Abstract
Primary cutaneous amyloidosis (PCA) is a localized skin disorder that is characterized by the abnormal deposition of amyloid in the extracellular matrix (ECM) of the dermis. The pathogenesis of PCA is poorly understood. The objective of the present study was to survey proteome changes in PCA lesions in order to gain insight into the molecular basis and pathogenesis of PCA. Total protein from PCA lesions and normal skin tissue samples were extracted and analyzed using the isobaric tags for relative and absolute quantitation technique. The function of differentially expressed proteins in PCA were analyzed by gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction analysis. The proteins that were most upregulated in PCA lesions were further analyzed by immunohistochemistry. A total of 1,032 proteins were identified in PCA lesions and control skin samples, with 51 proteins differentially expressed in PCA lesions, of which 27 were upregulated. In PCA lesions, the upregulated proteins were primarily extracellulary located. In addition, GO analysis indicated that the upregulated proteins were significantly enriched in the biological processes of epidermal development, collagen fiber organization and response to wounding (adjusted P<0.001). KEGG analysis indicated that the upregulated proteins were significantly enriched in the signaling pathways of cell communication, ECM receptor interaction and focal adhesion (adjusted P<0.001). Furthermore, the upregulated proteins were enriched in the molecular function of calcium ion binding, and the calcium binding proteins calmodulin-like protein 5, S100 calcium-binding protein A7 (S100A7)/fatty-acid binding protein and S100A8/A9 exhibited the highest levels of upregulation in PCA. This analysis of differentially expressed proteins in PCA suggests that increased focal adhesion, differentiation and wound healing is associated with the pathogenesis of PCA.
Keywords: primary cutaneous amyloidosis, iTRAQ, skin, amyloidosis
Introduction
Amyloidosis is a heterogeneous group of diseases characterized by the deposition of amyloid in the ECM, with pathological consequences on the affected tissues or organs (1). Primary cutaneous amyloidosis (PCA) is a localized amyloidosis that affects only the skin, resulting in discolored bumps or nodules, and can be further divided into lichen amyloidosis (LA), macular amyloidosis and nodular amyloidosis (2). The course of the PCA is prolonged; patients often experience severe itchiness, seriously affecting their daily lives (3). PCA has a higher incidence in Asian populations (4). Pathogenic factors for PCA include prolonged friction, and genetic and environmental causes; however, the precise molecular mechanisms underlying the pathogenesis of PCA remain unclear (5).
Numerous previous studies investigating amyloidosis, including PCA, have focused on the identification and classification of the precursor proteins in amyloid deposits, since the pathological process and effects of treatment are associated with the type of amyloid precursor (6). To date, >20 types of amyloid precursors have been identified, and cytokeratins (CKs), serum amyloid P, apolipoprotein E (ApoE), galectins and actins have been observed in PCA amyloid deposits (7–10). Previous studies on the genetic causes of PCA have identified mutations in several genes, including those encoding oncostatin M receptor and interleukin-31 receptor A (11,12). A previous study noted a reduction in the expression of several interfollicular keratinocyte stem cell markers, and differences in the expression of proteins involved in apoptosis and nerve conduction, in PCA samples (13). However, to the best of our knowledge, a survey of proteome changes in PCA has not yet been reported.
Comparative proteomics has been successfully used to identify differentially expressed proteins in various disease conditions and provide insights into the molecular mechanisms underlying these diseases, in addition to diagnostic candidates. For example, Lu et al (14) compared the proteome of islets between a type 2 diabetes mouse model and nondiabetes control mice, and identified differential proteins related to insulin-secretory defects and mutiple novel proteins related to diabetes. Also using a comparative proteomics stretegy, Masui et al (15) identified 29 proteins differentially expressed in metastatic and primary renal cell carcinoma, among which profilin-1, 14-3-3ζ and galectin-1 were demonstrated to be associated with poor prognosis. In the present study, a comparative proteomics study of PCA was performed, which identified numerous differentially expressed proteins, providing novel insights into the pathology of the disease.
Materials and methods
Tissue samples
Skin tissues from 10 patients diagnosed with LA (26–64 years old; 4 males and 6 females) and 10 healthy individuals (2–60 years old; 5 males and 5 females) were collected from January 2009 to December 2013 at Qilu Hospital (Shandong, China) for quantitative proteomics analysis. Additional skin samples from 29 patients diagnosed with LA (29–71 years old; 15 males and 14 females) and 17 healthy individuals (18–70 years old; 9 males and 8 females) were also collected from January 2009 to December 2013 at Qilu Hospital for immunohistochemistry (IHC) experiments. The diagnosis of LA was made by two physicians according to the diagnostic standards for PCA and histopathological examination (2). All PCA samples were also validated by crystal violet staining. For quantitative proteomics analysis, lesions with a size of 1×2 cm were obtained surgically, immediately frozen in liquid nitrogen and then stored at −80°C until required. For IHC analysis, skin tissue samples were fixed at room temperature with 4% formaldehyde for 24 h and formaldehyde (4%)-acetic acid (5%)-ethanol (45%) for a further 3 h, followed by stepwise dehydration with 80% ethanol for 2 h, 90% ethanol for 3 h, 95% ethanol overnight, 100% ethanol twice (20 min/treatment) and xylene twice (20 min/treatment). The fixed and dehydrated tissues were immersed in paraffin wax twice (30 min/treatment) and then embedded into paraffin blocks until use. The present study was approved by the Ethics Committee of Qilu Hospital. All patients provided written informed consent for the inclusion of their skin samples in the present study.
Sample preparation for mass spectrometry analysis
Tissue samples from each group were combined and ground into powder in liquid nitrogen. The powders were washed once with cold trichloroacetic acid/acetone at −20°C for 2 h, twice with cold acetone at −20°C for 30 min each, lyzed in lysis buffer (8 M urea, 30 mM hydroxyethyl piperazineethanesulfonic acid, 1 mM phenylmethylsulfonyl fluoride, 2 mM EDTA and 10 mM dithiothreitol) and sonicated. The proteins in the supernatants were precfigipitated with acetone and resuspended in resuspension buffer [50% triethylammonium bicarbonate (TEAB) and 0.1% SDS]. Total protein concentration was determined using the Bradford method (16). Proteins (100 µg) were digested with 3.3 µg of sequencing grade trypsin at 37°C for 24 h and with an additi onal 1 µg for a further 12 h. The digested proteins were frozen and lyophilized overnight in vacuum and resuspended in 61 µl TEAB solution. The digestion efficiency was assessed with UltraFlex MALDI-TOF/TOF (Bruker Corporation, Ettlingen, Germany) to see whether there were a large number of peaks with signal-to-noise ratio >10 and ≥20 peaks with intensities >1,000 within m/z of 500–3,000 Da.
Isobaric tags for relative and absolute quantitation (iTRAQ) labeling of peptides
Tryptic peptides from healthy and diseased groups were labeled with two different isobaric tags, using iTRAQ reagents 8-plex (SCIEX, Framingham, MA, USA) according to the manufacturer's protocol. Briefly, each iTRAQ reagent was dissolved in 70 µl isopropanol, then added to each specimen prior to a 2 h incubation at room temperature. The labeled peptides were then vacuum-dried.
Liquid chromatography tandem-mass spectrometry (LC-MS/MS) analysis
All iTRAQ-labeled tryptic peptides from the two groups were combined for relative quantification analysis by LC-MS/MS using a Q Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The combined samples were first fractionated into 30–40 fractions by strong cation exchange chromatography (Luna SCX, 4.6×250 mm, Phenomenex, Torrance, CA, USA) as described previously (17). After desalting on a C18 column, the relative amount of peptides in each fraction was assessed by MALDI/TOF, and neighboring fractions with lower signals were pooled, yielding 10 fractions. Each combined fraction was subjected to MS in the standard MS/MS data-dependent acquisition mode. The MS spectra were surveyed from 350–2,000 Da and the method of collision-induced dissociation was chosen.
Data analysis for peptide identification and quantification
Protein identification and quantification were performed using Mascot software (version 2.3.01; Matrix Science, Inc., Boston, MA, USA). The raw MS/MS data of the iTRAQ-labeled peptides was first converted into an msg file and then searched against the NCBI human reference sequence database (www.ncbi.nlm.nih.gov, downloaded on the 5th December 2011 with 216,686 sequences). The database-searching parameters were as follows: Carbamidomethyl (C) for fixed modifications, oxidation (M), Gln->Pyro-Glu (N-term Q), iTRAQ8plex (K), iTRAQ8plex (Y), and iTRAQ8plex (N-term) for variable modification, 1 for maximum missed cleavages, and 15 ppm and 20 mmu for peptide and MS/MS tolerance, respectively. The iTRAQ8plex modification rate of the spectrum was used for monitoring the labeling efficiency rate. For quantitative analysis, the protein ratio type was set at the median, the minimum unique peptides at 2 and the normalization method was the median ratio. P<0.05 was considered to indicate a statistically significant difference. The differential threshold was set at 1.5. Student's t-tests were performed using R software (version 3.2.4; https://www.r-project.org).
IHC analysis
Paraffin-embedded skin tissues were sliced into 3-µm-thick sections and mounted on glass slides. IHC was performed according to the protocols provided by the manufacturers of the primary antibodies (Abcam, Cambridge, UK) and PowerVision™ Two-Step Histostaining reagent (OriGene Technologies, Inc., Beijing, China). After de-waxing (placed in a 65°C incubator for 30 min, twice in dimethylbenzene at room temperature for 30 min), rehydration (5 min for each incubation at room temperature with descending ethanol from 100, 95, 85, to a final 75%), antigen retrieval and 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) blocking at room temperature for 1 h, the slides were incubated with primary antibodies (1:5,000 anti-S100A7 ab83543, 1:20,000 anti-S100A8 ab92331, and 1:20,000 anti-CALML5 ab154631) at 4°C overnight, followed by incubation with PowerVision™ Two-Step Histostaining reagent at room temperature for 20 min. The slides were then stained with 3,3′-diaminobenzidine for 5 min and hematoxylin for 2 min at room temperature. An Olympus BX-51 microscope (Olympus Corporation, Tokyo, Japan) equipped with a digital camera system was used to capture images of the slides. IHC staining on epidermal cells was quantified using ImageJ v1.50 software (National Institutes of Health, Bethesda, MD, USA). Threshold was determined for each target protein at the point where true negatives were about to show color. The percentage of the area that met the threshold was multiplied by the mean intensity of the area to generate an integrated score. Wilcoxon signed-rank tests were performed to compare IHC results between the two groups, using R software. P<0.05 was considered to indicate a statistically significant difference.
Bioinformatics analysis
Gene ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using a gene list enrichment analysis tool, Enrichr (http://amp.pharm.mssm.edu/Enrichr) (18). The output of Enrichr provides adjusted P values computed using the Fisher exact test, Z score computed by assessing the deviation from the expected rank, and combined score from multiplying log (adjusted P-value) and Z score. Adjusted P<0.001 was considered to indicate a statistically significant difference. Protein-protein interaction (PPI) analysis was performed using a gene interaction analysis tool, genes2FANs (http://actin.pharm.mssm.edu/genes2FANs) (19). A maximum path length of 2 and significance cutoff of 2.5 were used.
Results
Identification of differentially expressed proteins in PCA skin tissue
To identify proteins that are differentially expressed in PCA skin tissue, comparative proteomic analysis was performed on pooled diseased and normal skin tissues. Proteins extracted from skin tissues were trypsin-digested and labeled with iTRAQ reagents. The labeled samples from diseased and normal skin tissues were then combined prior to LC-MS/MS analysis. A total of 318,710 spectra were captured and 1,032 proteins were identified, with a false discovery rate of <1%. Among them, 533 proteins had at least two unique peptides and were subject to quantitative comparison. Using a strict 1.5-fold change and P<0.05 threshold, 51 proteins were identified to be differentially expressed between diseased and normal skin tissue (Table I). Among these, 27 were upregulated and 24 were downregulated in PCA lesions compared with normal skin tissue.
Table I.
Differentially expressed proteins identified in PCA.
| Accession no. | Gene abbreviation | Gene name | Expression ratioa | P-value |
|---|---|---|---|---|
| 4557581 | FABP5 | Fatty acid-binding protein, epidermal | 2.990 | 6.05×10−10 |
| 1195531 | KRT16 | Type I keratin 16 | 2.757 | 1.61×10−6 |
| 189053201 | S100A9 | S100 calcium-binding protein A9 | 2.602 | 9.96×10−26 |
| 223278387 | CALML5 | Calmodulin-like protein 5 (calmodulin-like skin protein) | 2.598 | 3.50×10−4 |
| 21614544 | S100A8 | S100 calcium-binding protein A8 | 2.557 | 1.32×10−6 |
| 12053626 | S100A7 | Psoriasin (S100 calcium-binding protein A7) | 2.315 | 2.11×10−24 |
| 194387918 | KRT6A | Keratin, type II cytoskeletal 6A | 2.088 | 4.70×10−7 |
| 553348 | TNC | Hexabrachion | 1.992 | 1.51×10−5 |
| 457464 | DSC1 | Dsc1a precursor (desmocollin 1) | 1.983 | 4.18×10−3 |
| 124056490 | COL3A1 | Collagen α1 (III) chain | 1.845 | 3.57×10−61 |
| 6912286 | CASP14 | Caspase-14 precursor | 1.834 | 2.08×10−12 |
| 49660012 | TPM1 | Sarcomeric tropomyosin κ | 1.821 | 2.05×10−3 |
| 1942977 | MIF | Chain a, macrophage migration inhibitory factor | 1.816 | 4.84×10−7 |
| 4503113 | CST6 | Cystatin-M precursor | 1.809 | 1.39×10−2 |
| 110349772 | COL1A1 | Collagen α1 (I) chain preproprotein | 1.744 | 0.00×100 |
| 115527066 | COL6A2 | Collagen α2 (VI) chain isoform 2C2a precursor | 1.734 | 1.52×10−3 |
| 178849 | APOE | Apolipoprotein E | 1.720 | 2.47×10−7 |
| 119585301 | COL7A1 | Collagen, type VII, α1 (epidermolysis bullosa, dystrophic, dominant and recessive), isoform CRA_e | 1.703 | 1.26×10−3 |
| 4505047 | LUM | Lumican precursor | 1.700 | 7.83×10−168 |
| 453157 | FMOD | Fibromodulin | 1.689 | 1.68×10−2 |
| 48255905 | TAGLN | Transgelin (SM22) | 1.683 | 1.10×10−18 |
| 183583553 | COL4A5 | Collagen α5 (VI) chain | 1.681 | 2.29×10−2 |
| 21708099 | DPT | Dermatopontin | 1.647 | 6.17×10−12 |
| 32451581 | COL1A2 | Collagen, type I, α2 | 1.640 | 1.03×10−138 |
| 4885165 | CSTA | Cystatin-A | 1.567 | 1.61×10−2 |
| 166343769 | gi|166343769 | Periostin isoform thy6 | 1.557 | 5.03×10−20 |
| 49456765 | SFN | Stratifin | 1.519 | 1.12×10−20 |
| 113414871 | KRT77 | Keratin 77 (type II) | 0.664 | 2.24×10−2 |
| 126035028 | HIST1H1E | Liver histone H1e | 0.646 | 2.35×10−4 |
| 119573007 | APOA2 | Apolipoprotein A-II, isoform CRA_d | 0.644 | 4.99×10−13 |
| 221045918 | TUBB4B | Tubulin β-4B chain | 0.643 | 1.77×10−5 |
| 167614485 | ACAA2 | 3-ketoacyl-coa thiolase, mitochondrial | 0.642 | 5.18×10−5 |
| 9857759 | gi|9857759 | Recombinant IgG4 heavy chain | 0.633 | 1.53×10−50 |
| 193785892 | ATP2A1 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 0.631 | 1.55×10−3 |
| 4502011 | AK1 | Adenylate kinase isoenzyme 1 | 0.620 | 2.98×10−3 |
| 221041416 | ACSL1 | Long-chain-fatty-acid-coA ligase 1 | 0.619 | 2.69×10−3 |
| 297304885 | FHL1 | PREDICTED: Four and a half LIM domains protein 1 | 0.613 | 3.48×10−3 |
| 194387956 | ADH1B | Alcohol dehydrogenase 1B | 0.593 | 6.27×10−17 |
| 61743954 | AHNAK | Neuroblast differentiation-associated protein AHNAK isoform 1 | 0.587 | 5.05×10−126 |
| 109731497 | MYH1 | Myosin, heavy chain 1, skeletal muscle, adult | 0.573 | 3.85×10−5 |
| 4506335 | PVALB | Parvalbumin α | 0.518 | 3.43×10−2 |
| 194374151 | ENO3 | β-enolase; 2-phospho-D-glycerate hydro-lyase | 0.504 | 9.95×10−4 |
| 34071 | KRT15 | Keratin 15 (type I) | 0.501 | 2.98×10−16 |
| 153791586 | MYH2 | Myosin-2 | 0.492 | 1.16×10−28 |
| 109111058 | TPM2 | PREDICTED: β-tropomyosin isoform | 0.445 | 1.22×10−2 |
| 825683 | TMSB4X | Thymosin β4 | 0.427 | 1.05×10−2 |
| 4507617 | TNNC2 | Troponin C, skeletal muscle | 0.421 | 3.93×10−7 |
| 28372499 | MYL2 | Myosin regulatory light chain 2, skeletal muscle isoform | 0.412 | 5.07×10−16 |
| 168985192 | MB | Myoglobin | 0.356 | 8.45×10−6 |
| 189053833 | CKM | Creatine kinase, muscle (CKM) | 0.354 | 1.04×10−11 |
| 17986273 | MYL1 | Myosin light chain 1/3, skeletal Muscle isoform isoform 1f | 0.304 | 2.95×10−15 |
Protein ratio of the PCA group vs. the control group. PCA, primary cutaneous amyloidosis.
Analysis of differentially expressed proteins in PCA lesions
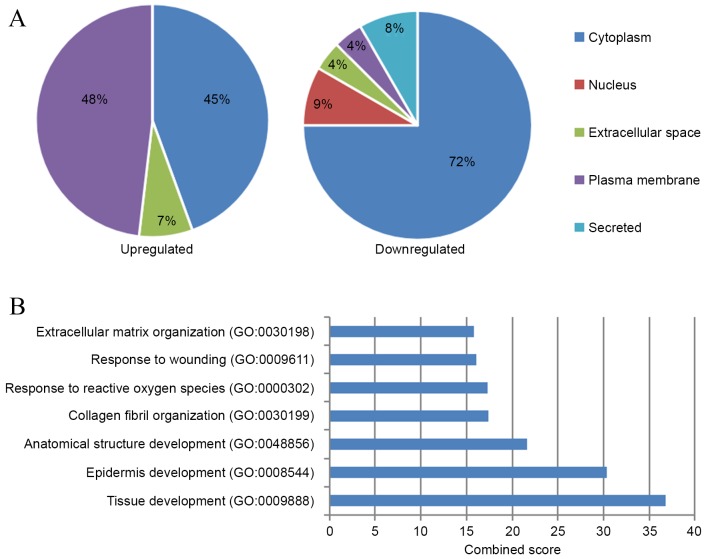
Subcellular distribution analysis of the differentially expressed proteins identified in PCA (Fig. 1A) revealed that the upregulated proteins were markedly enriched in the plasma membrane (48%), among which one known amyloid protein, ApoE, was identified (Table I). Only one plasma membrane protein was identified out of the downregulated proteins (4%), while the majority of downregulated proteins were cytoplasmic (72%).
Figure 1.
Subcellular distribution and functional analysis of differentially expressed proteins in PCA. (A) Subcellular distribution of differentially expressed proteins in PCA lesions. Pie charts indicate the percentage of various subcellular locations of upregulated (left) and downregulated (right) proteins. (B) Biological processes that the upregulated proteins in PCA were significantly enriched in (adjusted P<0.001). PCA, primary cutaneous amyloidosis; GO, gene ontology.
GO term enrichment analysis suggested that upregulated proteins in PCA (Fig. 1B) were significantly enriched in seven biological processes (adjusted P<0.001). The processes with the highest combined score were tissue development (GO:0009888) and epidermis development (GO:0008544), which both included S100 calcium-binding protein A7 (S100A7), collagen type III α1 chain (COL3A1), COL7A1, calmodulin-like protein 5 (CALML5), caspase-14 precursor and fatty acid-binding protein, epidermal (FABP5). The processes with the next highest scores were anatomical structure development (GO:0048856), collagen fibril organization (GO:0030199), response to reactive oxygen species (GO:0000302), response to wounding (GO:0009611) and ECM organization (GO:0030198). The only significantly enriched molecular function of the upregulated proteins in PCA was calcium ion binding (GO:0005509; adjusted P=0.004; data not shown). For downregulated proteins in PCA, the most enriched cellular process was regulation of muscle contraction (GO:0006937; adjusted P=0.003) and structural constituent of muscle (GO:0008307; adjusted P<0.001) (data not shown).
KEGG pathway enrichment analysis revealed that the upregulated proteins in PCA were enriched in three pathways as follows: Cell communication, ECM receptor interaction and focal adhesion (adjusted P<0.001; data not shown). The proteins involved in these three signaling pathways include COL3A1, COL6A2, COL1A2, COL1A1 and hexabrachion (TNC). No significantly enriched signaling pathways were identified for the downregulated proteins.
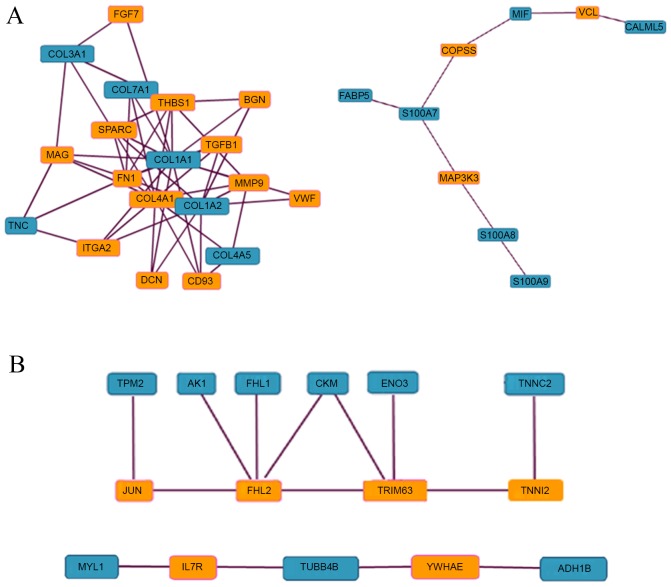
PPI analysis using a maximum path length of 2 and significance cutoff of 2.5 revealed two clusters of interaction among upregulated proteins (Fig. 2A). One of these included COL3A1, COL7A1, COL1A1, COL1A2, COL4A5 and TNC, the majority of which are involved in focal adhesion according to enrichment analysis. Within this cluster, COL1A1, COL1A2 and TNC directly interact with integrin α2 (ITGA2), the receptor in focal adhesion. The second cluster included S100A9, S100A8, S100A7, FABP5, macrophage migration inhibitory factor and CALML5, among which S100A9 and S100A8, and S100A7 and FABP5 interact directly. The majority of the proteins in this second cluster function in calcium ion binding according to enrichment analysis. For downregulated proteins, there were also two clusters of interaction (Fig. 2B). One cluster contained 6 downregulated proteins that formed a network through 4 intermediate proteins that interacted with one another. Troponin C interacts with troponin I fast skeletal muscle isoform 2, β-enolase and creatinine kinase muscle (CKM) interact with tripartite motif-containing protein 63; CKM, four and a half LIM domains protein 1 (FHL1) and adenylate kinase isoenzyme 1 all interact with FHL2; and β-tropomyosin interacts with JUN. In the second cluster, a downregulated protein tubulin β-4B chain interacted with another 2 downregulated proteins, myosin light chain 1/3 and alcohol dehydrogenase 1B, through the intermediate proteins interleukin 7 receptor and 14-3-3 protein ε, respectively.
Figure 2.
PPI analysis of differentially expressed proteins in PCA. Proteins that were (A) upregulated and (B) downregulated proteins in PCA were subjected to PPI analysis using a maximum path length of 2 and significance cutoff of 2.5. Blue and orange boxes indicate proteins that were and were not identified as differentially expressed proteins, respectively. PPI, protein-protein interaction; PCA, primary cutaneous amyloidosis.
Distribution of upregulated calcium ion binding proteins in PCA
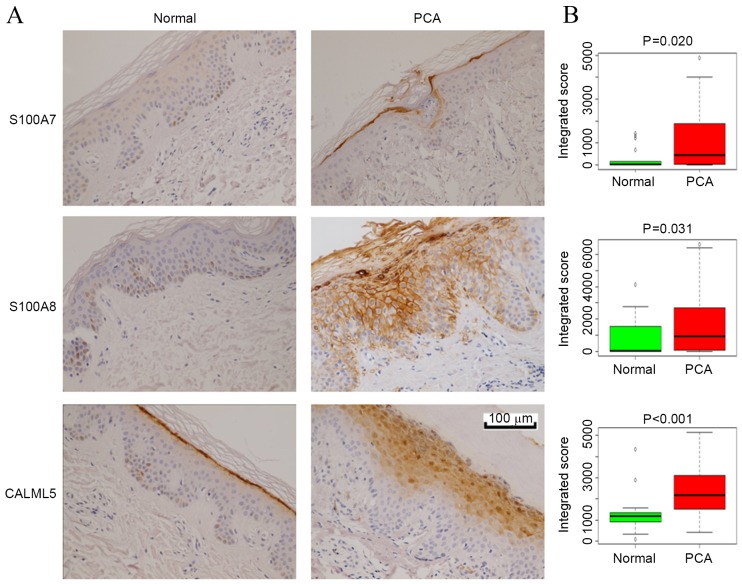
Calcium ion binding was identified as the most enriched molecular function associated with the upregulated proteins in PCA, and the calcium binding proteins CALML5, S100A7/FABP5 and S100A8/A9 were among the most upregulated proteins in the PCA samples, with fold changes >2. To validate these results and to observe the distribution of these proteins in epidermal layers, IHC analysis for CALML5, S100A7 and S100A8 was performed on skin tissues from 29 LA and 17 control samples. Fig. 3A shows representative IHC staining images and Fig. 3B presents quantification of the IHC results. The staining for all three proteins in PCA skin tissues was significantly stronger compared with normal skin tissues (P<0.05), which was consistent with the proteomic results. S100A7 and S100A8 were expressed in the cornified and granular layers in PCA lesions, while they were generally absent in normal skin. CALML5 was expressed primarily in the outermost cornified layer in normal skin, while it was expressed in the granular layers in PCA.
Figure 3.
IHC validation of selected differentially expressed proteins in PCA. (A) Representative images of IHC analysis. Skin samples from normal controls (n=17) and patients with PCA (n=29) were subjected to S100A7, S100A8 or CALML5 staining. (B) Box and whisker plots of integrated scores from the IHC staining of epidermal cells in the normal and PCA groups. S100A7/8, S100 calcium-binding protein A7/8; CALML5, calmodulin-like protein 5; IHC, immunohistochemistry; PCA, primary cutaneous amyloidosis.
Discussion
In the present study, comparative proteomics technology was used to identify 51 differentially expressed proteins in PCA lesions. Three of the upregulated proteins identified were also verified by IHC analysis. In PCA lesions, the majority of upregulated proteins were located extracellularly. It was demonstrated that the upregulated proteins were associated with the biological processes of epidermal development, collagen fiber organization and response to wounding, and the molecular function of calcium ion binding. The upregulated proteins were also significantly enriched in the signaling pathways of cell communication, ECM receptor interaction and focal adhesion.
Amyloid deposition in the ECM is the key feature of PCA as well as all other amyloidosis. Among all reported PCA amyloid components, only ApoE was demonstrated to be upregulated in PCA lesions in the present study. ApoE is an apolipoprotein that transports cholesterol and triglycerides in the blood (20). The function of ApoE in Alzheimer's disease, an amyloidosis of the brain, is to affect the fate of amyloid β (Aβ), which is deposited into the extracellular space after it undergoes a configuration change (21). However, the deposition of Aβ has not previously been reported in PCA. Whether ApoE serves a similar function to other amyloid proteins in PCA requires further study.
CKs are known to be the major amyloid proteins in PCA. The following four CKs have been reported to be present in amyloid deposits in descending order of abundance: CK5, CK1, CK14 and CK10 (22). In the present study, these four CKs were identified in PCA lesions; however, their abundance was not significantly different between the PCA and control samples (fold change, 0.82–1.22), which could be due to the fact that whole tissue lysates rather than isolated amyloid deposits were used.
In the present study, half of the upregulated extracellular proteins (6/12) in PCA lesions were collagens, which were identified to be enriched in the signaling pathways of cell communication, ECM receptor interaction and focal adhesion. Focal adhesions mediate a dynamic connection between cells and the ECM, functioning as a mechanical connection and biological signaling center. Integrins serve as mediators in focal adhesion, connecting extracellular collagens to proteins inside the cells. PPI analysis revealed that two upregulated collagens in PCA, COL1A1 and COL1A2, could directly bind to ITGA2. In addition to the six collagens, five additional upregulated extracellular proteins were associated with collagen function. TNC, a large ECM glycoprotein, is a component in focal adhesion and can bind to the ITGA2 (23). Lumican precursor (LUM), a dermatan sulfate proteoglycan, regulates the assembly of collagen microfibers (24). Fibromodulin (FMOD), a small leucine-rich glycoprotein, regulates the generation of collagen microfibers (25). Dermatopontin (DPT), a widely distributed small ECM protein, is primarily located on the surface of collagen fibers in skin where it exerts cell adhesion activity (26). Periostin, also called osteoblast-specific factor 2, regulates collagen fibrillogenesis (27). Periostin can also facilitate integrin-dependent cell adhesion and mobility through the binding of its C-terminal domain to ECM proteins, including collagens and fibronectin. Different isoforms of periostin have distinct effects on the formation of ECM fibers (28). A total of ~41% (11/27) of the upregulated proteins identified in PCA lesions in the present study were collagens or functional components in collagen fiber organization, suggesting that focal adhesion mediated by collagens serves a key role in PCA.
Calcium ion binding was identified as the top enriched molecular function associated with upregulated proteins in PCA in the present study, and the calcium binding proteins CALML5, S100A7/FABP5 and S100A8/A9 were among the top upregulated proteins, with a fold change >2. This may indicate the presence of a high Ca2+ concentration in PCA lesions. Calcium ions serve key functions in numerous cellular processes. Thus, as Ca2+ sensors and transductors, calcium-binding proteins are involved in numerous biological processes (29). For example, S100A7, a member of the S100 protein family, regulates essential cellular activities, including proliferation, differentiation, apoptosis, signal transduction and lipid metabolism. S100A7 is also involved in cell adhesion and migration as a substrate of transglutaminase, and is a precursor of cornified envelope formation (30). FABP5, the binding partner of S100A7, forms a complex with S100A7 in the cytoplasm in the absence of Ca2+ (31). Upon treatment with Ca2+, the complex moves to the outer region of the cell where it colocalizes with α-actinin and paxillin in a focal adhesion-like structure (32). S100A8 forms a heterodimer with S100A9 in a Ca2+-dependent manner, which is involved in inflammation, regulating cell growth and differentiation, inhibiting cell proliferation and inducing apoptosis (33). CALML5, also known as calmodulin-like skin protein, serves a function in keratinocyte differentiation (34). CALML5 is primarily located in the cornified and granular layers in the epidermis, and CALML5 positive cells are typically undergoing differentiation rather than proliferation (35). Using IHC in the present study, it was identified that CALML5 positive cells expanded from the outer cornified layer in normal skin to the granular layers in PLA lesions, which indicates an elevation of differentiation in PCA. An increase in differentiation in the epidermis of PCA lesions was also indicated in previous studies by the elevation of several keratinocyte differentiation markers, including desmocollin-1 (36), cystatin-M precursor (37) and Caspase-14 (38). Another indicator of increased differentiation is the downregulation of keratin 15 (KRT15), as observed in the present study, because its expression is normally limited to proliferating keratinocytes in the basal layer and is reduced when cells start to differentiate and move outwards (39).
A key pathogenic factor in PCA is prolonged and repeated friction, which may cause small wounds in the skin and trigger the wound healing process. GO analysis performed in the present study demonstrated that the upregulated proteins in PCA were significantly enriched in the process of response to wounding, suggesting that the wound healing process was triggered. The enriched proteins included S100A8, S100A9, TNC, COL3A1 and sarcomeric tropomyosin κ. Several additional proteins that have previously been reported to be involved in wound healing were identified to be upregulated in PCA in the present study, including TNC (40), LUM (24), FMOD (25) and DPT (26). In addition, the downregulation of KRT15 in keratinocytes has previously been reported during wound healing (41) and was demonstrated to be downregulated in PCA lesions in the present study. These results suggest a wound healing process occurs in PCA lesions. However, whether this is a causative association or not remains unclear.
In conclusion, the current study identified numerous proteins that are differentially expressed in PCA. Analysis of the upregulated proteins suggests a possible increase in focal adhesion, differentiation and wound healing activities, and a possible increase in Ca2+ concentration, which is consistent with the symptom of skin hyperplasia and the key pathogenic factor of prolonged friction. The present study expands the knowledge of the molecular basis of PCA and provides novel insights into the pathology of the disease.
Acknowledgements
The present study was supported by Shandong Province Science and Technology Development Projects (grant no. 2015GSF118060), and the Science Foundation of Qilu Hospital of Shandong University (grant no. 2015QLMS16).
Glossary
Abbreviations
- ECM
extracellular matrix
- GO
gene ontology
- IHC
immunohistochemistry
- iTRAQ
isobaric tags for relative and absolute quantitation
- LA
lichen amyloidosis
- PCA
primary cutaneous amyloidosis
- PPI
protein-protein interaction
References
- 1.Pinney JH, Hawkins PN. Amyloidosis. Ann Clin Biochem. 2012;49:229–241. doi: 10.1258/acb.2011.011225. [DOI] [PubMed] [Google Scholar]
- 2.Zhao B. The Chinese clinical dermatology. Phoenix Science press; Nanjing: 2010. pp. 1395–1401. (In Chinese) [Google Scholar]
- 3.Fang S, Shen X, Chen AJ, Li S, Shan K. Health-related quality of life in patients with primary cutaneous amyloidosis. PLoS One. 2015;10:e0120623. doi: 10.1371/journal.pone.0120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishna A, Nath B, Dhir GG, Kumari R, Budhiraja V, Singh K. Study on epidemiology of cutaneous amyloidosis in northern India and effectiveness of dimethylsulphoxide in cutaneous amyloidosis. Indian Dermatol Online J. 2012;3:182–186. doi: 10.4103/2229-5178.101814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka A, Arita K, Lai-Cheong JE, Palisson F, Hide M, McGrath JA. New insight into mechanisms of pruritus from molecular studies on familial primary localized cutaneous amyloidosis. Br J Dermatol. 2009;161:1217–1224. doi: 10.1111/j.1365-2133.2009.09311.x. [DOI] [PubMed] [Google Scholar]
- 6.Brambilla F, Lavatelli F, Merlini G, Mauri P. Clinical proteomics for diagnosis and typing of systemic amyloidosis. Proteomics Clin Appl. 2013;7:136–143. doi: 10.1002/prca.201200097. [DOI] [PubMed] [Google Scholar]
- 7.Apaydin R, Gürbüz Y, Bayramgürler D, Müezzinoglu B, Bilen N. Cytokeratin expression in lichen amyloidosis and macular amyloidosis. J Eur Acad Dermatol Venereol. 2004;18:305–309. doi: 10.1111/j.1468-3083.2004.00905.x. [DOI] [PubMed] [Google Scholar]
- 8.Furumoto H, Shimizu T, Asagami C, Muto M, Takahashi M, Hoshii Y, Ishihara T, Nakamura K. Apolipoprotein E is present in primary localized cutaneous amyloidosis. J Invest Dermatol. 1998;111:417–421. doi: 10.1046/j.1523-1747.1998.00294.x. [DOI] [PubMed] [Google Scholar]
- 9.Furumoto H, Hashimoto Y, Muto M, Shimizu T, Nakamura K. Apolipoprotein E4 is associated with primary localized cutaneous amyloidosis. J Invest Dermatol. 2002;119:532–533. doi: 10.1046/j.1523-1747.2002.18581.x. [DOI] [PubMed] [Google Scholar]
- 10.Miura Y, Harumiya S, Ono K, Fujimoto E, Akiyama M, Fujii N, Kawano H, Wachi H, Tajima S. Galectin-7 and actin are components of amyloid deposit of localized cutaneous amyloidosis. Exp Dermatol. 2013;22:36–40. doi: 10.1111/exd.12065. [DOI] [PubMed] [Google Scholar]
- 11.Saeedi M, Ebrahim-Habibi A, Haghighi A, Zarrabi F, Amoli MM, Robati RM. A novel missense mutation in oncostatin M receptor beta causing primary localized cutaneous amyloidosis. Biomed Res Int. 2014;2014:653724. doi: 10.1155/2014/653724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MW, Lee DD, Liu TT, Lin YF, Chen SY, Huang CC, Weng HY, Liu YF, Tanaka A, Arita K, et al. Novel IL31RA gene mutation and ancestral OSMR mutant allele in familial primary cutaneous amyloidosis. Eur J Hum Genet. 2010;18:26–32. doi: 10.1038/ejhg.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka A, Lai-Cheong JE, van den Akker PC, Nagy N, Millington G, Diercks GF, van Voorst Vader PC, Clements SE, Almaani N, Techanukul T, et al. The molecular skin pathology of familial primary localized cutaneous amyloidosis. Exp Dermatol. 2010;19:416–423. doi: 10.1111/j.1600-0625.2010.01083.x. [DOI] [PubMed] [Google Scholar]
- 14.Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of type 2 diabetes: A quantitative proteomics approach. Mol Cell Proteomics. 2008;7:1434–1451. doi: 10.1074/mcp.M700478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masui O, White NM, DeSouza LV, Krakovska O, Matta A, Metias S, Khalil B, Romaschin AD, Honey RJ, Stewart R, et al. Quantitative proteomic analysis in metastatic renal cell carcinoma reveals a unique set of proteins with potential prognostic significance. Mol Cell Proteomics. 2013;12:132–144. doi: 10.1074/mcp.M112.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Zhu Y, Wang Z, Zhu H, Pan Q, Su S, Dong Y, Li L, Zhang H, Wu L, et al. mTORC1 alters the expression of glycolytic genes by regulating KPNA2 abundances. J Proteomics. 2016;136:13–24. doi: 10.1016/j.jprot.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dannenfelser R, Clark NR, Ma'ayan A. Genes2FANs: Connecting genes through functional association networks. BMC Bioinformatics. 2012;13:156. doi: 10.1186/1471-2105-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 21.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YT, Liu HN, Wang WJ, Lee DD, Tsai SF. A study of cytokeratin profiles in localized cutaneous amyloidsis. Arch Dermatol Res. 2004;296:83–88. doi: 10.1007/s00403-004-0474-3. [DOI] [PubMed] [Google Scholar]
- 23.Midwood KS, Orend G. The role of tenascin-C in tissue injury and tumorigenesis. J Cell Commun Signal. 2009;3:287–310. doi: 10.1007/s12079-009-0075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XJ, Kong FZ, Wang YH, Zheng JH, Wan WD, Deng CL, Mao GY, Li J, Yang XM, Zhang YL, et al. Lumican accelerates wound healing by enhancing α2β1 integrin-mediated fibroblast contractility. PLoS One. 2013;8:e67124. doi: 10.1371/journal.pone.0067124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Z, Nguyen C, Zhang X, Khorasani H, Wang JZ, Zara JN, Chu F, Yin W, Pang S, Le A, et al. Delayed wound closure in fibromodulin-deficient mice is associated with increased TGF-β3 signaling. J Invest Dermatol. 2011;131:769–778. doi: 10.1038/jid.2010.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto O, Fujiwara S. Dermatopontin, a novel player in the biology of the extracellular matrix. Connect Tissue Res. 2006;47:177–189. doi: 10.1080/03008200600846564. [DOI] [PubMed] [Google Scholar]
- 27.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, et al. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morra L, Moch H. MPeriostin expression and epithelial-mesenchymal transition in cancer: A review and an update. Virchows Arch. 2011;459:465–475. doi: 10.1007/s00428-011-1151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Méhul B, Bernard D, Brouard M, Delattre C, Schmidt R. Influence of calcium on the proteolytic degradation of the calmodulin-like skin protein (calmodulin-like protein 5) in psoriatic epidermis. Exp Dermatol. 2006;15:469–477. doi: 10.1111/j.1600-0625.2006.00433.x. [DOI] [PubMed] [Google Scholar]
- 30.Ruse M, Lambert A, Robinson N, Ryan D, Shon KJ, Eckert RL. S100A7, S100A10, and S100A11 are transglutaminase substrates. Biochemistry. 2001;40:3167–3173. doi: 10.1021/bi0019747. [DOI] [PubMed] [Google Scholar]
- 31.Hagens G, Masouyé I, Augsburger E, Hotz R, Saurat JH, Siegenthaler G. Calcium-binding protein S100A7 and epidermal-type fatty acid-binding protein are associated in the cytosol of human keratinocytes. Biochem J. 1999;339:419–427. doi: 10.1042/bj3390419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Méhul B, Bernard D, Schmidt R. Calmodulin-like skin protein: A new marker of keratinocyte differentiation. J Invent Dermatol. 2001;116:905–909. doi: 10.1046/j.0022-202x.2001.01376.x. [DOI] [PubMed] [Google Scholar]
- 33.Kerkhoff C, Klempt M, Sorg C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9) Biochim Biophys Acta. 1998;1448:200–211. doi: 10.1016/S0167-4889(98)00144-X. [DOI] [PubMed] [Google Scholar]
- 34.Méhul B, Bernard D, Simonetti L, Bernard MA, Schmidt R. Identification and cloning of a new calmodulin-like protein from human epidermis. J Biol Chem. 2000;275:12841–12847. doi: 10.1074/jbc.275.17.12841. [DOI] [PubMed] [Google Scholar]
- 35.Watson PH, Leygue ER, Murphy LC. Psoriasin (S100A7) Int J Biochem Cell Biol. 1998;30:567–571. doi: 10.1016/S1357-2725(97)00066-6. [DOI] [PubMed] [Google Scholar]
- 36.Henkler F, Strom M, Mathers K, Cordingley H, Sullivan K, King I. Trangenic misexpression of the differentiation-specific desmocollin isoform 1 in basal keratinocytes. J Invest Dermatol. 2001;116:144–149. doi: 10.1046/j.1523-1747.2001.00234.x. [DOI] [PubMed] [Google Scholar]
- 37.Zeeuwen PL, van Vlijmen-Willems IM, Cheng T, Rodijk-Olthuis D, Hitomi K, Hara-Nishimura I, John S, Smyth N, Reinheckel T, Hendriks WJ, Schalkwijk J. The cystatin M/E- cathepsin L balance is essential for tissue homeostasis in epidermis, hair follicles, and cornea. FASEB J. 2010;24:3744–3755. doi: 10.1096/fj.10-155879. [DOI] [PubMed] [Google Scholar]
- 38.Brocklehurst K, Philpott MP. Cysteine proteases: Mode of action and role in epidermal differentiation. Cell Tissue Res. 2013;351:237–244. doi: 10.1007/s00441-013-1557-2. [DOI] [PubMed] [Google Scholar]
- 39.Bose A, Teh MT, Mackenzie IC, Waseem A. Keratin k15 as a biomarker of epidermal stem cells. Int J Mol Sci. 2013;14:19385–19398. doi: 10.3390/ijms141019385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa K, Ito M, Takeuchi K, Nakada A, Heishi M, Suto H, Mitsuishi K, Sugita Y, Ogawa H, Ra C. Tenascin-C is upregulated in the skin lesions of patients with atopic dermatitis. J Dermatol Sci. 2005;40:35–41. doi: 10.1016/j.jdermsci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Werner S, Munz B. Suppression of keratin 15 expression by transforming growth factor beta in vitro and by cutaneous injury in vivo. Exp Cell Res. 2000;254:80–90. doi: 10.1006/excr.1999.4726. [DOI] [PubMed] [Google Scholar]