Abstract
Endoplasmic reticulum (ER) sheet membranes are covered with ribosomes and RNAs that are involved in protein synthesis. A new study reveals that a calcium-activated endoribonuclease of the EndoU protein family promotes the formation of tubular ER networks, contributing to dynamic shaping of the ER in cells.
The endoplasmic reticulum (ER) is a continuous membrane system comprising the nuclear envelope, flat sheets often studded with ribosomes, and a polygonal network of mostly smooth tubules extending throughout the cell. Synthesis, modification, and transport of lipids and proteins as well as Ca2+ sequestration and protein quality control within the ER have been extensively investigated over many years, but mechanisms responsible for the distinctive morphology of the ER have only been uncovered more recently [1,2]. Several eukaryotic protein families, including reticulons and REEPs/DP1/Yop1p, harbor hydrophobic hairpin domains that partially insert into the lipid bilayer, shaping high-curvature ER tubules [3]. Members of the atlastin/RHD3/Sey1p family of large, membrane-bound GTPases mediate the formation of three-way junctions via homotypic membrane fusion, generating the reticulated tubular ER network [4,5]. Additional classes of tubular ER proteins, including some REEPs and the ATPase M1 spastin (which severs microtubules), interact with the cytoskeleton [6,7]. Flat ER sheets possess a different cadre of proteins, such as p180, CLIMP-63 and kinectin, which have been implicated in shaping, cisternal stacking and cytoskeletal interactions, with reticulons shaping the high-curvature edges [8]. Other proteins, including members of the Lunapark, SNARE, and Rab protein families, have also been suggested to have a role in shaping the ER network [1]. In a recent issue of the Journal of Cell Biology, Schwarz and Blower [9] identify a new and unexpected member of the cellular ER-shaping team — the Ca2+-activated ribonuclease XendoU (for Xenopus EndoU), previously studied mostly for its roles in processing intron-encoded small nucleolar RNAs and in viral replication [10–12].
Dramatic changes in intracellular organization and organelle structure occur during developmental differentiation, and this is certainly true for the ER network. Though published images of the ER tend to paint a static picture, the ER is in fact in constant motion, and numerous signaling pathways as well as interactions among cytoskeletal elements, the plasma membrane, and organelles cooperate to position and shape the ER dynamically. Striking morphological changes in the ER occur during cellular events, such as fertilization and cell division. For example, within minutes of fertilization, the ER in starfish eggs undergoes fragmentation, accompanied by Ca2+ release from internal stores [13]. Ca2+-induced, reversible ER fragmentation has also been reported in cell lines and neurons [14,15], prefiguring key roles for signaling pathways in the regulation of ER morphology.
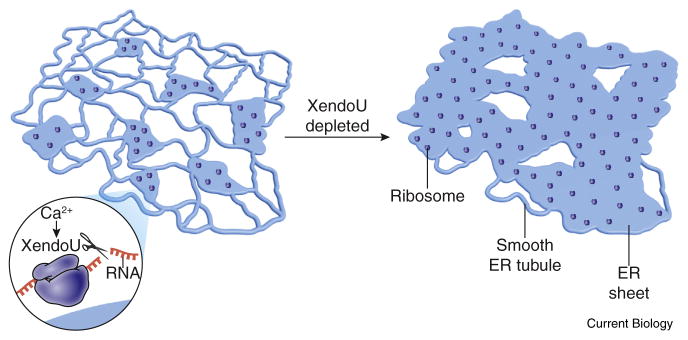
In the new work, Schwarz and Blower [9] set out to investigate the role of Ca2+, which is increased upon fertilization, in the developmental transition from oocyte to embryo. Starting with metaphase-arrested Xenopus egg extracts, they added Ca2+ to mimic the cytoplasmic Ca2+ influx that occurs from both intracellular and extracellular stores at fertilization. They then purified a Ca2+-dependent ribonuclease activity, identifying the protein as the XendoU ribonuclease. The authors further found that a subpopulation of XendoU is tightly membrane bound at the ER surface, where it functions in local RNA degradation. This Ca2+-dependent degradation results in the removal of ribosomes, ribonuclear proteins (RNPs) and RNAs from the ER surface (Figure 1), favoring ER tubule formation and thus helping to regulate the dynamic balance between ER sheets and tubules. Depletion of XendoU caused expansion of sheets at the expense of tubules, an alteration that could be rescued by XendoU in a catalysis-dependent manner. The authors concluded that Ca2+-dependent removal of RNA, ribosomes, and RNPs from the membrane by XendoU promotes ER remodeling and the formation of tubular ER [9].
Figure 1.
Schematic diagram of effects of XendoU on ER morphology.
Ca2+-activated XendoU favors ER tubule formation, with its depletion or loss of catalytic activity resulting in expansion of ER sheets. (Image drawn by Ethan Tyler.)
Mechanistically, there are a number of possibilities for how XendoU functions in regulating ER morphology. First, there could be sheet stability conferred by the presence of ribosomes. Thus, removing them would destabilize sheets and help promote tubule formation. In this regard, ER network formation in vitro using purified Xenopus egg membranes was inhibited by specific antibodies against XendoU, emphasizing the direct role of the membrane-bound subpopulation of XendoU. Schwarz and Blower [9] postulate that oligomerization of atlastin GTPases mediates membrane fusion and subsequent Ca2+ release through Ca2+ channels on the membrane. XendoU would then be activated by Ca2+ and degrade RNA locally, resulting in the release of ribosomes, RNPs, and RNA [9]. Studies of Xenopus, human, and viral EndoU orthologs (including XendoU, PP11, and Nsp15) have demonstrated that these Ca2+-dependent endonucleases are relatively nonspecific, cleaving RNAs in vitro after UU dinucleotides or a single U nucleotide. Interestingly, mild treatment of salt-washed vesicles with non-specific RNase A facilitated network formation, while an excess of RNAse A was very disruptive, emphasizing more generally the importance of RNA [9]. Similar findings were observed in mammalian HeLa cells, with effects on ER morphology also dependent on EndoU catalytic activity. Protein scaffolding could conceivably play a role, since a protein of the XendoU family produced by haemocytes of the moth Heliothis virescens can form large, amyloid-like fibrils at ER sheets, and these are released upon immune challenge [16].
Ca2+ is a ubiquitous signaling molecule implicated in a plethora of cellular pathways involved in organelle changes, including many that affect the ER. For instance, B lymphocyte activation is accompanied by increased Ca2+ signaling and expansion of the ER, with downregulation of EndoU suppressing activation-induced cell death in these cells [17]. Another example is found in neurons, where prominent dynamic changes in ER morphology occur in neuronal dendrites [18]. In this latter case, proteins including synaptic glutamate receptors rapidly diffuse within the continuous network of dendritic ER but are confined by increased ER complexity at branch points of dendrites and near dendritic spines. The spatial range of receptor mobility is rapidly restricted by phosphoinositide-linked metabotropic glutamate receptor signaling, which is linked to intracellular Ca2+ release via inositol (1,4,5) trisphophate (IP3) receptor channels, through a mechanism involving protein kinase C and the ER sheet protein CLIMP63. The morphological changes in local zones of ER have the effect of compartmentalizing ER export and also correspond to sites of new dendritic branches [18]. It will be particularly important to assess any role forXendoU in such Ca2+-dependent processes, and small-molecule inhibitors effective against XendoU in the low micromolar range provide additional tools for such studies [12].
In future studies, investigations of the function of EndoU proteins in vivo, particularly within cells in tissues such as the central nervous system, will be of particular interest. Though these proteins are known to be aberrantly expressed in human diseases, such as cancer, loss of function may similarly be related to disease. In fact, many proteins involved in shaping the ER network are mutated in neurological disorders, including hereditary spastic paraplegia and hereditary sensory neuropathy [19,20]. It will be interesting to probe any links of EndoU mutations to human neurological disease.
References
- 1.Goyal U, Blackstone C. Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta. 2013;1833:2492–2498. doi: 10.1016/j.bbamcr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol. 2013;5:a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 6.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlaitz AL, Thompson J, Wong CCL, Yates JR, 3rd, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell. 2013;26:313–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz DS, Blower MD. The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J Cell Biol. 2014;207:41–57. doi: 10.1083/jcb.201406037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioia U, Laneve P, Dlakić M, Arceci M, Bozzoni I, Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem. 2005;280:18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- 11.Ricagno S, Engloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. Crystal structure and molecular determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragno R, Gioia U, Laneve P, Bozzoni I, Mai A, Caffarelli E. Identification of small-molecule inhibitors of the XendoU endoribonucleases family. Chem Med Chem. 2011;6:1797–1805. doi: 10.1002/cmdc.201100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terasaki M, Jaffe LA, Hunnicutt GR, Hammer JA., 3rd Structural change of the endoplasmic reticulum during fertilization: evidence for loss of membrane continuity using the green fluorescent protein. Dev Biol. 1996;179:320–328. doi: 10.1006/dbio.1996.0263. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- 15.Kucharz K, Krogh M, Ng AN, Toresson H. NMDA receptor stimulation induces reversible fission of the neuronal endoplasmic reticulum. PLoS One. 2009;4:e5250. doi: 10.1371/journal.pone.0005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falabella P, Riviello L, Pascale M, Di Lelio I, Tettamanti G, Grimaldi A, Iannone C, Monti M, Pucci P, Tamburro AM, et al. Functional amyloids in insect immune response. Insect Biochem Mol Biol. 2012;42:203–211. doi: 10.1016/j.ibmb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Poe JC, Kountikov EI, Lykken JM, Natarajan A, Marchuk DA, Tedder TF. EndoU is a novel regulator of AICD during peripheral B cell selection. J Exp Med. 2014;211:57–69. doi: 10.1084/jem.20130648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hübner CA, Kurth I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain. 2014 doi: 10.1093/brain/awu287. http://dx.doi.org/10.1093/brain/awu287. [DOI] [PubMed]