Abstract
Mitochondria in cells comprise a tubulovesicular network shaped continuously by complementary fission and fusion events. The mammalian Drp1 protein plays a key role in fission, while Mfn1, Mfn2, and OPA1 are required for fusion. Shifts in the balance between these opposing processes can occur rapidly, indicating that modifications to these proteins may regulate mitochondrial membrane dynamics. We highlight posttranslational modifications of the mitochondrial fission protein Drp1, for which these regulatory mechanisms are best characterized. This dynamin-related GTPase undergoes a number of steps to mediate mitochondrial fission, including translocation from cytoplasm to the mitochondrial outer membrane, higher-order assembly into spirals, GTP hydrolysis associated with a conformational change and membrane deformation, and ultimately disassembly. Many of these steps may be influenced by covalent modification of Drp1. We discuss the dynamic nature of Drp1 modifications and how they contribute not only to the normal regulation of mitochondrial division, but also to neuropathologic processes.
Keywords: mitochondria, fission, phosphorylation, sumoylation, dynamin, GTPase
Introduction
The term mitochondria is derived from the Greek words mitos (thread) and khondrion (small grain), emphasizing their tubulovesicular appearance within cells. However, this description masks the dynamic nature of mitochondria, which undergo continuous cycles of division and fusion in cells.1 The steady state of these yin-yang processes is physiologically tuned during a number of important cellular events, such as development, apoptosis, autophagy, synaptic plasticity, and cell division.2 Perturbation of this balance has been implicated in a number of degenerative neurological disorders, both acquired and inherited, including Alzheimer disease, Parkinson disease, hereditary optic atrophy, and Charcot-Marie-Tooth neuropathy.3–5
The cellular machineries responsible for dynamic changes in mitochondrial morphology have been elucidated through studies in a large number of different experimental systems including the budding yeast Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila, mice, and mammalian cells in culture. Surprisingly, a relatively small number of proteins have been identified that play fundamental roles.6 Most notable are a number of different large, dynamin-related GTPases that function in mitochondrial fission (Drp1) or fusion (Mfn1, Mfn2, and OPA1).7 In addition, the mitochondrial outer membrane proteins Fis1 and Mff participate in fission.8–10 Though S. cerevisiae Mdv1 and the related Caf4 protein function as adaptors important for mitochondrial division in yeast, no clear orthologs for these proteins have been identified in mammalian cells. A number of other proteins have been implicated more recently in various facets of mitochondrial fission and fusion, but these may function in more specific contexts or play regulatory roles.11,12
The importance of carefully orchestrating the balance between mitochondrial fission and fusion has been emphasized in a large number of neurological disease studies. Consistent with the fundamental importance of the dynamin-related proteins in mitochondrial fusion, mutations in OPA1 and Mfn2 have been reported in patients with autosomal dominant optic atrophy type 1 and Charcot-Marie-Tooth type 2A neuropathy, respectively.13–15 In addition, a de novo middle domain mutation in the mitochondrial fissioning GTPase Drp1 that likely affects its higher-order assembly has been reported in a neonatally lethal syndrome of microcephaly, abnormal brain development, optic atrophy, and lactic acidemia. Cells derived from this patient showed aberrant elongation of mitochondria. Peroxisomes, which share components of the mitochondrial fission machinery, were also affected.16 Lastly, during programmed cell death there is increased recruitment of Drp1 to mitochondria,17 with the resulting increase in fission conspiring with a decrease in fusion to extensively fragment mitochondria. These fission events are also important for autophagic clearance of mitochondria that may be dysfunctional18,19 as well as for proper segregation of mitochondria into daughter cells during cell division. Thus, a properly regulated fission/fusion balance is critical, since inappropriate disruption of either fusion or fission can be deleterious.
The modulation of Drp1 function in particular has been a topic of great interest. Drp1 exists as small oligomers (dimers/tetramers) that can self-assemble into larger multimeric structures at the mitochondrial outer membrane (Fig. 1), where they mediate mitochondrial division through a guanosine triphosphate (GTP)-dependent conformational change. Reminiscent of other dynamin superfamily proteins, Drp1 has an N-terminal GTP-binding domain, a middle assembly domain, a small insert (insert B), and a C-terminal GTPase effector domain (GED; Fig. 1).7 One prominent difference is that the Drp1 protein does not contain the C-terminal proline-rich domain of dynamin. The GED domain of Drp1 is important for mediating both intra- and intermolecular interactions,20 and the middle domain is important for self-assembly into higher order structures. Based on structural studies of dynamin and the yeast Drp1 ortholog Dnm1,21,22 the basic building block of the Drp1 higher order spirals on the mitochondrial outer membrane may be a T-shaped dimer, with the GTP-binding domain at the head, and the middle and GED domains comprising the stalk.
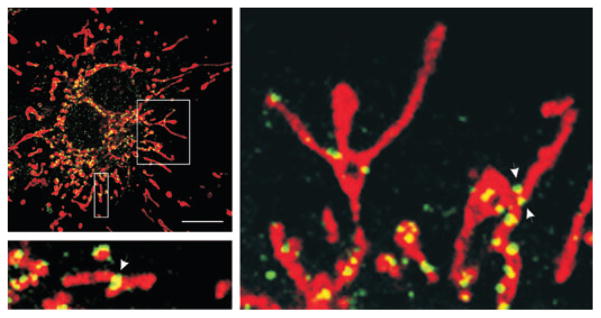
Figure 1.
Drp1 localization on mitochondria. Endogenous Drp1 puncta (green) are present in COS7 cells at discrete sites along mitochondrial tubules, which are visualized using MitoTracker Red CMXRos. Although some Drp1 foci are clearly evident at sites of fission (arrows in enlargements), not all foci represent sites of fission. Scale bar, 20 μm. Adapted from Zhu et al.20
Unlike the other three dynamin-related GTPases and Fis1/Mff that are involved in mitochondrial dynamics, Drp1 is mostly cytoplasmic, with only approximately 3% associated with the mitochondrial outer membrane under typical cellular conditions.23 Consequently, it seems unlikely that simply altering protein levels would significantly change mitochondrial fission, and many studies investigating wild-type Drp1 overexpressed in cells have not reported an increase in fission. Thus, regulation of Drp1 properties such as mitochondrial translocation, protein interactions, higher order assembly, or GTPase activity might be expected to be more important. Drp1 recruitment to mitochondria is increased prominently during events such as programmed cell death, with an associated increase in fission.17 Thus, a key level of regulation occurs at the mitochondrial recruitment step. Other opportunities for regulation occur during higher order Drp1 assembly/disassembly and the cycle of binding and hydrolysis of GTP. Any of these regulatory changes could conceivably occur though modifications of Drp1 and/or its interaction partners.
Posttranslational modifications of Drp1
Exciting insights have recently been published on the posttranslational modification and regulation of Drp1 by a variety of modifying enzymes.2,12,24,25 These covalent modifications include protein phosphorylation, sumoylation, ubiquitination, and S-nitrosylation (Fig. 2). Though these modifications will be discussed individually, it seems likely that they can occur in conjunction with one another, and that modification at one site may affect modifications at other sites.
Figure 2.
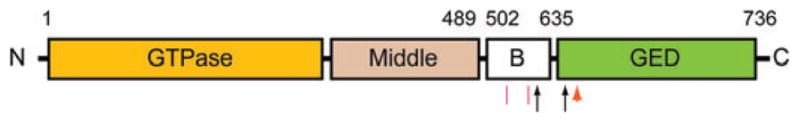
Schematic domain model of Drp1 showing identified sites of posttranslational modifications. Sumoylation sites are indicated by red line segments, and protein phosphorylation sites are identified by black arrows. An orange arrowhead identifies a site of S-nitrosylation. Sites of ubiquitination have not been identified. Boundary amino acid residues for the indicated domains are along the top. All amino acid numbering is based on the human Drp1 splice variant 1 sequence.
Protein phosphorylation
Protein phosphorylation is a posttranslational modification mediated by protein kinases with a wide range of substrate specificity that is widely used by cells to modulate protein function, with the advantage of integrating rapid functional changes with cellular signaling pathways. Thus, it was not surprising when this was initially identified as a regulatory mechanism for Drp1. The earliest reported phosphorylation was at Ser616 by Cdk1/cyclin B (residue numbers have been changed for consistency to reflect the position in human Drp1 variant 1).26 This mitotic phosphorylation promoted Drp1-dependent mitochondrial fission, accounting for the increase in mitochondrial fragmentation that occurs in cells undergoing mitosis. Since this modification does not directly affect GTPase activity,26 the increase in fission may be mediated by alterations in Drp1 interactions with other proteins.
A more widely studied phosphorylation site has been Drp1 Ser637, which is part of a consensus site highly conserved among metazoans. This was initially identified as a site for phosphorylation by protein kinase A (PKA), with dephosphorylation mediated by calcineurin.25,27–29 Subsequently, Ser637 was also identified as a site for phosphorylation by Ca2+/calmodulin-dependent protein kinase Iα (CaMKIα).30 Three studies reported that modification of this site inhibits mitochondrial division through a reduction in GTPase activity and/or inhibition of Drp1 translocation to mitochondria.27–29 However, a fourth study found that Ser637 phosphorylation by CaMKIα stimulates mitochondrial translocation of Drp1, perhaps through an increase in Drp1-binding affinity for Fis1, with an associated increase in mitochondrial fragmentation.30
This apparent discrepancy highlights the importance of where and at what stage of the Drp1 activity cycle phosphorylation occurs in the cell as well as the possibility that other proteins are concurrently modified by these kinases. For instance, the study that found an increase in mitochondrial fragmentation investigated CaMKIα, which is activated in response to increased intracellular Ca2+ concentrations; importantly, changes in Ca2+ concentrations can have a plethora of effects on other proteins. Furthermore, increased cAMP levels also can have a variety of cellular effects in addition to stimulating PKA-dependent phosphorylation of Drp1 Ser637. Though this example might portend a great degree of difficulty in understanding how phosphorylation at a given site might affect Drp1 function, it reinforces the point that physiological effects of a modification may differ depending on the cellular context in which it occurs.
A recent study found that calcineurin-dependent dephosphorylation increases Drp1 recruitment to mitochondria.29 At first this might seem incompatible with the low basal phosphorylation reported at this site in several studies,27,28 but there are a number of possible explanations. First, phosphorylation at this site may turn over very slowly, and thus not be evident in 32P-orthophosphate-labeling experiments, where labeling typically occurs for about 4 h. Arguing against this notion is that immunoblots with phosphopeptide antibodies also did not suggest significant basal phosphorylation.27,28 A second and more likely possibility is that there may be a small subpopulation of Drp1 protein that is phosphorylated and is specifically involved in this calcineurin-dependent process, emphasizing again the importance of the cellular context of a modification.
Sumoylation
The small ubiquitin-like modifier (Sumo) protein is also involved in Drp1 modification.31 Sumo attachment often alters subcellular localization of proteins or protects them from ubiquitin-mediated destruction. Interestingly, there is a Bax/Bak-dependent stable association of Drp1 at the mitochondrial membrane, with associated conjugation of Sumo to Drp1.32 Consistent with this modification of Drp1 occurring at the mitochondrial outer membrane, Drp1 is a substrate for at least one mitochondrial-anchored Sumo E3 ligase, MAPL,33 with a corresponding Sumo protease SenP5 that can desumoylate Drp1.34 Even so, the effects of sumoylation on Drp1 function remain unclear. A mutant Drp1 protein with a series of closely spaced Lys-to-Arg mutations that prevent detectable sumoylation of Drp1 does not exhibit differences in mitochondrial division or in mitochondrial recruitment of Drp1 during programmed cell death when expressed in cells, arguing that sumoylation of Drp1 itself is not required and that other proteins may play a role.35 The fact that sumoylation appears to occur within the B insert of Drp1 suggests that it may exert an effect on Drp1 interactions with the outer mitochondrial membrane or other proteins.
Ubiquitination
In addition to being sumoylated, Drp1 is also modified by a related process, ubiquitination. This modification often acts as a protein interaction motif and often targets protein for destruction. Recent studies have focused on the effects of the mitochondrial RING-CH E3 (MARCH-V, MARCH5, MITOL) ubiquitin ligase that resides in the mitochondrial outer membrane and ubiquitinates Drp1 as well as Fis1.36–38 Though originally proposed to promote mitochondrial fusion,36,37 more recent studies have provided evidence that it enhances fission, possibly by facilitating trafficking of Drp1 to mitochondrial sites of division.38 Ubiquitination might affect Drp1 assembly/disassembly also, since expression of a dominant-negative MARCH-V protein increases mitochondrial association of Drp1.38
S-Nitrosylation
A final Drp1 modification recently described is S-nitrosylation. This is a redox-related modification of thiols (such as in cysteine residues) by nitric oxide (NO), which transduces NO activity and affects a variety of proteins involved in a number of cellular processes.39 In fact, S-nitrosylation shares many properties with protein phosphorylation. Both modifications exhibit substrate specificity, strict spatial and temporal regulation, and reversibility. In some cases, as for the caspases involved in apoptosis, S-nitrosylation has been shown to inhibit enzyme activity. In other proteins, such as matrix metalloproteinases, S-nitrosylation appears to increase enzymatic activity.
NO functions as a signaling molecule, but in excess it can cause neuronal injury, in part via increasing mitochondrial fragmentation. A recent study has suggested that NO produced in response to β-amyloid protein, a key mediator of Alzheimer disease, triggers mitochondrial fission and subsequent synaptic loss and neuronal damage, in part via S-nitrosylation of Drp1 at Cys644.40 Preventing S-nitrosylation of Drp1 through a Cys644Ala mutation blocked the neurotoxicity.40 Thus, S-nitrosylation joins the others modifications already discussed in providing cells a repertoire of tools to regulate Drp1 function, with relevance for both normal and pathological processes.
Conclusion
The large number of distinct modifications of Drp1 already identified foretells an exciting new area of mitochondrial research. The studies reviewed here are likely just the beginning, since not only will additional modifications of Drp1 be revealed, but other proteins involved in regulating Drp1-mediated mitochondrial dynamics are likely also to be uncovered. A supporting cast of modifying enzymes coupled to a variety of signaling pathways dramatically increases the regulatory repertoire that a cell can dispatch to fine-tune Drp1-mediated mitochondrial fission.
We suggest that Drp1 is the predominant mediator of mitochondrial division, with a wide variety of interacting and modifying proteins providing the flexibility to regulate fission activity and to integrate such changes into a variety of cell processes. In this regard, an important point to emphasize is that effects of any protein modification are crucially dependent on where and when they occur within the cell, and what other proteins might be concurrently affected. Given their roles in the functional modulation of Drp1, it is not surprising that alterations in structure and modifications of Drp1 in inherited and acquired neurological disorders are pathologically significant. Whether the regulation of these posttranslational modifications can be harnessed therapeutically will be an important area for future investigation.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Bereiter-Hahn J, Vöth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 2.Soubannier V, McBride HM. Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta. 2009;1793:154–170. doi: 10.1016/j.bbamcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann NY Acad Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liesa M, Palacín M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 6.Westermann B. Molecular machinery of mitochondrial fusion and fission. J Biol Chem. 2008;283:13501–13505. doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- 7.Praefcke GJK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 8.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 10.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerveny KL, Tamura Y, Zhang Z, et al. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta. 2009;1792:1138–1144. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Züchner S, I, Mersiyanova V, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 14.Alexander C, Votruba M, Pesch UE, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 15.Delettre C, Lenaers G, Griffoin JM, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 16.Waterham HR, Koster J, van Roermund CWT, et al. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 17.Frank S, Gaume B, Bergmann-Leitner ES, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 18.Arnoult D, Rismanchi N, Grodet A, et al. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 19.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu PP, Patterson A, Stadler J, et al. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 22.Ingerman E, Perkins EM, Marino M, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santel A, Frank S. Shaping mitochondria: the complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- 25.Chang C-R, Blackstone C. Drp1 phosphorylation and mitochondrial regulation. EMBO Rep. 2007;8:1088–1089. doi: 10.1038/sj.embor.7401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi N, Ishihara N, Jofuku A, et al. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 27.Chang C-R, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 28.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cereghetti GM, Stangherlin A, Martins de Brito O, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han XJ, Lu YF, Li SA, et al. CaM kinase 1α-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zunino R, Schauss A, Rippstein P, et al. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 35.Figueroa-Romero C, Iñiguez-Lluhí JA, Stadler J, et al. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23:3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura N, Kimura Y, Tokuda M, et al. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yonashiro R, Ishido S, Kyo S, et al. A novel mitochondrial ubiquitin ligase plays a central role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho DH, Nakamura T, Fang J, et al. S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]