Abstract
Multiple clinical and experimental studies have suggested that epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) may be effective at treating advanced non-small cell lung cancer (NSCLC), however, the molecular basis of primary resistance to EGFR-TKIs in NSCLC remains unclear. In the current study, the insulin-like growth factor 1 receptor (IGF-1R) gene in the gefitinib-resistant human lung adenocarcinoma epithelial cell line A549 (A549/GR) was silenced using small interfering RNA (siRNA) in order to determine the role of microRNA (miRNA) in the development of resistance against epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) in lung adenocarcinoma. The relative gefitinib-resistant capacity in A549 and A549/GR cells was determined using a cell counting kit 8. A549/GR cells were transfected with chemically synthesized siRNA to silence the IGF-1R gene. A total of 48 h after siRNA transfection, IGF-1R expression in A549/GR cells was evaluated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting. miRNA expression in A549/GR cells and A549/GR cells with silenced IGF-1R was analyzed using a miRNA microarray. The microarray results of 10 miRNAs were then compared with the results of RT-qPCR. The results demonstrated that the gefitinib-resistance capacity of A549/GR cells was six times higher than that of A549 cells. Additionally, RT-qPCR and western blotting demonstrated that the IGF-1R gene in A549/GR cells was successfully silenced by siRNA. The highest silencing rate (72%) of the IGF-1R gene was obtained using siRNA-2. The microarray identified 72 miRNAs with significantly different expression in A549/GR cells with silenced IGF-1R compared with A549/GR cells. Of the 72 differentially expressed miRNAs, 13 miRNAs (including miR-497-3p and miR-1273c) were up-regulated and 59 miRNAs (including miR-361-3p and miR-345-3p) were down-regulated in A549/GR cells with silenced IGF-1R compared with A549/GR cells. The changes in the expression of 10 different miRNAs were confirmed by RT-qPCR. Thus, the present study successfully established an A549/GR cell line with silenced IGF-1R. The results suggest that a number of miRNAs associated with the IGF-1R signaling pathway, including miR-497-3p and miR-144-5p, were involved in the development of resistance against EGFR-TKIs in A549 cells. These miRNAs may provide novel targets to treat lung adenocarcinoma exhibiting resistance against EGFR-TKIs.
Keywords: non-small cell lung cancer, gefitinib, drug resistance, insulin-like growth factor receptor 1, microRNA
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide. Non-small-cell lung cancer (NSCLC) is the predominant form of lung cancer accounting for ~85% of all lung cancer cases (1). The primary use of systemic chemotherapy in the treatment of NSCLC is palliative and produces only modest pain relief (2). Multiple clinical and experimental studies have suggested that mutations in the epidermal growth factor receptor (EGFR) are an indicator of diseases, such as NSCLS (3) and colorectal cancer (4). Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are molecularly targeted drugs, able to target c-Src and Pak1, among others (5), that are effective at treating patients with advanced NSCLC (6). However, the development of resistance to EGFR-TKIs in tumors inhibits their efficacy. A number of potential mechanisms underlying the development of EGFR-TKI resistance in tumors, including mutations in the KRAS proto-oncogene and ALK receptor tyrosine kinase gene fusion, have been identified and an increasing number of studies are focusing on the regulation of EGFR bypass signaling pathways (7,8). Receptor tyrosine kinases (RTK) stand out among causal dominant oncogenes in NSCLC, and the ablation of RTK signaling has emerged as a novel tailored therapeutic strategy (9), and inhibition of mitogen-activated protein kinase (MEK) may decrease cell viability of NSCLC (10). The EGFR/AKT signaling pathway and the mTOR signaling pathway have demonstrated important roles in EGFR-TKI resistance (11). Other receptor tyrosine kinases (RTK) may activate the downstream phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt), Ras-Raf-mitogen-activated protein kinase 1 (MAPK) and signal transducer and activator of transcription (STAT)/STAT signaling pathways when the EGFR signaling pathway is blocked by EGFR-TKIs (12). As one of the most important components in the EGFR bypass signaling pathways, insulin-like growth factor 1 receptor (IGF-1R) is a potential target. IGF-1R is a transmembrane tetraploid that exhibits tyrosine kinase activity. The binding of IGF-1R to its ligands IGF-1 or IGF-2 induces the auto-phosphorylation of intracellular tyrosine kinase, which promotes the proliferation and metastasis of lung cancer cells by activating the PI3K/Akt and Ras-Raf-MAPK pathways (13).
microRNAs (miRNAs) make up only 1% of the human genome, however they regulate the expression of >30% of all genes (14). The abnormal expression of multiple miRNAs has been identified in the development, angiogenesis, metastasis and drug resistance of lung cancer. The expression of miRNAs associated with the IGF-1R bypass signaling pathways are altered following the inactivation of IGF-1R, which contributes to the proliferation of drug-resistant NSCLC cells (15). In the present study, the expression of IGF-1R in the gefitinib-resistant human lung adenocarcinoma epithelial cell line A549/GR was silenced using small interfering (si)RNA. A number of differentially expressed miRNAs were identified based on the results of a miRNA microarray. It was then determined which of these miRNAs may serve a role in the development of resistance to EGFR-TKIs in patients with NSCLC. The aim of the current study was to identify strategies to inhibit the development of primary resistance to EGFR-TKIs.
Materials and methods
Establishment of the gefitinib-resistant A549/GR cell line
Cells from the human lung carcinoma epithelial cell line A549 (CCL-185; ATCC, Manassas, VA, USA) were used. A569 cells were purchased from American Type Culture Collection (ATCC, Manassas, USA). Gefitinib-resistant A549 cells (A549/GR cells) were established by stepwise selection following exposure to increasing doses of gefitinib (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Briefly, A549 cells were gradually exposed to gefitinib of increasing concentrations of 5, 10, 20 and 30 µM, at room temperature. After 10 months, A549 cells grew stably at 37°C in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 100 U/ml penicillin and 100 g/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 30 µM gefitinib. The stability of the gefitinib-resistant cells was confirmed following culture in gefitinib-free medium for 6 months at room temperature.
Evaluation of A549 and A549/GR cell proliferation using the cell counting kit-8 (CCK-8)
A549 and A549/GR cells in the logarithmic growth phase were seeded onto a 96-well culture plate (2×103-3×103 cells/well), the DMEM was purchased from Gibco; Thermo Fisher Scientific, Inc. Different concentrations of gefitinib (2.5, 5, 10, 20, 30, 40, 60, 80 and 120 µM) were added to the DMEM culture medium (Gibco, MA, USA). For the negative control and blank groups, the same amount of culture medium and PBS, respectively, were added instead of gefitinib. For each group, the experiment was conducted in three wells. The cells were cultured in an incubator containing 5% CO2 at 37°C for 72 h. The medium was then replaced with 10 µl CCK-8 kit (catalogue no. 96992; Sigma-Aldrich; Merck KGaA) and 90 µl medium, and the cells were then cultured at room temperature for 3 h. The optical density (OD) value at 450 nm was determined to evaluate the vital rate (VR) of cells according to the following formula: VR(%)=(OD of the gefitinib group-OD of the blank group)/(OD of the negative control group-OD the blank group) ×100%. The half maximal inhibitory concentration (IC50) of gefitinib was calculated on the basis of the logarithm of the concentration of gefitinib and the linear regression of VR. The resistance index (RI) of gefitinib was calculated using the following formula: RI=IC50 (A549/GR)/IC50 (A549). The experiments were repeated 3 times with duplicate samples per group.
The use of siRNA to silence IGF-1R
To design the IGF-1R siRNA targets using short hairpin, the IGF-1R nucleotide sequence was obtained from NCBI GenBank databases (https://www.ncbi.nlm.nih.gov/gene/?term=NM_000875; Acc-ession number: NM_000875). The online siRNA design tool (http://www.mwg-biotech.com) was used to design three specific siRNAs: forward 5′-TTTCACAGGAGGCTCTCTC-3′ and reverse 5′-GAGAGAGCCTCCTGTGAAA-3′ (siRNA-1); forward 5′-TAGGACTGGATTATTCTCC-3′ and reverse 5′-GGAGAATAATCCAGTCCTA-3′ (siRNA-2) and forward 5′-TTGTACATTGTTGAGG-3′ and reverse 5′-CCATCAACAATGAGTACAA-3′ (siRNA-3), to target the IGF-1R gene in the current study. siRNA sequences, positive and negative controls were synthesized by Guangzhou RuiBo Biological Technology Co., Ltd (Guangzhou, China). siRNAs were stored at a −20°C, at a concentration of 20 pmol/µl. A549/GR cells were divided into six groups: i) siRNA-1 transfection group (cells transfected with siRNA-1); ii) siRNA-2 transfection group (cells transfected with siRNA-2); iii) siRNA-3 transfection group (cells transfected with siRNA-3); iv) blank group, (untransfected A549/GR cells); v) positive control group (A549/GR cells transfected with GAPDH) and vi) negative control group (A549/GR cells transfected with unrelated siRNA). A549/GR cells (0.5×106 per well) were seeded onto a 12-well culture plate. The transfection solution was prepared by mixing 4 µl siRNA-1, siRNA-2 and siRNA-3, respectively, 2 µl lipofectamine reagent (Gibco; Thermo Fisher Scientific, Inc.) and 200 µl DMEM without serum (Gibco; Thermo Fisher Scientific, Inc.). After standing at room temperature for 20 min, the transfection solution was added to each well. Cells were cultured with the transfection solution in an incubator containing 5% CO2 at 37°C for 5–6 h. The supernatant was then replaced with culture medium and cells were cultured at 37°C for 48 h.
Evaluation of IGF-1R mRNA using reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the cells of each group using TRIzol (Thermo Fisher Scientific, Inc., Waltham, MA USA) and the quality and concentration of total RNA was assessed on a UV spectrophotometer (Ultraspec 3300; Biochrom, Ltd., Cambridge, UK). Complementary DNA (cDNA) was obtained using a reverse transcription kit (DRR036A; Takara Bio, Inc., Otsu, Japan). The primers used in qPCR were as follows: IGF-1R, forward, 5′-AACAAGCCCACAGGGTATGG-3′ and reverse, 5′-GCTGACTTGGCAGGCTTGAG-3′; GAPDH, forward, 5′-GGACCTGACCTGCCGTCTAG-3′ and reverse, 5′-GAGGAGTGGGTGTCGCTGTT-3′. The PrimeScript RT Reagent kit (catalogue no. RR391A, Takara Bio, Inc.) was used for qPCR and SYBR Green I (catalogue no. RR086A; Takara Bio, Inc.) was used as the reporter fluorophore, which was conducted in a 20 µl reaction system with 10 µl 2x Mix buffer, forward and reverse primers (0.4 µl each), 1 µl cDNA, and 15.4 µl double distilled water. The reaction was performed as follows: 94°C for 5 min, 30 cycles of 94°C for 1 min, 45°C for 1 min and 72°C for 40 sec. The reaction was performed in a Bio-Rad CFX qPCR machine (Bio-Rad Laboratories, Inc., Hercules CA, USA). The 2−ΔΔCq method was applied for quantification of relative expression of target genes (16).
Evaluation of IGF-1R protein expression using western blotting
The expression of IGF-1R was evaluated using western blot analysis. Cells from each group (2 ml) were seeded into a 6-well plate (2×106 cells/well). Following 24 h incubation at 37°C, cells were collected and centrifuged at 12,000 × g for 10 min at 4°C. The protein was extracted by using cold RIPA buffer (Gibco; Thermo Fisher Scientific, Inc.) and centrifuged at 12,000 × g for 15 min at 4°C. Subsequently, IGF-1R protein was quantified using the Bradford protein quantification method, and by measuring absorbance at 595 nm using a microplate reader (Bio-Rad® Benchmark microplate reader; Bio-Rad Laboratories, Inc.). Proteins (50 µg/well) were separated by 12.5% SDS-PAGE and transferred onto a nitrocellulose membrane. Following blocking in 5% non-fat milk at room temperature for 1 h, the nitrocellulose membrane was rinsed with TBST at 4°C and then incubated with the primary antibodies for IGF-1R (1:200 dilution; catalogue no. #3027; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature overnight. Anti-GAPDH (1:500 dilution; catalogue no. ab8245; Abcam, Cambridge, MA, USA) was used as an internal control. After washing with PBST twice for 10 min, the membranes were incubated horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibodies (1:5,000; catalogue no. ab6721; Abcam) at room temperature for 1 h and visualized using enhanced chemiluminescence (ECL; catalogue no. WP20005; Invitrogen; Thermo Fisher Scientific, Inc.) according to manufacturers' protocol. Band intensity was determined using chemiluminescent (ECL) reagents (Pierce; Thermo Fisher Scientific, Inc.) and analyzed on Image J software version 14.1 (National Institutes of Health, Bethesda, MD, USA).
Microarray analysis of miRNA gene expression
A total of 2,565 miRNA probes were included on the RiBo Array™ miRNA Human Array (Shanghai Biotechnology Corporation, Shanghai, China). The probes were designed using the miRBase database (Sanger miRBase Release 21.0; http://www.mirbase.org). Following hybridization, the laser scanner GenePix 4000B (Molecular Devices, LLC, Sunnyvale, CA, USA) was used to obtain hybridization images. The results were analyzed using the Array-Pro 3.0 software (Axon Instruments, Inc., Union City, CA, USA). The Locally-Weighted Regression (LOWESS) method (17) was used to normalize the microarray results and calculate the fold changes and P-values for the paired t test. Fold changes of >1.5 or <-1.5 were determined to be statistically significant (P<0.05). Significant differences in the expression of miRNAs between the A549/GR-siRNA2 and the negative control group were identified using heat map clustering and scatter plots. The scatter plot was used to assess variation in miRNA expression between the siRNA2 and negative control group. The raw data from the microarray images were then extracted using Feature Extraction software version 11.0.1.1 (Agilent Technologies, Santa Clara, CA, USA), and the raw data was normalized using Locally-Weighted Regression (LOWESS; Silicon Genetics, Inc., Redwood, CA, USA). The values obtained for the X and Y axes in the scatter plot were the normalized signal values determined for the samples (log2 scaled) or the average normalized signal values obtained for the groups of samples (log2 scaled). Heat map clustering was performed using Cluster 3.0 software (http://rana.lbl.gov/EisenSoftware.htm; Lawrence Berkeley National Laboratory, Berkley, CA, USA). The colors in the map present the relative values of all tiles within two groups. Red represents up-regulated and green represents down-regulated miRNAs. The brightness of the color indicates the degree of up- or down-regulation.
Confirmation of microarray results: miRNA levels measured by RT-qPCR
A total of 10 miRNAs that exhibited the most significant differences in expression between the A549/GR cells and A549/GR cells with silenced IGF-1R, were selected for RT-qPCR. Total RNA was isolated from cells of each group using TRIzol reagent according to the manufacturer protocol (Thermo Fisher Scientific, Inc.) and the quality and concentration of total RNA was assessed on a UV spectrophotometer (Ultraspec 3300; Biochrom, Ltd., Cambridge, UK). cDNA was obtained using the One Step Prime Script™ RT-PCR kit (catalogue no. RR064A; Takara Bio, Inc.). The cDNA samples were then used for RT-qPCR measurement with miRNA-specific primers. SYBR Green I was used as the reporter fluorophore (Takara Bio, Inc.). The primers used in this study were as follows: i) miR-144-5p Forward 5′-CTGCACGGATATCATCATAC-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; ii) miR-550a-5p Forward 5′-CTGCACAGTGCCTGAGGGAG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; iii) miR-361-3p Forward 5′-CTGCACTCCCCCACCTG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; iv) miR-345-3p Forward 5′-CTGCACGCCCTGAACGAG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; v) miR-4450 Forward 5′-CTGCACTGGGGATTTGGAGAAG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; vi) miR-135a-3p Forward 5′-CTGCACTATAGGGATTGGAG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; vii) miR-124-3p Forward 5′-CTGCACTAAGGCACGCGGT-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; viii) miR-337-3p Forward 5′-CTGCACCTCCTATATGAT-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; viiii) miR-1273c Forward 5′-CTGTATGGCGACAAAACG-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′; x) miR-497-3p Forward 5′-CTGCGCCAAACCACACTGT-3′, Reverse 5′-GTGCAGGGTCCGAGGT-3′. U6 was used as an internal control, the primers were; Forward 5′-CTGCGCAAGGATGACACG-3′ and Reverse 5′-GTGCAGGGTCCGAGGT-3′. The Takara Kit (Takara Bio, Inc.) was used for qPCR, which was conducted in a 20 µl reaction system with 10 µl 2x Mix buffer, forward and reverse primers (0.4 µl each), 1 µl cDNA, and 15.4 µl double distilled water. The PCR parameters were as follows: 95°C for 1 min, followed by 40 cycles of 95°C for 10 sec and extension at 64°C for 25 sec. The 2−ΔΔCq method was used to determine the relative expression of miRNAs (16).
Statistical analyses
Statistical analyses were conducted using SPSS software 19.0 (IBM Corp., Armonk, NY, USA). Data were expressed as mean ± standard deviation. Comparisons between different groups were conducted using the t-test for two independent samples and P<0.05 was considered to indicate a statistically significant difference.
Results
A549/GR cells exhibit gerfitinib resistance
The results of CCK8 demonstrated that the viability of A549/GR and A549 cells decreased as concentrations of gerfitinib increased in the culture medium. However, the IC50 value of gerfitinib for A549/GR cells was 48.05±4.33 µmol/l, which is significantly higher than that of A549 cells (8.02±0.63 µmol/l; P<0.01; Fig. 1), this suggested that A549/GR exhibits significantly increased resistance to gerfitinib than A549 cells.
Figure 1.
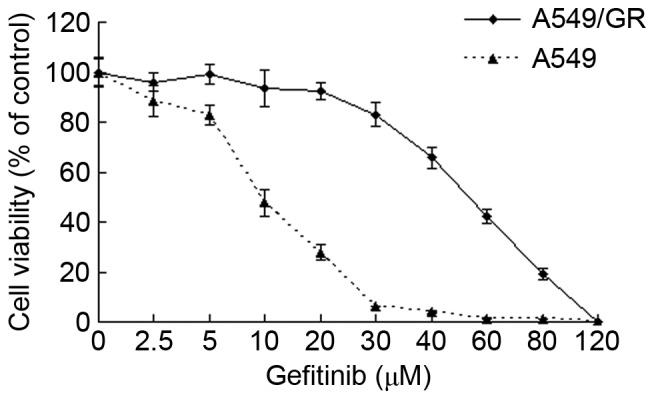
The viability of A549/GR and A549 cells decreased as concentrations of gerfitinib increased in the culture medium. The half maximal inhibitory concentration value of gerfitinib for A549/GR cells was 48.05±4.33 µmol/l, which is significantly greater than that of the A549 cells (8.02±0.63 µmol/l; P<0.01).
IGF-1R expression is successfully silenced by siRNAs
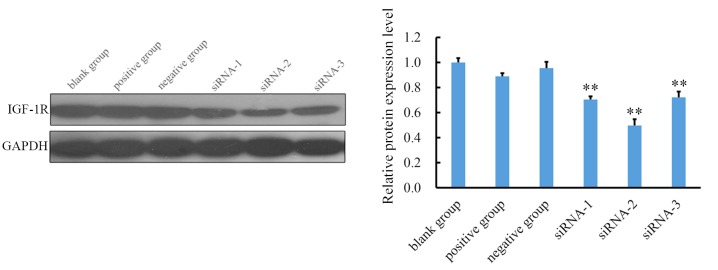
To examine the effect of siRNAs on the expression of IGF-1R, the expression of IGF-1R mRNA and protein was assessed using RT-qPCR and western blotting. The expression of IGF-1R mRNA was inhibited by siRNA-1, siRNA-2, and siRNA-3 (Fig. 2). A total of 48 h after transfection with siRNA, the relative Cq value of IGF-1R for the siRNA-1, siRNA-2, siRNA-3 groups were 0.55±0.05, 0.28±0.02, and 0.62±0.03, respectively. The expression of IGF-1R in all three siRNA groups was significantly lower than in the blank control group (P<0.05; Fig. 2A). These results demonstrate that siRNAs successfully inhibit the expression of IGF-1R mRNA. The siRNA silencing efficiency was calculated according to the following formula: siRNA silencing efficiency=(1-IGF-1R expression in the siRNA group/IGF-1R expression in the blank group) ×100%. siRNA-2 exhibited the highest efficiency (72%) of silencing (Fig. 2B). Western blot analysis demonstrated that in cells transfected with all three siRNAs, there was a significant decrease in IGF-1R expression (P<0.01) and that siRNA-2 exhibited the highest inhibition efficiency (Fig. 3). Therefore, siRNA-2 was used in subsequent experiments to silence IGF-1R and evaluate alterations in miRNA expression.
Figure 2.
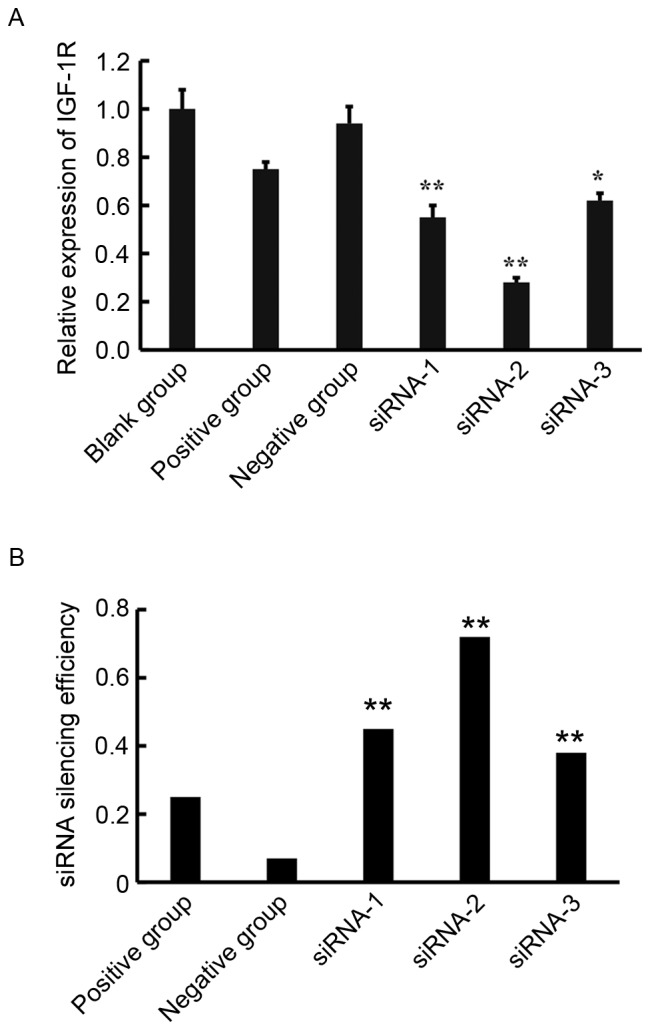
(A) Levels of IGF-1R mRNA in each group were detected by reverse transcription-quantitative polymerase chain reaction. IGF-1R expression was inhibited by transfection with siRNA-1, siRNA-2 and siRNA-3. Expression of IGF-1R mRNA in the three siRNA groups was significantly lower than that of the blank control group Data are expressed as the mean + standard deviation. *P<0.05, **P<0.01 vs. the blank control group. (B) The siRNA silencing efficiency. siRNA silencing efficiency=(1-IGF-1R expression in the siRNA group/IGF-1R expression in the blank group) ×100%. siRNA-2 exhibited the highest efficiency (72%) of silencing. **P<0.01 vs. the positive and negative groups. siRNA, small interfering RNA; IGF-1R, insulin-like growth factor receptor 1.
Figure 3.
Relative protein expression of IGF-1R in each group was detected by western blotting. Levels of IGF-1R in the three siRNA groups were lower than in the blank control group. Quantified data indicated that siRNA-2 exhibited the highest inhibition efficacy. All data are presented as the mean + standard deviation. **P<0.01 vs. blank control group. siRNA, small interfering RNA; IGF-1R, insulin-like growth factor receptor 1.
Differentially expressed miRNAs in A549/GR cells with silenced IGF-1R compared with A549/GR cells
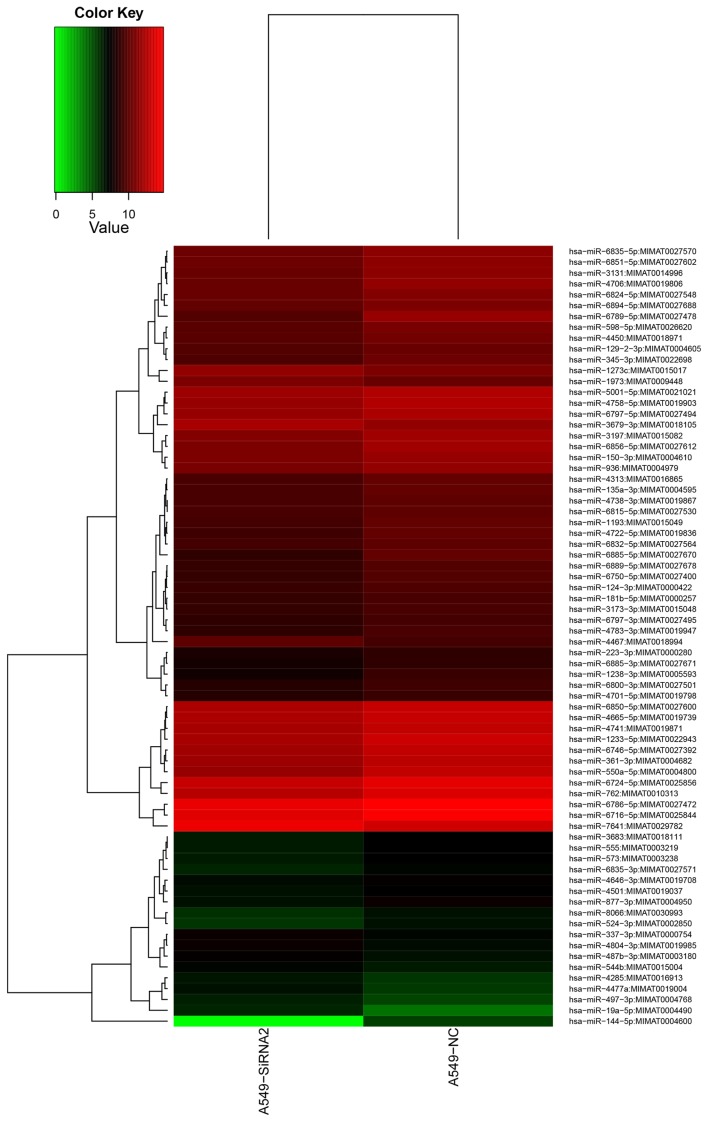
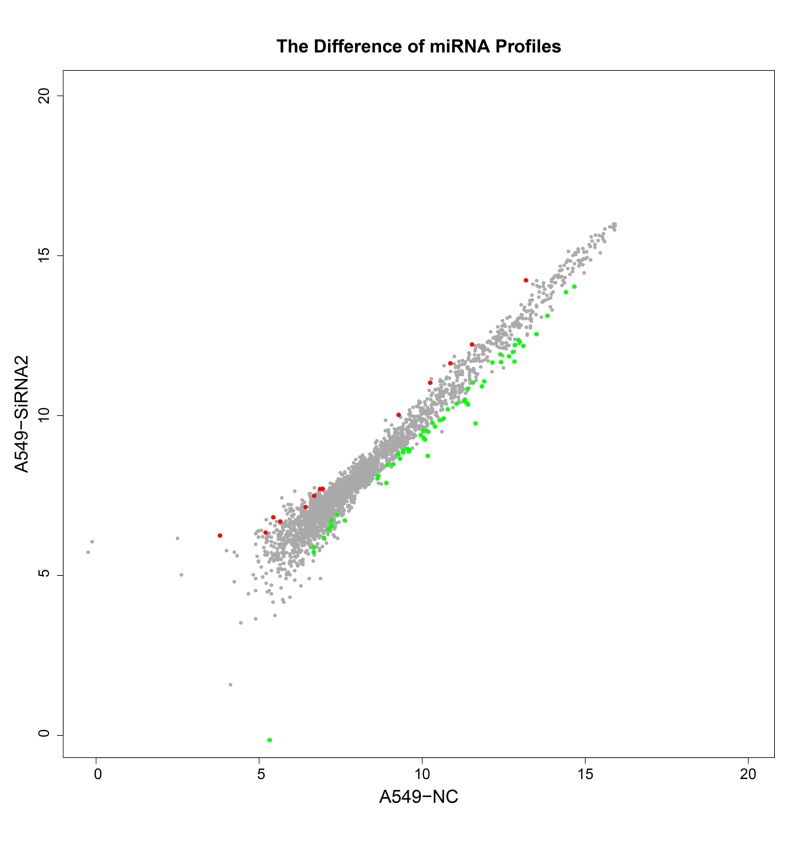
Based on the miRNA microarray, 72 differentially expressed miRNAs were identified in A549/GR cells that had undergone IGF-1R silencing compared with control A549/GR cells (P<0.05). Of the 72 differentially expressed miRNAs, 13 were up-regulated and 59 were down-regulated (Table I). The levels of expression of 15 miRNAs in A549/GR cells with IGF-1R silencing were two times higher or lower than in A549/GR cells. These differentially expressed miRNAs may serve important roles in the development of gerfitinib-resistance in A549/GR cells. Heat map clustering was performed based on ‘All Targets value-miRNAs’ using the siRNA-2 group and negative control group; the clustering results revealed distinct miRNA expression profiles in the two groups (Fig. 4). A scatter plot was constructed indicating the overlapping miRNA expression profiles of the siRNA-2 and negative control groups. The values obtained for the X and Y axes in the scatter-plot are the normalized signal values of the samples (log2 scaled) or the averaged normalized signal values of groups of samples (log2 scaled; Fig. 5). That there were differences in the expression profiles of 10 miRNAs between the siRNA-2 and negative control in A549 cells, indicating that these 10 miRNAs expression profiles were changed after transfection with IGF-1R siRNA-2 in A569 cells. The scatter plot was used to assess variation in miRNA expression between the A549 cells transfected with siRNA-2 and the negative control group. The axes of the scatter plot are the normalized signal values of the samples (the ratio scale), red color represents upregulated miRNAs, green color represents downregulated miRNAs, grey color represents equally-expressed miRNAs.
Table I.
Differentially expressed miRNAs in A549/GR cells with IGF-1R silencing compared with A549/GR cells.
| Number | miRNA name | Fold change | P-value |
|---|---|---|---|
| 1 | hsa-miR-19a-5p | 5.066666667 | 0.001502792 |
| 2 | hsa-miR-4477a | 2.419354839 | 0.006793443 |
| 3 | hsa-miR-497-3p | 2.050632911 | 0.006056884 |
| 4 | hsa-miR-76412 | 1.920393783 | 0.046714308 |
| 5 | hsa-miR-4285 | 1.907407407 | 0.025745132 |
| 6 | hsa-miR-4804-3p | 1.650793651 | 0.014660408 |
| 7 | hsa-miR-487b-3p | 1.621621622 | 0.017437008 |
| 8 | hsa-miR-1973 | 1.595256312 | 0.000733254 |
| 9 | hsa-miR-1273c | 1.58258483 | 0.002789136 |
| 10 | hsa-miR-337-3p | 1.571428571 | 0.025546335 |
| 11 | hsa-miR-4467 | 1.553892216 | 0.028963891 |
| 12 | hsa-miR-544b | 1.532608696 | 0.03936994 |
| 13 | hsa-miR-3679-3p | 1.505596721 | 0.016565788 |
| 14 | hsa-miR-144-5p | −47.63076917 | 0.046283511 |
| 15 | hsa-miR-6789-5p | −3.966396292 | 0.000284606 |
| 16 | hsa-miR-6885-5p | −2.902923977 | 0.043100598 |
| 17 | hsa-miR-550a-5p | −2.372917298 | 0.004168534 |
| 18 | hsa-miR-4706 | −2.249421742 | 0.012286458 |
| 19 | hsa-miR-1238-3p | −2.171066526 | 0.011922191 |
| 20 | hsa-miR-3131 | −2.11235119 | 0.040641938 |
| 21 | hsa-miR-762 | −2.094218057 | 0.006013498 |
| 22 | hsa-miR-524-3p | −2.075471698 | 0.040855564 |
| 23 | hsa-miR-1233-5p | −2.041464467 | 0.024882678 |
| 24 | hsa-miR-6856-5p | −2.034500649 | 0.033718572 |
| 25 | hsa-miR-877-3p | −2.028571429 | 0.032436555 |
| 26 | hsa-miR-4722-5p | −1.928336079 | 0.002188824 |
| 27 | hsa-miR-6851-5p | −1.925488089 | 0.001570519 |
| 28 | hsa-miR-3197 | −1.923085887 | 0.001736544 |
| 29 | hsa-miR-361-3p | −1.891474966 | 0.01367353 |
| 30 | hsa-miR-6835-5p | −1.884668508 | 0.041693405 |
| 31 | hsa-miR-6835-3p | −1.882758621 | 0.007597698 |
| 32 | hsa-miR-6746-5p | −1.862366121 | 0.020066861 |
| 33 | hsa-miR-8066 | −1.848739496 | 0.045020172 |
| 34 | hsa-miR-4758-5p | −1.8122311 | 0.010309085 |
| 35 | hsa-miR-598-5p | −1.808977035 | 0.010516834 |
| 36 | hsa-miR-1193 | −1.808841099 | 0.00103303 |
| 37 | hsa-miR-345-3p | −1.8 | 0.041455795 |
| 38 | hsa-miR-6724-5p | −1.770495483 | 0.006584465 |
| 39 | hsa-miR-6750-5p | −1.769516729 | 0.006643986 |
| 40 | hsa-miR-4313 | −1.741487144 | 0.045341542 |
| 41 | hsa-miR-573 | −1.74137931 | 0.048138838 |
| 42 | hsa-miR-6824-5p | −1.731818182 | 0.022809783 |
| 43 | hsa-miR-4450 | −1.727125068 | 0.046979788 |
| 44 | hsa-miR-6889-5p | −1.725773196 | 0.016773382 |
| 45 | hsa-miR-4665-5p | −1.722864198 | 0.030246907 |
| 46 | hsa-miR-3683 | −1.721925134 | 0.03417578 |
| 47 | hsa-miR-555 | −1.711229947 | 0.023664688 |
| 48 | hsa-miR-4783-3p | −1.706832298 | 0.040236296 |
| 49 | hsa-miR-4741 | −1.67372657 | 0.006643116 |
| 50 | hsa-miR-6800-3p | −1.673211781 | 0.016012437 |
| 51 | hsa-miR-6786-5p | −1.670338552 | 0.038049277 |
| 52 | hsa-miR-135a-3p | −1.638153428 | 0.048112573 |
| 53 | hsa-miR-124-3p | −1.635721493 | 0.039190116 |
| 54 | hsa-miR-6894-5p | −1.624092268 | 0.014021758 |
| 55 | hsa-miR-6885-3p | −1.620229008 | 0.006836833 |
| 56 | hsa-miR-6850-5p | −1.614990998 | 0.019169362 |
| 57 | hsa-miR-6832-5p | −1.608858859 | 0.004509677 |
| 58 | hsa-miR-3173-3p | −1.598476605 | 0.036559297 |
| 59 | hsa-miR-936 | −1.587800875 | 0.00122134 |
| 60 | hsa-miR-6716-5p | −1.576567456 | 0.036987695 |
| 61 | hsa-miR-129-2-3p | −1.565789474 | 0.011634879 |
| 62 | hsa-miR-223-3p | −1.563963964 | 0.011445976 |
| 63 | hsa-miR-150-3p | −1.546714456 | 0.017237636 |
| 64 | hsa-miR-6815-5p | −1.54072553 | 0.020109206 |
| 65 | hsa-miR-4738-3p | −1.538644876 | 0.004713573 |
| 66 | hsa-miR-181b-5p | −1.5375 | 0.029892403 |
| 67 | hsa-miR-4646-3p | −1.52742616 | 0.041419501 |
| 68 | hsa-miR-4501 | −1.523809524 | 0.008659241 |
| 69 | hsa-miR-6797-5p | −1.516079159 | 0.017211813 |
| 70 | hsa-miR-5001-5p | −1.50693994 | 0.037688515 |
| 71 | hsa-miR-6797-3p | −1.502837684 | 0.043167029 |
| 72 | hsa-miR-4701-5p | −1.50212766 | 0.02122897 |
miR, microRNA.
Figure 4.
Heat map clustering of miRNAs expressed in siRNA-2 and negative group. Clustering was performed using Cluster 3.0 software. The colors in the map present the relative values of all tiles within two groups. Red represents up-regulated and green represents down-regulated miRNAs. The brightness of the color indicates the degree of up- or down-regulation.
Figure 5.
Scatter plot. The scatter plot was used to assess variation in miRNA expression between the two groups. The values obtained for the X and Y axes in the scatter plot are the normalized signal values determined for the samples (log2 scaled) or the average normalized signal values obtained for the groups of samples (log2 scaled). miRNA, microRNA; siRNA, small interfering RNA; NC, negative control.
miRNA expression confirmed by RT-qPCR
The miRNA microarray results were further evaluated using RT-qPCR to evaluate the expression of 10 miRNAs, including miR-144-5p, miR-550a-5p, miR-361-3p, miR-345-3p, miR-4450, miR-135a-3p, miR-124-3p, hsa-miR-337-3p, hsa-miR-1273c and hsa-miR-497-3p, which exhibited the strongest hybridization signal. It was demonstrated that the results of RT-qPCR were consistent with those of the microarray, suggesting that the results of the miRNA microarray are reliable (Fig. 6).
Figure 6.
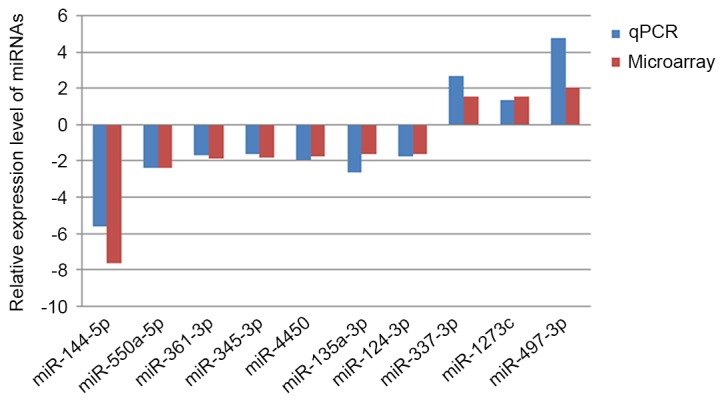
miRNA expression confirmed by RT-qPCR. The results of the miRNA microarray were further evaluated by performing RT-qPCR of 10 miRNAs, including miRNA-144-5p, miR-550a-5p, miR-361-3p, miR-345-3p, miR-4450, miR-135a-3p, miR-124-3p, hsa-miR-337-3p, hsa-miR-1273c and hsa-miR-497-3p. It was demonstrated that the results of RT-qPCR results were consistent with the microarray results. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miRNA, microRNA.
Discussion
IGF-1R and EGFR share two downstream signaling pathways, the Ras-Raf-MAPK and PI3K-Akt signaling pathways, which are involved in cell proliferation and apoptosis, as well as angiogenesis (18). In the present study, A549 cells were selected to identify why gefitinib exhibits poor efficacy in lung cancer cells. A549 cells are Ras-mutated, PI3K-mutated and EGFR wild-type and exhibit resistance to first-generation EGFR TKIs, such as gefitinib (19). Previous studies have demonstrated that EGFR-TKIs are effective against lung cancer in vitro and in vivo (20,21). Inhibition of EGFR by TKIs, including gefitinib and erlotinib, has provided hope for patients with NSCLC. However, a number of studies have reported that one of the EGFR TKIs is a more effective treatment for patients with NSCLC that exhibit EGFR mutations than in patients with wild-type EGFR, as patients with EGFR mutations are typically highly sensitive to EGFR TKIs (22,23). Due to acquired resistance of EGFR TKIs via EGFR-mutant NSCLC that arise through various molecular mechanisms including EGFR secondary mutations, MET proto-oncogene amplification and hepatocyte growth factor (HGF) overexpression, the anti-tumor effect of EGFR TKIs may also be weakened (24,25). Hence, the present study used the EGFR wild-type lung cancer cell line A569 as the cell model, to avoid the EGFR tyrosine kinase secondary mutation in the process of stepwise gefitinib selection.
A previous study by the current authors identified the increased expression of IGF-1R mRNA in A549/GR cells, which is associated with gerfitinib-resistance (26). The results of the current study suggest that IGF-1R activation contributes to the development of secondary resistance to gefitinib. Activated IGF-1R bypasses the EGFR pathway to directly activate the downstream Ras-Raf-MAPK and PI3K-Akt signaling pathways, which promote the proliferation and metastasis of tumor cells and secondary resistance to gefitinib (27).
Aberrant expression of miRNA has been identified in many tumors (28,29). The complementary binding between miRNAs and their target mRNAs induces the formation of RNA-induced silencing complexes (RISCs), which degrade mRNAs or inhibit the translation of mRNAs (30). During cancer development and metastasis, miRNAs acts as a proto-oncogene and a tumor suppressor gene. One miRNA may target multiple mRNAs and several miRNAs may coordinate to regulate the expression of a single mRNA (31). In addition, the transcription of miRNA is regulated by transcription factors, which form a complicated regulatory network (32). The characterization of miRNAs involved in the IGF-1R pathway may facilitate the identification of novel targets to treat different types of cancer that are resistant to gefitinib. As an effective tool for gene knockout, siRNA interference has been widely used in biological research. Following siRNA transfection into host cells, the siRNA duplex melts and integrates into the RISC. The reverse strand of siRNA guides the complementary binding between RISC and mRNA, leading to the efficient and specific degradation of intracellular mRNA and gene silencing (33). Silenced expression of a gene may inactivate the downstream signaling pathway, which facilitates the identification and characterization of the regulatory miRNAs involved in the signaling pathway (34). To efficiently silence a specific gene, a number of siRNAs must be designed and their silencing efficiency compared under the same conditions (35). In the present study, three siRNAs were designed on the basis of the nucleotide sequence of IGF-1R gene and a pair of unrelated nucleotides was used as a negative control.
It has been demonstrated that exogenous double-stranded RNA >50 base pairs long may activate the interferon pathway, thus inducing apoptosis (36). However, the transfection of double-stranded RNA of 19–23 nucleotides long can efficiently and rapidly silence target genes without affecting the interferon pathway. Therefore, in the current study, siRNAs 19 nucleotides long were used to avoid interference with the interferon pathway. In addition, numerous studies have demonstrated that siRNAs containing 30–50% guanine and cytosine (GC) are more efficient at gene silencing than siRNAs containing 52% GC (37). In the present study, the GC contents of the three pairs of siRNA used for IGF-1R silencing were 52, 42 and 37%. Compared with the blank control cells, the levels of IGF-1R mRNA in A549/GR cells significantly decreased following siRNA transfection. Of the three pairs of siRNAs, siRNA2 exhibited the highest (72%) efficiency of IGF-1R gene silencing, which is slightly higher than the 70% efficiency reported in a previous study (38). It has been demonstrated that chemically synthesized siRNAs exhibit a lower impact on downstream components of the signaling pathway than vector-based siRNAs. Therefore, synthesized siRNAs were used in the present study to silence IGF-1R, facilitating the study of the role and mechanisms of IGF-1R in the development of gefitinib-resistance in A549/GR cells.
In the present study, a microarray containing 2,565 miRNA probes from the Sanger miRBase database Release 21.0 was used to identify human miRNAs involving the IGF-1R signaling pathway. Based on analysis of the hybridization results of A549/GR cells and A549/GR cells in which the IGF-1R gene was silenced by siRNA2, a total of 72 differentially expressed miRNA were identified. Of the 72 differentially expressed miRNAs, 59, including miR-19a-5p and miR-4477a, exhibited decreased expression and 13, including miR-144-5p, miR-6789-5p, exhibited increased expression. The expression profiles of 10 miRNAs including miR-144-5p, miR-550a-5p, miR-361-3p, miR-345-3p, miR-4450, miR-135a-3p, miR-124-3, hsa-miR-497-3p, hsa-miR-1273c and hsa-miR-337-3 were measured using RT-qPCR and were consistent with those of the microarray results, suggesting that the miRNA microarray is a reliable tool to use to identify differentially expressed miRNAs. The expression of miR-497-3p was significantly increased in A549/GR cells with silenced IGF-1R. miR-497-3p, a member of the miR-15/16/195/424/497 family, is located on the short arm of chromosome 17 (17p13.1). As with other members of the miR-15/16/195/424/497 family, miR-497-3p is involved in mammalian meiosis by regulating the expression of cell cycle-related genes (39). It has been demonstrated that miR-497 transfection leads to G0/G1 cell cycle arrest in host cells (39). Zhao et al (40) identified the down-regulation of miR-497 in NSCLC tissue, as well as A549, SPC-A1, H1299 and H460 cells. Another study demonstrated that miR-497 promotes the apoptosis of NSCLC cells through the hepatoma-derived growth factor (HDGF) (40). Han et al (41) reported that miR-497 and miR-34a inhibited the proliferation and metastasis of lung cancer cells by binding to the 3′-untranslated region (UTR) of the cyclin E1 gene and inhibiting its expression, the synergistic actions of miR-497 and miR-34a partly correlated with cyclin E1 levels, cyclin E1 is downregulated by both miR-497 and miR-34a, which synergistically retard the growth of human lung cancer cells. In the present study, the expression of miR-497-3p in A549/GR cells with silenced IGF-1R was significantly different than that in A549 cells, suggesting that miR-497-3p is involved in the development of gefitinib resistance in NSCLC cells through the IGF-1R bypass pathway.
Of the 71 differentially expressed miRNAs, miR-144-5p exhibited the most significant difference in expression between A549/GR cells and A549/GR cells with silenced IGF-1R. However, the role of miR-144-5p in the development and progression of tumors is controversial. Studies have suggested that miR-144-5p acts as a tumor suppressor gene during the development of osteosarcoma. Ye et al (42) reported that miR-144-5p promotes the progress of colon cancer. Zha et al (43) determined that the expression of miR-144-5p in NSCLC tissues was significantly lower than in adjacent tissues (P<0.01) and that the expression of miR-144-5p in A549 cells was significantly lower than in normal bronchial epithelial cells (P<0.01). Increased expression of the apoptotic marker apoptosis related protein 3 (ARP3) and cytochrome C has been detected in A549 cells overexpressing miR-144-5p (44). In addition, it has been demonstrated that miR-144-5p inhibits the growth and promotes the apoptosis of A549 cells (45). Therefore, it was speculated that miR-144-5p serves a tumor suppressor role in NSCLC and that zinc finger proteins may be involved in the underlying mechanism. Xiang et al (46) demonstrated that miR-144-5p inhibits the proliferation and invasion of A549 and H460 cells through the TP53-inducible glycolysis and apoptosis regulator. Furthermore, Han et al (47) demonstrated that the down-regulation of miR-223 may activate the IGF-1R/phosphatidylinositol 3-kinase (PI3K)/Akt pathway in lung cancer PC9/ER and PC9/CD133+ cells, suggesting that miR-233 is a potential therapeutic target for overcoming resistance to EGFR-TKIs.
In the present study, the results from the microarray and RT-qPCR identified significant differences in the expression of miR-497-3p and miR-144-5p in A549/GR cells with silenced IGF-1R compared with A549/GR cells. This suggests that miR-497-3p and miR-144-5p stimulate the development of gefitinib-resistance in A549 cells through the IGF-1R bypass pathway. However, the underlying molecular mechanisms have not yet been reported and further studies are therefore required.
In conclusion, the current study identified 72 differentially expressed miRNAs using a miRNA microarray in A549/GR cells in which the IGF-1R gene was silenced using siRNA. The results of the current study provide a basis for investigating the roles of miRNAs in the development of resistance to gefitinib in lung adenocarcinoma. Given that miRNAs involved in the IGF-1R bypass pathway serve important roles in the proliferation of drug-resistant NSCLC cells, the identification of these miRNAs may establish novel targets to treat gefitinib-resistant NSCLC.
Acknowledgements
This study was supported by the Guangzhou Science Technology and Innovation Commission (Grant no. 2011J4100027).
References
- 1.Carbone DP, Gandara DR, Antonia SJ, Zielinski C, Paz-Ares L. Non-Small-Cell Lung Cancer: Role of the immune system and potential for immunotherapy. J Thorac Oncol. 2015;10:974–984. doi: 10.1097/JTO.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Eastern cooperative oncology group: Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Bagheri-Yarmand R, Wang RA, Adam L, Papadimitrakopoulou VV, Clayman GL, El-Naggar A, Lotan R, Barnes CJ, Hong WK, Kumar R. The epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) suppresses c-Src and Pak1 pathways and invasiveness of human cancer cells. Clin Cancer Res. 2004;10:658–667. doi: 10.1158/1078-0432.CCR-0382-03. [DOI] [PubMed] [Google Scholar]
- 6.Kosaka T, Yamaki E, Mogi A, Kuwano H. Mechanisms of resistance to EGFR TKIs and development of a new generation of drugs in non-small-cell lung cancer. J Biomed Biotechnol. 2011;2011:165214. doi: 10.1155/2011/165214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voena C, Di Giacomo F, Panizza E, D'Amico L, Boccalatte FE, Pellegrino E, Todaro M, Recupero D, Tabbò F, Ambrogio C, et al. The EGFR family members sustain the neoplastic phenotype of ALK+ lung adenocarcinoma via EGR1. Oncogenesis. 2013;2:e43. doi: 10.1038/oncsis.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozano MD, Labiano T, Echeveste J, Gurpide A, Martín-Algarra S, Zhang G, Sharma A, Palma JF. Assessment of EGFR and KRAS mutation status from FNAs and core-needle biopsies of non-small cell lung cancer. Cancer Cytopathol. 2015;123:230–236. doi: 10.1002/cncy.21513. [DOI] [PubMed] [Google Scholar]
- 9.Sutterlüty H, Mayer CE, Holzmann K, Miksche M, Berger W. The role of Sprouty proteins as negative regulators of RTK signaling in non-small cell cancer lung cancer (NSCLC) Cancer Res. 2005;65 [Google Scholar]
- 10.Lundholm L, Hååg P, Juntti T, Lewensohn R, Viktorsson K. Phosphoprotein analysis reveals MEK inhibition as a way to target non-small cell lung cancer tumor initiating cells. Int J Radiat Biol. 2014;90:718–726. doi: 10.3109/09553002.2014.905725. [DOI] [PubMed] [Google Scholar]
- 11.Xiang XD, Yu J, Li GF, Xie L, Li H. In vitro study on blocking mTOR signaling pathway in EGFR-TKI resistance NSCLC. Asian Pac J Trop Med. 2014;7:394–397. doi: 10.1016/S1995-7645(14)60063-8. [DOI] [PubMed] [Google Scholar]
- 12.Kudo M. Signaling pathway and molecular-targeted therapy for hepatocellular carcinoma. Dig Dis. 2011;29:289–302. doi: 10.1159/000327562. [DOI] [PubMed] [Google Scholar]
- 13.Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretić L, Lim D, Wang L, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20:1027–1034. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peled N, Wynes MW, Ikeda N, Ohira T, Yoshida K, Qian J, Ilouze M, Brenner R, Kato Y, Mascaux C, Hirsch FR. Insulin-like growth factor-1 receptor (IGF-1R) as a biomarker for resistance to the tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. Cell Oncol (Dordr) 2013;36:277–288. doi: 10.1007/s13402-013-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Ma JZ, Li MD. Normalization of cDNA microarray data using wavelet regressions. Comb Chem High Throughput Screen. 2004;7:783–791. doi: 10.2174/1386207043328274. [DOI] [PubMed] [Google Scholar]
- 18.Zhao R, Chen M, Jiang Z, Zhao F, Xi B, Zhang X, Fu H, Zhou K. Platycodin-D induced autophagy in non-small cell lung cancer cells via PI3K/Akt/mTOR and MAPK signaling pathways. J Cancer. 2015;6:623–631. doi: 10.7150/jca.11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, Fujii T, Kuwano M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–472. [PubMed] [Google Scholar]
- 20.Xie M, He CS, Wei SH, Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur J Cancer. 2013;49:3559–3572. doi: 10.1016/j.ejca.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–289. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JY, Wu SG, Yang CH, Chang YL, Chang YC, Hsu YC, Shih JY, Yang PC. Comparison of gefitinib and erlotinib in advanced NSCLC and the effect of EGFR mutations. Lung Cancer. 2011;72:205–212. doi: 10.1016/j.lungcan.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Li T, Wang X, Ye M, Cai J, Xu Y, Wu B. Gene-guided gefitinib switch maintenance therapy for patients with advanced EGFR mutation-positive non-small cell lung cancer: An economic analysis. BMC Cancer. 2013;13:39. doi: 10.1186/1471-2407-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nurwidya F, Takahashi F, Murakami A, Kobayashi I, Kato M, Shukuya T, Tajima K, Shimada N, Takahashi K. Acquired resistance of non-small cell lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Respir Investig. 2014;52:82–91. doi: 10.1016/j.resinv.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Hata A, Katakami N, Yoshioka H, Takeshita J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119:4325–4332. doi: 10.1002/cncr.28364. [DOI] [PubMed] [Google Scholar]
- 26.Ning LL, Wei MA, Liu J, Wang HP, Yu-Jun LI, Respiration DO. Establishment of a gefitinib-resistant cell line of human lung adenocarcinoma and its drug resistant mechanism. China Tropical Med. 2014;14:1045–1049. [Google Scholar]
- 27.Luo M, Fu LW. Redundant kinase activation and resistance of EGFR-tyrosine kinase inhibitors. Am J Cancer Res. 2014;4:608–628. [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang Y, Zhao J, McCrae MA, Zhuang H. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118:2431–2442. doi: 10.1002/cncr.26566. [DOI] [PubMed] [Google Scholar]
- 29.Manikandan M, Rao DM, Munirajan A. Analyzing the expression of candidate microRNAs in primary tumors of oral squamous cell carcinoma. Mol Cytogenetics. 2014;7:P7. doi: 10.1186/1755-8166-7-S1-P7. [DOI] [Google Scholar]
- 30.Kawasaki H, Fukuda Y, Taira K. Small RNA-mediated chromatin modification and transcriptional gene silencing. Gene Therapy Reg. 2004;2:351–367. doi: 10.1163/1568558043967463. [DOI] [Google Scholar]
- 31.Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW, Zhu MX, Lin HC, Chu DD, Yao M, Yan MX. MicroRNA-148a suppresses invasion and metastasis of human non-small-cell lung cancer. Cell Physiol Biochem. 2015;37:1847–1856. doi: 10.1159/000438546. [DOI] [PubMed] [Google Scholar]
- 32.Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: A combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288:77–87. doi: 10.1007/s00438-013-0734-z. [DOI] [PubMed] [Google Scholar]
- 33.Pham JW, Sontheimer EJ. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- 34.Leung AW, Hung SS, Backstrom I, Ricaurte D, Kwok B, Poon S, McKinney S, Segovia R, Rawji J, Qadir MA, et al. Combined Use of gene expression modeling and siRNA screening identifies genes and pathways which enhance the activity of cisplatin when added at no effect levels to non-small cell lung cancer cells in vitro. PLoS One. 2016;11:e0150675. doi: 10.1371/journal.pone.0150675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sledz CA, Williams BR. RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans. 2004;32:952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 36.Weise F. Review of siRNA/shRNA applications in cell-based microarrays. Springer; New York: 2014. pp. 17–31. [DOI] [Google Scholar]
- 37.Pan M, Ni J, He H, Gao S, Duan X. New paradigms on siRNA local application. BMB Rep. 2015;48:147–152. doi: 10.5483/BMBRep.2015.48.3.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Ou M, Keough E, Roberts J, Koeplinger K, Lyman M, Fauty S, Carlini E, Stern M, Zhang R, et al. Quantitation of physiological and biochemical barriers to siRNA liver delivery via lipid nanoparticle platform. Mol Pharm. 2014;11:1424–1434. doi: 10.1021/mp400584h. [DOI] [PubMed] [Google Scholar]
- 39.Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: Evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao WY, Wang Y, An ZJ, Shi CG, Zhu GA, Wang B, Lu MY, Pan CK, Chen P. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem Biophys Res Commun. 2013;435:466–471. doi: 10.1016/j.bbrc.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Han Z, Zhang Y, Yang Q, Liu B, Wu J, Zhang Y, Yang C, Jiang Y. miR-497 and miR-34a retard lung cancer growth by co-inhibiting cyclin E1 (CCNE1) Oncotarget. 2015;6:13149–13163. doi: 10.18632/oncotarget.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye JJ, Cao J. MicroRNAs in colorectal cancer as markers and targets: Recent advances. World J Gastroenterol. 2014;20:4288–4299. doi: 10.3748/wjg.v20.i15.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8:e74175. doi: 10.1371/journal.pone.0074175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson CA, Burcham PC. Protein alkylation, transcriptional responses and cytochrome c release during acrolein toxicity in A549 cells: Influence of nucleophilic culture media constituents. Toxicol In Vitro. 2008;22:844–853. doi: 10.1016/j.tiv.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Zha WJ, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8:e74175. doi: 10.1371/journal.pone.0074175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang C, Cui SP, Ke Y. MiR-144 inhibits cell proliferation of renal cell carcinoma by targeting MTOR. J Huazhong Univ Sci Technolog Med Sci. 2016;36:186–192. doi: 10.1007/s11596-016-1564-0. [DOI] [PubMed] [Google Scholar]
- 47.Han J, Zhao F, Zhang J, Zhu H, Ma H, Li X, Peng L, Sun J, Chen Z. miR-223 reverses the resistance of EGFR-TKIs through IGF1R/PI3K/Akt signaling pathway. Int J Oncol. 2016;48:1855–1867. doi: 10.3892/ijo.2016.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]