). White band lesions are defined by bands that appear centrally or peripherally and advance on healthy tissue, whereas the patch disease shows distinct white patches expanding on healthy crusts (Quéré et al., 2015a). Both diseases result in tissue loss, which often leads to the death of the affected patch, with subsequent colonization by endophytic algae (Quéré et al., 2015b). Although these syndromes have been well-described in field surveys, standard histopathological techniques have failed to identify the potential infectious agents (Quéré et al., 2015a). Further molecular characterization of the microbial communities is needed to complete the diagnostic picture.
The microbes that are associated with macro-organisms play an important role in the host’s health and metabolism (Rosenberg and Zilber-Rosenberg, 2016). Microbial communities associated with CCA remain poorly understood (Webster et al., 2011; Sneed et al., 2015; Hester et al., 2016), but there are indications that they can induce settlement and metamorphosis of coral larvae (Negri et al., 2001). CCA bacterial communities can be altered by environmental changes such as temperature (Webster et al., 2011), pH (Webster et al., 2013), or disease (Kimes et al., 2010), but the identification of the causative pathogens associated with epizootics in CCA is still unknown. Recently, the development of “omic” studies has shown the limits of the Koch and Hill’s postulate “one microbe – one disease” and brought forward the pathobiome concept (Vayssier-Taussat et al., 2014), which has been proposed for the study of coral diseases (Sweet and Bulling, 2017). The pathobiome is defined as the pathogenic agent integrated within its biotic environment (Vayssier-Taussat et al., 2014). Pathogens interact with other microbes, and these complex interactions influence the outcome of the disease.
Here, we investigated both healthy and diseased CCA tissues to test the hypothesis that tissue lesions indicative of CWBS and CWPD are characterized by distinct pathobiomes.
Materials and Methods
Field Collection
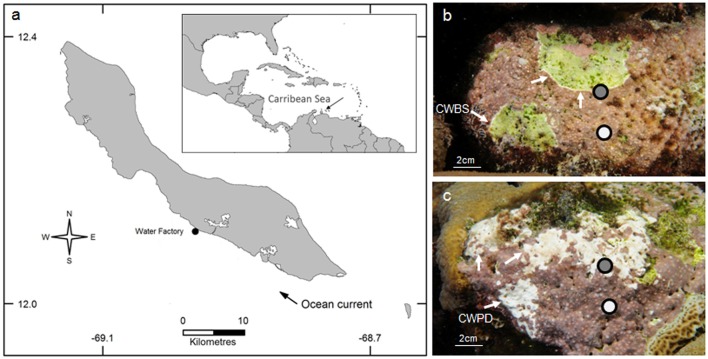
The common CCA species Neogoniolithon mamillare was sampled on 25 October and 15 November 2014 at the diving site Water Factory on the leeward coast of Curaçao, Southern Caribbean (12°N, 69°W) (Figure 1a). Fragments (ca. 2–5 cm2) of the CCA species were collected using hammer and chisel on the reef terrace at 5–10 m depth. Two different symptoms were targeted: CWBS and the CWPD (Figures 1b,c). Samples were gathered from five healthy-looking individuals, five CWBS and five CWBD individuals. Fragments of diseased individuals included both healthy-looking and diseased tissues. Healthy and diseased individuals were placed in individual collection bags to avoid contamination and transported in the dark to the laboratory.
FIGURE 1.
Geographical location of the Water Factory sampling site in Curaçao in 2014. The dark gray areas indicate urbanized area (a). Neogoniolithon mamillare affected by coralline white band syndrome (CWBS) (b) and coralline white patch disease (CWPD) (c). Healthy and diseased tissues sampled for each pathology are indicated in white and gray shading respectively. Arrows point to the white bands in CWBS and the white patches in CWPD.
DNA Extraction and Sequencing
Back in the laboratory, the CCA fragments were dissected with sterilized instruments by scraping the surface of the samples. One sample was chipped from each of the five healthy-looking individuals (controls), whereas a sample of healthy-looking tissue and a sample of diseased-looking tissue was chipped from each of the 10 diseased individuals, making a total of 25 samples. Samples of diseased-looking tissue included the boundary between healthy and diseased tissues (Figures 1b,c). Each sample was fixed separately in ethanol 96% v/v. Lysis of CCA was realized by mechanic action on a FastPrep Instrument with a Y Matrix (MP Biomedical, Santa Ana, CA, United States), after removing the ethanol and washing with DNase free water, followed by incubation with proteinase K at 57°C during 1 h. DNA was extracted using Maxwell® Blood DNA Purification Kit LEV (Promega, Madison, WI, United States) on Maxwell 16 MDx Instrument (Promega) following the manufacturer instructions. DNA concentrations were measured by spectrophotometry (Nanodrop ND-1000, Thermo Fisher Scientific, Inc., Waltham, MA, United States), and the quality was assessed by electrophoresis migration on a 1% agarose gel. DNA was diluted to 15 ng μL-1 and 30 ng were used for PCR amplification. The V1–V3 region of the bacterial 16S rRNA genes were amplified using bacteria specific primers 27F (AGRGTTTGATCMTGGCTCAG) and 519R (GTNTTACNGCGGCKGCTG) with the barcode on the forward primer. All samples were then pooled together in equal proportions based on their DNA concentrations. Pooled samples were purified using calibrated Agencourt® AMPure® XP (Beckman Coulter, Brea, CA, United States). Then the pooled and purified PCR product was used to prepare a DNA library by following Illumina TruSeq DNA library preparation protocol. All samples were sequenced on the same Miseq Illumina sequencer run (Illumina, San Diego, CA, United States) using Miseq reagent kit V3 (Illumina) producing 2 × 300-bp long reads in a commercial laboratory (MR DNA, Lubbock, TX, United States). All sequences were deposited in GenBank under SRA accession SRP113196.
Sequence Analyses
R1 and R2 reads were joined with PANDAseq (Masella et al., 2012). The read quality filtering and length trimming, data set partitioning based on barcodes, de-replication, clustering at 97% sequence identity and taxonomic classification were performed with PyroTagger (Kunin and Hugenholtz, 2010). The quality of the sequences was controlled by removing all the reads that had a mismatch with the 16S rRNA primers, contained ambiguous nucleotides (N) or were <300 bp long beyond the forward primer. A stringent quality trimming criteria was applied to remove reads that had ≥10% of bases with Phred values <27. This procedure is recommended to ensure that when clustering at 97%, the influence of erroneous reads is minimized (Kunin et al., 2010). The sequences were then de-replicated and clustered at a 97% threshold using UCLUST (Edgar, 2010). The sequences from each operational taxonomic unit (OTU) were classified by comparing them with those from the SILVA 119 (Quast et al., 2013). The taxonomic affiliations of the OTUs of interest were further verified against sequences from the NCBI databases using BLAST with default parameters. Sequences annotated as belonging to algal chloroplasts were removed. Putatively chimeric sequences were identified as sequences having a best Blast alignment <90% of the trimmed read length to the reference sequence in the database, ≥90% sequence identity to the best Blast match and OTU size ≤2. One sample from healthy patch disease tissues was dominated by a chimeric sequence and was thus removed from the dataset.
Data Analysis
A total of 6,083,434 bacterial 16S rRNA genes sequences were retained after removing poor quality reads and putative algal chloroplastic hits. A total of 10,818 OTUs were identified at a 97% sequence similarity cutoff. All samples were randomly re-sampled to match the size of the sample containing the fewest sequences (n = 11,695). The Shannon diversity index (H′) was calculated at the OTU level on the re-sampled dataset with the PAST software (Hammer et al., 2001). Differences in OTU abundance between two groups of samples were tested using the White’s non-parametric t-test using the STAMP v2.1.3 software (Parks et al., 2014). OTUs were defined as pathology-specific when their representative sequence abundance was significantly higher in the diseased tissues compared to control tissue.
A multidimensional scaling ordination (MDS) based on Bray–Curtis similarity was conducted to visualize similarities in community composition between samples. The MDS was computed with the R package phyloseq (McMurdie and Holmes, 2013). An analysis of similarity (one-way ANOSIM) was conducted to assess the significance of the MDS grouping using PAST v3.10 software. A t-test was used to assess the differences in community diversities were significant using Statistica v10 software after testing for normality and equal variance.
To test if N. mamillare had a species specific bacterial community, OTU sequence representatives were blasted against 454 sequences previously found in four different species of CCA sampled in Belize: Titanoderma prototypum, Porolithon pachydermum, Paragoniolithon solubile, and Hydrolithon boergesenii (SRA database number SRP056487) (Sneed et al., 2015).
Results and Discussion
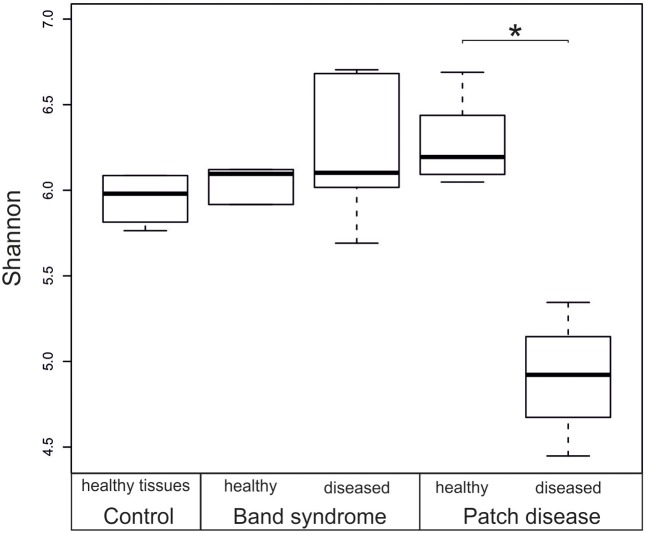
A comparison of the bacterial community composition was conducted using the Bray–Curtis similarity index and MDS analysis. The MDS ordination of 16S tag sequencing data separated the N. mamillare samples into three main groups (Figure 2). One group corresponded to the CCA sampled from diseased CWPD tissues, a second group included CWBS diseased tissues and the third group had healthy tissues from both control and diseased individuals. There was only one exception with one diseased CWPD sample similar to a healthy community. There was more variability within CWBS disease tissue communities than within CWPD communities (Figure 2). In summary, our results show that bacterial communities associated with CWBS were significantly different from the ones found in CWPD diseased tissues (Table 1) and they were different from control tissues (ANOSIM, p < 0.01), demonstrating that the two syndromes corresponded to different pathobiomes. The pathobiome diversity in patch diseased samples was significantly lower than the diversity of the bacterial community in healthy tissues (Figure 3). This finding, together with the observed unchanged diversity of the band syndrome community, is in marked contrast to the patterns that are commonly observed for disease associated microbial communities. Higher diversity has been observed in diseased sponge tissues (Blanquer et al., 2016), and in corals, the yellow band or white plague diseases are associated with an increased number of bacterial taxa (Closek et al., 2014; Roder et al., 2014). These studies have hypothesized that a diverse array of opportunistic bacteria from the surrounding environment colonizes the compromised coral tissues. In the case of CCA, the lower diversity of the CWPD diseased tissues suggests the occurrence of a few specialized pathogens.
FIGURE 2.
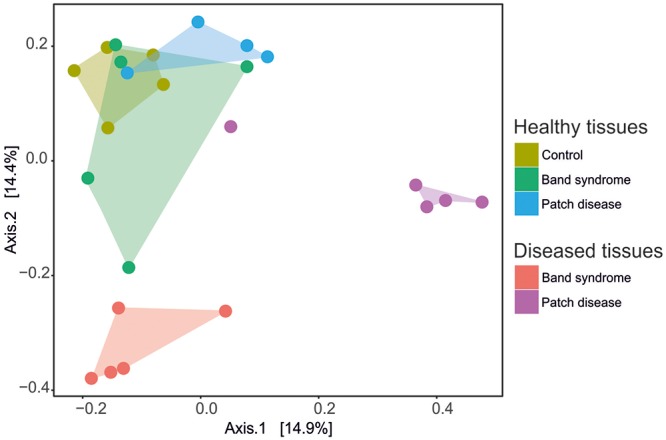
Multi-dimensional scaling plot (MDS) based on the Bray–Curtis similarity index showing the similarity between bacterial community compositions for healthy and diseased tissues in CCA affected by CWBS and CWPD.
Table 1.
Operational taxonomic units (OTUs) that showed a significant difference in abundance between healthy and diseased tissues (White’s non-parametric t-test).
| Sample | OTU | Class | Order | Best match | Similarity | Origin or bacterial strain | Reference |
|---|---|---|---|---|---|---|---|
| Healthy | OTU 62 | Alphaproteobacteria | Rhodobacterales | HE574911 | 98% | Euprymna scolopes (squid) | Collins et al., 2012 |
| OTU 185 | Alphaproteobacteria | Rhodobacterales | NR_118329 | 100% | Loktanella litorea | Yoon et al., 2013 | |
| OTU 102 | Alphaproteobacteria | Rhodobacterales | AB611495 | 97% | Sea water | Yoshida-Takashima et al., 2012 | |
| OTU 362 | Chloroflexi | Ardenticatenia | KU688646 | 96% | Cystoseira compressa (brown algae) | Mancuso et al., 2016 | |
| OTU 380 | Alphaproteobacteria | Parvularculales | EF123358 | 99% | Siderastrea siderea (coral) | Sekar et al., 2008 | |
| Band syndrome | OTU 23 | Alphaproteobacteria | Rhodobacterales | GU118282 | 98% | Diploria strigosa (coral) | Sunagawa et al., 2010 |
| OTU 172 | Alphaproteobacteria | Rhodobacterales | AY038533 | 98% | Black band diseased coral | Frias-Lopez et al., 2002 | |
| OTU 239 | Alphaproteobacteria | Rhodobacterales | KY577415 | 98% | Black band diseased Orbicella faveolata (coral) | Unpublished | |
| Patch disease | OTU 20 | Alphaproteobacteria | Rickettsiales | FJ930418 | 98% | Porites lobata (coral) | Unpublished |
| OTU 665 | Gammaproteobacteria | Vibrionales | KP329558 | 100% | Vibrio tubiashii Strain T33 | Mohamad et al., 2015 |
Operational taxonomic units are given together with their best match in GenBank.
FIGURE 3.
Box plot showing the diversity (Shannon index) based on 16S rRNA sequences from healthy and diseased N. mamillare tissues. The mean value and the standard deviation are represented by a line and a box respectively, and minimum and maximum values are represented by whiskers. ∗p < 0.05.
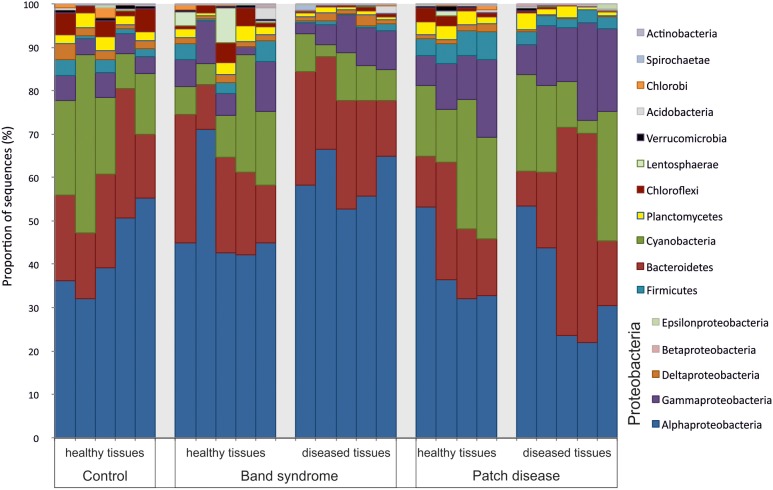
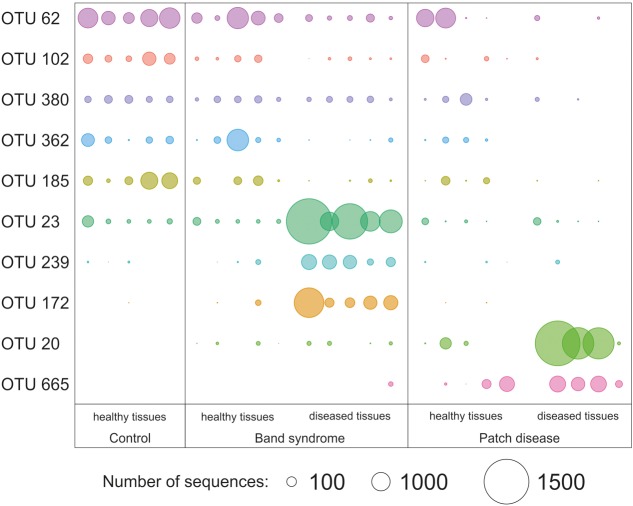
Diseased CWPD tissues were characterized by a higher proportion of Bacteroidetes and Gammaproteobacteria (White’s non-parametric t-test between control and diseased tissues, p < 0.05) (Figure 4). At a higher taxonomic resolution, the CWPD pathobiomes showed a significantly higher abundance of a Rickettsiales (OTU 20), similar to an uncultured bacterium isolated from Porites coral mucus (White’s non-parametric t-test, Table 1 and Figure 5), but without match to any cultured microorganisms. OTU 20 represented up to 13% of the sequences in diseased PD tissues. The CWPD pathobiome also showed a significantly higher abundance of a Vibrio sp. OTU (OTU 665, Figure 5), which was similar to Vibrio tubiashii (Table 1), a known pathogen of mollusks often associated with larval vibriosis (Beaz-Hidalgo et al., 2010). This OTU was observed in all the CWPD diseased tissues (Figure 5) and represented up to 2% of the sequences in diseased CWPD tissues. High abundances of Vibrio were previously found in another CCA species, P. pachydermum (21% of sequences), but without correlation to CCA diseases (Sneed et al., 2015). In our study, the vibrio OTU was present in three of the five healthy tissues of the CWPD individuals but was never detected in control individuals (Figure 5). This vibrio could thus be the putative agent causing CWPD.
FIGURE 4.
Bacterial community composition at the phylum level in healthy and diseased N. mamillare tissues based on 16S rRNA sequences. The most abundant phylum are shown (>0.1% of the sequences).
FIGURE 5.
Relative abundance of operational taxonomic units (OTUs) that showed a significant difference in abundance between healthy and diseased tissues (White’s non-parametric t-test). Circle sizes are proportional to the number of sequences contained in an OTU.
Vibrios are ubiquitous and abundant in the aquatic environment (Thompson et al., 2004). Host endogen factors such as reproduction could play a crucial role in triggering vibriosis (Travers et al., 2009). However, the growth, behavior (Garren et al., 2016) and virulence (Kimes et al., 2012) of pathogenic vibrios in the natural environment are largely dictated by the temperature. The abundance of vibrios has therefore been proposed to represent a microbial barometer of climate change (Baker-Austin et al., 2017). The association between vibriosis and temperature supports our hypothesis of putative pathogenicity of vibrios because CWPD has recently been described as being likely to be temperature-dependent in Curaçao (Quéré et al., 2015b). Together, these results strengthen the hypothesis that CWPD is a temperature dependent disease.
The CWBS pathobiome was dominated by OTUs that belong to the Alphaproteobacteria class (Figure 4), most of which were Rhodobacterales (Table 1). The three OTUs, OTU 23, OTU 172 and OTU 239, represented up to 13, 6, and 1.8%, respectively, of the sequences obtained from diseased CWBS tissues (Figure 5). We did not identify any potential pathogens in the CWBS community, such as cyanobacteria known to be a causative agent of the black band disease in corals (Casamatta et al., 2012), but Rhodobacterales OTUs had close similarity (98%) to sequences found in corals affected by microbial originating white plague or black band disease (Frias-Lopez et al., 2002; Sunagawa et al., 2009). Thus, they could represent opportunist bacteria thriving on dying tissues. They may invade the depleted, diseased CCA cells, which have been shown to contain higher abundances of bioeroders by histological analysis, including boring sponges, helminths, and cyanobacteria (Quéré et al., 2015a). Such as in coral affected by white syndromes disease (Pollock et al., 2017), opportunist bacteria could play a role in pathogenesis and/or serve as a diagnostic criterion for disease differentiation. Further investigations of the pathobiome and/or viral component of the CWBS are required to reveal the causative agent of the disease.
The microbiome associated with control CCA might be specific to the species N. mamillare. Only 0.005% of the sequences found in N. mamillare were similar to those found in four different CCA species sampled in Belize, including Titanoderma textitprototypum and Hydrolithon boergesenii, which facilitate coral larval settlement, and P. solubile and P. pachydermum, which cause lower levels of coral settlement (Ritson-Williams et al., 2014). N. mamillare is not known to induce high coral settlements (Quéré et al., 2015b). The presence of a high proportion of cyanobacteria (average of 20% on healthy tissues, Figure 2) in N. mamillare, P. solubile, and P. pachydermum feeds the hypothesis that a high abundance of cyanobacteria in certain CCA species could inhibit coral settlements (Sneed et al., 2015). Benthic cyanobacteria, especially those in the order Oscillatoriales, are known to produce potent allelopathic compounds and inhibit coral settlements (Kuffner and Paul, 2004; Kuffner et al., 2006).
In summary, the two different CCA diseases appear to have different causes. The patch disease could be of bacterial origin as shown by the presence of Vibrio tubiashii, whereas the origin (i.e., bacterial, viral, or environmental stress) of band syndrome, characterized by opportunistic bacteria, remains unknown.
Author Contributions
A-LM conceived and designed the experiments, analyzed the data and wrote the paper. PG contributed reagents/material/analysis tools, analyzed the data and wrote the paper. MN contributed materials and wrote the paper. GQ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PG is supported by the Agence Nationale de la Recherche (ANR) through the project EUREKA (ANR-14-CE02-0004-01). MN acknowledges support for the CNRS Chaire d’Excellence and the PEPS ExoMod program.
References
- Baker-Austin C., Trinanes J., Gonzalez-Escalona N., Martinez-Urtaza J. (2017). Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25 76–84. 10.1016/j.tim.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R., Balboa S., Romalde J. L., Figueras M. J. (2010). Diversity and pathogenecity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep. 2 34–43. 10.1111/j.1758-2229.2010.00135.x [DOI] [PubMed] [Google Scholar]
- Blanquer A., Uriz M. J., Cebrian E., Galand P. E. (2016). Snapshot of a bacterial microbiome shift during the early symptoms of a massive sponge die-offin the western Mediterranean. Front. Microbiol. 7:752 10.3389/fmicb.2016.00752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamatta D., Stanić D., Gantar M., Richardson L. L. (2012). Characterization of Roseofilum reptotaenium (Oscillatoriales, Cyanobacteria) gen. et sp. nov. isolated from Caribbean black band disease. Phycologia 51 489–499. 10.2216/11-10.1 [DOI] [Google Scholar]
- Closek C. J., Sunagawa S., DeSalvo M. K., Piceno Y. M., DeSantis T. Z., Brodie E. L., et al. (2014). Coral transcriptome and bacterial community profiles reveal distinct yellow band disease states in Orbicella faveolata. ISME J. 8 2411–2422. 10.1038/ismej.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. J., LaBarre B. A., Wong Won B. S., Shah M. V., Heng S., Choudhury M. H., et al. (2012). Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl. Environ. Microbiol. 78 4200–4208. 10.1128/AEM.07437-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Frias-Lopez J., Zerkle A. L., Bonheyo G. T., Fouke B. W. (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68 2214–2228. 10.1128/AEM.68.5.2214-2228.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M., Son K., Tout J., Seymour J. R., Stocker R. (2016). Temperature-induced behavioral switches in a bacterial coral pathogen. ISME J. 10 1363–1372. 10.1038/ismej.2015.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D. (2001). Past: paleontological statistics software package for education and data analysis.Paleontol. Electron. 4 9. [Google Scholar]
- Harvell C. D., Kim K., Burkholder J. M., Colwell R. R., Epstein P. R., Grimes D. J., et al. (1999). Emerging marine diseases-climate links and anthropogenic factors. Science 285 1505–1510. 10.1126/science.285.5433.1505 [DOI] [PubMed] [Google Scholar]
- Harvell C. D., Mitchell C. E., Ward J. R., Altizer S., Dobson A. P., Ostfeld R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296 2158–2162. 10.1126/science.1063699 [DOI] [PubMed] [Google Scholar]
- Harvell D., Aronson R., Baron N., Connell J., Dobson A., Ellner S., et al. (2004). The rising tide of ocean diseases: unsolved problems and research priorities. Front. Ecol. Environ. 2 375–382. 10.1890/1540-92952004002[0375:TRTOOD]2.0.CO;2 [DOI] [Google Scholar]
- Hester E. R., Barott K. L., Nulton J., Vermeij M. J. A., Rohwer F. L. (2016). Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 10 1157–1169. 10.1038/ismej.2015.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes N. E., Grim C. J., Johnson W. R., Hasan N. A., Tall B. D., Kothary M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6 835–846. 10.1038/ismej.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes N. E., Van Nostrand J. D., Weil E., Zhou J., Morris P. J. (2010). Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 12 541–556. 10.1111/j.1462-2920.2009.02113.x [DOI] [PubMed] [Google Scholar]
- Kuffner I. B., Paul V. J. (2004). Effects of the benthic cyanobacterium Lyngbya majuscula on larval recruitment of the reef corals Acropora surculosa and Pocillopora damicornis. Coral Reefs 23 455–458. 10.1007/s00338-004-0416-8 [DOI] [Google Scholar]
- Kuffner I. B., Walters L. J., Becerro M. A., Paul V. J., Ritson-Williams R. (2006). Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar. Ecol. Prog. Ser. 323 107–117. 10.3354/meps323107 [DOI] [Google Scholar]
- Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12 118–123. 10.1111/j.1462-2920.2009.02051.x [DOI] [PubMed] [Google Scholar]
- Kunin V., Hugenholtz P. (2010). PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J. 1–8. [Google Scholar]
- Mancuso F. P., D’Hondt S., Willems A., Airoldi L., De Clerck O. (2016). Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweed Cystoseira compressa (Esper) Gerloff and Nizamuddin. Front. Microbiol. 7:476 10.3389/fmicb.2016.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13:31 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J., Holmes S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad N. I., Yin W.-F., Chan K.-G. (2015). Whole-genome sequence of quorum-sensing Vibrio tubiashii strain T33. Genome Announc. 3:e01362-14. 10.1128/genomeA.01362-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri A. P., Webster N. S., Hill R. T., Heyward A. J. (2001). Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar. Ecol. Prog. Ser. 223 121–131. 10.3354/meps223121 [DOI] [Google Scholar]
- Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock F. J., Wada N., Torda G., Willis B. L., Bourne D. G. (2017). White syndrome-affected corals have a distinct microbiome at disease lesion fronts. Appl. Environ. Microbiol. 83:e02799-16. 10.1128/AEM.02799-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéré G., Meistertzheim A.-L., Steneck R. S., Nugues M. M. (2015a). Histopathology of crustose coralline algae affected by white band and white patch diseases. PeerJ 3:e1034 10.7717/peerj.1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéré G., Steneck R. S., Nugues M. M. (2015b). Spatiotemporal and species-specific patterns of diseases affecting crustose coralline algae in Curaçao. Coral Reefs 34 259–273. 10.1007/s00338-014-1225-3 [DOI] [Google Scholar]
- Rasser M., Riegl B. (2002). Holocene coral reef rubble and its binding agents. Coral Reefs 21 57–72. 10.1007/s00338-001-0206-5 [DOI] [Google Scholar]
- Ritson-Williams R., Arnold S. N., Paul V. J., Steneck R. S. (2014). Larval settlement preferences of Acropora palmata and Montastraea faveolata in response to diverse red algae. Coral Reefs 33 59–66. 10.1007/s00338-013-1113-2 [DOI] [Google Scholar]
- Roder C., Arif C., Bayer T., Aranda M., Daniels C., Shibl A., et al. (2014). Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J. 8 31–39. 10.1038/ismej.2013.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Zilber-Rosenberg I. (2016). Microbes drive evolution of animals and plants: the hologenome concept. MBio 7:e01395 10.1128/mBio.01395-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R., Kaczmarsky L. T., Richardson L. L. (2008). Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar. Ecol. Prog. Ser. 362 85–98. 10.3354/meps07496 [DOI] [Google Scholar]
- Sneed J. M., Ritson-Williams R., Paul V. J. (2015). Crustose coralline algal species host distinct bacterial assemblages on their surfaces. ISME J. 9 2527–2536. 10.1038/ismej.2015.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., DeSantis T. Z., Piceno Y. M., Brodie E. L., DeSalvo M. K., Voolstra C. R., et al. (2009). Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3 512–521. 10.1038/ismej.2008.131 [DOI] [PubMed] [Google Scholar]
- Sunagawa S., Woodley C. M., Medina M. (2010). Threatened corals provide underexplored microbial habitats. PLoS ONE 5:e9554 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet M. J., Bulling M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4:9 10.3389/fmars.2017.00009 [DOI] [Google Scholar]
- Thompson F. L., Iida T., Swings J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68 403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers M.-A., Basuyaux O., Le Goïc N., Huchette S., Nicolas J.-L., Koken M., et al. (2009). Influence of temperature and spawning effort on Haliotis tuberculata mortalities caused by Vibrio harveyi: an example of emerging vibriosis linked to global warming. Glob. Chang. Biol. 15 1365–1376. 10.1111/j.1365-2486.2008.01764.x [DOI] [Google Scholar]
- Vayssier-Taussat M., Albina E., Citti C., Cosson J. F., Jacques M.-A., Lebrun M.-H., et al. (2014). Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 4:29 10.3389/fcimb.2014.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Soo R., Cobb R., Negri A. P. (2011). Elevated seawater temperature causes a microbial shift on crustose coralline algae with implications for the recruitment of coral larvae. ISME J. 5 759–770. 10.1038/ismej.2010.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Uthicke S., Botté E. S., Flores F., Negri A. P. (2013). Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob. Chang. Biol. 19 303–315. 10.1111/gcb.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Jung Y. T., Lee J. S. (2013). Loktanella litorea sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 63 175–180. 10.1099/ijs.0.039198-0 [DOI] [PubMed] [Google Scholar]
- Yoshida-Takashima Y., Nunoura T., Kazama H., Noguchi T., Inoue K., Akashi H., et al. (2012). Spatial distribution of viruses associated with planktonic and attached microbial communities in hydrothermal environments. Appl. Environ. Microbiol. 78 1311–1320. 10.1128/AEM.06491-11 [DOI] [PMC free article] [PubMed] [Google Scholar]