ABSTRACT
Vibrio vulnificus is an estuarine bacterium and potent opportunistic human pathogen. It enters the food chain by asymptomatically colonizing a variety of marine organisms, most notably oysters. Expression of the brp-encoded extracellular polysaccharide, which enhances cell-surface adherence, is regulated by cyclic di-GMP (c-di-GMP) and the activator BrpT. The Vibrio cholerae and Vibrio parahaemolyticus homologs VpsT and CpsQ, directly bind c-di-GMP via a novel W[F/L/M][T/S]R motif, and c-di-GMP binding is absolutely required for activity. Notably, BrpT belongs to a distinct subclass of VpsT-like regulators that harbor a proline in the third position of the c-di-GMP binding motif (WLPR), and the impact of this change on activity is unknown. We show that the brp locus is organized as two linked operons with BrpT specifically binding to promoters upstream of brpA and brpH. Expression data and structural modeling suggested that BrpT might be less dependent on c-di-GMP binding for activity than VpsT or CpsQ. We show that the affinity of BrpT for c-di-GMP is low and that signal binding is not a requisite for BrpT function. Furthermore, a BrpT mutant engineered to carry a canonical WLTR motif (BrpTP124T) bound c-di-GMP with high affinity and its activity was now c-di-GMP dependent. Conversely, introduction of the WLPR motif into VpsT suppressed its dependence on c-di-GMP for activity. This is the first demonstration of reduced dependence on signal association for regulator function within this motif family. Thus, BrpT defines a new class of VpsT-like transcriptional regulators, and the WLPR motif variant may similarly liberate the activity of other subclass members.
IMPORTANCE A Vibrio genome may encode nearly 100 proteins that make, break, and bind c-di-GMP, underscoring its central role in the physiology of these bacteria. The activity of the biofilm regulators VpsT of V. cholerae and CpsQ of V. parahaemolyticus is regulated by the direct binding of c-di-GMP via a novel W[F/L/M][T/S]R motif. The V. vulnificus homolog, BrpT, bears an unusual WLPR variant and remains active at low intracellular c-di-GMP levels. This suggests that the WLPR motif may also liberate the activity of other members of this subclass. A single point mutation at the 3rd position of the motif was sufficient to moderate dependence on c-di-GMP binding for activator function, highlighting the simplicity with which complex bacterial signaling networks can be rewired.
KEYWORDS: biofilms, c-di-GMP, foodborne pathogens, regulation of gene expression
INTRODUCTION
In nature, the majority of microorganisms live as surface-associated communities (biofilms) that are sheathed in a hydrated matrix of DNA, proteins, and polysaccharides produced by the resident microorganisms (1). Compared with their planktonic counterparts, biofilm consortia are highly resilient, allowing bacteria to withstand environmental stress, the onslaught of antimicrobials, and immune responses of the host.
The second messenger cyclic di-GMP (c-di-GMP) is a key signaling molecule used by bacteria to regulate biofilm development (2–6). Once triggered, the signaling pathway directs diguanylate cyclases (DGC) and phosphodiesterases (PDE), which synthesize and degrade, respectively, c-di-GMP to alter the internal c-di-GMP levels (7, 8). Downstream effector proteins sense this change and transduce the signal (9). The domains and motifs within effector proteins that bind c-di-GMP include the RXXD motif, the PilZ, EAL, and HD-GYP domains, and the novel binding sites identified within MshE and CuxR (9–12). The circuit culminates with a phenotypic response driven by modified protein activity and/or differential expression of target genes. In general, increases in intracellular c-di-GMP concentrations enhance the expression of adhesion factors and biofilm formation, while repressing motility and virulence gene expression.
Vibrio vulnificus is a potent opportunistic human pathogen. It is unique among Vibrio species in that it is best known for causing septicemia (13). Infection typically occurs through open wounds or via the food chain, in which V. vulnificus is known to colonize bivalves such as oysters. Disease progression is rapid, and the fatality rate of septicemic patients is greater than 50%, the highest death rate of any foodborne disease agent (14). Biofilm formation is important for the ecology and transmission of V. vulnificus; however, our understanding of its regulation is lacking. Greater insight could lead to the development of approaches to decrease the Vibrio load in filter feeders and lower the incidence of invasive disease.
We previously demonstrated that increased intracellular c-di-GMP levels induced biofilm formation in V. vulnificus (15). Among the plethora of genes with increased transcription were four that clustered to one region of the chromosome that was later designated the brp (biofilm and rugose polysaccharide) locus (16). The locus contained nine genes (brpABCDFHIJK), and disruption of brpF and brpI prevented c-di-GMP-induced biofilm, aggregate, and rugose colony formation. Screening of a transposon mutant library for clones that failed to form a biofilm in response to elevated c-di-GMP levels led to the identification of two regulators of brp expression, designated brpR and brpT. Both BrpR and BrpT contained a helix-turn-helix (HTH) DNA binding domain and shared 79% and 33% amino acid identity with the transcriptional regulators VpsR and VpsT of Vibrio cholerae (17). VpsR binds c-di-GMP by an unknown mechanism and drives vpsT expression (18). VpsT in turn binds c-di-GMP (19) and activates transcription of two linked operons, vpsI and vpsII, which are part of the Vibrio polysaccharide synthesis (vps) locus (20).
The crystal structure of VpsT revealed that it interacts with c-di-GMP via a novel W[F/L/M][T/S]R motif. T133 and R134 of the WLTR motif participate in H bonding with the c-di-GMP dimer, and signal binding is required for activity. The Vibrio parahaemolyticus homolog CspQ, which activates capsular polysaccharide (cps) expression, harbors a WLSR motif and binds c-di-GMP. A CpsQR134A (WLSA) mutant failed to bind c-di-GMP and consequently could not activate cps expression (21). BrpT belongs to a distinct subclass of VpsT-like regulators that deviate from the canonical c-di-GMP binding sites of VpsT and CpsQ in that a proline, rather than a serine or threonine, is found in the third position of the motif (WLPR). Not only is proline conformationally rigid, but the lack of an amide hydrogen precludes it from donating to H bonding. As a consequence, proline is not commonly found in the binding or active sites of proteins (22). As such, its presence in the putative BrpT c-di-GMP binding site is unusual and its effect on signal binding by and subsequent activity of BrpT is unknown.
Here, we show that the brp locus is organized as two linked operons that are separately transcribed. Expression of the locus is BrpT dependent, and purified BrpT specifically bound to promoters upstream of brpA and brpH. Expression data and structural modeling suggested that BrpT could potentially activate transcription without binding c-di-GMP. We show that, in contrast to VpsT and CpsQ, purified BrpT was able to bind target promoters and drive brp expression under low-c-di-GMP conditions. By engineering a canonical WLTR motif into BrpT and the proline variant into VpsT via single point mutations, we were able to alter the degree to which signal binding was required for activator function.
RESULTS
The activities of DcpA and CdgJ alter the intracellular c-di-GMP concentrations.
To begin examining the mechanism by which elevated c-di-GMP levels induce brp expression, we sought to controllably manipulate the intracellular c-di-GMP concentration. We previously identified a putative V. vulnificus DGC (DcpA) that, when overexpressed, increased brp expression and biofilm formation. To demonstrate that DcpA expression resulted in elevated intracellular c-di-GMP concentrations, we expressed it in V. vulnificus and measured the change in the level of the second messenger relative to a control strain. Following DcpA induction, c-di-GMP levels increased 14-fold, while expression of the V. cholerae PDE, CdgJ, decreased c-di-GMP levels 15-fold (see Fig. S1, top, in the supplemental material). Similar effects were observed in Escherichia coli, in which DcpA expression boosted the intracellular c-di-GMP concentration nearly 114-fold, while CdgJ expression decreased levels 3-fold. We also tested the phenotypic effect of DcpA and CdgJ expression on V. vulnificus morphology (Fig. S1, bottom). Wild-type cells were flat and featureless on solid medium and remained so following expression of CdgJ alone. DcpA expression induced a rugose-colony development that was punctuated by a dramatic wrinkled phenotype, while simultaneous expression of DcpA and CdgJ reverted the rugose phenotype. These results confirmed that we could use DcpA and CdgJ to manipulate intracellular c-di-GMP concentrations from the millimolar (high) to low nanomolar (low) range.
c-di-GMP regulates brp locus and brpT expression.
Our previous studies suggested that elevated intracellular c-di-GMP levels induced biofilm formation in V. vulnificus and that the enhanced biofilm phenotype was dependent on BrpT (15, 16). We monitored the expression of PbrpA in V. vulnificus using an ectopically integrated transcriptional reporter (Fig. 1A). PbrpA-gfp expression was observed in wild-type (WT) cells only when intracellular c-di-GMP levels were increased. No PbrpA-gfp expression was observed in the brpT-null strain under unaltered or elevated c-di-GMP conditions. Moreover, ectopic expression of brpT in the null mutant (ΔbrpT-C) restored PbrpA-gfp expression when intracellular c-di-GMP levels were elevated. This confirmed that brp expression was regulated by c-di-GMP and dependent on BrpT. To determine if the expression of brpT was also regulated by c-di-GMP and/or BrpR, we measured the activity of a PbrpT-gfp reporter in WT and ΔbrpR strains under unaltered and elevated intracellular c-di-GMP conditions (Fig. 1B). PbrpT-gfp expression was low in WT cells but increased 4.5-fold when intracellular c-di-GMP was elevated. However, only a 1.5-fold increase in PbrpT-gfp expression was observed in the ΔbrpR strain. This suggested that brpT expression was regulated by BrpR and c-di-GMP and BrpT in turn regulated brp expression.
FIG 1.

Transcription of brpA and brpT is regulated by c-di-GMP. (A) Relative PbrpA-gfp expression in the WT, ΔbrpT, and complemented (ΔbrpT-C) strains is shown in relative fluorescence units (RFU). (B) Relative PbrpT-gfp expression in the WT and ΔbrpR strains. Plots show the means, and error bars represent the standard deviations. Significance: ns, not significant (P > 0.05); *, P < 0.001.
BrpT directly regulates brp expression.
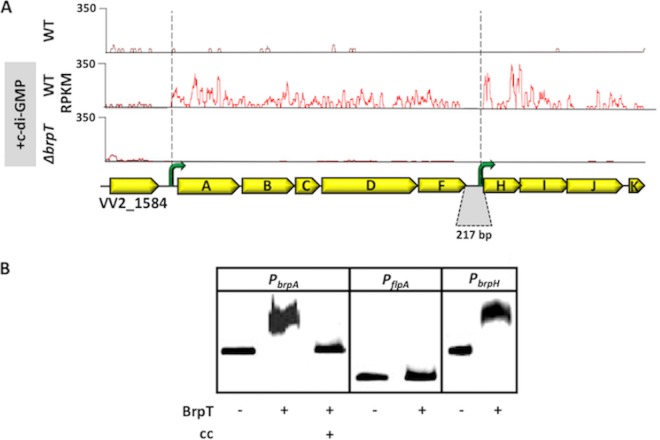
We previously showed that increased c-di-GMP levels triggered the expression of four of the nine brp genes. However, it was still unclear if all of the genes were expressed or how their expression was organized. We used transcriptome sequencing (RNA-seq) to analyze c-di-GMP-dependent brp expression to better define its transcriptional architecture. Few transcripts mapping to the locus were detected in wild-type cells carrying the empty expression vector, but transcripts were readily detected in cells following DcpA expression (Fig. 2A, top and middle panels). Expression of all nine brp genes ceased in a ΔbrpT strain, even when DcpA was expressed (Fig. 2A, bottom panel).
FIG 2.
Architecture of the brp locus and DNA binding by BrpT to the brpA and brpH promoter regions. (A) Reads per kilobase per million mapped sequence reads (RPKM) to the brp locus. The top, middle, and bottom panels show the traces for the WT and ΔbrpT strains under unaltered (top panel) and elevated (middle and bottom panels) intracellular c-di-GMP conditions (i.e., expressing DcpA). Yellow arrows denote the respective brp genes. Green arrows, putative promoters upstream of brpA and brpH. The genes brpF and brpH are separated by 217 bp (gray region). (B) BrpT was purified and incubated with labeled DNA fragments spanning the brpA promoter (PbrpA) or the brpH promoter (PbrpH) regions. Unlabeled PbrpA was used as a cold competitor (cc) for DNA binding. The flpA promoter (PflpA) was used as a control for specific binding.
The brpF and brpH genes are separated by 217 bp, and we did not identify any transcripts that mapped to this intergenic region. This raised the possibility of the brp locus being expressed as two separate transcripts, the first initiating upstream of brpA and the second initiating upstream of brpH. To determine if BrpT specifically bound to promoter regions upstream of brpA and brpH, we purified it (see Fig. S2A in the supplemental material) and used it in gel mobility shift assays (Fig. 2B). BrpT was able to bind and shift a labeled 325-bp DNA fragment corresponding to the brpA promoter (PbrpA), and binding was prevented when excess unlabeled PbrpA (cold competitor) was added to the reaction mixture. Conversely, BrpT was unable to bind to a labeled 100-bp DNA fragment spanning the promoter region of flpA (PflpA), which codes for a type IVb pilin of V. vulnificus. BrpT was also capable of shifting a 325-bp DNA fragment mapping to the brpH promoter region (PbrpH). These results suggested that BrpT specifically binds to target promoters upstream of brpA and brpH to regulate brp expression and that the brp locus consists of two operons, brpABCDF and brpHIJK, that are separately transcribed.
brp expression is temperature dependent.
We previously demonstrated that the rugose phenotype in V. vulnificus was dependent on expression of the brp locus (15, 16). During the course of those studies, we noticed that rugosity was striking at 25°C and almost imperceptible at 37°C. This led us to speculate that brp expression might be temperature dependent. To investigate this, we monitored the expression of brpA and wzc (required for capsular polysaccharide [CPS] transport to the bacterial surface [23]) as a function of temperature in a V. vulnificus rugose isolate. There was a clear correlation between the level of rugosity, brpA expression, and temperature. Rugosity was more pronounced and biofilm formation was 4.6-fold greater at 25°C than 37°C (Fig. 3A), and while the expression of wzc was unaffected by the growth temperature, the expression of brpA was 4.9-fold higher at 25°C than 37°C (Fig. 3B). These results suggested that the loss of rugosity at 37°C was due to decreased brp expression and that brp expression was temperature dependent.
FIG 3.
PbrpA expression is temperature dependent. Colony morphology (top) and biofilm formation (bottom) (A) and RT-PCR analysis (B, top and bottom) of rugose V. vulnificus grown at 37°C or 25°C. Representative colony morphology and agarose gel images are shown. The transcript levels for wzc and brpA were normalized relative to the level of rplT (ribosomal L20) transcript in the same sample. Assays were performed in triplicate on three biological samples. Plots show the means, and error bars represent the standard deviations. White and gray bars indicate transcript levels at 37°C and 25°C, respectively. Significance: ns, not significant (P > 0.05); *, P < 0.001.
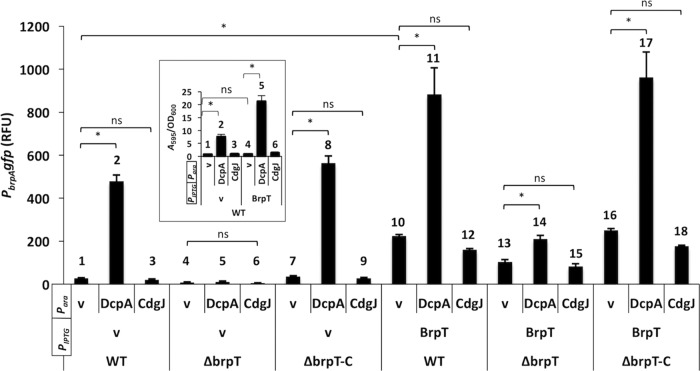
BrpT activates transcription at low intracellular c-di-GMP concentrations.
VpsT and CpsQ harbor c-di-GMP binding motifs (WLTR and WLSR, respectively) that conform to the W[F/L/M][T/S]R consensus. BrpT is different; a proline occupies the third position of the motif (WLPR), and the impact of this change on BrpT function is not known. We monitored PbrpA-gfp expression and biofilm formation of the same cells under low, unaltered, and elevated c-di-GMP levels (Fig. 4). Wild-type cells harboring empty vector or expressing CdgJ exhibited very low PbrpA-gfp expression (Fig. 4, bars 1 and 3) and biofilm formation (Fig. 4, inset, bars 1 and 3). PbrpA-gfp levels increased 18-fold following DcpA expression (Fig. 4, bar 2), and biofilm formation increased 11-fold (inset, bar 2). PbrpA-gfp expression was entirely dependent on BrpT, as DcpA was unable to induce expression in a ΔbrpT mutant (bars 4 to 6). Expression was restored when brpT was ectopically expressed from its native promoter (bars 7 to 9).
FIG 4.
BrpT mediates the effect of c-di-GMP on brp locus expression and biofilm formation. Wild type (WT), ΔbrpT, and complemented (ΔbrpT-C) V. vulnificus strains expressing either dcpA or cdgJ from an arabinose-inducible promoter (Para) and/or brpT from an IPTG-inducible promoter (PIPTG) were grown in LB under inducing conditions. Vector control strains contained the empty expression vectors. The relative level of PbrpA-gfp expression is shown in relative fluorescence units (RFU). Inset, biofilm formation by the indicated WT strains. BrpT mediates the effect of c-di-GMP on brp expression and biofilm formation. Plots show the means, and error bars represent the standard deviations. Significance: ns, not significant (P > 0.05); *, P < 0.001. Refer to the text for bar numbering.
To minimize the potential influence of altered c-di-GMP levels on brpT transcription from its native promoter, we expressed brpT from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. While the expression of DcpA alone increased biofilm formation in WT cells (Fig. 4, inset, compare bars 1 and 2), the expression of BrpT alone did not (inset, compare bars 1 and 4). This suggested that in addition to stimulating brpT and ultimately brp expression, c-di-GMP may also regulate the function of additional factors required for biofilm development that cannot be overcome by simply overexpressing brpT. However, IPTG-induced expression of BrpT increased PbrpA-gfp levels in each background (Fig. 4, bars 10 to 18), including when coexpressed with CdgJ (bars 12, 15, and 18). In fact, only a slight decrease in reporter expression was observed in strains coexpressing CdgJ relative to the corresponding vector controls (Fig. 4, bars 10, 13, and 16). This hinted at BrpT being capable of driving PbrpA-gfp expression at low intracellular c-di-GMP concentrations.
To confirm that BrpT could activate gene expression under limiting c-di-GMP conditions, PbrpA-gfp levels were monitored in E. coli cells expressing BrpT under unaltered (4 nM), low (1 nM), and high (400 nM) intracellular c-di-GMP concentrations. Under low conditions, the intracellular c-di-GMP levels were approaching the limit of our detection capability (∼1 nM) (Fig. S1). We did not detect PbrpA-gfp expression under any condition in the absence of BrpT (Fig. 5, bars 1 to 3). This is likely due to the lower overall intracellular c-di-GMP levels in E. coli than in V. vulnificus (Fig. S1) and the dependence of PbrpA-gfp expression on BrpT. Induction of BrpT resulted in a similar increase (>230-fold) in PbrpA-gfp expression under low, unaltered, and high intracellular c-di-GMP concentrations, with a 1.5-fold difference at most among the three strains (Fig. 5, bars 4 to 6). This suggested that BrpT can specifically bind to target promoters and activate gene expression over a wide range of intracellular c-di-GMP concentrations (nanomolar to micromolar).
FIG 5.
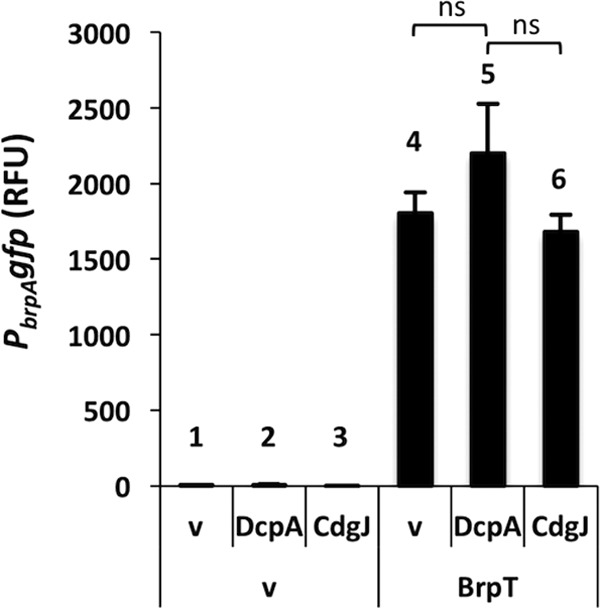
BrpTWT-driven PbrpA expression in E coli. Plots show the means, and error bars represent the standard deviations. Significance: ns, not significant (P > 0.05). RFU, relative fluorescence units. Refer to the text for bar numbering.
The BrpT WLPR motif suppresses the requirement for c-di-GMP binding for activation.
To delve further into the impact of the proline of the WLPR motif on BrpT activity, we mutated it to a T (BrpTP124T) as found in the canonical VpsT c-di-GMP binding site. Structural modeling of the BrpTP124T motif (WLTR) suggested that it would mirror the arrangement of the c-di-GMP binding pocket of VpsT (see Fig. S3 in the supplemental material). The R in the 4th position of the motif is essential for c-di-GMP binding by, and function of, VpsT and CpsQ, so the analogous change was introduced into BrpT (BrpTR125A). However, the BrpTR125A motif (WLPA) maintains the 3rd-position proline, so a BrpTP124T/R125A double mutant was also generated. Each mutant was assayed for its ability to drive PbrpA expression in a ΔbrpT background under low (CdgJ), unaltered (empty vector), or high (DcpA) intracellular c-di-GMP concentrations.
Reporter expression in control cells was 2-fold higher with BrpTP124T than observed with BrpTWT (Fig. 6A, bar 1; compare with Fig. 4, bar 13). Although a similar increase (2.5-fold) was observed when BrpTP124T was coexpressed with DcpA (Fig. 6A, bars 1 and 2; compare with Fig. 4, bars 13 and 14), overall PbrpA-gfp expression was twice the level observed with BrpTWT. Importantly, PbrpA-gfp expression was 4-fold lower when CdgJ was coexpressed with BrpTP124T than with BrpTWT (Fig. 6A, bar 3; compare with Fig. 4, bar 15). This suggested that BrpTP124T was more active and more sensitive to changes in the intracellular c-di-GMP concentrations than BrpTWT. The BrpTR125A mutant, which bears a proline in the 3rd position of the motif, could still activate PbrpA-gfp expression, and its activity was largely insensitive to the intracellular c-di-GMP level (Fig. 6A, bars 4 to 6). Conversely, only a low level of promoter activity was detected with the BrpTP124T/R125A double mutant under each condition (Fig. 6A, bars 7 to 9). This decrease suggested that the activity of BrpTP124T was now dependent on R125A (Fig. 6A, compare bars 1 to 3 with bars 7 to 9).
FIG 6.
PbrpA expression driven by BrpT WLPR motif mutants. PbrpA-gfp expression profiles in a V. vulnificus ΔbrpT strain (A) and E coli (B). RFU, relative fluorescence units. Plots show the means, and error bars represent the standard deviations. Significance: ns, not significant (P > 0.05); *, P < 0.001. Refer to the text for bar numbering.
The same overall trend was observed when the reporter assays were conducted in E. coli (Fig. 6B). BrpTP124T-driven PbrpA-gfp expression was highly dependent on the intracellular c-di-GMP concentration (bars 1 to 3). While only a 1.5-fold change in PbrpA-gfp expression was observed between high and low intracellular c-di-GMP concentrations for BrpTWT (Fig. 5, bars 5 and 6), a 17-fold drop in PbrpA-gfp expression occurred for BrpTP124T under the same conditions (Fig. 6B, bars 2 and 3). This again suggested that BrpTP124T was more active but more sensitive than BrpTWT to changes in the intracellular c-di-GMP concentrations. Although PbrpA-gfp expression driven by BrpTR125A (Fig. 6B, bars 4 to 6) was 2- to 3-fold lower than that driven by BrpTWT, the overall expression pattern appeared to be largely independent of the intracellular c-di-GMP concentration. Finally, the BrpTP124T/R125A double mutant could not support PbrpA-gfp expression under any condition tested (Fig. 6B, bars 7 to 9). Since both BrpTWT and BrpTR125A bear a proline in the third position of their respective WLP[R/A] motif (and each variant was stably expressed to comparable levels in vivo [Fig. S2]) and both still drove PbrpA-gfp expression when coexpressed with CdgJ, this suggested that a proline at the third position of the BrpT motif was sufficient to support BrpT activity at low intracellular c-di-GMP levels. Mutating the proline to a threonine rendered BrpT more active than BrpTWT at high intracellular c-di-GMP levels but less active at unaltered and low intracellular c-di-GMP levels, and this activity was now dependent on the arginine in the 4th position of the motif.
BrpT has a low affinity for c-di-GMP.
BrpT could specifically bind to promoters upstream of brpA and brpH. Since only nanomolar amounts of c-di-GMP were detected in our purified BrpT preparations, this suggested that DNA binding could occur at low intracellular c-di-GMP concentrations. Structural modeling of the WLPR motif of BrpT suggested that the proline would likely affect the H bond at that position and potentially at the neighboring arginine (R125) as well (Fig. S3). Empirically, the ability of BrpT to stably bind c-di-GMP seemed suspect. Isothermal titration calorimetry (ITC) was used to measure the c-di-GMP binding potential of purified BrpT (see Fig. S4 in the supplemental material). We were unable to saturate BrpTWT with c-di-GMP at any molar ratio (from 1:4 to 18:1). The data indicated minimal protein-molecule association, and the affinity of BrpT for c-di-GMP (apparent dissociation constant [Kd] = 135.4 μM with n = 1, ΔS = 408.2 J/mol/K) was >40 times weaker than reported for VpsT (Kd = 3.2 μM). This result suggested that BrpT interacted poorly with c-di-GMP. The ability of BrpT to activate PbrpA expression under low intracellular c-di-GMP concentrations together with its low affinity for c-di-GMP suggested that signal binding was not absolutely required for BrpT to function.
We next tested the ability of BrpTP124T to bind c-di-GMP. BrpTP124T was purified to homogeneity, and as with BrpTWT, only a low concentration (nanomolar) of c-di-GMP was detected in the preparation (Fig. S2). ITC data suggested that BrpTP124T bound c-di-GMP with a Kd of ∼1.4 μM and a stoichiometry of 1:1 (Fig. S4), metrics very similar to those of c-di-GMP binding by VpsT. Electromobility shift assays (EMSAs) with BrpTP124T were conducted to determine if promoter binding was now c-di-GMP dependent (Fig. 7). BrpTP124T was unable to shift the PbrpA fragment (Fig. 7, compare lanes 1 and 2) unless exogenous c-di-GMP was added to the reaction (lanes 1 to 3). Binding was specific, since BrpTP124T was not able to bind PflpA (lanes 4 and 5). These results suggested that the P124T substitution altered the DNA binding behavior of BrpT such that stable complex formation was now c-di-GMP dependent. Together, these data support our contention that BrpTWT likely does not directly sense c-di-GMP and so can remain active at low intracellular c-di-GMP concentrations.
FIG 7.

c-di-GMP dependent binding of BrpTP124T to the brpA promoter. BrpTP124T was incubated with a labeled 325-bp fragment spanning the brpA promoter (PbrpA) or a 100-bp fragment of the flpA promoter (PflpA). c-di-GMP was added where indicated.
To verify the importance of the third position of the motif, we next introduced the reciprocal change into VpsT (T133P), in anticipation that this would render it less dependent on intracellular c-di-GMP levels for activation than VpsTWT. VpsTWT and VpsTT133P were expressed alone or together with CdgJ in a V. cholerae vpsT-null mutant derived from a rugose parental strain (FY-Vc618ΔvpsT). This mutant appears smooth on solid medium (Fig. 8, top), and the development of rugosity was used as a visual indicator of activator function. VpsTWT, when expressed alone, induced rugose-colony formation but failed to do so when coexpressed with CdgJ (Fig. 8, middle). FY-Vc618ΔvpsT cells expressing VpsTT133P were also rugose. Although it was subtle, the cells exhibited a partially rugose phenotype when coexpressed with CdgJ (Fig. 8, bottom). This suggested that VpsTT133P, unlike VpsTWT, was able to suppress to some extent the CdgJ-induced smooth phenotype. This demonstrated that the VpsTT133P allele remained functional at intracellular c-di-GMP concentrations too low to support VpsTWT activity and supported the notion that the WLPR variant of the c-di-GMP binding motif plays an important role in moderating c-di-GMP-dependent regulator function.
FIG 8.
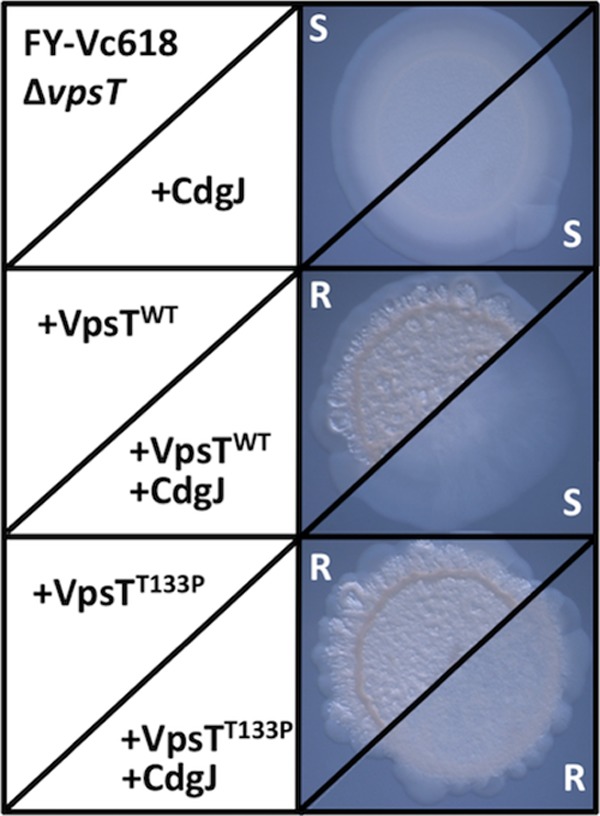
The VpsTT133P is active at lower intracellular c-di-GMP concentrations than VpsTWT. VpsTWT and VpsTT133P were expressed alone or together with CdgJ in strain FY-Vc618ΔvpsT. Colony morphology was monitored for a switch from the smooth (S) to the rugose (R) phenotype as an indicator of VpsT activity.
DISCUSSION
In V. vulnificus, increased intracellular c-di-GMP concentrations trigger biofilm development, a process that includes expressing the brp-encoded EPS (15, 16). We have shown that the brp locus is organized as two linked operons of five and four genes, respectively, and that its c-di-GMP dependent expression is mediated by BrpT, which binds to promoters upstream of brpA and brpH. We also showed that brp expression was temperature dependent, similar to the expression of the vps locus in V. cholerae, in which it has been reported that intracellular c-di-GMP concentrations increase with decreasing growth temperature (24). Although we did not demonstrate that the intracellular c-di-GMP concentration of V. vulnificus varied with temperature, the change in rugosity may be of ecological and pathological relevance. V. vulnificus is most prevalent in oysters at estuary temperatures just below 20°C (25–27), a range that supports brp expression, biofilm formation, and host colonization. The CPS is an essential virulence factor in V. vulnificus (28–30). Decreased brp expression at 37°C, such as during disease progression in the human body, might help to ensure that important cellular resources for polysaccharide synthesis are invested in CPS production rather than being diverted to a competing Brp-EPS biosynthesis pathway.
BrpT belongs to the subclass of VpsT-type regulators that bear a proline in the 3rd position of the canonical W[F/L/M][T/S]R c-di-GMP binding motif (19). We demonstrated that the WLPR variant of the BrpT c-di-GMP binding motif diminished its dependence on signal binding for activator function. Thus, BrpT defines a new class of VpsT-like transcriptional regulators, and we anticipate that other members of the WLPR motif subclass may behave similarly. In support of this notion, a VpsT mutant bearing a WLPR motif (VpsTT133P) also exhibited activity at intracellular c-di-GMP concentrations too low to support the activity of wild-type VpsT. These data suggest that the 3rd position of the WLTR motif plays a key role in moderating the degree to which activator function is regulated by direct signal binding. Obtaining a detailed structure of BrpTWT and BrpTP124T (free and c-di-GMP bound) should shed light on the manner in which BrpT activates gene expression.
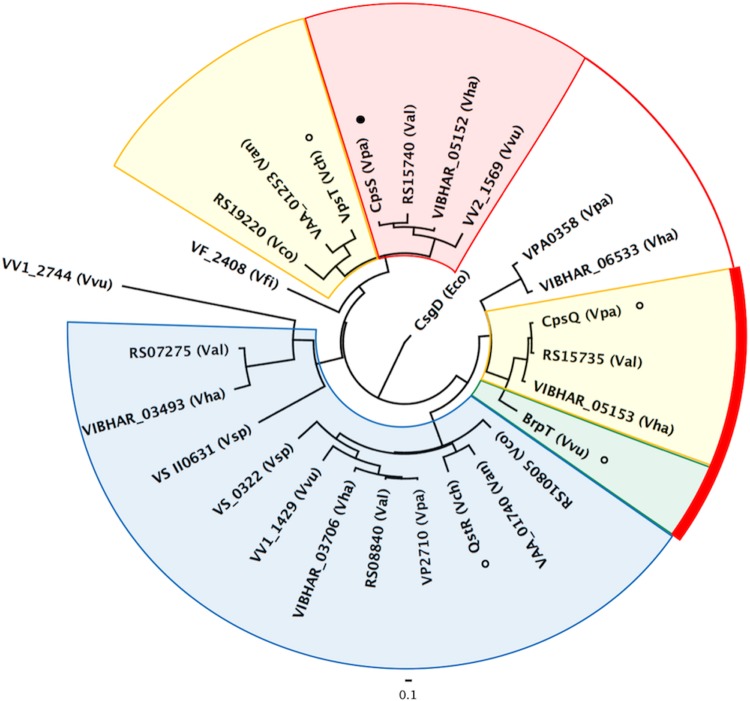
VpsT-type regulators bearing a 3rd-position proline motif were identified in 8 of 9 Vibrio species examined. The exception was the squid symbiont Vibrio fischeri, whose genome coded for only one VpsT-type regulator and no identifiable brp-type locus. Estimation of the maximum likelihood phylogeny for VpsT-type regulators from these species revealed that regulators with a 3rd-position proline grouped separately from other W[F/L/M][T/S]R motif members (Fig. 9). Notably, BrpT clustered with CpsQ on a separate branch from VpsT. This is particularly interesting since BrpT is not absolutely dependent on signal binding for activity and the apo form of CpsQ is reported to bind c-di-GMP so tightly that, once bound, it may in effect be constitutively active (21). As a result, both V. parahaemolyticus and V. vulnificus would likely require additional systems to decrease expression of their respective eps loci, since the activity of CpsQ and BrpT would be challenging to quickly moderate via further direct interactions with c-di-GMP. V. parahaemolyticus codes for additional VpsT-type regulators not found in V. cholerae. One of these, CpsS, is more closely related to VpsT than the CpsQ activator (Fig. 9, red shading). CpsS functions to repress cps expression, although it is not known if its activity is c-di-GMP dependent. V. vulnificus also codes for additional VpsT-type regulators not found in V. cholerae. It is thus conceivable that c-di-GMP also regulates the activity of systems that counter BrpT activity to keep brp expression low under low intracellular c-di-GMP conditions.
FIG 9.
Inferred evolutionary relationship of VpsT-type transcriptional regulators in 9 species of Vibrio. Phylogenetic tree of VpsT-type regulators bearing a W[F/L/M][T/S/P]R motif. The protein name or locus tag is shown followed by the host species. The V. harveyi (Vha) genome coded for five VpsT homologs, while V. vulnificus (Vvu), V. parahaemolyticus (Vpa), and V. alginolyticus (Val) each encoded four. Two representatives were identified in V. cholerae (Vch), V. anguillarum (Van), V. coralliilyticus (Vco), and V. splendidus (Vsp), and a single homolog was found in V. fischeri (Vfi). Eco, Escherichia coli. Shading is as follows: blue, regulators with a 3rd-position proline; yellow, host strains that also exhibit homology/synteny in their eps loci (see Fig. S5); green, BrpT shares aspects of the adjacent blue and yellow subgroups; red, putative c-di-GMP-regulated repressors encoded by the genomes of strains with activators that cluster with BrpT and CpsQ (connected by a red line and bar).
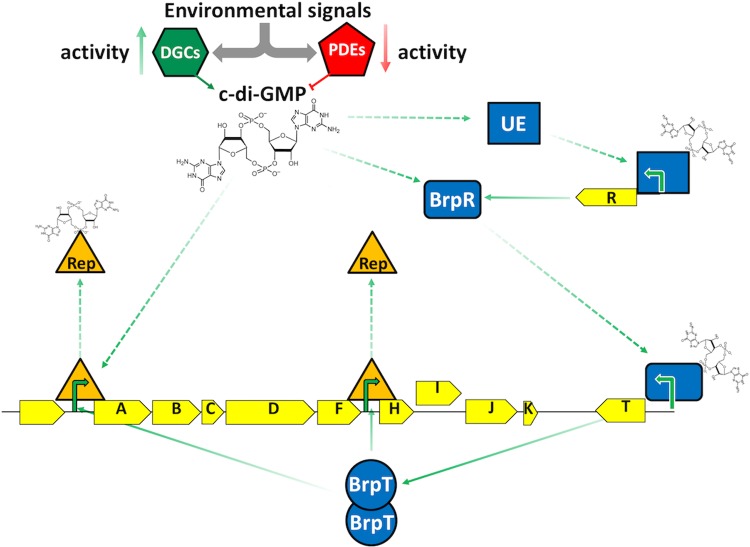
The synteny and homology of the eps loci from species encoding a similar number of VpsT-type regulators were conserved (see Fig. S5 in the supplemental material). We suspect that species coding for only two VpsT-type regulators (V. cholerae, V. anguillarum, V. coralliilyticus, and V. splendidus) moderate activator function by direct molecule binding, whereas the regulatory network of those with four or more VpsT-type regulators may use additional VpsT-type repressors to counter activator function. This notion is supported by our observation that although the expression of BrpT alone in V. vulnificus induced PbrpA expression, it was not sufficient to promote biofilm formation. Maximal and timely brp expression would thus be dependent on the simultaneous activation and relief of brp repression (Fig. 10).
FIG 10.
Model for BrpT- and c-di-GMP-dependent regulation of brp expression. Environmental cues activate DGCs and/or PDEs that alter the internal c-di-GMP concentration. Increased c-di-GMP levels invoke an effector(s) that activates brpR expression, or c-di-GMP may bind directly to BrpR already present in the cell. BrpR drives expression of PbrpT, and functional BrpT oligomers bind to promoters upstream of brpA and brpH to activate brp expression. Concomitant derepression of brp expression by specific repressors occurs, possibly via physical interaction of the repressor with c-di-GMP or its displacement from the DNA by BrpT. UE, unknown effector; Rep, repressor; green arrows, known (solid) and unknown (dashed) steps in the activation of brp expression.
VpsT and BrpT share only 33% identity (57% similarity [Fig. S3]), yet a single nucleotide transversion was sufficient to produce the proline/threonine mutation that shifted their dependence on c-di-GMP binding for activity. It is not known which activity (signal-dependent or liberated) is ancestral or if they evolved independently; however, both pathways remain dependent on an increased intracellular c-di-GMP concentration to initiate biofilm formation; in V. vulnificus, the expression of brpT itself is c-di-GMP regulated. The third-position proline variant of the c-di-GMP binding motif simply suppresses the barrier of regulator-signal association before downstream target promoter activation occurs. An obvious advantage is that, once expressed, BrpT is competent for activation. However, a tenet of this arrangement is that the activity of BrpT would require counterbalancing by repressors, whereas the activity of VpsT can be directly mitigated. Direct signal binding by VpsT-type regulators may also permit them to behave as c-di-GMP sentinels, promoting their roles from specific to global regulators that are capable of influencing multiple networks. Conversely, the liberated activity of BrpT likely restricts its impact to a few biofilm-related pathways.
MATERIALS AND METHODS
Media and strains.
LB and LB agar were purchased from BD Difco. Antibiotics and additives were purchased from Sigma and used at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 25 μg/ml; gentamicin (Gm), 10 (E. coli) or 35 (V. vulnificus) μg/ml; IPTG, 100 μM; X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 20 μg/ml; l-arabinose, 0.1%. Strains and plasmids are listed in Table S1 in the supplemental material.
Colony morphology, CV assays, and RT-PCR.
Colony morphology and crystal violet (CV) assays were conducted as previously described (16). Colony morphology images were captured on a Leica MS5 dissecting scope equipped with a Leica DC300F camera. The following reverse transcription (RT)-PCR primer pairs (data not shown) were used to monitor expression of wzc, brpA, and rplT: wzcseq2 and wzcR, wcrART1 and wcrART2, and vvuL20RT1 and vvuL20RT2, respectively. PCR products were visualized by agarose gel electrophoresis and quantified by densitometry using the ImageLab gel analysis software package (Bio-Rad). The transcript levels for wzc and brpA were normalized relative to the level of rplT (ribosomal L20) transcript in the same sample. Assays were performed in triplicate on three biological samples. Plots show the means, and error bars represent the standard deviations. P values were calculated using the Student t test.
Construction of the ΔbrpT mutant.
PCR fragments (3 kb) upstream and downstream of brpT were fused together by Gibson assembly (New England BioLabs), such that only the start and stop codons of the brpT coding region remained (data not shown). The fragment was cloned into pRE112 (31) and conjugated to V. vulnificus. Cointegrants were selected on LB Cm plates, gridded onto LB 10% sucrose plates for sacB counterselection, and confirmed to have a deletion in brpT by PCR.
Construction of reporter plasmids.
A 325-bp PbrpA PCR product was fused to gfp or the E. coli lacZ (lacZEc) and inserted into pTX1KebgA by Gibson assembly. Plasmids were maintained in E. coli S17.1λπ and conjugated to V. vulnificus for integration into ebgA (a cryptic lacZ). The same approach was used to fuse brpTWT or brpTP124T to gfp for cloning into the IPTG-inducible pC2X6HT expression plasmid for subcellular localization studies. In order to monitor β-galactosidase activity in V. vulnificus without interference due to expression of its endogenous lacZ (lacZVv), pTX1KebgA-PbrpAlacZEc was conjugated to a V. vulnificus strain bearing a markerless deletion of the promoter for lacZVv that was created using the pRE112 sucrose counterselection plasmid (32).
Fluorescence and reporter fusion analysis.
Strains were grown overnight, diluted in fresh medium containing inducer, and inoculated in triplicate into 96-well plates. Optical density at 600 nm (OD600) and fluorescence measurements were taken at 30-min intervals on a Biotek Synergy H1 microplate reader (excitation wavelength, 485 nm; emission wavelength, 528 nm). Data for three technical replicates from three biological replicates were collected for all experiments. Plots show the means, and error bars represent the standard deviations. P values were calculated using the Student t test.
Protein expression and purification.
His-tagged PCR products of the wild-type and mutated brpT genes were cloned into pACYC-Duet (Novagen) by Gibson assembly and transformed into E. coli BL21(DE3) (Novagen). Cells were grown overnight at 37°C with shaking (200 rpm) in LB containing Cm. The cultures were diluted 1:100 into 4 liters fresh LB Cm and grown to an OD600 of 0.5 at 37°C with shaking. IPTG was added to 250 μM, and incubation was continued at 25°C with shaking for 6 h. The cells were harvested by centrifugation, and pellets were frozen at −80°C for later purification.
Cell pellets were thawed on ice, resuspended in lysis buffer (20 mM imidazole, 550 mM NaCl, 1 mg/ml lysozyme, and 50 mM NaH2PO4 [pH 7.5]), and disrupted by sonication. Lysates were clarified by centrifugation at 12,000 × g for 15 min at 4°C and then passed through a 0.22-μm filter. The cleared lysates were loaded onto a 1-ml HisTRAP HP column (GE Healthcare) connected to an AKTA FLPC (Amersham Pharmacia) with a flow rate of 1 ml/min. The column was washed with 20 column volumes of lysis buffer minus lysozyme, and bound proteins were eluted using a linear imidazole gradient from 20 mM to 500 mM over 20 column volumes. The eluted fractions were separated by SDS-PAGE (12%), and those containing greater than 95% His-tagged BrpT (judged by Coomassie blue staining) were pooled. The pooled sample was applied to a 5-ml HiTrap desalting column and eluted in 50 mM NaH2PO4 (pH 7.5) and 550 mM NaCl to exchange the buffer. Purified protein was recovered in the flowthrough. Protein purity was analyzed by SDS-PAGE. If necessary, the protein was concentrated using an Amicon Ultracel 10K column (Milipore) and stored at 4°C for up to 14 days.
To demonstrate that purified BrpT (Fig. S2A) was, in effect, free of c-di-GMP for subsequent ITC experiments, it was extracted and quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Bovine serum albumin (BSA) spiked with a known quantity of c-di-GMP was treated in parallel and served as a control for recovery of the signaling molecule. Nearly 86% of the c-di-GMP added to the BSA sample was recovered (Fig. S2B), proving the efficacy of the extraction method. The amount of c-di-GMP detected in our purified BrpT was in the low nanomolar range and approached the limit of detection. Western blots were probed with Sigma monoclonal anti-polyhistidine-peroxidase primary conjugate antibody and detection was performed by using the Invitrogen Novex ECL HRP chemiluminescent substrate reagent kit.
Quantification of the intracellular c-di-GMP concentration.
For extraction of intracellular c-di-GMP from whole cells, strains carrying pSU38GT-dcpA, pSU38GT-cdgJ, or the empty vector were grown overnight in LB with Gm. The next day, cells were diluted 1:100 in fresh medium and grown to an OD600 of 0.5. Cells were again diluted to an OD600 of 0.05 into fresh LB containing Gm with 0.2% l-arabinose and grown to an OD600 of 1.0. Six milliliters of each sample was centrifuged at 4°C for 10 min at 12,000 × g. The pellet was resuspended in 200 μl of 40% (vol/vol) acetonitrile–40% (vol/vol) methanol–0.1 N formic acid. This suspension was incubated at −20°C for 30 min and then centrifuged at 4°C for 5 min. The supernatant was recovered, and 10 μl of each sample was analyzed by LC-MS/MS on a Quattro Premier XE mass spectrometer (Waters) coupled with an Acquity Ultra Performance LC system (Waters). c-di-GMP was detected with electrospray ionization using multiple-reaction monitoring in the negative-ion mode at m/z 689.16→344.31. The MS parameters were as follows: capillary voltage, 3.5 kV; cone voltage, 50 V; collision energy, 34 V; source temperature, 110°C; desolvation temperature, 350°C; cone gas flow (nitrogen), 50 liters/h; desolvation gas flow (nitrogen), 800 liters/h; collision gas flow (nitrogen), 0.15 ml/min; and multiplier voltage, 650 V. Chromatography separation was reverse phase using a Waters BEH C18 2.1- by 50-mm column with a flow rate of 0.3 ml/min with a gradient of 10 mM tributylamine plus 15 mM acetic acid in 97:3 water-methanol (solution A) to methanol (solution B) with the following parameters: t = 0 min; A, 99%, and B, 1%, t = 2.5 min; A, 80%, and B, 20%, t = 7.0 min; A, 35%, and B, 65%, t = 7.5 min; A, 5%, and B, 95%, t = 9.01 min; A, 99%, and B, 1%, t = 10 min. A c-di-GMP standard curve for calculating the c-di-GMP concentration in each extract was generated by dissolving synthesized c-di-GMP (Axxora) in extraction buffer to 250 nM followed by 2-fold serial dilutions to 0.975 nM.
Isothermal titration calorimetry.
Apparent dissociation constants (Kd) and interaction stoichiometry of BrpTWT and BrpTP124T with c-di-GMP were measured by isothermal titration calorimetry (ITC) using a Nano ITC instrument from TA Instruments. Calorimetric titrations were done using 2 μM c-di-GMP and 3-μl injections spaced 180 s apart for BrpT and using 300 μM c-di-GMP with 2-μl injections spaced 180 s apart for BrpTP124T. The cuvette contained 30 μM protein, and reactions were conducted at 25°C in 50 mM NaH2PO4 (pH 7.5), 550 mM NaCl, and 0.3 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). The data were analyzed by integrating heat effects normalized to the amount of injected c-di-GMP. Curves were fit based on a single-site binding model using the TA Instruments NanoAnalyze data analysis software (v 3.4.0), and the Kd was derived from the data using standard methods.
EMSAs.
EMSAs were performed using purified BrpTWT and BrpTP124T and infrared 700 (IR700)-labeled probes (IDT). IR700-labeled primers (data not shown) were used to generate PbrpA, PbrpH, and PflpA probes from V. vulnificus genomic DNA. Reaction mixtures contained 5 μM BrpTWT or BrpTP124T, 0.2 mg/ml BSA, 0.1 μg/ml poly(dI-dC) in binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 75 mM NaCl, 10 mM MgCl2, 5% glycerol, 1 mM dithiothreitol [DTT], and 0.1 mM EDTA). When indicated, c-di-GMP was added to a final concentration of 50 μM. After a 30-min incubation at room temperature in the dark, reaction mixtures were loaded onto a 5% Tris-borate-EDTA (TBE)-polyacrylamide gel with 10% glycerol and run in 0.5× TBE. Gels were prerun for 1 h at 50 V before loading, and samples were resolved at 50 V for 2 h. Gels were visualized on an Odysee FC imaging system (Licor).
Structural modeling, phylogenetic analysis, and genomic comparisons.
The crystal structure of VpsT (PDB 3KLO) was used as a scaffold to build structural models for wild-type and mutant BrpT using PHYRE2 (33) and MODELLER (34). Chimera (35) was used for molecular visualization. Five homology models were generated for each protein, and the model with the best overall agreement score was analyzed further. Protein sequences were aligned using T-Coffee (36), and maximum likelihood trees were generated with PhyML (37). Mauve alignments (38) were viewed using Geneious (Biomatters).
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Dann and members of his lab for assistance with ITC and Carl Bauer and Ankur Dalia for insightful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00344-17.
REFERENCES
- 1.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 2.D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 3.Romling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz FH. 2008. Cyclic dimeric GMP signaling and regulation of surface-associated developmental programs. J Bacteriol 190:781–783. doi: 10.1128/JB.01852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha DG, O'Toole GA. 2015. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol Spectr 3:MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hengge R, Grundling A, Jenal U, Ryan R, Yildiz F. 2016. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J Bacteriol 198:15–26. doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner JG, Zamorano-Sanchez D, Park JH, Sondermann H, Yildiz FH. 2017. The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr Opin Microbiol 36:20–29. doi: 10.1016/j.mib.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, Galperin MY, Yildiz FH, Lee VT. 2015. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog 11:e1005232. doi: 10.1371/journal.ppat.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaper S, Steinchen W, Krol E, Altegoer F, Skotnicka D, Sogaard-Andersen L, Bange G, Becker A. 2017. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc Natl Acad Sci U S A 114:E4822–E4831. doi: 10.1073/pnas.1702435114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YC, Chin KH, Tu ZL, He J, Jones CJ, Sanchez DZ, Yildiz FH, Galperin MY, Chou SH. 2016. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun 7:12481. doi: 10.1038/ncomms12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda JM, Newton AE, Bopp CA. 2015. Vibriosis. Clin Lab Med 35:273–288. doi: 10.1016/j.cll.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Todd EC. 1989. Costs of acute bacterial foodborne disease in Canada and the United States. Int J Food Microbiol 9:313–326. doi: 10.1016/0168-1605(89)90099-8. [DOI] [PubMed] [Google Scholar]
- 15.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl Environ Microbiol 74:4199–4209. doi: 10.1128/AEM.00176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Rowe-Magnus DA. 2010. Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus. Infect Immun 78:1390–1402. doi: 10.1128/IAI.01188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol 189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A 96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira RB, Chodur DM, Antunes LC, Trimble MJ, McCarter LL. 2012. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J Bacteriol 194:914–924. doi: 10.1128/JB.05807-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betts MJ, Russell RB. 2003. Amino acid properties and consequences of substitutions, p 289–316. In Barnes MR, Gray IC (ed), Bioinformatics for geneticists. Wiley, Hoboken, NJ. [Google Scholar]
- 23.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 24.Townsley L, Yildiz FH. 2015. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ Microbiol 17:4290–4305. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koenig KL, Mueller J, Rose T. 1991. Vibrio vulnificus. Hazard on the half shell. West J Med 155:400–403. [PMC free article] [PubMed] [Google Scholar]
- 26.Motes ML, DePaola A, Cook DW, Veazey JE, Hunsucker JC, Garthright WE, Blodgett RJ, Chirtel SJ. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl Environ Microbiol 64:1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly MT. 1982. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl Environ Microbiol 44:820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright AC, Powell JL, Kaper JB, Morris JG Jr. 2001. Identification of a group 1-like capsular polysaccharide operon for Vibrio vulnificus. Infect Immun 69:6893–6901. doi: 10.1128/IAI.69.11.6893-6901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuppardo AB, Siebeling RJ. 1998. An epimerase gene essential for capsule synthesis in Vibrio vulnificus. Infect Immun 66:2601–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun 75:5550–5558. doi: 10.1128/IAI.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 32.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eswar N, Eramian D, Webb B, Shen MY, Sali A. 2008. Protein structure modeling with MODELLER. Methods Mol Biol 426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Taly JF, Magis C, Bussotti G, Chang JM, Di Tommaso P, Erb I, Espinosa-Carrasco J, Kemena C, Notredame C. 2011. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc 6:1669–1682. doi: 10.1038/nprot.2011.393. [DOI] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.