ABSTRACT
One of the first identified and best-studied toxin-antitoxin (TA) systems in Escherichia coli is the F-plasmid-based CcdAB system. This system is involved in plasmid maintenance through postsegregational killing. More recently, ccdAB homologs have been found on the chromosome, including in pathogenic strains of E. coli and other bacteria. However, the functional role of chromosomal ccdAB genes, if any, has remained unclear. We show that both the native ccd operon of the E. coli O157 strain (ccdO157) and the ccd operon from the F plasmid (ccdF), when inserted on the E. coli chromosome, lead to protection from cell death under multiple antibiotic stress conditions through formation of persisters, with the O157 operon showing higher protection. While the plasmid-encoded CcdB toxin is a potent gyrase inhibitor and leads to bacterial cell death even under fully repressed conditions, the chromosomally encoded toxin leads to growth inhibition, except at high expression levels, where some cell death is seen. This was further confirmed by transiently activating the chromosomal ccd operon through overexpression of an active-site inactive mutant of F-plasmid-encoded CcdB. Both the ccdF and ccdO157 operons may share common mechanisms for activation under stress conditions, eventually leading to multidrug-tolerant persister cells. This study clearly demonstrates an important role for chromosomal ccd systems in bacterial persistence.
IMPORTANCE A large number of free-living and pathogenic bacteria are known to harbor multiple toxin-antitoxin systems, on plasmids as well as on chromosomes. The F-plasmid CcdAB system has been extensively studied and is known to be involved in plasmid maintenance. However, little is known about the function of its chromosomal counterpart, found in several pathogenic E. coli strains. We show that the native chromosomal ccd operon of the E. coli O157 strain is involved in drug tolerance and confers protection from cell death under multiple antibiotic stress conditions. This has implications for generation of potential therapeutics that target these TA systems and has clinical significance because the presence of persisters in an antibiotic-treated population can lead to resuscitation of chronic infection and may contribute to failure of antibiotic treatment.
KEYWORDS: toxin-antitoxin, persistence
INTRODUCTION
In the early 1940s it was reported that a small fraction of staphylococcal cells, about one in a million, survived prolonged penicillin treatment (1). These cells were termed persisters. Later it was established that persisters are phenotypic variants in a given population that show antibiotic tolerance due to probable differences in their physiology under a given set of conditions. Upon subsequent culturing, they give rise to a population that retains antibiotic sensitivity. They are different from resistant cells in which resistance is genetically encoded and is passed on from one generation to the next. In contrast, persisters are transient and rare, ranging from 10−2 to 10−6 in frequency. After their initial discovery by Joseph Bigger, various studies have probed the conditions and genes involved in the generation of persisters (2–4). Exposure to two different classes of antibiotics, namely, beta-lactams and fluoroquinolones, has been shown to produce a fraction of persister cells as large as 10−1 in the case of hip mutants (5). HipAB belongs to the family of toxin-antitoxin (TA) systems, with HipB being the antitoxin for the toxin HipA (6).
Many bacterial genomes harbor a class of genes referred to as TA genes. These genes were initially discovered on low-copy-number plasmids such as the F plasmid of Escherichia coli, and they are known to be involved in plasmid maintenance in the bacterial population by a process known as postsegregational killing (7). TA systems comprise of a pair of genes organized in an operon encoding a stable toxin and a labile antitoxin that antagonizes it (7). In most cases, the first gene encodes an antitoxin, while the second gene encodes a toxin that targets an essential cellular function. The antitoxin usually acts as a transcriptional repressor by binding to operator sites within its own promoter. Therefore, transcription of both genes is typically autoregulated by their encoded protein products. This negative feedback loop helps in maintaining steady-state levels of both toxin and antitoxin such that the toxin/antitoxin ratio is <1 (8). Under conditions of stress, the labile antitoxin is degraded. The resulting increase in the toxin/antitoxin ratio leads to transient derepression of the operon and fresh synthesis of both antitoxin and toxin (9). In the case of the ccdAB system on F-plasmid-containing cells, the toxin CcdB is expressed along with its unstable antidote, CcdA, forming a TA complex. In cells lacking the F plasmid, CcdA is degraded, and in the absence of fresh CcdA synthesis, CcdB kills the cell, ensuring F-plasmid maintenance. Homologs of plasmid-based TA systems have recently been discovered on the chromosomes of a large number of bacteria, many of which are pathogenic (10).
The biological function of TA loci when present on a plasmid is often clear, but the roles of many chromosomal TA loci are unclear. There are different models that have been proposed to explain their presence on the chromosome. For the chromosomal ccd systems, these include the antiaddiction model (11), which states that the chromosomally encoded ccd system (ccdEch) protects the cell against postsegregational killing mediated by its F-plasmid ccd (ccdF) homolog, whereas for the ccdO157 system, it has been suggested that this system is devoid of any biological role in the E. coli species (12). Models proposed for other chromosomal TA systems include the programmed cell death (PCD) model, the growth modulation model, the persistence model, and the development model (13). TA loci have been proposed as general stress response elements in prokaryotes (14–17). Despite a plethora of studies on TA systems, in most cases their functions remain to be elucidated. A common difficulty encountered during study of a specific TA system is functional redundancy with other TA systems present in the same organism. Hence, study of the in vivo role of a particular TA locus requires the selective activation of that specific locus in the background of other TA loci. Ectopic overexpression of the active, toxin component often leads to cell death, further complicating elucidation of TA system function. Since many bacteria contain multiple homologous TA systems with redundant functions, multiple TA systems may need to be knocked out before there is an observable phenotype (18).
Various chromosomal TA systems as well as stress-responsive pathways in bacteria have been shown to be overexpressed in transcriptomic studies done on isolated persister cells. The pathogenic E. coli O157:H7 strain harbors the ccd operon in its genome. A global analysis of the transcriptional response to nutritional stress in this strain (19) revealed an increased expression of the dinJ-yafQ, chpBS, and ccdAB TA loci upon entry into stationary phase, indicating that these systems are actively transcribed in the stationary phase (19). However, their contribution to cell survival still remains to be elucidated.
Most chromosomal TA systems are thought to be acquired by horizontal transmission from their plasmidic counterparts, and there is conservation of both target and antitoxin binding residues in the chromosomal CcdB toxin relative to its F-plasmid counterpart. It was recently shown that the ccd operon of the F plasmid can also function as a transmissible persistence factor (20). Hence, it becomes important to test whether the ccd operon in the chromosomal context has a role to play in bacterial drug tolerance that is similar to or enhanced compared to that of its F-plasmid counterpart.
This issue is clinically significant, as the presence of persisters in an antibiotic-treated population can lead to resuscitation of chronic infection and may contribute to failure of antibiotic treatment (2, 21). It has been previously demonstrated that prolonged antimicrobial therapy selects for high-persistence variants. Given that many of these TA systems are found on the chromosomes of pathogenic strains of E. coli, it is important to study the contribution of these chromosomally encoded TA systems toward persister generation under antibiotic stress conditions. This can have implications for generation of potential therapeutics that target these TA systems. We have therefore probed the role of the chromosomally encoded CcdAB TA system. We found that both the native ccdO157 and the F-plasmid-carried ccd operon, when in a chromosomal context, confer protection from cell death under antibiotic stress, with the ccdO157 chromosomal form showing better protection and weaker DNA gyrase poisoning activity.
RESULTS
Activation of ccd of E. coli O157 by heat or antibiotic stress.
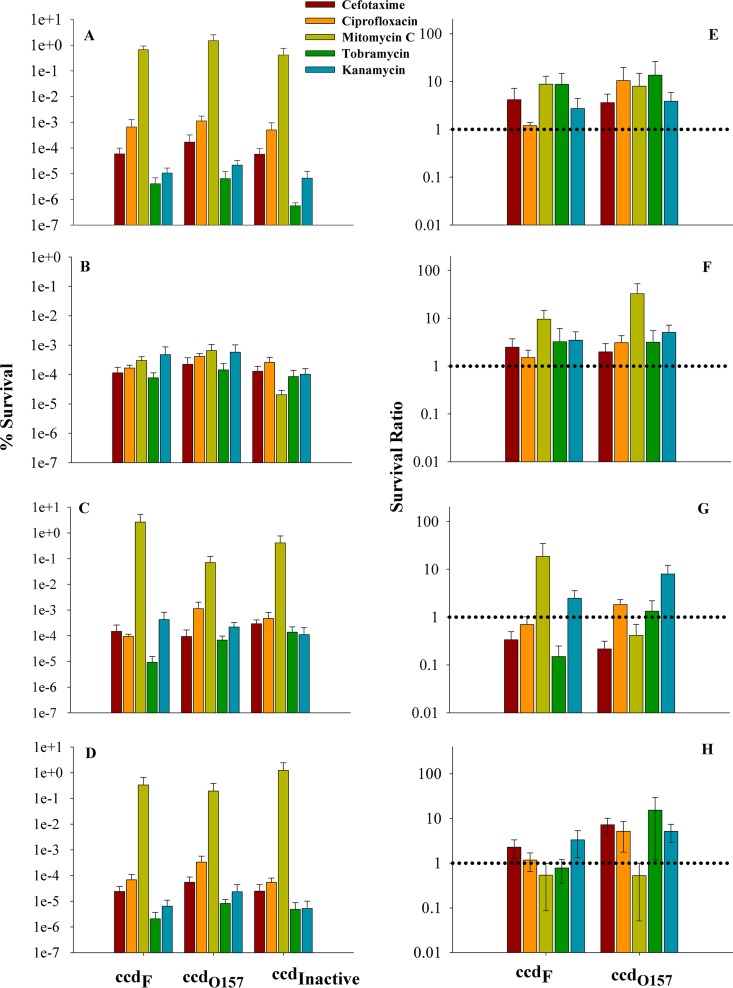
A DNA fragment containing the complete ccd operon from pathogenic E. coli O157:H7 was synthesized and inserted into the 77-bp intergenic region between the folA and apaH genes of E. coli strain BW25113 using lambda red recombination (22). We chose the Escherichia coli BW25113 strain because it is the parent strain of the Keio collection with a known genotype [F− Δ(araD-araB)567 lacZ4787(Δ)::rrnB3 LAM− rph1 Δ(rhaD-rhaB)568 hsdR514] and the complete genome is sequenced (23). Bioinformatics analysis has shown that most known chromosomal ccd operons are located in this locus in the E. coli chromosome. The resulting strains were BW25113 ccdO157 (contains the ccd operon from E. coli O157:H7 strain), BW25113 ccdF (contains the F-plasmid-borne ccd operon), and BW25113 ccdInactive (control strain containing the inactive, truncated ccd operon from a serogroup O51 E. coli O157:H7 strain). All three strains were first exposed to a prestress that involved either heat (48°C for 20 mins) or a sublethal dose of ampicillin (2.5 μg/ml for 1 h) or ciprofloxacin (0.008 μg/ml for 1 h). Multidrug-tolerant persisters are known to increase upon exposure to sublethal levels of antibiotic, at least in case of the clinically relevant bacterium Staphylococcus aureus (24). The prestress was followed by exposure of the cells to different antibiotics at lethal doses of 10× the MIC (cefotaxime, 100 μg/ml; ciprofloxacin, 0.4 μg/ml; mitomycin C, 10 μg/ml; kanamycin, 50 μg/ml; and tobramycin, 25 μg/ml) for 4 h. We used these antibiotics because prior studies have shown that persisters isolated under different conditions show various degrees of tolerance to different antibiotics, but typically only 2 or 3 antibiotics were examined (18, 25). In order to carry out a more comprehensive study we used multiple different antibiotics while studying the contribution of the chromosomal ccdAB operon to bacterial drug tolerance. We took at least one representative from each major class of antibiotics, based on their mechanism of action: kanamycin and tobramycin as aminoglycoside-type bactericidal antibiotics; cefotaxime, a member of the cephalosporin class of antibiotics that act on actively growing cells by inhibiting cell wall synthesis; mitomycin C, which selectively inhibits DNA synthesis; and ciprofloxacin, which is a fluoroquinolone and inhibits DNA gyrase, similarly to the CcdB toxin. Cells were then washed, plated on LB medium, and grown for 14 to 18 h in order to count the number of viable colonies. Figure 1A to D show the percent survival of the E. coli strains BW25113 ccdO157, BW25113 ccdF, and BW25113 ccdInactive in the presence of heat, a sublethal dose of ampicillin or ciprofloxacin, and without prestress, respectively. The survival ratios shown in Fig. 1E to H for the respective prestresses for the E. coli strains BW25113 ccdO157 and BW25113 ccdF were calculated as percent survival of the strain containing active ccd/percent survival of the reference strain containing inactive truncated ccd (BW25113 ccdInactive). A survival ratio of greater than one indicates that protection from the antibiotic is due to specific expression of the Ccd toxin. There was a general nonspecific increase in survival for all three strains upon exposure to prestress. When the survivals of strains BW25113 ccdO157 and BW25113 ccdF were compared with that of BW25113 ccdInactive, 2- to 10-fold higher survival was seen for both BW25113 ccdO157 and BW25113 ccdF (Fig. 1). Survival of BW25113 ccdO157 was higher than that of BW25113 ccdF against cefotaxime, ciprofloxacin, and tobramycin (Fig. 1), especially under conditions of heat prestress. When ciprofloxacin prestress was given, no enhancement in survival relative to that of BW25113 ccdInactive was seen, except toward mitomycin C (15-fold) in strain BW25113 ccdF. Surprisingly a similar protection from antibiotic killing was seen even in the absence of a prestress for the ccdO157 strain (Fig. 1D and H). These data show that persisters surviving treatment with different antibiotics are not identical and may be induced by different mechanisms. The difference in the survival ratios upon different antibiotic stresses could potentially indicate the shutdown of only selected metabolic processes that are targets for different antibiotics, for example, cell wall synthesis for ampicillin and cefotaxime. A similar experiment was carried out in the E. coli MG1655 strain. The plasmids with the ccd operons cloned in them (see Table S2 in the supplemental material) were individually transformed into the MG1655 strain, and relative survival was monitored under different antibiotic stress conditions. Results similar to those with the BW25113 strain were obtained (see Fig. S1 in the supplemental material).
FIG 1.
Survival of E. coli strains containing the ccd operon upon antibiotic exposure. The BW25113 strains BW25113 ccdO157, BW25113 ccdF, and BW25113 ccdInactive were first exposed to a prestress that involved either heat or a sublethal dose of ampicillin or ciprofloxacin. Survival of these strains was monitored in the presence of various antibiotics. Percent survival was calculated for each strain as the ratio of CFU after antibiotic exposure to CFU prior to antibiotic exposure. The survival ratio was calculated as the ratio of the percent survival of a strain containing active ccd to the percent survival of the reference strain containing inactive truncated ccd (BW25113 ccdInactive). (A to D) Percent survival of E. coli strains BW25113 ccdO157, BW25113 ccdF, and BW25113 ccdInactive in the presence of heat or a sublethal dose of ampicillin or ciprofloxacin or without prestress, respectively. (E to H) Survival ratios of E. coli strains BW25113 ccdO157 and BW25113 ccdF in the presence of heat or a sublethal dose of ampicillin or ciprofloxacin or without prestress, respectively. The y axis is shown on a log scale, and error bars indicate the standard error from 4 independent experiments. Each experiment was done using three biological replicates. The survival ratio for strain BW25113 ccdO157 in the absence of prestress is higher than that for strain BW25113 ccdF against cefotaxime, ciprofloxacin, tobramycin, respectively. Survival of both strains BW25113 ccdO157 and BW25113 ccdF is greater than that of strain BW25113 ccdInactive for most antibiotics, as indicated by the dotted lines.
Estimating cell survival using flow cytometry.
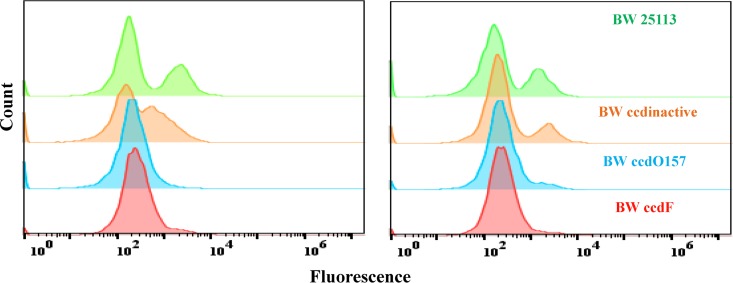
In addition to the viable cell count on plates, we verified our results on cell survival using a flow cytometry-based approach. The forward scatter (FSC)-side scatter (SSC) plot gives a measure of cell size and granularity. Differences in these parameters can be seen between live and dead cells. Log-phase cultures were subjected to antibiotic stress followed by flow cytometric analysis. Further, the cells were stained with propidium iodide (PI) after antibiotic treatment to measure the relative levels of cell survival for different strains. This dye is known to stain dead cells due to increased selective permeability of dead cells but is generally excluded by persisters (26). A larger fraction of cells showed fluorescence due to PI staining in the case of both strains BW25113 and BW25113 ccdInactive than in the case of either BW25113 ccdF or BW25113 ccdO157 cells (Fig. 2; see Table S4 in the supplemental material).
FIG 2.
Monitoring cell survival following antibiotic exposure using flow cytometry. Cells were subjected to antibiotic stress with 25 μg/ml tobramycin (left panel) and 100 μg/ml cefotaxime (right panel) for 4 h. Cells were pelleted, washed in saline, and grown in the presence of 0.01% propidium iodide (PI) solution for 2 h for staining of dead cells. PI fluorescence for a given volume of cells was recorded in a BDAccuri flow cytometer. A rightward shift in the population shows that a larger fraction of cells showed fluorescence due to PI staining in the case of both BW25113 and BW25113 ccdInactive than in the case of either BW25113 ccdF or BW25113 ccdO157.
Monitoring growth kinetics and assessing metabolic activity.
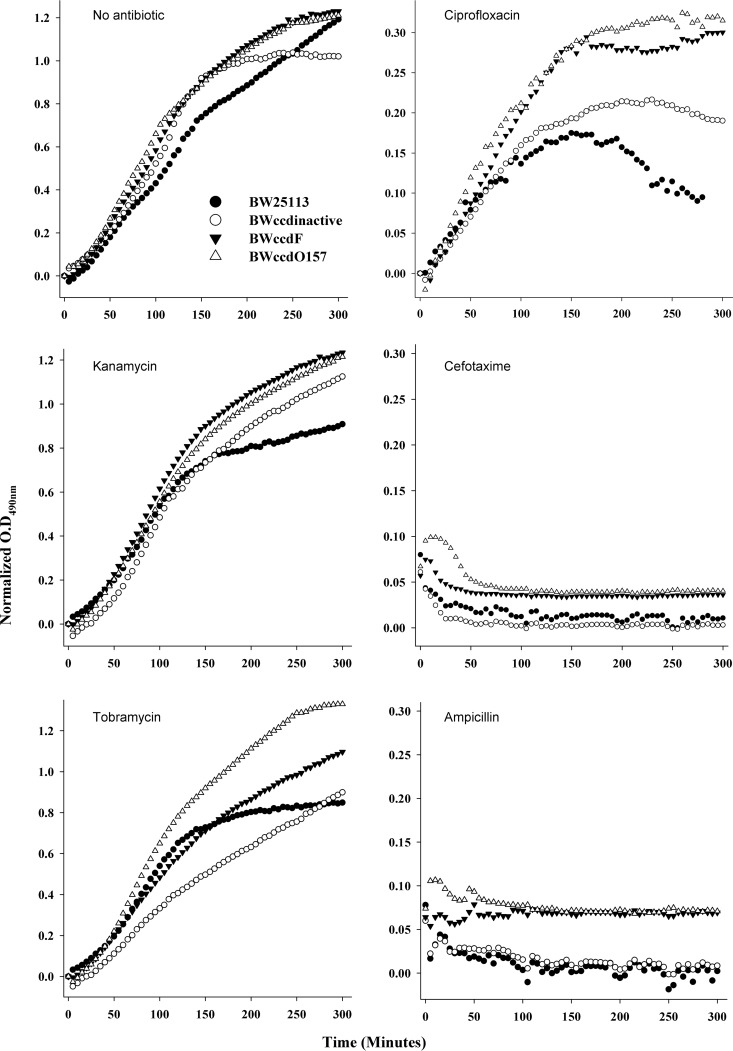
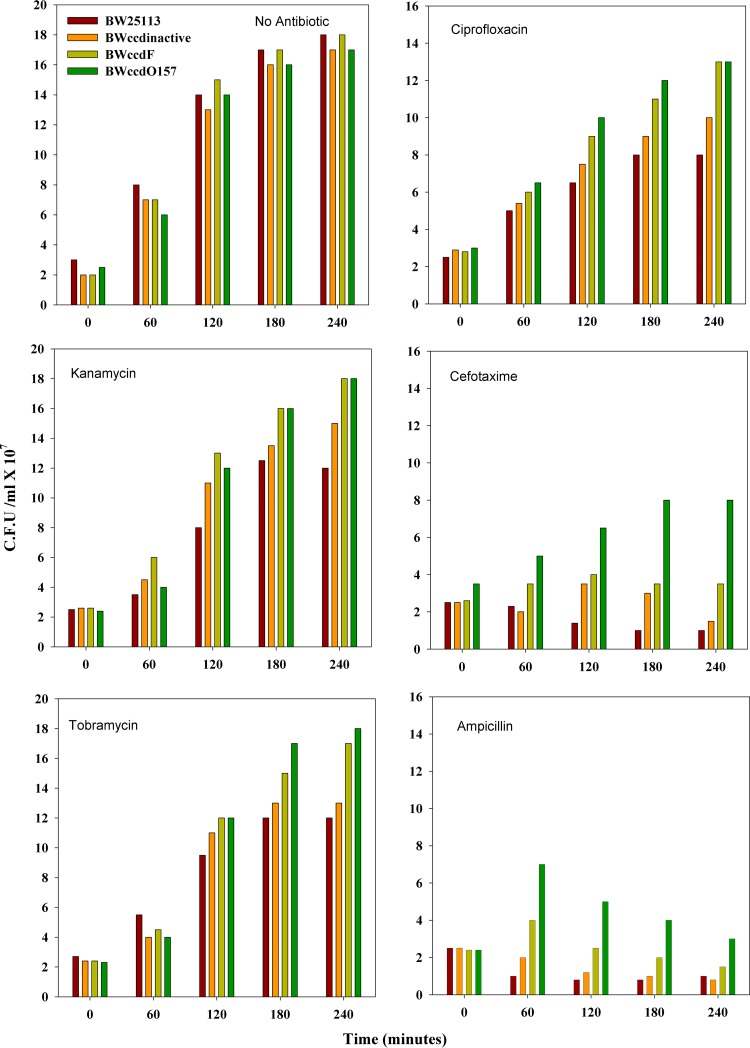
Most persister cells that are tolerant to antibiotics are metabolically active, but there are known exceptions (27, 28). Many may exhibit biphasic killing (29). The second phase arises due to killing of the colonies formed from persister cells in cases where the persister cells continue to divide but at a rate lower than normal. We employed the 5-cyano-2,3,-ditolyl tetrazolium chloride (CTC) redox dye to monitor growth and assess the metabolic activity of the different strains spectrophotometrically (30). The optical density measured at 490 nm (OD490) was plotted against time to follow the growth kinetics. In the absence of antibiotic, all the strains showed comparable growth, although the wild-type (WT) BW25113 strain attained the stationary phase earlier than the other strains (Fig. 3). The aminoglycoside-type bactericidal antibiotics kanamycin and tobramycin, which act by inhibiting protein synthesis, did not cause much reduction in growth, although both the BW25113 and BW25113 ccdInactive strains showed lower growth rates than the BW25113 ccdF and BW25113 ccdO157 strains. Lower cell death with these antibiotics may be due to the use of comparatively low concentrations (50 μg/ml and 25 μg/ml for kanamycin and tobramycin, respectively). Cefotaxime, a member of the cephalosporin class of antibiotics, and ampicillin, which is in the penicillin group of beta-lactam antibiotics, both showed typical killing curves for cultures treated with antibiotic. Both act on actively growing cells by inhibiting cell wall synthesis. In both cases, phenotypically sensitive cells showed marked cell death within 1 h of antibiotic treatment, and a plateau of persister cells was observed. Both ccd operon-containing strains showed similar protection, higher than that for the two control strains. Upon treatment with ciprofloxacin, strains containing the ccd operon showed better growth than the control strains, and the cells are metabolically more active than after treatment with other antibiotics (Fig. 3). Overall, the ccdO157-containing cells showed growth in liquid culture similar to or better than that of ccdF-containing cells under most antibiotic stress conditions. Similar results were obtained when we monitored growth using measurements of OD600 (see Fig. S2 in the supplemental material) or with viable cell counts (Fig. 4).
FIG 3.
Growth kinetics and metabolic activity of different strains under antibiotic stress. Mid-log-phase cells were grown in either the absence or presence of various antibiotics (cefotaxime, 100 μg/ml; ciprofloxacin, 0.4 μg/ml; ampicillin, 10 μg/ml; kanamycin, 50 μg/ml; and tobramycin, 25 μg/ml) in a 96-well microtiter plate, and growth was monitored using the absorbance of tetrazolium dye (0.02%) at 490 nm. Growth kinetics were monitored for 5 h, and the OD was plotted against time. The OD490 was normalized to zero with respect to the first reading. The average readings from three independent replicates were plotted. Both strains BW25113 ccdF and BW25113 ccdO157 show protection from cell death upon antibiotic exposure compared to the control strains, BW25113 ccdInactive and BW25113.
FIG 4.
Viable cell counts for different strains under antibiotic stress. Mid-log-phase cells were grown in either the absence or presence of various antibiotics (cefotaxime, 100 μg/ml; ciprofloxacin, 0.4 μg/ml; ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and tobramycin, 25 μg/ml) in a 96-well microtiter plate. Viable cell counts were measured at intervals of 30 min for 5 h and plotted against time. The average readings from three independent replicates are shown. For clarity, only data at hourly intervals are presented.
Low-level expression of CcdBO157 leads to reversible growth inhibition.
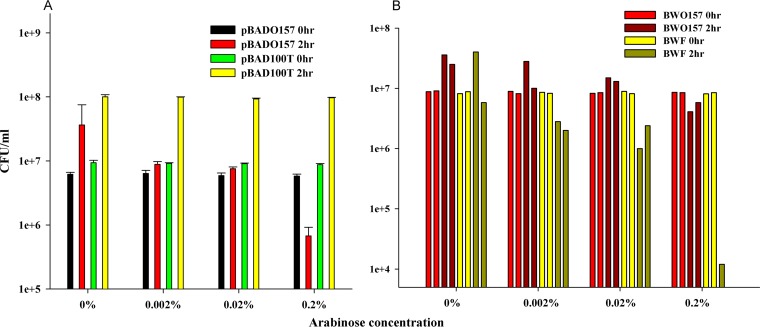
We have previously shown (31) that even very low levels of CcdBF expression, for example, under highly repressed conditions (0.2% glucose) under the control of the PBAD promoter, lead to cell death in sensitive E. coli strains such as BW25113. In contrast, we show here that low levels of CcdBO157 expression under identical conditions do not cause significant cell death (Fig. 5A). A previous study has shown that the ccdF system is able to mediate postsegregational killing in an E. coli strain harboring the ccdO157 system on its chromosome. This shows that the plasmid ccdF system is functional even in the presence of its chromosomal counterpart (32). This requires that the CcdAO157 antitoxin either binds weakly or does not bind to the CcdBF toxin. To explore this further, we employed a previously described methodology (20) to specifically activate a given TA system in the background of others using an active-site inactive mutant toxin. We observed that overexpression of the active-site inactive mutant toxin, CcdBF G100T, is nontoxic in cells lacking WT CcdB but leads to cell death at very low levels of induction (0.002%) when transformed in cells having the ccdF operon inserted in the chromosome (Fig. 5). This is expected, as the binding of G100T to the cognate antitoxin CcdAF releases WT CcdBF, which then binds to gyrase, causing cell death. Interestingly, we also saw reduced cell survival at higher levels of induction of the mutant when transformed in cells having the ccdO157 operon inserted in the chromosome (Fig. 5B). This shows that there is weak cross activity between CcdBF and CcdAO157. CcdBF can bind to CcdAO157 but likely with reduced affinity relative to CcdAF, as inferred from the inducer levels at which cells start to die. This is in contrast to a previous study where CcdBF and CcdAO157 were shown to be noninteracting when overexpressed from two different heterologous promoters (32). This is possibly because in the latter case, the CcdAO157 antitoxin is unable to neutralize overexpressed CcdBF completely, and even a small amount of CcdBF can bind to gyrase and cause cell death. In contrast, in our experiments, we scored the expression and binding of CcdBO157 toxin in its native context. Also, we controlled the expression of the active-site inactive mutant by varying the inducer and repressor levels. This finding also demonstrates that exposure of cells to low levels of CcdBO157 causes growth inhibition rather than cell death.
FIG 5.
Low-level expression of CcdBO157 leads to reversible growth inhibition. (A) Plasmids pBAD24ccdBF, pBAD24ccdBO157, and pBAD24ccdB100T were individually transformed in E. coli strain BW25113 and grown in LB medium containing 0.2% glucose. Cells were induced with various concentrations of arabinose at an OD600 of 0.2. The x axis indicates various concentrations of arabinose used for transient expression of the construct. Induced cells were grown for 2 h, washed twice with LB medium, diluted, and plated on 0.2% glucose-containing medium to repress further expression. CcdBF expression leads to complete cell death, even under highly repressed conditions. CcdBO157 leads to cell death at very high expression levels (0.2% arabinose) but leads to growth inhibition when the expression is low (0.002% to 0.02% arabinose). The y axis is shown on a log scale. Results from two independent experiments are plotted as two different bars. Overexpression of the G100T mutant is nontoxic to E. coli Top10 cells. WT CcdBF is toxic to cells even under highly repressed conditions, and no transformants are obtained under any condition. (B) To score the expression of ccdBO157 from its own promoter, the active-site inactive mutant of CcdBF, G100T, was transformed into the E. coli BW25113 ccdF (having chromosomal CcdF) and BW25113 ccdO157 (having chromosomal CcdF) strains individually. Expression of G100T was induced with various concentrations of arabinose (as described above) at an OD600 of 0.2. Cells were grown for 2 h, washed twice, diluted, and plated on 0.2% glucose-containing medium to repress further expression. Overexpressed G100T will titrate out cellular CcdA, leading to release of WT CcdB as well as derepression of the ccdAB operon. The reduction in cell survival at higher levels of induction (0.2% arabinose) of the mutant in BW25113 ccdO157 cells shows that there is cross activity between the CcdBF and CcdAO157 TA systems. Low expression levels of the mutant (0.002% to 0.02% arabinose) result in reduced toxicity and cause growth inhibition.
Estimates of the relative activities of CcdBF and CcdBO157.
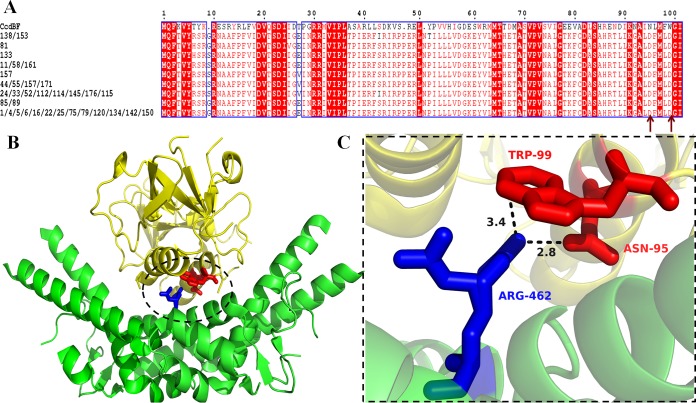
CcdBF, when expressed from the F plasmid, and CcdBO157, when expressed from the E. coli chromosome, bind to the same target, gyrase A. However, they have been implicated in different functional roles (12, 32, 33). While the F-plasmid-carried ccd operon is involved in plasmid maintenance by postsegregational killing, no clear role has been attributed to the chromosomal ccd counterpart. To explore the molecular mechanisms involved in the possibly differential functions of CcdBF and CcdBO157, we monitored their activities in vivo as well as in vitro. The in vivo results showed that CcdBO157 is much less efficient than CcdBF in causing cell death, as can be seen from significant growth under inducing conditions for CcdBO157 and no growth even under highly repressed conditions for CcdBF (Fig. 5A). In vitro gyrase binding of CcdBF and CcdBO157 using surface plasmon resonance (SPR) showed that CcdBO157 has 5-fold-lower affinity for its target, GyrA (see Fig. S3 and Table S3 in the supplemental material). Although the dissociation rates for the two proteins were somewhat similar, there was a significant decrease in the association rates. These results are consistent with the experimentally observed lower toxicity of CcdBO157 in vivo. Sequence comparison of the two proteins showed that though most of the residues known to be involved in gyrase binding are conserved, two important interacting residues are not conserved. Analysis of the CcdBF-gyrase bound structure (PDB no. 1X75) indicated that substitutions N95D and W99D observed in CcdBO157 are likely to disrupt gyrase binding (Fig. 6). Our previous study involving saturation mutagenesis on the CcdBF protein also indicates that both the N95D and W99D mutations have an inactive phenotype (34).
FIG 6.
Sequence and structural basis for differential activity of CcdBF and CcdBO157. (A) Sequence alignment of CcdBF and CcdBO157 serotypes. Sequenced variants of CcdBO157 from 32 different isolates, which tested positive for toxicity (12), were aligned with the CcdBF sequence using the Clustal Omega multiple-sequence alignment tool. Residues that are fully conserved either are active-site residues or form a part of the hydrophobic core (34). (B) CcdBF-gyrase A14 bound crystal structure (PDB no. 1X75) (40). The key region of the interaction between CcdBF and gyrase A14 is circled. (C) Residues in the interaction between CcdB (in red) and gyrase A14 (in blue) that are not identical in the CcdBF and CcdBO157 sequences (marked with arrows in the multiple-sequence alignment in panel A). CcdBO157 contains the mutations N95D and W99D relative to CcdF.
DISCUSSION
One of the first identified and best-studied toxin-antitoxin systems in E. coli is the F-plasmid-based CcdAB system. This is involved in plasmid maintenance through postsegregational killing. More recently, ccdAB homologs have been found on chromosomes, including those in pathogenic strains of E. coli and other bacteria. However, their functional role, if any, has remained unclear. We show that both the native ccd operon of the E. coli O157 strain and the ccd operon from the F plasmid, when inserted on the E. coli chromosome, lead to protection from cell death under multiple antibiotic stress conditions through formation of persisters. The frequency of these persisters ranges from 10−1 (for antibiotics such as mitomycin) to <10−5 (for tobramycin). Under many antibiotic stress conditions, the frequency of persisters is increased up to 10-fold compared to that for the isogenic strain having an inactive ccd operon at the same locus in the BW25113 genome, showing that the protection from antibiotics is due to specific expression of the Ccd toxin (Fig. 1). Strains with chromosomal ccdO157 show greater drug tolerance, particularly in the presence of heat stress and even in the absence of prestress, conditions that are probably more physiologically relevant. Both the ccdF and ccdO157 operons may share common mechanisms for activation under stress conditions and also display weak cross activation. CcdBO157 has weaker affinity for GyrA than the plasmidic counterpart but can still cause some cell death. This suggests that inhibitors of the CcdO157 toxin-antitoxin interaction can cause cell death and therefore that the system might represent a drug target. Alternatively, inhibitors of CcdO157 might also decrease the fraction of persisters, which could have clinical significance.
The ccdAB system when present on the F plasmid has a role in plasmid maintenance. In F-plasmid-containing cells, CcdB is expressed along with an excess of its unstable antidote, CcdA. Hence, CcdB is present almost entirely as a complex with CcdA. In cells lacking the F plasmid, CcdA is degraded, and in the absence of fresh CcdA synthesis, CcdB kills the cell. This process is known as postsegregational killing. There are other reports which suggest activation of the ccd operon during nutritional stress (35). Homologs of the ccd operon have been discovered on the chromosomes of a variety of bacterial strains, including many pathogenic strains such as those of Shigella dysenteriae, E. coli O157:H7, and Vibrio cholerae. The chromosomal ccd operon cloned on a plasmid shows a lower degree of postsegregational killing than ccdABF (32). When the ccd operon present on the F plasmid is activated by stress, such as heat or a sublethal dose of antibiotics, an increased frequency of drug-tolerant persisters is observed (20). A previous study of E. coli O157:H7 suggests that CcdBO157 is also active and that when it is expressed alone in the absence of its cognate antidote, CcdA, it causes cell death by poisoning DNA gyrase (32). The ccd operon in this bacterium is located in an intergenic region between two metabolic genes, folA and apaH, which encode dihydrofolate reductase and diadenosine tetraphosphatase, respectively. This operon is absent from the E. coli K-12 strains, and a 77-bp intergenic region is present at the same locus. When a bioinformatics analysis was carried out on various serogroups of this strain, it was suggested that the intergenic region between the folA and apaH genes is a site for transposon insertion. The promoter activity of the CcdO157 system was found to be lower than that of the CcdF system, although at the protein level, some of the crucial active-site residues involved in target binding were found to be conserved (32).
One model that was proposed to explain the presence of TA modules on the genomes of bacteria is the stabilization module; i.e., chromosomally encoded TA modules could act against large-scale deletion of neighboring genomic regions. Given that many of these ccd TA modules have been consistently found to be integrated between the repeat regions spanned by the folA and apaH metabolic genes, this could also be one explanation for the role of the ccdAB operon on the chromosome.
The antiaddiction model suggests that the presence of TA modules on the chromosome which can cross-react with a plasmidic or phage TA module could potentially give selective advantage to the plasmid-free cells by inhibiting postsegregational killing of cells that lose the plasmid (11). Mutations that are able to evade the antiaddiction will be selected by the plasmidic TA system, and vice versa for the chromosomally encoded TA system. Analysis of the CcdAB complex structure (PDB no. 3HPW) (36) indicates that the presence of an N69Y mutation in CcdAO157 with respect to CcdAF and of a Y8R mutation in CcdBO157 with respect to CcdBF (see Fig. S4 in the supplemental material) will likely hamper the binding of the noncognate Ccd toxin and antitoxin. This implies that the chromosomal ccd operon would be unable to cause a strong antiaddiction phenotype in the presence of the plasmidic F plasmid, consistent with previous experimental data (32).
In the present study, we found that there is weak cross talk between the plasmidic toxin and chromosomal antitoxin, since overexpression of the active-site inactive CcdF toxin mutant causes cell death in CcdO157-containing cells. A previous study indicated that chromosomally carried ccd operons undergo a decay process within the E. coli species and therefore are likely to lose their antiaddictive property, with about 29% of the strains harboring an inactive toxin (12). The present work suggests a probable explanation for the presence of an active ccd operon in more than 69% of the E. coli strains studied, namely, that they functionally integrate into the host regulatory network, leading to enhanced tolerance to multiple antibiotics through the formation of persisters. It is probable that the chromosomal TA systems, such as the ccd operon, are acquired from the plasmidic counterparts and that mutations that lead to lower toxicity but increased drug tolerance have been selected for in the course of evolution.
The ccd operon of the F plasmid can function as a transmissible persistence factor (20). Under stress conditions, the antitoxins of multiple TA systems undergo degradation by cellular proteases such as Lon, leading to activation of multiple TA systems, which eventually leads to multidrug tolerance, as previously suggested (20). CcdB poisons the gyrase-DNA complex, blocking the passage of polymerases and leading to double-strand breaks. Additionally, the GyrA subunit has also been found as an inactive binary complex with CcdB when overexpressed (37). Although lethal effects of CcdB are probably due to poisoning of the gyrase-DNA complex, the inactivation pathway may lead to growth inhibition and prevent cell death, as is possible in the case of CcdBO157, which shows lower toxicity in vivo. Under conditions where there is specific activation of the CcdAB system, the SOS response to DNA damage caused by the CcdB toxin may be the specific trigger for activation of other TA systems. This response is mediated through the DNA repair enzyme RecA and Lon protease. It is very likely that similar pathways operate when the ccd operon in placed in a chromosomal context, leading to enhanced persister generation, as previously shown (20) for the plasmidic CcdF TA system. Future experiments aimed at exploring the molecular mechanisms underlying the drug-tolerant phenotype of E. coli cells carrying the ccd operon in the chromosome would further aid in designing targeted therapeutics against such antibiotic-tolerant persisters.
MATERIALS AND METHODS
Construction of the BW25113 ccdO157, BW25113 ccdInactive, and BW25113 ccdF strains.
The ccd operons of E. coli strain O157:H7 (based on reference strain EDL933) and the F plasmid were both synthesized at GenScript. To enable insertion of the operon at a specific locus in the E. coli genome, these constructs had homology at the 5′ end with sequences downstream of the folA gene of the E. coli K-12 chromosome and at the 3′ end with a cat gene from a pKD3 plasmid. The synthesized fragments were named ccdO157 and ccdF, respectively. As a control, the ccd operon from serogroup O51 of strain O157:H7 was also synthesized at GenScript and named ccdInactive The ccdInactive operon has a truncated ccdB gene, resulting in an inactive CcdB protein, although the antitoxin sequence and the regulatory regions are intact. To construct ccd-cat fragments for all synthesized ccd operons for the purpose of selection, a cat gene containing FLP recombination target (FRT) sites was amplified separately from a pKD3 plasmid by using the cat (for2) and apaH (rev3) primers, respectively. This fragment has homology at the 3′ end with upstream sequences of the apaH gene of the E. coli K-12 chromosome. All three ccd operons were fused with the PCR-amplified cat gene by overlap PCR. Each ccd-cat cassette has sequences at the 5′ and 3′ ends that are similar to the downstream and upstream sequences of the folA and apaH genes, respectively. The three final constructs, ccdO157-cat, ccdF-cat, and ccdInactive-cat, were inserted at a 77-bp intergenic region between the folA and apaH genes of an E. coli strain K-12 locus in E. coli strain BW25113 using lambda red recombination (38). A list of all strains and plasmids used in this study is provided in Table S1 in the supplemental material. Sequences for all of these constructs are listed in Table S2 in the supplemental material. Recombinants were screened on chloramphenicol (25 μg/ml)-containing LB plates, and the presence of the ccd operon was confirmed by both PCR and DNA sequencing. The cat gene was later removed by transforming each strain individually with plasmid pCP20 expressing the FRT-FLP recombinase, as described previously (38).
Activation of the ccd operon by heat or antibiotic stress.
Overnight cultures of E. coli strains BW25113 ccdO157, BW25113 ccdF, and BW25113 ccdInactive were diluted 100-fold and grown to an OD600 of 0.3. Subsequently, cells were subjected to three different types of sublethal prestresses, namely, exposure of cells to a high temperature (48°C for 20 min) or to a sublethal dose of either ampicillin (2.5 μg/ml for 1 h) or ciprofloxacin (0.008 μg/ml for 1 h). This was followed by exposure to lethal doses of various antibiotics with different modes of action (cefotaxime, 100 μg/ml; ciprofloxacin, 0.4 μg/ml; mitomycin C, 10 μg/ml; kanamycin, 50 μg/ml; and tobramycin, 25 μg/ml) for 4 h with growth under shaking conditions (180 rpm) at 37°C. Similar antibiotic concentrations were used for all experiments unless otherwise specified. The cultures were washed twice, appropriately diluted, and plated on LB agar plates at 37°C to determine the viable counts after antibiotic treatment, and the percent survival for each E. coli strain was calculated. Percent survival was defined as the ratio of CFU after antibiotic exposure to CFU prior to antibiotic exposure. For the calculation of the survival ratio with respect to the control strain, the percent survival of the ccd strain (containing the ccd operon from either the E. coli O157:H7 strain or the F plasmid) was normalized to the percent survival of a reference strain containing the truncated ccd operon, ccdInactive.
Estimating cell survival using flow cytometry.
Primary cultures were inoculated in LB medium and grown overnight. Thirty microliters of the overnight culture was inoculated in 3 ml of LB and grown for 2 h to an OD600 of 0.4. Cells were subjected to antibiotic stresses, namely, 25 μg/ml tobramycin and 100 μg/ml cefotaxime individually for 4 h. Three milliliters of cell culture was pelleted, washed in saline, and grown in the presence of 0.01% propidium iodide (PI) solution for 2 h for staining of dead cells. One milliliter of cell culture was again pelleted and washed with saline, and PI fluorescence for a 100-μl volume of cells was recorded in a BDAccuri flow cytometer using the FL-4 channel.
Monitoring growth kinetics and assessing metabolic activity.
Two hundred microliters of mid-log-phase cells (OD600 of 0.4) was grown in the presence of various antibiotics (as mentioned above) in a microtiter plate, and growth was monitored for 5 h by conventional OD600 measurements as well as by using 5-cyano-2,3,-ditolyl tetrazolium chloride (CTC) redox dye (0.02%), which upon formation of formazan reduces to give a purple color having an absorption maximum at 490 nm (30). OD measurements at both 490 nm and 600 nm were made at an interval of every 5 min using a SpectraMax Plus microplate spectrophotometer. In addition, this OD490 is indicative of the survival of the bacterial population as a function of time of exposure to a given antibiotic. Five microliters of sample was taken from each of the wells every 30 min and plated onto LB medium at three different dilutions to obtain the viable cell count.
Cloning and expression of CcdBF and CcdBO157 to monitor their relative activities. (i) Cloning, expression, and purification.
The ccdB genes (both ccdBF and ccdBO157) were cloned under the control of the PBAD promoter in plasmid pBAD24 as described previously (39). pBADCcdBF and pBADCcdBO157 clones were individually transformed into the E. coli CSH501 strain, which is resistant to the action of CcdB. This strain harbors the gyrA462 mutation in its chromosomal DNA, which prevents gyrase from binding to CcdB (33). A 200-ml cell culture in LB was induced with 0.2% (wt/vol) arabinose at an OD600 of 0.6 and grown for 8 h at 25°C. Cells were harvested by centrifugation at 1,800 × g for 10 min at 4°C. The pellet was resuspended in HEG buffer (10 mM HEPES, 50 mM EDTA, 10% glycerol), pH 7.4, and sonicated, followed by centrifugation at 11,000 × g for 30 min at 4°C. The supernatant was loaded onto a GyrA14 affinity column (3-ml bed volume) and incubated at 4°C for 4 h, followed by removal of unbound proteins by washing the column with coupling buffer (5 times the bed volume). Elution of CcdB was carried out with 0.2 M glycine (pH 2.5) into a tube containing an equal volume of 400 mM HEPES (pH 8.5) at 4°C. The eluted fractions were subjected to 15% SDS-PAGE, and the concentration was determined.
(ii) In vitro gyrase binding by SPR.
Binding studies were performed with a ProteOn XPR 36 system (Bio-Rad). GyrA14 was immobilized onto the ProteOn GLC chip surface by standard amine coupling. A sensor surface (without GyrA14) that had been activated and deactivated served as a negative control for each binding interaction. Three different concentrations of CcdB were passed across each sensor surface in running buffer (phosphate-buffered saline [PBS; pH 7.4] containing 0.01% P20 surfactant). Protein concentrations ranged from 50 nM to 500 nM. Both binding and dissociation were measured at a flow rate of 60 μl/min. In all cases, the sensor surface was regenerated between binding reactions with 4 M MgCl2. Each binding curve was corrected for nonspecific binding by subtraction of the signal obtained with the channel where only buffer instead of the analyte was passed. The kinetic parameters were obtained by fitting the data to a simple 1:1 Langmuir interaction model.
(iii) In vivo activity estimation.
E. coli strain BW25113 was individually transformed with the pBADCcdBF, pBADCcdBO157, and pBADCcdB100T (control) plasmids. Cells were transiently induced with various concentrations of arabinose (0%, 0.002%, 0.02%, and 0.2%) at an OD600 of 0.2. The induced cells were grown for 2 h, washed twice, diluted, and plated on 0.2% glucose-containing medium to repress further expression. Activity was assayed by plating the transformation mix on LB-ampicillin plates at 37°C in the presence of the following concentrations of glucose (repressor) or arabinose (inducer): 0.2% glucose, 0.02% glucose, 0.002% glucose, 0% glucose/arabinose, 0.002% arabinose, 0.02% arabinose, and 0.2% arabinose.
Overexpression of a CcdB active-site inactive mutant to probe the specificity and cross-reactivity between the CcdF and CcdO157 gene products.
An active-site inactive mutant of CcdBF (G100T) was cloned under control of a heterologous arabinose-inducible promoter as described previously (31). This mutant cannot bind to the toxin target (gyrase) and is therefore inactive, but it retains binding to the cognate antitoxin, CcdA. Therefore, overexpression of G100T will lead to cell death only if it binds to the chromosomally expressed CcdA antitoxin, thus releasing the WT CcdB toxin, which is capable of binding to the target. To probe the specificity and cross-reactivity between CcdF and CcdO157, the G100T mutant of CcdBF was transformed individually into the BW25113 ccdF and BW25113 ccdO157 strains. A single colony was picked, inoculated in LB-ampicillin medium containing 0.2% glucose, and grown at 37°C overnight. Expression of G100T was induced with various concentrations of arabinose (0%, 0.002%, 0.02%, and 0.2%) at an OD of 0.2 after washing the cells twice with LB medium. Cells were grown for 2 h, washed twice, diluted, and plated on 0.2% glucose-containing medium to repress further expression. Surviving colonies were counted for each strain.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a grant to R.V. from the Department of Biotechnology (grant number NO.BT/COE/34/SP15219/2015, DT.20/11/2015), Government of India. We also acknowledge funding for infrastructural support from the following programs of the Government of India: DST-FIST, UGC Centre for Advanced Study, the Ministry of Human Resource Development (MHRD), and the DBT-IISc Partnership Program.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We acknowledge Sivasankar Devanarayanan for help with FACS data acquisition.
We declare that no competing interests exist.
K.G., A.T., and R.V. designed the experiments. K.G., A.T., A.S., and R.V. performed the experiments. K.G., A.T., and R.V. analyzed the results and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00397-17.
REFERENCES
- 1.Bigger JW. 1944. The bactericidal action of penicillin on staphylococcus pyogenes. Irish J Med Sci (1926-1967) 19:585–595. doi: 10.1007/BF02948462. [DOI] [Google Scholar]
- 2.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 3.Coussens NP, Daines DA. 2016. Wake me when it's over—bacterial toxin-antitoxin proteins and induced dormancy. Exp Biol Med 241:1332–42. doi: 10.1177/1535370216651938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y. 2014. Persisters, persistent infections and the Yin-Yang model. Emerg Microbes Infect 3:e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob Agents Chemother 34:1938–1943. doi: 10.1128/AAC.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 8.Magnuson R, Yarmolinsky MB. 1998. Corepression of the P1 addiction operon by Phd and Doc. J Bacteriol 180:6342–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afif H, Allali N, Couturier M, Van Melderen L. 2001. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol Microbiol 41:73–82. doi: 10.1046/j.1365-2958.2001.02492.x. [DOI] [PubMed] [Google Scholar]
- 10.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saavedra De Bast M, Mine N, Van Melderen L. 2008. Chromosomal toxin-antitoxin systems may act as antiaddiction modules. J Bacteriol 190:4603–4609. doi: 10.1128/JB.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mine N, Guglielmini J, Wilbaux M, Van Melderen L. 2009. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species. Genetics 181:1557–1566. doi: 10.1534/genetics.108.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Melderen L, Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol 46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 16.Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 332:809–819. doi: 10.1016/S0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Bergholz TM, Wick LM, Qi W, Riordan JT, Ouellette LM, Whittam TS. 2007. Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol 7:97. doi: 10.1186/1471-2180-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi A, Dewan PC, Barua B, Varadarajan R. 2012. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc Natl Acad Sci U S A 109:12497–12502. doi: 10.1073/pnas.1121217109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 22.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete Genome Sequence of Escherichia coli BW25113. Genome Announc 2(5):e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechner S, Lewis K, Bertram R. 2012. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J Mol Microbiol Biotechnol 22:235–244. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofsteenge N, van Nimwegen E, Silander OK. 2013. Quantitative analysis of persister fractions suggests different mechanisms of formation among environmental isolates of E. coli. BMC Microbiol 13:25. doi: 10.1186/1471-2180-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leszczynska D, Matuszewska E, Kuczynska-Wisnik D, Furmanek-Blaszk B, Laskowska E. 2013. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS One 8:e54737. doi: 10.1371/journal.pone.0054737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orman MA, Brynildsen MP. 2013. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 29.Canas-Duarte SJ, Restrepo S, Pedraza JM. 2014. Novel protocol for persister cells isolation. PLoS One 9:e88660. doi: 10.1371/journal.pone.0088660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhupathiraju VK, Hernandez M, Landfear D, Alvarez-Cohen L. 1999. Application of a tetrazolium dye as an indicator of viability in anaerobic bacteria. J Microbiol Methods 37:231–243. doi: 10.1016/S0167-7012(99)00069-X. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj K, Dewan PC, Chakrabarti P, Goswami D, Barua B, Baliga C, Varadarajan R. 2008. Structural correlates of the temperature sensitive phenotype derived from saturation mutagenesis studies of CcdB. Biochemistry 47:12964–12973. doi: 10.1021/bi8014345. [DOI] [PubMed] [Google Scholar]
- 32.Wilbaux M, Mine N, Guerout AM, Mazel D, Van Melderen L. 2007. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J Bacteriol 189:2712–2719. doi: 10.1128/JB.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard P, Couturier M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226:735–745. doi: 10.1016/0022-2836(92)90629-X. [DOI] [PubMed] [Google Scholar]
- 34.Tripathi A, Gupta K, Khare S, Jain PC, Patel S, Kumar P, Pulianmackal AJ, Aghera N, Varadarajan R. 2016. Molecular determinants of mutant phenotypes, inferred from saturation mutagenesis data. Mol Biol Evol 33:2960–2975. doi: 10.1093/molbev/msw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre-Ramirez M, Ramirez-Santos J, Van Melderen L, Gomez-Eichelmann MC. 2006. Expression of the F plasmid ccd toxin-antitoxin system in Escherichia coli cells under nutritional stress. Can J Microbiol 52:24–30. doi: 10.1139/w05-107. [DOI] [PubMed] [Google Scholar]
- 36.De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, Loris R. 2009. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell 35:154–163. doi: 10.1016/j.molcel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M. 1999. Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem 274:10936–10944. [DOI] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakshusmathi G, Mondal K, Lakshmi GS, Singh G, Roy A, Ch RB, Madhusudhanan S, Varadarajan R. 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc Natl Acad Sci U S A 101:7925–7930. doi: 10.1073/pnas.0402222101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dao-Thi MH, Van Melderen L, De Genst E, Afif H, Buts L, Wyns L, Loris R. 2005. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 348:1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.