Abstract
A secretory defect causes specific transcriptional repression of both ribosomal protein and ribosomal RNA genes, suggesting the coupling of plasma membrane and ribosome syntheses. We previously reported that the rap1-17 allele, which produced C-terminally truncated Rap1p, derepressed transcription of ribosomal protein genes when the secretory pathway was blocked. In this paper, we demonstrate that the rap1-17 mutation also leads to significant attenuation of transcriptional repression of rRNA genes due to a secretory defect. In contrast, the rap1-2 temperature-sensitive allele containing a unique missense mutation in the middle of the coding sequence has only a weak effect on repression. These results suggest that the C-terminal silencing domain of Rap1p is required for transcriptional repression of rDNA in response to a secretory defect. We also demonstrated that transcriptional regulation of ribosomal protein genes in response to nitrogen limitation was not affected by the rap1-17 allele, suggesting that the mechanism of nitrogen response is distinct from that of the secretory response.
INTRODUCTION
The yeast ribosome consists of four rRNAs and 78 ribosomal proteins (RP), which are encoded by 100–200 copies of rRNA genes and 137 genes for 78 RP per haploid cell. Nevertheless, the transcription of these genes appears to be coordinately regulated in response to various kinds of stresses and environmental changes (1). Recent genome-wide analysis also suggested that almost all RP genes were coordinately regulated in response to heat shock (2). A defect in the secretory pathway causes transcriptional repression of the three sets of genes, rRNA genes by RNA polymerase I (3), RP genes by RNA polymerase II (3) and 5S rRNA genes and tRNA genes by RNA polymerase III (4). This coupling of the secretion pathway and ribosome synthesis apparently arises through feedback from stretch sensors within the plasma membrane/cell wall, as the plasma membrane is stressed by the increase in cell mass without the possibility of a concomitant increase in plasma membrane (4). As a candidate member of the signaling pathway, we cloned RRS1 and demonstrated that Rrs1p has a role in 25S rRNA maturation and 60S ribosomal subunit assembly (5). This suggested that the molecular mechanism of signaling is closely related to that of normal ribosome biogenesis.
We previously demonstrated that the C-terminal region of Rap1p was also essential for transcriptional repression of RP genes due to a secretory defect; a C-terminally truncated allele of RAP1, rap1-17, caused substantial attenuation of repression (6). Rap1 is a relatively abundant, multifunctional protein in yeast. It is involved in both transcriptional activation and silencing. Transcription of many genes are regulated by Rap1p: the majority of RP genes, glycolytic genes, such as PGK1 and PYK1, and others. The C-terminal region of Rap1 interacts with Rif proteins and Sir proteins and is implicated in both silencing and telomere length (7–9). Recent studies revealed that Rap1p has a function for chromatin opening (10). Transcription of most RP genes is dependent on Rap1p, although a few RP genes, such as RPL3, have an Abf1p binding site but not a Rap1p-binding site upstream. Nevertheless, we showed that Rap1p is responsible for the repression of both sets of RP genes due to a secretory defect, suggesting that the repression does not necessarily require a Rap1p binding site within the upstream activating sequence (UAS) (6).
Here we demonstrate that Rap1p is responsible for the repression of rRNA genes as well, indicating yet another role for this remarkable protein.
MATERIALS AND METHODS
Yeast strains, media and plasmids
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose)-rich medium, synthetic complete medium containing 2% glucose (SC) or SC dropout medium depending on the plasmid markers (11). Nitrogen-starvation medium (SC–N) contained 2% glucose and 0.17% Yeast Nitrogen Base without amino acids and without ammonium sulfate. Yeast transformation was performed by a lithium acetate procedure (12). For epitope-tagging, an NheI site was introduced by PCR into pRAP1 (6) just after the initiation codon. The DNA cassette encoding three copies of the nine-amino acid influenza virus hemaggulutinin (HA) epitope was obtained by the digestion of pYT11 (13) with NheI and was inserted in-frame into the NheI site of pRAP1. The digested DNA fragment including epitope-tagged RAP1 was subcloned into pRS313 (14), designated pRS313-RAP1-HA. pRS313-rap1-17-HA was constructed by replacing the SacI–SphI region of pRS313-rap1-17 (6), including the upstream region and part of the ORF, with the corresponding region of pRS313-RAP1-HA. A plasmid containing the rap1-2 allele in pRS413 was generously provided by H. Uemura (National Institute of AIST, Japan).
Table 1. Yeast strains used in this study.
| Strain |
Genotype |
Source or reference |
| W303a | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 | R.Rothstein |
| KM011 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 sly1 [pRS313-RAP1] | (6) |
| KM013 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 sly1 [pRS313-rap1-17] | (6) |
| KM014 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-RAP1] | (6) |
| KM016 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-rap1-17] | (6) |
| KM017 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-RAP1-HA] | This study |
| KM019 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 sly1 [pRS313-RAP1-HA] | This study |
| KM326 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-rap1-17-HA] | This study |
| KM327 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 sly1 [pRS313-rap1-17-HA] | This study |
| KM309 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS413-rap1-2] | This study |
| KM328 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-RAP1, pRS414-ADE2-URA3] | This study |
| KM329 | MATa his3-11,15 ade2-1 ura3-1 leu2-3, 112 trp1-1 can1-100 rap1::LEU2 [pRS313-rap1-17, pRS414-ADE2-URA3] | This study |
Northern blot analysis
Northern blot analysis was carried out using 1.5% agarose gels as described previously (3,15). 32P-labeled probes for mRNAs were prepared by random priming and a 32P-labeled oligonucleotide probe for snoRNA U3 was prepared using polynucleotide kinase. Northern blots were hybridized with 32P-labeled probes and radioactivity of each band was quantified using BAS-1800 (Fuji Photo Film Co.). The radioactivity was normalized by the level of snoRNA U3.
[Methyl-3H]methionine pulse-chase analysis
[Methyl-3H]methionine pulse-chase analysis was carried out as previously described (16). Total RNA was prepared and 20 µg of each sample was analyzed by electrophoresis and blotted to a Nytran membrane. The blot of the upper portion of the gel was sprayed with En3Hance (NEN) and exposed to a film for 4 days. The blot of the lower portion was probed for snoRNA U3. Incorporation of [3H] into RNA was determined by counting the radioactivity of TCA precipitates in a liquid scintillator.
Western blot analysis
Western blotting followed standard techniques, and signals were visualized by Enhanced Chemiluminescence (Amersham).
RESULTS
Transcriptional repression of rDNA appears not to be secondary event following that of RP genes in response to a secretory defect
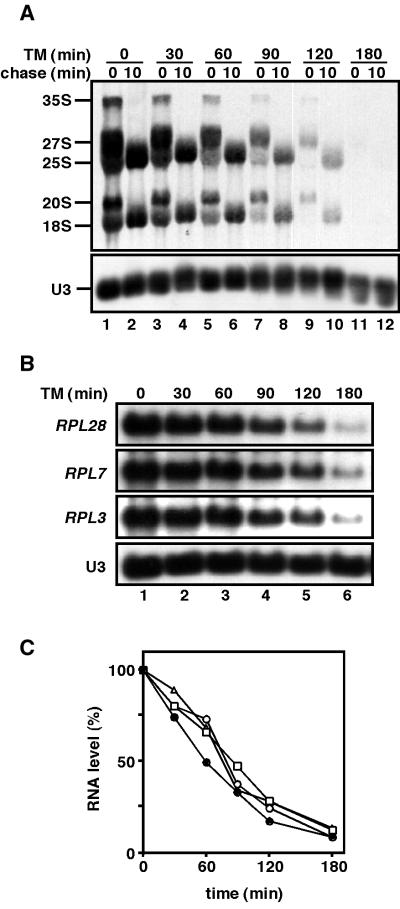
A secretory defect causes transcriptional repression of both rRNA and RP genes (3). In order to elucidate the relationship between these two sets of the repression, the time-courses of transcriptional repression of rRNA genes and that of RP genes were compared with each other when the secretory pathway was blocked by addition of tunicamycin. We detected the synthesis and processing of rRNA by [methyl-3H]methionine pulse-chase analysis, since newly synthesized precursor rRNA is methylated immediately (17). Northern analysis and [methyl-3H]methionine pulse-chase analysis showed that transcription of RP genes and that of rRNA genes were gradually repressed after addition of tunicamycin (Fig. 1A and B), consistent with previous data (3). Comparison of data from northern blot and [methyl-3H]methionine pulse suggest that transcription of rDNA was repressed at the same time or slightly earlier than that of RP genes (Fig. 1C, see Discussion).
Figure 1.
Transcription of rDNA is repressed apparently earlier than that of RP genes in response to a secretory defect. Wild-type cells (W303a) were grown to early log phase in SC–Met medium at 30°C and treated with tunicamycin (2.5 µg/ml). Each culture at the indicated times was taken for (A) pulse-chase analysis with [methyl-3H] methionine or (B) northern analysis. (C) Closed circle, incorporation of [3H] into RNA [as in (A)]; square, open circle and triangle, levels of mRNA for RPL28, RPL7 and RPL3, respectively [as in (B)]. Nomenclature of RP genes follows Mager et al. (18).
The rap1-17 mutation leads to attenuation of transcriptional repression of rDNA as well as that of RP genes due to a secretory defect
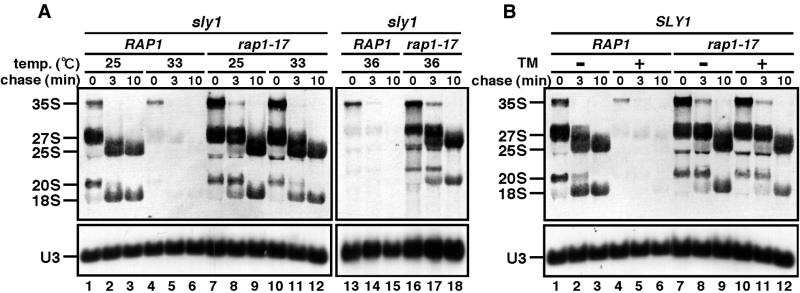
We previously reported that the C-terminal silencing domain of Rap1p is required for transcriptional repression of RP genes in response to a secretory defect. This is true for both Rap1p-dependent and -independent RP genes, suggesting that the Rap1p-binding element of the RP gene upstream is not required (6). Thus, we examined whether the rap1-17 mutation, a C-terminally truncated allele of RAP1, had any effect on the repression of rDNA as well. As shown in Figure 2A, a secretory defect by shifting a temperature-sensitive sly1 mutant, which is defective in endoplasmic reticulum-to-Golgi trafficking (3,19), to the restrictive temperature led to strong repression of rDNA transcription (lanes 4–6 and 13–15), consistent with the previous data (3). On the other hand, in rap1-17 mutant cells, a secretory defect did not cause repression of rDNA transcription (lanes 10–12 and 16–18), indicating that the rap1-17 mutation also affects the signaling pathway from a secretory defect to the repression of rDNA transcription. A similar result was obtained after inhibiting the secretory pathway with tunicamycin (Fig. 2B), indicating that the effect of rap1-17 is not specific to the sly1 allele.
Figure 2.
The rap1-17 mutation affects the transcriptional repression of rDNA due to a secretory response. (A) Strains KM011 (sly1 RAP1, lanes1–6 and 13–15) and KM013 (sly1 rap1-17, lanes 7–12 and 16–18) were grown to log phase (OD600 = 0.5–0.6) in SC–Met medium at 25°C. One-third of the culture was shifted to 33 or 36°C for 90 min. (B) Strains KM014 (SLY1 RAP1, lanes 1–6) and KM016 (SLY1 rap1-17, lanes 7–12) were grown in SC–Met medium at 25°C. Half of the culture was treated with tunicamycin at a final concentration of 1 µg/ml for 4 h at 25°C. Each culture was pulsed with [methyl-3H]methionine and chased with non-radioactive methionine. Samples were taken at the time of addition of cold methionine (t = 0) and after a chase time of 3 or 10 min to prepare total RNA.
Protein concentration of neither wild-type nor C-truncated form of Rap1p changes due to a secretory defect
In order to compare the expression between wild-type and mutant Rap1p when the secretory pathway is blocked, three tandem copies of a sequence encoding an epitope from the influenza virus HA protein was inserted just after the initial codon of RAP1 or rap1-17. Each of the fused genes was subcloned into a CEN-plasmid, pRS313. The plasmid containing either RAP1-HA or rap1-17-HA could complement the lethality of the rap1 null mutation (data not shown). The HA-tagged wild-type protein and C-truncated protein, designated Rap1-HA and rap1-17-HA, were detected as 104- and 87-kDa bands on SDS–polyacrylamide gel, respectively (data not shown). These are considerably larger than the predicted molecular weights of the proteins as previously described for Rap1p without tag (6). The protein level of neither Rap1-HA nor rap1-17-HA changed in response to a secretory defect, indicating that the effect of the rap1-17 mutation was not due to a change in concentration of the products (data not shown). It is noteworthy that the stability of the truncated protein appears to be similar to that of the wild-type protein.
The rap1-2 mutation has only a weak effect on transcriptional repression of both rRNA and RP genes due to a secretory defect
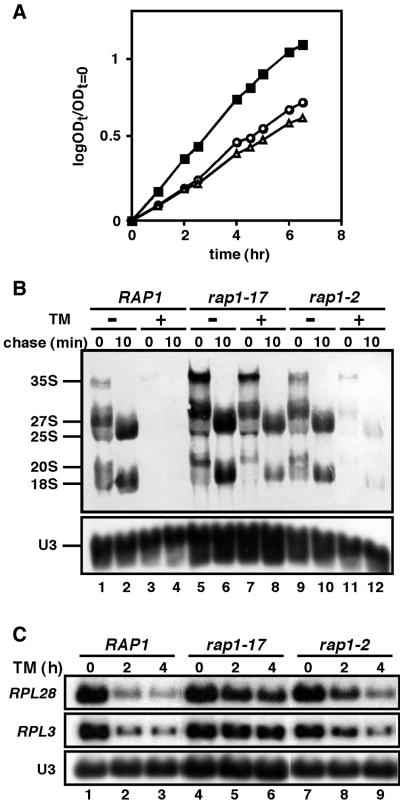
To determine whether the effect of the rap1 mutation is specific to the rap1-17 allele or not, we used the rap1-2 mutant, in which DNA-binding activity was heat labile, leading to a temperature-sensitive phenotype (20). The rap1-2 allele has a missense mutation, A458T. Although the rap1-17 cells are not temperature-sensitive, they grow more slowly than wild-type cells. We set up experimental conditions under which the two kinds of rap1 mutant cells have similar growth defect. As at 33°C, rap1-2 and rap1-17 strains grew at a similar rate (Fig. 3A) and thus the cells were cultured at 33°C to examine the secretory response. Induction of a secretory defect by addition of tunicamycin shows that, compared to the effect of the rap1-17 allele, the rap1-2 allele had only a weak effect on the repression of both rDNA and RP gene transcription in response to a secretory defect (Fig. 3B and C). These results suggest that the C-terminal region of Rap1p is important for the signaling.
Figure 3.
The rap1-2 allele has only weak effect on the secretory response. Strains KM014 (RAP1, square), KM016 (rap1-17, triangle), and KM309 (rap1-2, circle) were grown in SC–Met medium at 30°C overnight, diluted and shifted to 33°C. (A) Growth curves at 33°C. (B) [Methyl-3H]methionine pulse-chase analysis before and after the secretory block. The cells were grown for 2.5 h at 33°C and half of the culture was treated with 2.5 µg/ml tunicamycin for 4 h. Each culture was pulsed for 3 min with [methyl-3H]methionine and chased with non-radioactive methionine. Samples were taken at the time of addition of cold methionine (t = 0) and after a chase time of 10 min to prepare total RNA. (C) Northern analysis before and after the secretory block. The cells were grown for 2.5 h at 33°C and half of the culture was treated with 2.5 µg/ml tunicamycin. Samples were taken to prepare total RNA.
The rap1-17 allele does not affect transcriptional regulation of RP genes in response to N-starvation
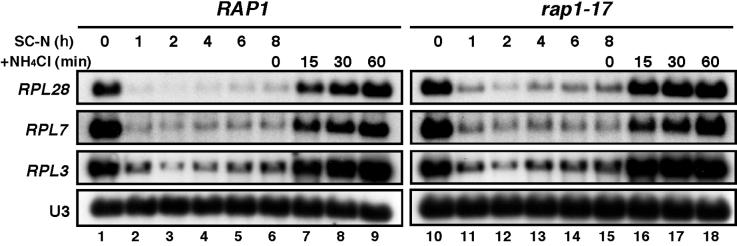
We previously demonstrated that the rap1-17 allele affects the secretory response but not heat shock response (6). As it is known that ribosome synthesis is also regulated by limitation and re-addition of nitrogen source (21), we next examined the effect of rap1-17 on N-starvation and ammonium chloride addition to a N-starved cells. As shown in Figure 4, the rap1-17 allele had little effect on the repression of RP gene transcription in response to N-starvation as well as induction by addition of ammonium chloride. These results suggest that the signaling pathway in response to a secretory defect is distinct from the nitrogen regulation.
Figure 4.
The rap1-17 allele has little effect on N-starvation response. Strains KM328 (RAP1) and KM329 (rap1-17) were grown to OD600 = 0.3–0.4 in SC medium at 25°C, harvested by filtration and resuspended in SC–N medium. The culture was incubated at 25°C until >95% of the cells were unbudded (for 8 h), at which time ammonium chloride was added to a concentration of 5 g/l. Samples were taken 1, 2, 4, 6 and 8 h after shifting of the cells to SC–N medium prior and at 15, 30 and 60 min after nitrogen addition to prepare total RNA. Northern blots were hybridized to the probes specific for ribosomal proteins (RPL28, RPL7 and RPL3) or snoRNA U3.
DISCUSSION
As a large number of ribosomes are synthesized in growing cells, its regulation is essential for the normal growth of the cells, especially in microorganisms (22). Indeed, ribosome biosynthesis in yeast is regulated in response to various kinds of environmental changes such as carbon-source upshift (23,24), mild heat shock (25,26), amino acid starvation (27), nitrogen limitation (21) and a secretory defect (3,28). Recently we described that Rrs1p, which was an essential nuclear protein required for pre-rRNA processing/ribosomal subunit assembly, had an important role in the signaling due to a secretory defect (5). This suggests that pre-rRNA processing/ribosomal subunit assembly is related to regulation of rRNA synthesis. Fath et al. (29) isolated nucleolar subcomplexes active for both transcription and processing of rRNA and proposed the idea that synthesis and processing of rRNA are coupled. The idea is consistent with our model and suggests that transcriptional repression of rRNA genes might be a primary event in secretory signaling. This urged us to determine the timing of these two sets of transcriptional repression. rRNA synthesis measured by [methyl-3H]methionine pulse declined apparently earlier than mRNA level of RP genes by 15–30 min (Fig. 1). Considering that the half-life of RP mRNA is 5–9 min (30), it is estimated that transcription of rRNA synthesis is repressed at the same time or slightly earlier than that of RP genes.
Most RP genes are driven by Rap1-binding site(s) (31). A few RP genes including RPL3 have no site for Rap1, but have a single Abf1-binding site instead (32–34). Nevertheless, the transcription of all the RP genes appears to be coordinated in the several experimental conditions. A secretory defect also causes the coordinated repression of RP genes (3). We previously showed that transcriptional repression of both RPL28 (Rap1-driven) and RPL3 (Abf1-driven) was diminished by the C-terminally truncated rap1-17 mutation (6). Furthermore, not only the Rap1p binding sites, but also promoter proximal regions of a RP gene can mediate repression of transcription in response to a secretory defect (30). These results suggest that Rap1p is essential for the repression in response to a secretory defect, but that Rap1p-binding sites are not necessary as cis-acting elements for repression. In this paper, we have demonstrated that the rap1-17 mutation attenuated transcriptional repression of both genes encoding RP and rRNA. It is interesting that different classes of promoters were defined by their transcriptional requirement for TATA-binding protein (TBP)-associated factors (TAFs) and many RP genes were classified to have TAF-dependent promoter (35).
It has been suggested that transcriptional repression of pol I, II and III genes due to a secretory defect requires the cell integrity pathway including the Wsc family of putative plasma membrane sensors and protein kinase C (Pkc1p) (4,36). We have shown that RAP1 is required for the repression of transcription of both rRNA and RP genes. In contrast, it was reported that the rap1-17 mutation did not affect repression of transcription of pol III genes (4), suggesting that the signaling pathway due to a secretory defect diverges downstream of Pkc1p. Also, the target of Pkc1p in this signaling pathway remains to be elucidated; it was suggested that the signaling pathway was not mediated by the downstream mitogen-activated protein kinase cascade (36). Rap1p is one of the candidates that are phosphorylated by Pkc1p in response to a secretory defect; it was suggested that phosphorylation might affect its binding to and/or transcriptional activity of Rap1p (37,38). However, simple modification of Rap1 activity cannot explain the repression of RP transcription, because not only RP genes but also other genes such as glycolytic genes are controlled by Rap1p and because RP genes without Rap1 sites upstream are repressed in response to a secretory defect as mentioned above.
The rap1-2 allele had only a weak effect on the repression of rDNA transcription due to a secretory defect, suggesting that the C-terminal silencing domain of Rap1p is important for repression (Fig. 3). Rap1p has a direct role in silencing at HM mating-type loci and telomeres. A balance between silencing at HM loci and telomeres is maintained by telomere length and by interactions between the C-terminal domain of Rap1p and Sir-proteins (39). The rap1-17 allele affects silencing of HM loci and telomere length. As Sir-proteins appeared not to be implicated in this repression (30), it is unlikely that a defect in interaction between Rap1p and Sir-proteins in rap1-17 is responsible for its effect on the repression. However, we cannot deny the possibility that some factor whose activity or localization is influenced by the status of the telomeres may be involved in the repression.
There is increasing evidence for acetylation/deacetylation of histones on conserved lysine residues within N-terminal tails modifying activities to specific promoters. Esa1p, essential histone acetylase in yeast, is the catalytic subunit of the NuA4 (nucleosome acetyl-transferase of histone H4) complex that acetylates histone H4 and H2A (40). It has recently been reported that Esa1p is specifically recruited to RP promoters, suggesting that Esa1p recruitment to RP promoters correlates with coordinate regulation of RP genes in response to growth stimuli. Specifically, the decrease in RP transcription that occurs under condition of heat shock or amino acid starvation is accompanied by a corresponding decrease in Esa1p occupancy at RP promoters (41). It remains to be seen whether Esa1p is implicated in the secretory response.
The detailed mechanism of transcriptional repression of both rRNA and RP genes in response to a secretory defect remains to be elucidated. However, we showed that regulation of rRNA transcription is important in the mechanism and that Rap1p has an essential role. As far as we know, this is the first demonstration of the role of Rap1p on Pol I transcription.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Shore (University of Geneva) and H. Uemura for providing the plasmid and E. Tsuchiya, D. Hirata and H. Uemura for valuable discussion. We are grateful to J. R. Warner for critical reading of the manuscript. This research was supported by grants from the Ministry of Education, Science and Culture of Japan and Special Coordination Funds for Promoting Science and Technology of the Science and Technology Agency of the Japanese Government.
References
- 1.Woolford J.L.,Jr and Warner,J.R. (1991) The ribosome and its synthesis. In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 587–626.
- 2.Eisen M.B., Spellmen,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuta K. and Warner,J.R. (1994) Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol., 14, 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Moir,R.D., Sethy-Coraci,I.K., Warner,J.R. and Willis,I.M. (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol., 20, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuno A., Miyoshi,K., Tsujii,R., Miyakawa,T. and Mizuta,K. (2000) RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuta K., Tsujii,R., Warner,J.R. and Nishiyama,M. (1998) The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res., 26, 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shore D. (1994) RAP1: a protein regulator in yeast. Trends Genet., 10, 408–412. [DOI] [PubMed] [Google Scholar]
- 8.Kyrion G., Boakye,K.A. and Lustig,A.J. (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Mao,X. and Lustig,A.J. (1994) Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics, 138, 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L. and Morse,R.H. (1999) Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol ., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takita Y., Ohya,Y. and Anraku,Y. (1995) The CLS2 gene encodes a protein with multiple membrane-spanning domains that is important Ca2+ tolerance in yeast. Mol. Gen. Genet., 246, 269–281. [DOI] [PubMed] [Google Scholar]
- 14.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng F.J. and Warner,J.R. (1991) Structural basis for the regulation of splicing of a yeast messenger RNA. Cell, 65, 797–804. [DOI] [PubMed] [Google Scholar]
- 16.Tsujii R., Miyoshi,K., Tsuno,A., Matsui,Y., Toh-e,A., Miyakawa,T. and Mizuta,K. (2000) Ebp2p, yeast homologue of a human protein that interacts with Epstein–Barr virus Nuclear Antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells, 5, 543–553. [DOI] [PubMed] [Google Scholar]
- 17.Udem S.A. and Warner,J.R. (1972) Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol., 65, 227–242. [DOI] [PubMed] [Google Scholar]
- 18.Mager W.H., Planta,R.J., Ballesta,J.-P.G., Lee,J.C., Mizuta,K., Suzuki,K., Warner,J.R. and Woolford,J. (1997) A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res., 25, 4872–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ossig R., Dascher,C., Trepte,H.-H., Schmitt,H.D. and Gallwitz,D. (1991) The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol. Cell. Biol., 11, 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurtz S. and Shore,D. (1991) RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5, 616–628. [DOI] [PubMed] [Google Scholar]
- 21.Neuman-Silberberg F.S., Bhattacharya,S. and Broach,J.R. (1995) Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell. Biol., 15, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- 23.Kief D.R. and Warner,J.R. (1981) Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol., 1, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herruer M.H., Mager,W.H., Woudt,L.P., Nieuwint,R.T., Wassenaar,G.M., Groeneveld,P. and Planta,R.J. (1987) Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res., 15, 10133–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herruer M.H., Mager,W.H., Raué,H.A., Vreken,P., Wilms,E. and Planta,R.J. (1988) Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res., 16, 7917–7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim C.H. and Warner,J.R. (1983) Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol. Cell. Biol., 3, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner J.R. and Gorenstein,C. (1978) Yeast has a true stringent response. Nature, 275, 338–339. [DOI] [PubMed] [Google Scholar]
- 28.Li B. and Warner,J.R. (1996) Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J. Biol. Chem., 271, 16813–16819. [DOI] [PubMed] [Google Scholar]
- 29.Fath S., Milkereit,P., Podtelejnikov,A.V., Bischler,N., Schultz,P., Bier,M., Mann,M. and Tschochner,H. (2000) Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J. Cell Biol., 149, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B., Nierras,C.R. and Warner,J.R. (1999) Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol., 19, 5393–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lascaris R.F., Mager,W.H. and Planta,R.J. (1999) DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics, 15, 267–277. [DOI] [PubMed] [Google Scholar]
- 32.Dorsman J.C., Doorenbosch,M.M., Maurer,C.T.C., de Winde,J.H., Mager,W.H., Planta,R.J. and Grivell,L.A. (1989) An ARS/silencer binding factor also activates two ribosomal protein genes in yeast. Nucleic Acids Res., 17, 4917–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamil K.G., Nam,H.G. and Fried,H.M. (1988) Constitutive transcription of yeast ribosomal protein gene TCM1 is promoted by uncommon cis- and trans-acting elements. Mol. Cell. Biol., 8, 4328–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herruer M.H., Mager,W.H., Doorenbosch,T.M., Wessels,P.L.M., Wassenaar,T.M. and Planta,R.J. (1989) The extended promoter of the gene encoding ribosomal protein S33 in yeast consists of multiple protein binding elements. Nucleic Acids Res., 17, 7427–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuras L., Kosa,P., Mencia,M. and Struhl,K. (2000) TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science, 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- 36.Nierras C.R. and Warner,J.R. (1999) Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274, 13235–13241. [DOI] [PubMed] [Google Scholar]
- 37.Tsang J.S.H., Henry,Y.A.L., Chambers,A., Kingsman,A.J. and Kingsman,S.M. (1990) Phosphorylation influences the binding of the yeast RAP1 protein to the upstream activating sequence of the PGK gene. Nucleic Acids Res., 18, 7331–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein C. and Struhl,K. (1994) Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol., 14, 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck S.W. and Shore,D. (1995) Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev., 9, 370–384. [DOI] [PubMed] [Google Scholar]
- 40.Allard S., Utley,R.T., Savard,J., Clarke,A., Grant,P., Brandl,C.J., Pillus,L., Workman,J.L. and Cote,J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATF-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid J.L., Iyer,V.R., Brown,P.O. and Struhl,K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell, 6, 1297–1307. [DOI] [PubMed] [Google Scholar]