ABSTRACT
The coordination of group behaviors in bacteria is accomplished via the cell-cell signaling process called quorum sensing. Vibrios have historically been models for studying bacterial communication due to the diverse and remarkable behaviors controlled by quorum sensing in these bacteria, including bioluminescence, type III and type VI secretion, biofilm formation, and motility. Here, we discuss the Vibrio LuxR/HapR family of proteins, the master global transcription factors that direct downstream gene expression in response to changes in cell density. These proteins are structurally similar to TetR transcription factors but exhibit distinct biochemical and genetic features from TetR that determine their regulatory influence on the quorum sensing gene network. We review here the gene groups regulated by LuxR/HapR and quorum sensing and explore the targets that are common and unique among Vibrio species.
KEYWORDS: quorum sensing, LuxR, HapR, SmcR, gene regulation, Vibrio, Vibrio cholerae, Vibrio harveyi
INTRODUCTION
Quorum sensing is a type of cell-cell communication used by a wide variety of bacteria. Through the detection of small signaling peptides or molecules termed autoinducers, cells monitor and respond to changes in the surrounding bacterial population and coordinate group behaviors. The first evidence of quorum sensing was the observation that a bacterial phenotype correlated with changes in population density (1–3). We now know that a plethora of bacterial activities are regulated by quorum sensing, ranging from bioluminescence and motility to protease production and toxin secretion (4, 5).
Owing to the easily monitored bioluminescent quorum sensing phenotype of several Vibrio species, vibrios have become major model organisms for studies of bacterial communication. In Gram-negative bacteria, most known quorum sensing gene regulatory systems are controlled through direct binding of an autoinducer to a cytosolic transcription factor. These quorum sensing systems are named LuxI/LuxR systems after the Vibrio fischeri system that was originally discovered (6, 7). The LuxI protein is the autoinducer synthase, and the LuxR protein is the transcriptional regulator that binds autoinducer as a ligand, allowing it to dimerize and bind DNA to control quorum sensing-regulated genes (7–9). LuxI/LuxR proteins are found in a wide variety of Gram-negative bacteria, and many are involved in quorum sensing signaling (1). However, V. fischeri seems to be the exception rather than the rule in the Vibrio genus. Most other vibrios for which quorum sensing has been characterized have systems with membrane-bound autoinducer receptors (Fig. 1) (10–19). These histidine kinase receptors initiate a signaling cascade that culminates in the expression of a TetR-type transcription factor, which regulates quorum sensing genes (20, 21). The members of the Vibrio TetR family of master quorum sensing regulators are highly conserved and include HapR in Vibrio cholerae, SmcR in Vibrio vulnificus, LitR in V. fischeri, OpaR in Vibrio parahaemolyticus, VanT in Vibrio anguillarum, and VtpR in Vibrio tubiashii (22–27). The TetR-type master regulator of quorum sensing genes in Vibrio harveyi was also named LuxR. However, it is structurally, biochemically, and genetically distinct from the V. fischeri LuxR protein that binds an autoinducer as part of the LuxI/LuxR regulatory system (20, 28–31). V. harveyi LuxR and its Vibrio homologs do not require a ligand to dimerize or bind DNA (28, 32, 33). In fact, V. fischeri encodes a LuxR/HapR homolog called LitR in its central quorum sensing system in addition to the LuxI/LuxR system (21, 34). Further, LuxR, HapR, and SmcR can cross-complement activity in the respective strains, thus supporting the grouping of these proteins as functional homologs (22, 35).
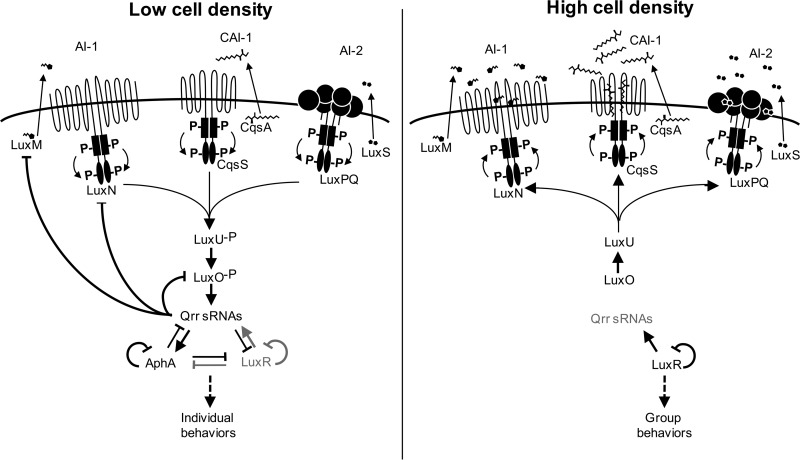
FIG 1.
Model of the V. harveyi quorum sensing system. The autoinducer synthases LuxM, CqsS, and LuxS produce AI-1, CAI-1, and AI-2, respectively. At LCD, autoinducers are at low concentrations in the external environment. LuxO∼P activates qrr gene expression. The Qrr sRNAs activate the expression of AphA and repress the expression of LuxR, LuxM, and LuxO. High levels of AphA and low levels of LuxR together regulate individual behavior genes. AphA autorepresses its expression and feeds back to repress the expression of the qrr genes and luxR. LuxR autorepresses its expression, represses aphA, and activates the qrr genes. At HCD, autoinducer concentrations are high, and AI-1, CAI-1, and AI-2 bind to the LuxN, CqsS, and LuxPQ receptors, respectively. The receptors dephosphorylate LuxU. Thus, LuxU does not phosphorylate LuxO, and the Qrrs are not expressed. LuxR is expressed at high levels, and AphA is not expressed. LuxR regulates group behavior genes. LuxR autorepresses its own expression and feeds back to repress aphA transcription and to activate qrr transcription.
THE VIBRIO QUORUM SENSING PATHWAY CULMINATES IN EXPRESSION OF AphA AND LuxR/HapR
V. harveyi was the first of the vibrios shown to have membrane-bound receptors that bind autoinducers (16). Thus, V. harveyi has become an ideal model organism for studying quorum sensing pathways in other vibrios where similar quorum sensing system architectures have been identified involving membrane-bound autoinducer receptors, small regulatory RNAs (sRNAs), and LuxR homologs acting as master regulators. These include V. cholerae, V. parahaemolyticus, V. vulnificus, and Vibrio alginolyticus (15, 22–24, 34, 36–41). In V. harveyi, the autoinducers are produced and diffuse through the cell membrane (Fig. 1). When Vibrio cells are growing at low cell density (LCD), there is an insufficient concentration of autoinducers in the extracellular environment to bind the membrane receptors (10, 11, 16, 40, 42), and they function as kinases to phosphorylate the phosphotransfer protein LuxU (43). LuxU transfers phosphate to the response regulator LuxO, and phosphorylated LuxO (LuxO∼P) activates the transcription of the sRNAs termed quorum regulatory RNAs (Qrrs) (44, 45). The Qrrs both positively and negatively posttranscriptionally regulate the expression of quorum sensing genes (46). At LCD, translation of the transcription factor AphA is activated by the Qrrs (47), while production of the transcription factor LuxR is negatively regulated by the Qrrs (45). Thus, at LCD, AphA is at its highest level and LuxR is at its lowest level (48). As autoinducers accumulate in the surrounding environment at high cell density (HCD), the membrane-bound receptors bind the autoinducers and switch to acting as phosphatases, removing the phosphates from the regulatory circuit. LuxO is dephosphorylated and the Qrrs are no longer expressed (44). Thus, AphA is no longer produced, and LuxR expression is high (48, 49). LuxR and AphA are the two master transcription factors that control 99% of the quorum sensing regulons: AphA is the LCD regulator, and LuxR is the HCD regulator (47, 48). The remainder of genes are posttranscriptionally controlled by the Qrrs (50). Even at LCD when LuxR levels are lowest, LuxR controls >80 genes and AphA controls >100 genes (48). At HCD, LuxR regulates hundreds of genes.
The roles of LuxR and AphA as the master regulators of quorum sensing gene expression are conserved across the Vibrio genus, even when other factors within the signaling cascade differ (12, 15, 47). For example, V. harveyi has three cognate membrane-bound receptors (Fig. 1), LuxN, LuxPQ, and CqsS, and each binds a specific autoinducer (autoinducer 1 [AI-1], AI-2, and cholera autoinducer 1 [CAI-1], respectively) (41). V. cholerae has four autoinducer receptors that feed into the same circuit to regulate HapR, one of which is unlike canonical Vibrio receptors in that it is not membrane bound (11–13). A new autoinducer-receptor pair was identified in V. cholerae; binding of the autoinducer 3,5-dimethylpyrazin-2-ol (DPO) is required for dimerization and function of the cytosolic transcription factor VqmA, which indirectly inhibits biofilm formation (51). The autoinducer-receptor systems in V. parahaemolyticus are likely similar to those in V. harveyi, based on phylogenetic analyses and signaling assays (10, 14, 16, 42), whereas only the LuxPQ system that detects AI-2 has been identified in V. vulnificus (40). The number of Qrrs in different Vibrio species varies from one to five among vibrios, and they act either additively or redundantly (15, 45, 52–56). Further, the Qrr regulon in V. harveyi (∼20 genes) does not appear to include the type VI secretion genes, which are regulated by the Qrrs in V. cholerae (50, 57).
There are several feedback loops in the quorum sensing circuit (Fig. 1). The Qrrs positively regulate AphA and negatively regulate LuxO, LuxM, and LuxR (46). LuxR represses aphA expression, and AphA represses luxR expression (47, 58). Further, because AphA and LuxR are autorepressors and regulate the transcription of the Qrrs, the concentrations of LuxR and AphA in the cell are highly controlled (29, 45, 59). Collectively, these feedback loops ensure precise expression levels of LuxR and AphA, drive transitions between LCD and HCD, determine the effective range of autoinducer concentrations, and prevent small fluctuations in autoinducer concentration from having a large impact on gene expression (59–62).
AphA, THE LCD MASTER REGULATOR
AphA is the LCD master quorum sensing regulator. It belongs to the MarR family of transcriptional regulators, and like many members of this family, it has a conserved winged helix DNA binding motif at its N terminus. AphA has a unique antiparallel coiled-coil dimerization domain at the C terminus (63, 64). The V. harveyi AphA protein shares 86% identity with V. cholerae and 96% with V. parahaemolyticus and has a consensus binding sequence of ATATGCAN6TGCATAT (65). In addition to positive regulation by the Qrrs, V. cholerae AphA expression is activated by binding of two transcriptional regulators, Lrp and VpsR, to the aphA promoter (47, 66). AphA is induced by high cyclic di-GMP levels in V. cholerae through binding of cyclic di-GMP to VpsR, which impacts the expression of downstream genes (67). Importantly, AphA gene regulation is critical for pathogenesis (64, 68). At the V. cholerae tcpPH operon, AphA directly interacts with coactivator AphB to positively regulate the expression of cholera toxin and the toxin-coregulated pilus to initiate the virulence cascade (69). In V. harveyi, AphA directly represses the expression of aphA, luxR, and the qrr genes (47). Although it is not known whether an AphA interaction with AphB is required for regulation of all these genes, at least for qrr4 repression, AphB is not required (47). Further, quorum sensing regulatory genes have not been found to be part of the AphB regulon under conditions that have been tested (47, 70). In V. harveyi, AphA controls 170 genes at LCD, and among these are >40 type III secretion system (T3SS) genes that are repressed by AphA (47, 48). Repression of T3SS genes at LCD by AphA and at HCD by LuxR results in the expression of T3SS genes at mid-cell density, and these genes are upregulated >1,000-fold during infection (48, 71). Thus, AphA plays a critical role in virulence gene regulation at LCD in vibrios.
THE VIBRIO LuxR/HapR FAMILY: DISTINCT TetR PROTEINS
LuxR shares high amino acid identity with other LuxR-type proteins: 71% with HapR (V. cholerae), 96% with OpaR (V. parahaemolyticus), 93% with SmcR (V. vulnificus), and 60% with LitR (V. fischeri). Structural determination of the LuxR homologs HapR and SmcR revealed that these proteins are TetR family transcription factors with a characteristic helix-turn-helix DNA binding motif in the N-terminal domain (Fig. 2A) (32, 33). Although the shared amino acid identity of the Vibrio LuxR homologs and other TetR family proteins is low, a high level of conservation exists at the secondary and tertiary levels. For example, SmcR shares just 27% similarity with the TetR-type regulator from Staphylococcus aureus, QacR, yet it is clear from examining the root mean square deviation (RMSD) values that these proteins are structural orthologs (Fig. 2A).
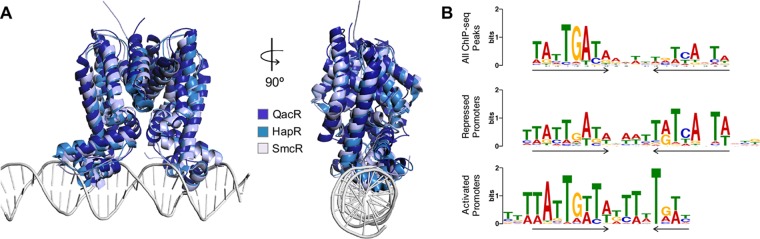
FIG 2.
LuxR/HapR homolog structural and DNA binding properties. (A) Superimposed crystal structures of the TetR proteins QacR, HapR, and SmcR (32, 33, 84). Structures were superimposed with DaliLite (142) and the figures created with PyMOL. The DNA-bound QacR dimer superimposed with HapR or SmcR both resulted in a root mean square deviation (RMSD) of 3.1. Superimposition of HapR to SmcR results in an RMSD of 1.5. HapR is shown in cyan (PDB: 2PBX), SmcR is shown in light blue (PDB: 3KZ9), and for the QacR-DNA structure, QacR is shown in dark blue and the DNA in gray (PDB: 1JT0) (32, 33, 84). (B) LuxR DNA binding motifs from ChIP-seq data grouped by the location of the binding site peak. Top, all LuxR DNA binding peaks from ChIP-seq; middle, peaks in promoters of repressed genes; bottom, peaks in promoters of activated genes. The arrows indicate dyad symmetry in the binding site. Image reproduced with permission (74).
Although LuxR proteins function as dimers and exhibit TetR structural features (Fig. 2A) (72), they are distinct from most other TetR proteins in numerous ways. First, LuxR family proteins act as both activators and repressors, whereas TetR-type proteins typically act only as repressors (22, 47, 49, 72–76). Second, LuxR proteins regulate hundreds of genes (48, 77–79), while TetR-type proteins generally regulate 1 to 2 genes (72). For example, the V. harveyi LuxR regulon is 625 genes, and the V. cholerae HapR regulon is 100 genes (48, 80). Third, LuxR proteins bind to multiple binding sites within the promoter regions of some genes, unlike most TetR-type proteins that have a single binding site per promoter (49, 73, 74, 78, 81). The presence of multiple LuxR binding sites in a single promoter tends to correlate with genes that are activated, but this is not strictly the case. Regarding activated promoters, there are eight LuxR binding sites in the luxCDABE locus, which drives the expression of the bioluminescence genes, and two LuxR binding sites in the betIBA-proXWV promoter, which controls the expression of the osmotic stress genes (82, 83). However, there are two LuxR binding sites in the luxR promoter that are autorepressed by LuxR (29, 31, 82). Based on chromatin immunoprecipitation sequencing (ChIP-seq) data, there are 227 LuxR binding sites in the promoters of 115 genes, yielding an average of ∼2 sites per promoter (74). Of note, some TetR-type proteins, such as QacR, bind cooperatively as dimers of dimers on opposite sides of the DNA helix (84–86). While the mechanism of DNA binding by LuxR proteins is unclear in the absence of a DNA-bound structure, it is possible that LuxR proteins bind DNA in this manner.
A fourth difference between LuxR proteins and typical TetR proteins is in regard to the conservation of the binding site sequence. LuxR proteins bind to a 20- to 22-bp consensus binding motif with dyad symmetry, which is a typical binding site organization and length for TetR proteins (Fig. 2B) (72). However, for each of the Vibrio LuxR-type proteins that has been examined, the palindrome is asymmetric, with a preference for one side of the palindrome (Fig. 2B) (73, 74, 78, 81). The asymmetry of the consensus palindrome is produced from the combination of sites from activated and repressed promoters: repressed binding sites generally retain symmetrical inverted repeats, while activated binding sites contain only half of the palindrome (Fig. 2B) (74). The asymmetric nature of the LuxR binding motif may be an artifact of the locations of the binding sites in activated and repressed promoters. Alternatively, LuxR recognition of various DNA sequences may be connected to the structure of LuxR and possibly to bending of the DNA helix. Some TetR proteins alter DNA structure: TetR induces a 17° bend, whereas QacR bends DNA 3° and widens the DNA major groove (84, 87). Indeed, some of the alignment differences between SmcR/HapR and QacR occur in the helix-turn-helix region (Fig. 2A). This may be due to superimposition of the apo structures of HapR/SmcR onto the DNA-bound structure of QacR. Thus, even structural homologs may interact with DNA in different ways, which remains to be explored for the Vibrio LuxR family of regulators.
Finally, LuxR proteins exhibit various binding affinities for their binding sites, ranging from 0.5 nM to >100 nM (74). The strength of LuxR binding to its consensus binding site is postulated to have a strong impact on the timing of LuxR gene expression (49, 82). Because LuxR expression increases 10-fold in a gradient between LCD and HCD (49, 88), the range of LuxR concentrations at different quorum sensing phases likely influences gene expression at different times during population growth based on the affinity of LuxR for the binding site(s) in promoters. LuxR-type proteins also interact with other proteins to activate transcription via synergistic DNA binding and possibly direct protein-protein interactions (76, 82, 89). LuxR binds DNA synergistically with integration host factor (IHF) in V. harveyi, which is necessary for the precise timing of bioluminescence gene expression during quorum sensing (82). IHF also plays a role in bending DNA at the vvpE promoter in V. vulnificus to facilitate an interaction between SmcR and RNA polymerase for transcription activation (76). These many features of LuxR generate a complex transcription profile: genes are activated and repressed both at LCD and HCD, and the changes range from 2- to 200-fold (48, 77–80, 82).
LuxR/HapR REGULATION OF GROUP BEHAVIORS IN VIBRIOS
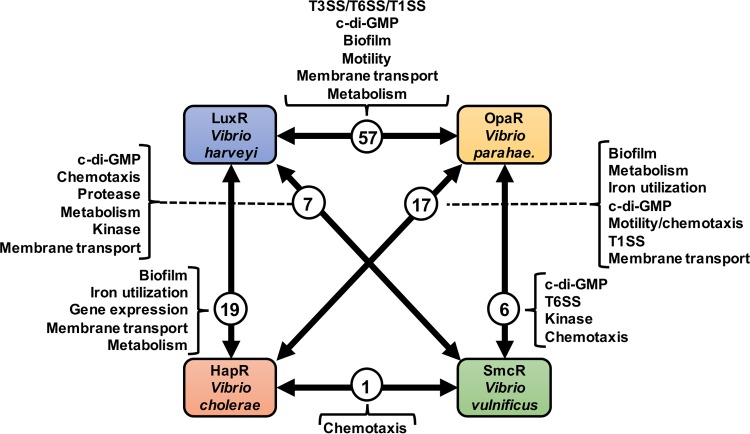
LuxR and its homologs were named according to the genes they were initially discovered to regulate. For example, HapR is the regulator of the hemagglutinin/protease A (hapA) in V. cholerae (22), V. parahaemolyticus OpaR is the regulator of opacity (e.g., capsule production), and LuxR is the regulator of bioluminescence (e.g., lux) (24, 28, 89). However, it has become clear that these proteins control transcription on a global scale, influencing the expression of hundreds of genes (4, 5). The advent of global transcriptomic techniques, like microarrays and RNA sequencing (RNA-seq), uncovered the regulons of LuxR, HapR, SmcR, and OpaR, as well as the entire quorum sensing regulons (including the AphA and sRNA-regulated genes) of V. cholerae and V. harveyi (50, 74, 77–80, 82, 90). These experiments supply a comprehensive view of genes controlled by LuxR homologs, including directly and indirectly regulated genes. As a global transcription factor, LuxR directly regulates 115 promoters out of a total of 625 genes (74). The SmcR regulon experiment identified only direct regulatory targets, of which there were 121 genes (78). Conversely, for HapR and OpaR, only the global regulons (direct and indirect combined) were identified and contain 100 genes and 267 genes, respectively (77, 80). The regulon data for HapR, OpaR, SmcR, and LuxR were collected using different approaches, including microarrays (HapR and LuxR), RNA-seq (LuxR and OpaR), and DNA pulldowns combined with bioinformatics (SmcR). The samples were collected at various cell densities and on different media. However, even though methods to gather these data are quite different, there are striking similarities among the LuxR, HapR, SmcR, and OpaR regulons. We examined these existing regulon data sets to identify genes with similar functions that are regulated by quorum sensing in multiple Vibrio species. The global gene regulons were analyzed for LuxR, HapR, and OpaR, while the SmcR direct regulon was used. Genes with similar function between pairs of Vibrio species were revealed by performing pairwise reciprocal BLAST searches (Fig. 3). Genes were considered “shared” between species if reciprocal protein BLAST searches yield the same best hit. Shared genes can be listed as associated with one pair or with multiple paired comparisons. Clearly, many gene classes are common among quorum sensing regulons, yet some gene groups and modes of regulation are extraordinarily different. Of particular interest are the pathways found in multiple Vibrio LuxR regulons, which include the group behaviors of motility, metabolism, production of public goods, cyclic di-GMP signaling, biofilm production, and secretion.
FIG 3.
Overlapping regulons of Vibrio LuxR-type regulators. The LuxR, HapR, SmcR, and OpaR regulons, which were previously published (77, 78, 80, 82), were analyzed to identify mutual gene constituents via pairwise reciprocal BLAST searches (143). The numbers of homologous genes identified between pairs of regulons are shown in circles and connected by arrows. Genes associated with major cellular processes are listed in the brackets extending from each circled number. Lists are not comprehensive; the paired lists contain genes encoding hypothetical proteins with no annotated function. Vibrio parahae., Vibrio parahaemolyticus.
Motility.
Although regulation of motility and/or chemotaxis is common to Vibrio regulons (Fig. 3), the regulation of these genes is remarkably different among Vibrio species (77, 78, 80, 82). In V. harveyi and V. cholerae, LuxR and HapR activate the expression of swimming motility genes, producing cells that display the highest level of motility at HCD (80, 91). This phenotype is also apparent in V. vulnificus and V. tubiashii, as smcR and vtpR mutants show reduced swimming motility (26, 38). Conversely, in V. parahaemolyticus, quorum sensing appears to modulate motility in the opposite fashion. OpaR inhibits swarming motility by repressing lateral flagellar genes, and AphA activates swimming and swarming behaviors at LCD. Thus, V. parahaemolyticus displays the highest levels of motility at LCD (68, 92). Similarly, motility is repressed by quorum sensing in the more distantly related species V. fischeri (18). In addition, multiple chemotaxis genes are found in the LuxR regulons, and several of them are common between vibrios (77, 78, 80, 82). In V. harveyi, LuxR positively regulates chemotaxis genes, whereas in V. parahaemolyticus and V. cholerae, OpaR and HapR negatively regulate homologous genes. These regulatory differences may be explained by the different niches inhabited by these bacteria. In V. cholerae, evidence suggests that activation of motility genes at HCD via quorum sensing promotes the detachment of V. cholerae from host epithelial cells and propels the bacteria into the intestinal lumen for shedding back into the environment (93). Conversely, in V. fischeri, swimming motility is essential to initiate the symbiosis with Euprymna scolopes (94–96). Following colonization, V. fischeri downregulates motility genes at HCD in a quorum sensing-dependent manner, suggesting that motility is not important during maintenance of the symbiosis (18).
Metabolism.
The expression of several metabolic enzymes is regulated by LuxR homologs (Fig. 3) (77, 80, 82), suggesting a key role for quorum sensing control of metabolism. Indeed, the Qrrs in V. cholerae repress the expression of the AlsSDO pathway, which is important for pyruvate metabolism (97). As the V. cholerae population grows, more pyruvate is fluxed through the AlsSDO pathway, which produces neutral metabolites instead of toxic organic acids, thereby allowing the population to remain stable in stationary phase. Similarly, in V. fischeri, as the population grows, acetate accumulates in the medium, which effectively lowers the local pH and can toxify the environment (98). Quorum sensing via LitR activates the production of acetyl coenzyme A (acetyl-CoA) synthase (Acs), which converts acetate into acetyl-CoA and counteracts the acidification (98). In addition, the quorum sensing network in V. parahaemolyticus represses 64 genes associated with the transport and metabolism of amino acids, carbohydrates, and lipids (99). A ΔluxO mutant exhibits significant fitness defects, while an ΔopaR mutant shows fitness advantages when grown on a variety of different carbon sources (e.g., glucose, gluconate, mannose, ribose, and arabinose). These results suggest that OpaR balances metabolic flux in V. parahaemolyticus in a manner similar to HapR in V. cholerae (99).
Public goods.
Quorum sensing within the Vibrio clade appears to regulate a number of genes associated with importing/exporting shared molecules that benefit the both the producers and nonproducers in the local community, which are termed “public goods” (Fig. 3) (77, 78, 80, 82). In V. harveyi, light production and metalloprotease production are considered public goods and are directly regulated by LuxR (31, 100, 101). Additionally, the metalloproteases VtpA/VtpB and EmpA are activated by VtpR in V. tubiashii and VanT in V. anguillarum, and the alkaline serine protease ProA is activated by LuxRval in V. alginolyticus (25–27). Quorum sensing also represses siderophore production in both V. harveyi and V. vulnificus (102, 103). The production of extracellular polysaccharide (EPS) for biofilms is modulated by quorum sensing and LuxR proteins (see section below) (104), and chitin metabolism genes are controlled at various levels by quorum sensing in V. cholerae (105–107).
Because public goods usually comprise biomolecules that can be utilized by both producers and nonproducers, social “cheaters” can arise within bacterial populations that are at quorum. Cheaters often originate from mutations that yield defective quorum sensing systems, such as in V. cholerae, in which ∼50% of natural isolates contain mutations in hapR (108). These mutants can outcompete wild-type cells in vitro, suggesting that such mutations could easily spread in the environment. A similar effect is observed for V. fischeri litR and luxO mutants (34, 109). However, bacterial populations can police themselves against cheaters via a number of mechanisms, including toxin production and metabolite exclusion (110, 111). In V. harveyi, constitutive cooperators (ΔluxO mutant) and cheaters (ΔluxR mutant) have reduced growth yields or are outcompeted by the wild type under various conditions (112). Thus, a functional quorum sensing system can act as a controlling mechanism for the cooperative expression of global behaviors that are fine-tuned for growth performance.
Cyclic di-GMP signaling.
Quorum sensing in vibrios regulates the production of cyclic di-GMP (c-di-GMP), which is an important intracellular signaling molecule in prokaryotes (54, 113). Levels of c-di-GMP are modulated by two classes of cellular enzymes, diguanylate cyclases (which contain GGDEF motifs) and cyclic diguanylate phosphodiesterases (which contain EAL/HD-GYP motifs) (114, 115). In V. cholerae, V. vulnificus, V. parahaemolyticus, and likely other vibrios, c-di-GMP is critical for coordinating biofilm formation and motility (116–120). LuxR homologs regulate the expression of a number of genes encoding diguanylate cyclases and cyclic diguanylate phosphodiesterases (Fig. 3). However, in V. harveyi and V. parahaemolyticus, some of the genes encoding these enzymes are upregulated, while others are downregulated by quorum sensing. V. cholerae is known to possess 62 genes that encode proteins capable of modulating the concentration of c-di-GMP inside the cell (121), and some of these control motility in opposite manners (122). Intracellular c-di-GMP signaling functions via a high-specificity model, in which individual diguanylate cyclase c-di-GMP production rates and levels drive distinct changes in transcription and downstream phenotypes (e.g., biofilm formation) (123). This is in contrast to a low-specificity model, in which total cellular c-di-GMP concentrations control transcriptional responses. High-specificity c-di-GMP signaling may enable cells to independently control pathways through these segregated c-di-GMP microdomains. Quorum sensing controls the expression of some diguanylate cyclases in vibrios, but not all, which in turn regulate downstream transcription via high-specificity c-di-GMP signaling. This type of network design likely enables fine-tuning of the population-wide quorum sensing response toward specific pathway control via c-di-GMP specificity for phenotypic outputs.
Biofilm production.
A plethora of research has shown that quorum sensing and LuxR-type proteins control the expression of biofilm genes (24, 67, 104, 124–130). Accordingly, multiple genes involved in EPS biosynthesis are common among the LuxR regulons examined (Fig. 3). In V. cholerae, HapR represses the VpsR and VpsT activators of biofilm formation, which results in biofilm formation at LCD and repression of biofilm genes at HCD (113, 121, 124). Conversely, SmcR and OpaR activate biofilm gene expression at HCD (24, 127). As discussed above, quorum sensing also indirectly affects the expression of biofilm genes through the modulation of c-di-GMP levels (128). The opposite biofilm lifestyles of these vibrios likely are connected to niche adaptation and/or environmental signals. V. cholerae biofilm production at LCD is thought to promote persistence in natural aqueous environments by providing protection from stresses (93). Biofilms also appear to protect cells from acid stresses during infection and colonization of the host (129). Following attachment, there are “biofilm-like” microcolonies that form (131), although it is not clear if these are similar to biofilms formed on abiotic surfaces (132).
Secretion systems.
The type III and type VI secretion systems (T3SS and T6SS, respectively) are intimately linked to quorum sensing in several Vibrio species (Fig. 3) (41, 49, 57, 71, 90, 133). T3SS and T6SS are complex syringe-like structures that are capable of penetrating proximal cellular membranes to deliver effector proteins that interfere with various cellular processes to cause cell death (134). T3SS are generally used to breach eukaryotic membranes, whereas T6SS can target both eukaryotic and prokaryotic membranes. In V. harveyi and V. parahaemolyticus, LuxR and OpaR repress expression of the T3SS operons, including those encoding the structural proteins, effector proteins, and the transcription factors in the system (41, 49, 135). Not only is the expression of T3SS vastly different between LCD and HCD in culture, but these operons are also highly upregulated during infection in a quorum sensing-dependent manner (71). While other vibrios, such as V. cholerae, contain T3SS, the regulatory connection to quorum sensing remains to be explored (136). In addition, the expression of T6SS is activated by quorum sensing in vibrios. LuxR/HapR and the Qrrs activate and repress the expression of T6SS operons in V. cholerae and V. harveyi, respectively (57, 90, 133). In the case of V. anguillarum, the LuxR homolog VanT represses the expression of hcp, which encodes a T6SS structural protein that is necessary for delivering effector proteins into the target cell (137, 138). HapR also activates qstR, which encodes the transcription factor that controls the expression of T6SS and competence genes (139, 140). Although the mechanism of T6SS regulation has not yet been examined in V. vulnificus, the SmcR regulon includes hcp, and thus, T6SS is controlled by quorum sensing (78). Interestingly, a type II secretion system has been recently revealed in V. cholerae, and this gene cluster is regulated by quorum sensing (141).
CONCLUDING REMARKS AND FUTURE PERSPECTIVE
For decades, Vibrio species have served as excellent model systems for studying quorum sensing, cell-cell interactions, and pathogenesis. Many of the intricacies of quorum sensing networks have been uncovered and yielded a fundamental understanding of the role of LuxR/HapR-type proteins. It is clear that LuxR protein expression is regulated through a phosphocascade circuit that responds to different autoinducers produced by various members of the Vibrio clade. The balance between AphA and LuxR protein concentrations is at the center of most Vibrio quorum sensing networks and directs changes in downstream gene expression. Furthermore, the Qrrs are critical for managing these concentrations, allowing rapid transitions from LCD to HCD or vice versa. Ultimately, the concentration of LuxR determines the regulation of hundreds of genes downstream. The mechanism by which this is accomplished is unclear, and many questions remain. For example, what are the multiple binding sites required for activation? How does LuxR affect transcription initiation? Do LuxR protein-protein interactions play roles in activation and/or repression? Does LuxR binding alter the DNA structure? These questions can be answered with biochemical, biophysical, and structural assays with LuxR proteins to gain a better understanding of the regulatory mechanisms of these transcription factors.
As global regulators, LuxR homologs regulate the expression of hundreds of genes to produce group behaviors. The elucidation of the LuxR regulons by transcriptomics in multiple Vibrio species revealed that these master regulators likely control a variety of cellular processes in addition to those discussed above. These findings have prompted the question: what other group behaviors are controlled by LuxR? LuxR homologs control biofilm formation, secretion, and c-di-GMP signaling, but there are likely others. It was only recently revealed that quorum sensing is connected to metabolism and the osmotic stress response. Indeed, hundreds of genes regulated by LuxR proteins in vibrios are annotated as hypothetical proteins with unknown function. Future research should focus on careful examination of these genes and patterns of expression that may reveal their function. Further experimental and bioinformatics examination of Vibrio quorum sensing regulons may guide the field toward important functional genes. Our simple search for orthologs in pairs of Vibrio quorum sensing transcriptomes revealed interesting classes of genes, such as chemotaxis and membrane transport genes. Because these four transcriptomes were determined under various assay conditions and methods, a side-by-side comparison of transcriptomes from vibrios under the same assay conditions would likely yield more comprehensive results. These types of transcriptomic and bioinformatics studies in Vibrio species reveal gene functions that benefit different habitats or lifestyles and may be particularly beneficial in identifying genes that are critical for pathogenesis.
Following the field's deep focus on bacterial cell-cell signaling molecules, a breadth of knowledge now exists on autoinducer synthesis, structures, receptors, and signal transduction components, as well as the regulatory mechanisms of the sRNAs, LuxR, and AphA. The downstream genes regulated by quorum sensing signaling (e.g., motility and biofilm formation) are also fairly well studied, although, as discussed above, there are likely many gene classes regulated by quorum sensing that have yet to be discovered. Thus, on the surface, it appears that the general signaling scheme in vibrios is elucidated: high concentrations of autoinducer signals drive changes in LuxR and AphA concentrations that up- or downregulate gene groups. However, there are major gaps in connecting signaling to behavior. What gene expression changes occur during transitions from LCD to HCD or from HCD to LCD? Certainly, these transitions occur under various conditions as cells grow in communities, move around, infect hosts, and become dispersed in the environment. What happens at midlevel concentrations of autoinducers or in the presence of one dominant autoinducer? A few research studies have hinted that there are discrete groups of genes controlled by various levels of autoinducers, suggesting that there is a specific series of events that occur over the growth of a bacterial culture as autoinducers accumulate. Because many quorum sensing studies are performed in planktonic monoculture, it is difficult to assess the effects of signal concentrations on the transcriptome or to determine how to separate these effects from those resulting from concurrent changes in nutrient availability or the accumulation of toxic by-products. As more autoinducer-receptor pairs are uncovered in vibrios, the web of signaling circuits becomes complex. With strong depth in the field and ever-improving technologies, we are now poised to ask questions such as: how does autoinducer signaling affect the timing of gene expression? How is this timing affected by nutrient availability during planktonic or biofilm growth? Do different autoinducers confer distinct levels of gene expression? How does the growth of mixed species cultures affect quorum sensing gene expression, and does this impact the physiology and/or ecology of these bacteria? It will be illuminating to explore the complex connections between autoinducer signaling and the control of group behavior genes in vibrios.
ACKNOWLEDGMENTS
We thank Douglas Rusch and Ram Podicheti from the Indiana University Center for Genomics and Bioinformatics for analyses of the quorum sensing regulons. We thank Susanne Ressl for analyses of LuxR protein structures. We also thank Matthew Bochman and Ankur Dalia for comments on the manuscript.
REFERENCES
- 1.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomasz A. 1965. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155–159. doi: 10.1038/208155a0. [DOI] [PubMed] [Google Scholar]
- 4.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 5.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engebrecht J, Nealson K, Silverman M. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 7.Engebrecht J, Silverman M. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A 81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi SH, Greenberg EP. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci U S A 88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi SH, Greenberg EP. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol 174:4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henke JM, Bassler BL. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303–314. doi: 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 12.Jung SA, Hawver LA, Ng WL. 2016. Parallel quorum sensing signaling pathways in Vibrio cholerae. Curr Genet 62:255–260. doi: 10.1007/s00294-015-0532-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung SA, Chapman CA, Ng WL. 2015. Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog 11:e1004837. doi: 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Qiu Y, Tan Y, Guo Z, Yang R, Zhou D. 2012. Transcriptional regulation of opaR, qrr2–4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One 7:e34622. doi: 10.1371/journal.pone.0034622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassler BL, Wright M, Showalter RE, Silverman MR. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 17.Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J Bacteriol 186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupp C, Ruby EG. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J Bacteriol 187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A 102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto CM, Chatterjee J, Swartzman E, Szittner R, Meighen EA. 1996. The role of lux autoinducer in regulating luminescence in Vibrio harveyi; control of luxR expression. Mol Microbiol 19:767–775. doi: 10.1046/j.1365-2958.1996.417948.x. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto CM, Dunlap PV, Ruby EG, Meighen EA. 2003. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol Microbiol 48:537–548. doi: 10.1046/j.1365-2958.2003.03453.x. [DOI] [PubMed] [Google Scholar]
- 22.Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol 26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 23.McDougald D, Rice SA, Kjelleberg S. 2000. The marine pathogen Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene 248:213–221. doi: 10.1016/S0378-1119(00)00117-7. [DOI] [PubMed] [Google Scholar]
- 24.McCarter LL. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol 180:3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croxatto A, Chalker VJ, Lauritz J, Jass J, Hardman A, Williams P, Camara M, Milton DL. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J Bacteriol 184:1617–1629. doi: 10.1128/JB.184.6.1617-1629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa H, Hase CC. 2009. TetR-type transcriptional regulator VtpR functions as a global regulator in Vibrio tubiashii. Appl Environ Microbiol 75:7602–7609. doi: 10.1128/AEM.01016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rui H, Liu Q, Ma Y, Wang Q, Zhang Y. 2008. Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol Lett 285:155–162. doi: 10.1111/j.1574-6968.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 28.Showalter RE, Martin MO, Silverman MR. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol 172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee J, Miyamoto CM, Meighen EA. 1996. Autoregulation of luxR: the Vibrio harveyi lux-operon activator functions as a repressor. Mol Microbiol 20:415–425. doi: 10.1111/j.1365-2958.1996.tb02628.x. [DOI] [PubMed] [Google Scholar]
- 30.Swartzman E, Meighen EA. 1993. Purification and characterization of a poly(dA-dT) lux-specific DNA-binding protein from Vibrio harveyi and identification as LuxR. J Biol Chem 268:16706–16716. [PubMed] [Google Scholar]
- 31.Swartzman E, Silverman M, Meighen EA. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2007. Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J Bacteriol 189:5683–5691. doi: 10.1128/JB.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y, Kim BS, Park YJ, Choi WC, Hwang J, Kang BS, Oh TK, Choi SH, Kim MH. 2010. Crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription. J Biol Chem 285:14020–14030. doi: 10.1074/jbc.M109.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 35.Shao CP, Hor LI. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J Bacteriol 183:1369–1375. doi: 10.1128/JB.183.4.1369-1375.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C, Jing-Jing Z, Chun-Hua R, Chao-Qun H. 2010. Deletion of valR, a homolog of Vibrio harveyi luxR generates an intermediate colony phenotype between opaque/rugose and translucent/smooth in Vibrio alginolyticus. Biofouling 26:595–601. doi: 10.1080/08927014.2010.499511. [DOI] [PubMed] [Google Scholar]
- 37.Ye J, Ma Y, Liu Q, Zhao DL, Wang QY, Zhang YX. 2008. Regulation of Vibrio alginolyticus virulence by the LuxS quorum-sensing system. J Fish Dis 31:161–169. doi: 10.1111/j.1365-2761.2007.00882.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Rhee JE, Park U, Ju HM, Lee BC, Kim TS, Jeong HS, Choi SH. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J Microbiol Biotechnol 17:325–334. [PubMed] [Google Scholar]
- 39.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Kim SY, Lee SE, Kim YR, Kim CM, Ryu PY, Choy HE, Chung SS, Rhee JH. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol Microbiol 48:1647–1664. doi: 10.1046/j.1365-2958.2003.03536.x. [DOI] [PubMed] [Google Scholar]
- 41.Henke JM, Bassler BL. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol 186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassler BL, Greenberg EP, Stevens AM. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol 179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freeman JA, Bassler BL. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol 181:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 45.Tu KC, Bassler BL. 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng L, Rutherford ST, Papenfort K, Bagert JD, van Kessel JC, Tirrell DA, Wingreen NS, Bassler BL. 2015. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell 160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. 2013. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J 32:2158–2171. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papenfort K, Silpe JE, Schramma KR, Cong JP, Seyedsayamdost MR, Bassler BL. 2017. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat Chem Biol 13:551–557. doi: 10.1038/nchembio.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svenningsen SL, Tu KC, Bassler BL. 2009. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J 28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen Y, Kim IH, Kim KS. 2016. Iron- and quorum-sensing signals converge on small quorum-regulatory RNAs for coordinated regulation of virulence factors in Vibrio vulnificus. J Biol Chem 291:14213–14230. doi: 10.1074/jbc.M116.714063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gode-Potratz CJ, McCarter LL. 2011. Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol 193:4224–4237. doi: 10.1128/JB.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez-Reytor D, Plaza N, Espejo RT, Navarrete P, Bastias R, Garcia K. 2016. Role of non-coding regulatory RNA in the virulence of human pathogenic vibrios. Front Microbiol 7:2160. doi: 10.3389/fmicb.2016.02160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao Y, Bassler BL. 2014. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol Microbiol 92:921–930. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovacikova G, Skorupski K. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 59.Tu KC, Waters CM, Svenningsen SL, Bassler BL. 2008. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol 70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svenningsen SL, Waters CM, Bassler BL. 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev 22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teng SW, Schaffer JN, Tu KC, Mehta P, Lu W, Ong NP, Bassler BL, Wingreen NS. 2011. Active regulation of receptor ratios controls integration of quorum-sensing signals in Vibrio harveyi. Mol Syst Biol 7:491. doi: 10.1038/msb.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. 2010. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell 37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J Biol Chem 280:13779–13783. doi: 10.1074/jbc.M413781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 65.Sun F, Zhang Y, Wang L, Yan X, Tan Y, Guo Z, Qiu J, Yang R, Xia P, Zhou D. 2012. Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One 7:e44210. doi: 10.1371/journal.pone.0044210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin W, Kovacikova G, Skorupski K. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol 64:953–967. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- 67.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Ling Y, Jiang H, Qiu Y, Qiu J, Chen H, Yang R, Zhou D. 2013. AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int J Food Microbiol 160:245–251. doi: 10.1016/j.ijfoodmicro.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruwandeepika HA, Karunasagar I, Bossier P, Defoirdt T. 2015. Expression and quorum sensing regulation of type III secretion system genes of Vibrio harveyi during infection of gnotobiotic brine shrimp. PLoS One 10:e0143935. doi: 10.1371/journal.pone.0143935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. 2008. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol 70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. 2013. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4(4):e00378-13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang M, Frey EM, Liu Z, Bishar R, Zhu J. 2010. The virulence transcriptional activator AphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect Immun 78:697–703. doi: 10.1128/IAI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong HS, Kim SM, Lim MS, Kim KS, Choi SH. 2010. Direct interaction between quorum-sensing regulator SmcR and RNA polymerase is mediated by integration host factor to activate vvpE encoding elastase in Vibrio vulnificus. J Biol Chem 285:9357–9366. doi: 10.1074/jbc.M109.089987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kernell Burke A, Guthrie LT, Modise T, Cormier G, Jensen RV, McCarter LL, Stevens AM. 2015. OpaR controls a network of downstream transcription factors in Vibrio parahaemolyticus BB22OP. PLoS One 10:e0121863. doi: 10.1371/journal.pone.0121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee DH, Jeong HS, Jeong HG, Kim KM, Kim H, Choi SH. 2008. A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J Biol Chem 283:23610–23618. doi: 10.1074/jbc.M801480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog 2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. 2009. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res 37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA, van Kessel JC. 2016. Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101:823–840. doi: 10.1111/mmi.13425. [DOI] [PubMed] [Google Scholar]
- 83.van Kessel JC, Rutherford ST, Cong JP, Quinodoz S, Healy J, Bassler BL. 2015. Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J Bacteriol 197:73–80. doi: 10.1128/JB.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J 21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeng WY, Ko TP, Liu CI, Guo RT, Liu CL, Shr HL, Wang AH. 2008. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res 36:1567–1577. doi: 10.1093/nar/gkm1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhukya H, Bhujbalrao R, Bitra A, Anand R. 2014. Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2) Nucleic Acids Res 42:10122–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol 7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 88.Teng SW, Wang Y, Tu KC, Long T, Mehta P, Wingreen NS, Bassler BL, Ong NP. 2010. Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell. Biophys J 98:2024–2031. doi: 10.1016/j.bpj.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jeong HS, Lee MH, Lee KH, Park SJ, Choi SH. 2003. SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J Biol Chem 278:45072–45081. doi: 10.1074/jbc.M308184200. [DOI] [PubMed] [Google Scholar]
- 90.Bagert JD, van Kessel JC, Sweredoski MJ, Feng L, Hess S, Bassler BL, Tirrell DA. 2016. Time-resolved proteomic analysis of quorum sensing in Vibrio harveyi. Chem Sci 7:1797–1806. doi: 10.1039/C5SC03340C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Q, Defoirdt T. 2015. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ Microbiol 17:960–968. doi: 10.1111/1462-2920.12420. [DOI] [PubMed] [Google Scholar]
- 92.Jaques S, McCarter LL. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J Bacteriol 188:2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 94.Ruby EG, Lee KH. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol 64:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millikan DS, Ruby EG. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hawver LA, Giulietti JM, Baleja JD, Ng WL. 2016. Quorum sensing coordinates cooperative expression of pyruvate metabolism genes to maintain a sustainable environment for population stability. mBio 7(6):e01863-16. doi: 10.1128/mBio.01863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Studer SV, Mandel MJ, Ruby EG. 2008. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol 190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kalburge SS, Carpenter MR, Rozovsky S, Boyd EF. 2017. Quorum sensing regulators are required for metabolic fitness in Vibrio parahaemolyticus. Infect Immun 85:e00930-16. doi: 10.1128/IAI.00930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anetzberger C, Schell U, Jung K. 2012. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiol 12:209. doi: 10.1186/1471-2180-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mok KC, Wingreen NS, Bassler BL. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J 22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lilley BN, Bassler BL. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 103.Wen Y, Kim IH, Son JS, Lee BH, Kim KS. 2012. Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J Biol Chem 287:26727–26739. doi: 10.1074/jbc.M112.374165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 105.Sun S, Tay QX, Kjelleberg S, Rice SA, McDougald D. 2015. Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J 9:1812–1820. doi: 10.1038/ismej.2014.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Defoirdt T, Darshanee Ruwandeepika HA, Karunasagar I, Boon N, Bossier P. 2010. Quorum sensing negatively regulates chitinase in Vibrio harveyi. Environ Microbiol Rep 2:44–49. doi: 10.1111/j.1758-2229.2009.00043.x. [DOI] [PubMed] [Google Scholar]
- 107.Watve SS, Thomas J, Hammer BK. 2015. CytR is a global positive regulator of competence, type VI secretion, and chitinases in Vibrio cholerae. PLoS One 10:e0138834. doi: 10.1371/journal.pone.0138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katzianer DS, Wang H, Carey RM, Zhu J. 2015. “Quorum non-sensing”: social cheating and deception in Vibrio cholerae. Appl Environ Microbiol 81:3856–3862. doi: 10.1128/AEM.00586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kimbrough JH, Stabb EV. 2015. Antisocial luxO mutants provide a stationary phase survival advantage in Vibrio fischeri ES114. J Bacteriol 198:673–687. doi: 10.1128/JB.00807-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Travisano M, Velicer GJ. 2004. Strategies of microbial cheater control. Trends Microbiol 12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 111.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. 2015. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci U S A 112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bruger EL, Waters CM. 2016. Bacterial quorum sensing stabilizes cooperation by optimizing growth strategies. Appl Environ Microbiol 82:6498–6506. doi: 10.1128/AEM.01945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 115.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol 188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Conner JG, Zamorano-Sanchez D, Park JH, Sondermann H, Yildiz FH. 2017. The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr Opin Microbiol 36:20–29. doi: 10.1016/j.mib.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boles BR, McCarter LL. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol 184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl Environ Microbiol 74:4199–4209. doi: 10.1128/AEM.00176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol 60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 121.Beyhan S, Odell LS, Yildiz FH. 2008. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol 190:7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. 2010. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol 192:4541–4552. doi: 10.1128/JB.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A 109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol 53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 125.Shikuma NJ, Fong JC, Odell LS, Perchuk BS, Laub MT, Yildiz FH. 2009. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol 191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee KJ, Kim JA, Hwang W, Park SJ, Lee KH. 2013. Role of capsular polysaccharide (CPS) in biofilm formation and regulation of CPS production by quorum-sensing in Vibrio vulnificus. Mol Microbiol 90:841–857. doi: 10.1111/mmi.12401. [DOI] [PubMed] [Google Scholar]
- 127.Enos-Berlage JL, McCarter LL. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J Bacteriol 182:5513–5520. doi: 10.1128/JB.182.19.5513-5520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell 5:647–656. doi: 10.1016/S1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 130.Vance RE, Zhu J, Mekalanos JJ. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect Immun 71:2571–2576. doi: 10.1128/IAI.71.5.2571-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Silva AJ, Benitez JA. 2016. Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl Trop Dis 10:e0004330. doi: 10.1371/journal.pntd.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4(1):VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Waters CM, Wu JT, Ramsey ME, Harris RC, Bassler BL. 2010. Control of the type 3 secretion system in Vibrio harveyi by quorum sensing through repression of ExsA. Appl Environ Microbiol 76:4996–5004. doi: 10.1128/AEM.00886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 137.Weber B, Hasic M, Chen C, Wai SN, Milton DL. 2009. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol 11:3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 138.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 140.Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zielke RA, Simmons RS, Park BR, Nonogaki M, Emerson S, Sikora AE. 2014. The type II secretion pathway in Vibrio cholerae is characterized by growth phase-dependent expression of exoprotein genes and is positively regulated by sigmaE. Infect Immun 82:2788–2801. doi: 10.1128/IAI.01292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]