Abstract
Background
The widespread presence of antibiotics resistance genes in pathogenic bacteria can cause enormous problems. Food animals are one of the main reservoirs of intestinal pathogens that pose a potential risk to human. Analyzing the epidemiological characteristics and resistance patterns of Shigella flexneri in calves is necessary for animal and human health.
Methods and results
A total of 54 Shigella flexneri isolates, including six serotypes (1a, 2a, 2b, 4a, 6 and Xv), were collected from 837 fecal samples obtained from 2014 to 2016. We performed pulsed-field gel electrophoresis (PFGE) and applied the restriction enzyme NotI to analyze the genetic relatedness among the 54 isolates and to categorize them into 31 reproducible and unique PFGE patterns. According to the results of antimicrobial susceptibility tests, all 26 Shigella flexneri 2a serotypes were resistant to cephalosporin and/or fluoroquinolones. The genes bla TEM-1, bla OXA-1, and bla CTX-M-14 were detected in 19 cephalosporin-resistant S. flexneri 2a isolates. Among 14 fluoroquinolone-resistant isolates, the aac(6′)-Ib-cr gene was largely present in each strain, followed by qnrS (5). Only one ciprofloxacin-resistant isolate harbored the qepA gene. Sequencing the quinolone resistance determining regions (QRDRs) of the fluoroquinolone-resistant isolates revealed two point mutations in gyrA (S83 L, D87N/Y) and a single point mutation in parC (S80I). Interestingly, two gyrA (D87N/Y) strains were resistant to ciprofloxacin.
Conclusions
The current study enhances our knowledge of Shigella in cattle, although continual surveillance is necessary for the control of shigellosis. The high level of cephalosporin and/or fluoroquinolone resistance in Shigella warns us of a potential risk to human and animal health.
Keywords: Shigella flexneri, Antimicrobial susceptibility, Resistant
Background
The majority of Enterobactericeae family bacteria, including Salmonella, E. coli and Shigella spp., the major etiological agent of diarrheal disease, are a global public health burden, particularly in low-income countries [1–3]. Shigella is phylogenetically distinct from several independent E. coli strains and has evolved through convergent evolution [4]. The genus Shigella consists of four subgroups differentiated according to their biochemical and serological properties: A (S. dysenteriae), B (S. flexneri), C (S. boydii), and D (S. sonnei). All four species of Shigella cause shigellosis, but S. flexneri is the predominant subgroup found in developing countries, whereas S. sonnei is found in industrialized countries [5]. The first Shigella species identified was S. dysenteriae, followed by S. flexneri at the end of the 19th century. Shigellosis became a notorious and widespread epidemic during World War 1 with the transmission of S. flexneri strain NCTC1, a 2a lineage [6, 7]. Based on the differing structural characteristics of the antigenic determinants of the O antigen, S. flexneri is divided into no fewer than 20 serotypes: 1a, 1b, 1c, 1d, 2a, 2b, 2v, 3a, 3b, 4a, 4av, 4b, 4c, 5a, 5b, X, Xv, Y, Yv, 6, and 7b [8, 9].
Given that shigellosis is a global public health burden, previous studies have focused on the human gastrointestinal pathogens but have ignored animal groups. Shigella spp. infect and also cause corresponding clinical symptoms in monkeys, cows, pigs, chickens and other animals [10–13]. Indeed, animals that live in environments characterized by poor sanitary hygiene, restricted access to clean drinking water and long-term exposure to contaminated food are prone to dysentery [14, 15]. Many antibiotics are used to control disease and promote growth during the breeding process, leading to the widespread dissemination of antibiotic resistance genes (ARGs). The spread of drug resistance among pathogenic bacteria in humans and animals may be disastrous.
The present study investigated the Shigella epidemic in cows in the northwest region of China. S. flexneri 2a was first isolated from a yak with diarrhea in Tibet in 2014. In this study, we used pulsed-field gel electrophoresis (PFGE) to analyze the relationships among S. flexneri isolates and tested for antimicrobial susceptibility patterns. Our results will help prevent diarrhea in calves and will assist in the selection of effective antibiotics against Shigella.
Methods
Bacterial isolation and identification
Fresh stool samples were isolated from 2014 to 2016 in Northwest China (Gansun, Shanxi, Qinghai, Xinjiang and Tibet) from calves (3 to 20 days) with diarrhea. Samples were stored in transport medium, cultured directly on Salmonella-Shigella (SS) agar and incubated at 37 °C for 24 h to select for Shigella. Resultant colonies (colorless, semitransparent, smooth, and moist circular) [16] were picked and grown at 37 °C for 24 h on MacConkey (MAC) Agar to verify identity. Colonies were selected and cultured in brain heart infusion broth at 37 °C for 5 h with shaking at 250 rpm. All isolates were confirmed using API20E test strips (bioMerieux Vitek, Marcy-l’ Etoile, France) according to the manufacturer’s recommendations. Shigella was serotyped using a commercially available kit (Denka Seiken, Tokyo, Japan) and confirmed by PCR [17].
Antimicrobial susceptibility testing
The antimicrobial susceptibility of S. flexneri isolates was determined via the Kirby–Bauer disc-diffusion method in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [18].
The antibiotic discs (OXOID, UK) included penicillin G (P, 10 μg), ampicillin (AMP, 10 μg), amoxicillin/clavulanic acid (AMC, 30 μg), cephalothin (KF, 30 μg), cephazolin (KZ, 30 μg), cefamandole (MA, 30 μg), cefoxitin (FOX, 30 μg), ceftriaxone (CRO, 30 μg), cefotaxime (CTX, 30 μg), cefoperazone (CFP, 75 μg), cefepime (FEP, 30 μg), meropenem (MEM, 10 μg), imipenem (IPM, 10 μg), norfloxacin (NOR, 10 μg), enrofloxacin (ENR, 5 μg), levofloxacin (LEV, 5 μg), ciprofloxacin (CIP, 5 μg), erythromycin (E, 15 μg), chloramphenicol (C, 30 μg), tetracycline (TE, 30 μg), streptomycin (S, 10 μg), gentamicin (CN, 10 μg), and amikacin (AK, 30 μg). E. coli strain ATCC25922 was used as a quality control strain in each test batch.
PCR amplification of ARGs
We performed PCR assays that targeted 24 different ARGs using the primers described in Table 1. To determine the underlying resistance mechanism of β-lactam antibiotics, we amplified extended-spectrum β-lactamase (ESBL) genes, specifically bla CTX-M, bla SHV, bla TEM, and bla OXA, as well as ampC genes, specifically bla MOX, bla FOX, bla DHA, bla CIT, bla ACC, and bla MIR [19–21]. Plasmid-mediated quinolone resistance (PMQR) determinant genes, including qnrA, qnrB, qnrD, qnrS, qepA and aac(6′)-Ib-cr and four quinolone resistance determining region (QRDR) genes as well as DNA gyrase (gyrA,gyrB) and topoisomerase IV (parC,parE) were amplified to determine the underlying mechanism of quinolone resistance [16, 21–23]. The PCR fragments were sequenced after purification and compared to sequences in GenBank.
Table 1.
Primers for the detection of antibiotic resistance genes
| Target | Primer sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|
| β-lactamase | |||
| bla CTX-M-1 | F: GGTTAAAAAATCACTGCGTC | 873 | Cui et al., 2015 [16] |
| R: TTACAAACCGTCGGTGACGA | |||
| bla CTX-M-2 | F: CGACGCTACCCCTGCTATT | 552 | Zong et al., 2008 [17] |
| R: CCAGCGTCAGATTTTTCAGG | |||
| bla CTX-M-8 | F: TCGCGTTAAGCGGATGATGC | 689 | Zong et al., 2008 [17] |
| R: AACCCACGATGTGGGTAGC | |||
| bla CTX-M-9 | F: AGAGTGCAACGGATGATG | 868 | Cui et al., 2015 [16] |
| R: CCAGTTACAGCCCTTCGG | |||
| bla CTX-M-25 | F: TTGTTGAGTCAGCGGGTTGA | 497 | Liu et al., 2015 [18] |
| R: GCGCGACCTTCCGGCCAAAT | |||
| bla SHV | F: CGCCGGGTTATTCTTATTTGTCGC | 1015 | Zong et al., 2008 [17] |
| R: TCTTTCCGATGCCGCCGCCAGTCA | |||
| bla TEM | F: ATGAGTATTCAACTTTCCG | 876 | This study |
| R: CCAATGCTTAATCAGTGAG | |||
| bla OXA | F: ATTAAGCCCTTTACCAAACCA | 890 | Cui et al., 2015 [16] |
| R: AAGGGTTGGGCGATTTTGCCA | |||
| bla MOX | F: GCTGCTCAAGGAGCACAGGAT | 520 | Cui et al., 2015 [16] |
| R: CACATTGACATAGGTGTGGTGC | |||
| bla FOX | F: AACATGGGGTATCAGGGAGATG | 190 | Cui et al., 2015 [16] |
| R: CAAAGCGCGTAACCGGATTGG | |||
| bla DHA | F: AACTTTCACAGGTGTGCTGGGT | 405 | Cui et al., 2015 [16] |
| R: CCGTACGCATACTGGCTTTGC | |||
| bla CIT | F: TGGCCAGAACTGACAGGCAAA | 462 | Cui et al., 2015 [16] |
| R: TTTCTCCTGAACGTGGCTGGC | |||
| bla ACC | F: AACAGCCTCAGCAGCCGGTTA | 346 | Cui et al., 2015 [16] |
| R: TTCGCCGCAATCATCCCTAGC | |||
| bla MIR | F: TCGGTAAAGCCGATGTTGCGG | 302 | Cui et al., 2015 [16] |
| R: CTTCCACTGCGGCTGCCAGTT | |||
| PMQRs | |||
| qnrA | F: ATTTCTCACGCCAGGATTTG | 516 | Colobatiu et al.,2015 [19] |
| R: GATCGGCAAAGGTTAGGTCA | |||
| qnrB | F: GATCGTGAAAGCCAGAAAGG | 476 | Colobatiu et al.,2015 [19] |
| R: ACGATGCCTGGTAGTTGTCC | |||
| qnrD | F: CGAGATCAATTTACGGGGAATA | 656 | Cui et al.,2015 [13] |
| R: AACAAGCTGAAGCGCCTG | |||
| qnrS | F: ACGACATTCGTCAACTGCAA | 417 | Colobatiu et al.,2015 [19] |
| R: TAAATTGGCACCCTGTAGGC | |||
| aac(6′)-Ib-cr | F: CCCGCTTTCTCGTAGCA | 544 | Colobatiu et al.,2015 [19] |
| R: TTAGGCATCACTGCGTCTTC | |||
| qepA | F: CGTGTTGCTGGAGTTCTTC | 403 | Colobatiu et al.,2015 [19] |
| R: CTGCAGGTACTGCGTCATG | |||
| QRDR | |||
| gyrA | F: TACACCGGTCAACATTGAGG | 648 | Hu et al.,2007 [20] |
| R: TTAATGATTGCCGCCGTCGG | |||
| gyrB | F: TGAAATGACCCGCCGTAAAGG | 309 | Hu et al.,2007 [20] |
| R: GCTGTGATAACGCAGTTTGTCCGGG | |||
| parC | F: GTACGTGATCATGGACCGTG | 531 | Hu et al.,2007 [20] |
| R: TTCGGCTGGTCGATTAATGC | |||
| parE | F: ATGCGTGCGGCTAAAAAAGTG | 290 | Hu et al.,2007 [20] |
| R: TCGTCGCTGTCAGGATCGATAC | |||
PFGE
Genotypes and transmission patterns were determined by performing PFGE according to the method described in a previous study [19]. S. flexneri isolates were digested with the restriction enzyme NotI (TaKaRa, Japan) at 37 °C for 3 h to generate a DNA fingerprinting profile. Salmonella enterica serotype Braenderup strain H9812 was digested with XbaI (TaKaRa, Japan) and used as a molecular size standard. Electrophoresis was performed on the CHEF Mapper XA system (Bio-Rad) with a 1% agarose SeaKem Gold gel (Lonza, USA). Electrophoretic parameters were determined by performing multiple screening runs and included switching times of 2.16 to 54.17 s, a voltage of 6 v/cm, a 120° angle and a run time of 21 h. PFGE images were obtained using a Universal Hood II (Bio-RAD, USA) and analyzed using BioNumerics software version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium). A clustering tree that indicated relative genetic similarity was constructed using UPGMA (Unweighted Pair Group Method with Arithmetic Mean) and the Dice-predicted similarity value with a 1.0% pattern optimization and 1.5% band position tolerance.
Results
Bacterial isolation and identification
During our epidemiological survey of Shigella, we collected 873 fecal samples from calves with diarrhea and obtained 54 S. flexneri isolates from five provinces in northwest China from 2014 to 2016. Isolate information is shown in detail in Table 2. Among the 54 S. flexneri isolates, there were six serotypes: five (9.26%) isolates were 1a, twenty-six (48.15%) isolates were 2a, four (7.41%) isolates were 2b, six (11.11%) isolates were 4a, eight (14.81%) isolates were 6, and five (9.26%) isolates were Xv (Fig. 1). Our surveillance of the Gansu isolates identified all of the serotypes, except 4a. All 4a serotypes were isolated from Shanxi, while all Xv and 1a serotypes were from Gansu. Additionally, serotype 2a was widely isolated from each province, with the exception of Xinjiang, and serotype 6 was found only in yaks. Interestingly, Shigella was primarily isolated in the first quarter and fourth quarter, accounting for 54% (29/54) and 30% (16/54), respectively (Fig. 2).
Table 2.
Strain information of S. flexneri isolates from diarrheal calves, 2014 to 2016
| Strain name | Serotype | Isolation year | Origin | Province |
|---|---|---|---|---|
| TYSF1412001 | 2a | 2014 | Yak | Tibet |
| GBSF1412056 | 2a | 2014 | Beef cattle | Gansu |
| GBSF1501026 | 2a | 2015 | Beef cattle | Gansu |
| GBSF1501071 | Xv | 2015 | Beef cattle | Gansu |
| GYSF1501076 | 6 | 2015 | Yak | Gansu |
| QYSF1501088 | 6 | 2015 | Yak | Qinghai |
| XBSF1501093 | 2b | 2015 | Beef cattle | Xinjiang |
| GBSF1501105 | 2a | 2015 | Beef cattle | Gansu |
| SBSF1501123 | 4a | 2015 | Beef cattle | Shanxi |
| QYSF1502130 | 6 | 2015 | Yak | Qinghai |
| GBSF1502176 | 2a | 2015 | Beef cattle | Gansu |
| GYSF1502197 | 6 | 2015 | Yak | Gansu |
| SBSF1502219 | 4a | 2015 | Beef cattle | Shanxi |
| XBSF1502236 | 2b | 2015 | Beef cattle | Xinjiang |
| GBSF1503241 | 2a | 2015 | Beef cattle | Gansu |
| GYSF1503270 | 1a | 2015 | Yak | Gansu |
| GBSF1503288 | 1a | 2015 | Beef cattle | Gansu |
| GBSF1505314 | 2a | 2015 | Beef cattle | Gansu |
| SBSF1505331 | 2a | 2015 | Beef cattle | Shanxi |
| GBSF1506340 | Xv | 2015 | Beef cattle | Gansu |
| GBSF1507358 | 1a | 2015 | Beef cattle | Gansu |
| GBSF1509369 | 2a | 2015 | Beef cattle | Gansu |
| GBSF1510375 | 2a | 2015 | Beef cattle | Gansu |
| GBSF1510390 | 2a | 2015 | Beef cattle | Gansu |
| QYSF1511395 | 2a | 2015 | Yak | Qinghai |
| GBSF1511401 | 2a | 2015 | Beef cattle | Gansu |
| GYSF1511409 | 2a | 2015 | Yak | Gansu |
| SBSF1512413 | 4a | 2015 | Beef cattle | Shanxi |
| GBSF1512419 | 2b | 2015 | Beef cattle | Gansu |
| GBSF1512425 | 2a | 2015 | Beef cattle | Gansu |
| GBSF1512433 | 2a | 2015 | Beef cattle | Gansu |
| GBSF1601015 | Xv | 2016 | Beef cattle | Gansu |
| GBSF1601024 | Xv | 2016 | Beef cattle | Gansu |
| TYSF1601031 | 2b | 2016 | Yak | Tibet |
| GBSF1601064 | 2a | 2016 | Beef cattle | Gansu |
| GYSF1601073 | 6 | 2016 | Yak | Gansu |
| GBSF1602082 | 2a | 2016 | Beef cattle | Gansu |
| QYSF1602094 | 6 | 2016 | Yak | Qinghai |
| GBSF1602098 | 2a | 2016 | Beef cattle | Gansu |
| GBSF1602103 | 2a | 2016 | Beef cattle | Gansu |
| SBSF1603115 | 4a | 2016 | Beef cattle | Shanxi |
| SBSF1603121 | 4a | 2016 | Beef cattle | Shanxi |
| GBSF1603138 | 2a | 2016 | Beef cattle | Gansu |
| GBSF1603149 | 2a | 2016 | Beef cattle | Gansu |
| QYSF1603158 | 6 | 2016 | Yak | Qinghai |
| SBSF1604173 | 2a | 2016 | Beef cattle | Shanxi |
| SBSF1604195 | 4a | 2016 | Beef cattle | Shanxi |
| GBSF1605203 | Xv | 2016 | Beef cattle | Gansu |
| GBSF1608241 | 2a | 2016 | Beef cattle | Gansu |
| GYSF1610256 | 2a | 2016 | Yak | Gansu |
| GYSF1610266 | 6 | 2016 | Yak | Gansu |
| GBSF1610275 | 1a | 2016 | Beef cattle | Gansu |
| GBSF1611283 | 1a | 2016 | Beef cattle | Gansu |
| GBSF1611290 | 2a | 2016 | Beef cattle | Gansu |
Fig. 1.
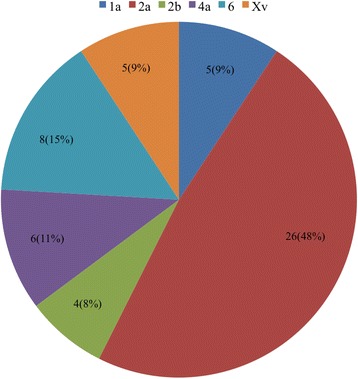
S. flexneri serotypes collected from 2014 to 2016
Fig. 2.
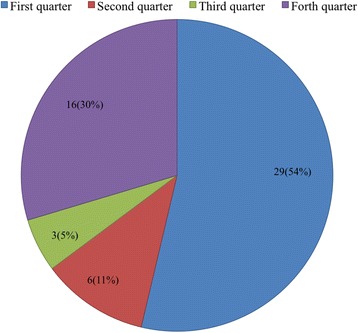
Number of S. flexneri isolated from different quarters
Antimicrobial susceptibility testing
The 54 S. flexneri isolates were examined for susceptibility to 23 antibiotics. More than 50% of isolates were resistant to 8 antibiotics. Among them, resistance to P (54/54, 100%), AMP (51/54, 94.44%) and TE (49/54, 90.74) was most common, followed by E (46/54, 85.19%), S (38/54, 70.37%), KZ (34/54, 62.96%), KF (29/54, 53.70%) and CN (29/54, 53.70%). None of the isolates were resistant to IMP, MEM and the fourth-generation cephalosporin FEP. In addition, although a certain number of isolates were resistant to second- and third-generation cephalosporins (MA, FOX, CRO, CTX and CFP) and fluoroquinolones (CIP, NOR, ENR and LEV), these comprised no more than 30% of the total number of isolates, and the resistance rate was lower than those of other antibiotics (Table 3, Fig. 3).
Table 3.
Statistical analysis of the results of antimicrobial susceptibility to 23 antibiotics for 54 S. flexneri
| Antibiotic | Antimicrobial resistance rate No. (%) | |||||
|---|---|---|---|---|---|---|
| Total (n = 54) | Gansu (n = 37) | Shanxi (n = 8) | Xinjiang (n = 2) | Qinghai (n = 5) | Tibet (n = 2) | |
| Penicillin G (P) | 54 (100%) | 37 (100%) | 8 (100%) | 2 (100%) | 5 (100%) | 2 (100%) |
| Ampicillin (AMP) | 51 (94.44%) | 37 (100%) | 8 (100%) | 2 (100%) | 3 (60%) | 1 (50%) |
| Amoxycillin/Clavulanic acid (AMC) | 5 (9.62%) | 3 (8.11%) | 1 (12.5%) | 1 (50%) | 0 | 0 |
| Cephalothin (KF) | 29 (53.70%) | 19 (51.35%) | 5 (62.5%) | 2 (100%) | 2 (40%) | 1 (50%) |
| Cephazolin (KZ) | 34 (62.96%) | 21 (56.76%) | 7 (87.5%) | 2 (100%) | 3 (60%) | 1 (50%) |
| Cefamandole (MA) | 16 (29.63%) | 12 (32.43%) | 2 (25%) | 1 (50%) | 1 (20%) | 0 |
| Cefoxitin (FOX) | 3 (5.56%) | 2 (5.41%) | 1 (12.5%) | 0 | 0 | 0 |
| Ceftriaxone (CRO) | 12 (22.22%) | 9 (24.32%) | 2 (25%) | 1 (50%) | 0 | 0 |
| Cefotaxime (CTX) | 14 (25.93%) | 10 (27.03%) | 2 (25%) | 1 (50%) | 1 (20%) | 0 |
| Cefoperazone (CFP) | 6 (11.11%) | 6 (16.22%) | 0 | 0 | 0 | 0 |
| Cefepime (FEP) | 0 | 0 | 0 | 0 | 0 | 0 |
| Meropenem (MEM) | 0 | 0 | 0 | 0 | 0 | 0 |
| Imipenem (IPM) | 0 | 0 | 0 | 0 | 0 | 0 |
| Norfloxacin (NOR) | 16 (29.63%) | 12 (32.43%) | 3 (37.5%) | 1 (50%) | 0 | 0 |
| Enrofloxacin (ENR) | 13(24.07%) | 11 (29.73%) | 2 (25%) | 0 | 0 | 0 |
| Levofloxacin (LEV) | 14 (25.93%) | 11 (29.73%) | 2 (25%) | 1 (50%) | 0 | 0 |
| Ciprofloxacin (CIP) | 2 (3.70%) | 2 (5.41%) | 0 | 0 | 0 | 0 |
| Erythromycin (E) | 46 (85.19%) | 35 (94.59%) | 6 (75%) | 2 (100%) | 3 (60%) | 0 |
| Tetracycline (TE) | 49 (90.74%) | 35 (94.59%) | 8 (100%) | 2 (100%) | 3 (60%) | 1 (50%) |
| Chloramphenicol (C) | 17 (31.48%) | 10 (27.03%) | 6 (75%) | 1 (50%) | 0 | 0 |
| Streptomycin (S) | 38 (70.37%) | 30 (81.08%) | 4 (50%) | 2 (100%) | 2 (40%) | 0 |
| Gentamicin (CN) | 29 (53.70%) | 23 (62.16%) | 4 (50%) | 2 (100%) | 0 | 0 |
| Amikacin (AK) | 3 (5.56%) | 3 (8.11%) | 0 | 0 | 0 | 0 |
Fig. 3.
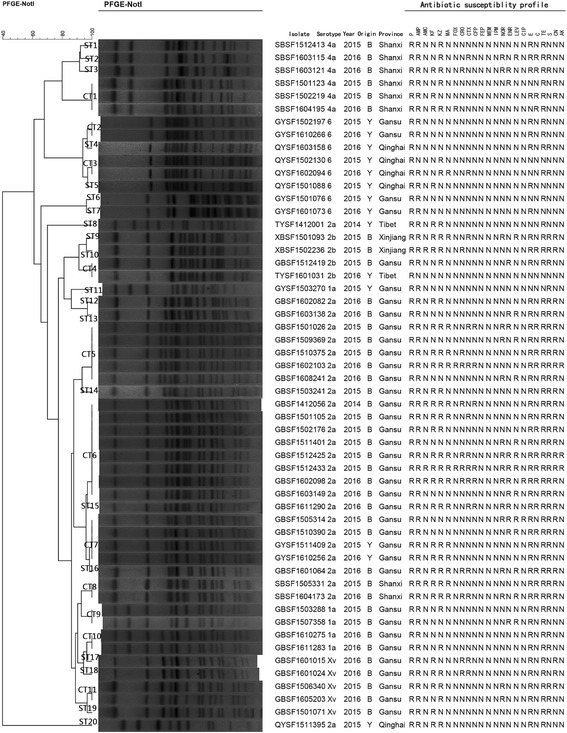
PFGE dendrogram and antibiotic susceptibility profile of 54 NotI-digested S. flexneri. B = Beef cattle; Y = Yak. R = Resistance; N = Sensitive and Intermediary
Remarkably, all 26 S. flexneri 2a isolates demonstrated varying degrees of resistance to cephalosporins and/or fluoroquinolones and exhibited multidrug resistance (MDR). The S. flexneri 2a isolates were resistant to 14 diverse cephalosporins/fluoroquinolones. Among them, 73.06% (19/26) of isolates were resistant to cephalosporin, 53.85% (14/26) of isolates were resistant to fluoroquinolones, and 26.92% (7/26) of isolates were resistant to both cephalosporin and fluoroquinolones. Furthermore, isolate GBSF1512433 was resistant to all cephalosporins (with the exception of FEP) and fluoroquinolones (with the exception of CIP). Compared with the S. flexneri 2a isolates collected from beef calves, the 4 yak isolates were sensitive to most cephalosporins and fluoroquinolones but resistant to KF, KZ and MA (Table 4).
Table 4.
Statistical analysis of the cephalosporin and/or fluoroquinolone susceptibility for 26 S. flexneri 2a
| Cephalosporin and/or Fluorquinolones resistance spectrum | Cephalosporin and/or Fluorquinolones resistance rate No. (%) | ||||
|---|---|---|---|---|---|
| Total (n = 26) | Gansu (n = 22a) | Shanxi (n = 2) | Qinghai (n = 1a) | Tibet (n = 1a) | |
| KF/KZ | 5 (19.23%) | 3 (13.64%) | 0 | 1a (100%) | 1a (100%) |
| KF/KZ/MA | 3 (11.54%) | 2a (9.09%) | 1 (50%) | 0 | 0 |
| KF/KZ/MA/CRO | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/CTX | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/CRO/CFP | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/FOX/CRO/CTX/CFP | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| NOR/LEV | 3 (11.54%) | 3 (13.64%) | 0 | 0 | 0 |
| ENR/LEV | 3 (11.54%) | 3 (13.64%) | 0 | 0 | 0 |
| NOR/ENR/LEV/CIP | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/CRO/CTX/NOR/LEV | 2 (7.69%) | 1 (4.55%) | 1 (50%) | 0 | 0 |
| KF/KZ/MA/CRO/CTX/NOR/ENR/LEV | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/CTX/CFP/CIP/NOR/ENR | 2 (7.69%) | 2 (9.09%) | 0 | 0 | 0 |
| KF/KZ/MA/CRO/CTX/CFP/NOR/ENR/CIP | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
| KF/KZ/MA/FOX/CRO/CTX/CFP/NOR/ENR/LEV | 1 (3.85%) | 1 (4.55%) | 0 | 0 | 0 |
aa yak origin S. flexneri 2a isolate
ARGs analysis of cephalosporin- and/or fluoroquinolone-resistant S. flexneri 2a isolates
In this study, only three β-lactamase gene types (bla OXA-1, bla TEM-1 and bla CTX-M-14) were identified among the 19 cephalosporin-resistant S. flexneri 2a isolates (Table 5). All isolates harbored bla TEM-1 type ARGs (100%), 15 isolates harbored bla OXA-1 (15/19, 78.95%), and 14 harbored bla CTX-M-14 (14/19, 73.68%). In total, 63.16% (12/19) of isolates harbored three β-lactamase gene types. All S. flexneri 2a isolates from yaks were negative for bla CTX-M type ARGs.
Table 5.
Antimicrobial spectrum and ARGs analysis of S. flexneri 2a with resistance to cephalosporin
| Strain name | Antimicrobial spectrum | ARGs in plasmid | ||
|---|---|---|---|---|
| TEM | OXA | CTX-M-9 | ||
| TYSF1412001 | KF/KZ | TEM-1 | OXA-1 | ---- |
| QYSF1511395 | KF/KZ | TEM-1 | ---- | ---- |
| GBSF1503241 | KF/KZ | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1502176 | KF/KZ | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1602082 | KF/KZ | TEM-1 | ---- | CTX-M-14 |
| SBSF1505331 | KF/KZ/MA | TEM-1 | OXA-1 | CTX-M-14 |
| GYSF1511409 | KF/KZ/MA | TEM-1 | ---- | ---- |
| GYSF1610256 | KF/KZ/MA | TEM-1 | OXA-1 | ---- |
| GBSF1510375 | KF/KZ/MA/CRO | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1501105 | KF/KZ/MA/CTX | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1412056 | KF/KZ/MA/CRO/CFP | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1602103 | KF/KZ/MA/FOX/CRO/CTX/CFP | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1601064 | KF/KZ/MA/CRO/CTX/NOR/LEV | TEM-1 | ---- | CTX-M-14 |
| SBSF1604173 | KF/KZ/MA/CRO/CTX/NOR/LEV | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1611290 | KF/KZ/MA/CTX/CFP/NOR/ENR | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1501026 | KF/KZ/MA/CTX/CFP/NOR/ENR | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1512425 | KF/KZ/MA/CRO/CTX/NOR/ENR/LEV | TEM-1 | OXA-1 | ---- |
| GBSF1602098 | KF/KZ/MA/CRO/CTX/CFP/NOR/ENR/CIP | TEM-1 | OXA-1 | CTX-M-14 |
| GBSF1512433 | KF/KZ/MA/FOX/CRO/CTX/CFP/NOR/ENR/LEV | TEM-1 | OXA-1 | CTX-M-14 |
Both PMQR genes and SNPs in QRDRs were identified for 14 quinolone-resistant isolates (Table 6). According to the PCR results, all quinolone-resistant isolates were positive for aac(6′)-Ib-cr but negative for qepA, except strain GBSF1602098. Only five (5/14, 35.71%) strains isolated from Gansu harbored qnrS, and no isolate harbored all three ARGs simultaneously. The point mutations in the QRDR genes play important roles in determining quinolone and/or fluoroquinolone resistance [24]. In the present study, we successfully amplified all four QRDR genes and compared them to reference sequences. We found two point mutations in gyrA and one point mutation each in gyrA and parC (Table 6). All quinolone-resistant strains carried mutations that altered the amino acid sequences of gyrA (S83 L) and parC (S80I). In addition, each strain carried the mutation 87 (D → N or Y) in gyrA, with the exception of GBSF1510390. Interestingly, GBSF1505314 and GBSF1602098 harbored the gyrA D87Y mutation, which confers resistance to ciprofloxacin.
Table 6.
Antimicrobial spectrum and amino acid types in QRDR and PMQRs genes analysis of S. flexneri 2a with resistance to fluoroquinolones
| Strain name | Antimicrobial spectrum | QRDR | ARGs in plasmid | ||||
|---|---|---|---|---|---|---|---|
| gyrA | parC | aac(6′)-Ib-cr | qnrS | qepA | |||
| 83 | 87 | 80 | |||||
| GBSF1509369 | NOR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1511401 | NOR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1608241 | NOR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1510390 | ENR/LEV | S83 L | D87D | S80I | + | + | − |
| GBSF1603138 | ENR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1603149 | ENR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1505314 | NOR/ENR/LEV/CIP | S83 L | D87Y | S80I | + | + | − |
| GBSF1601064 | KF/KZ/MA/CRO/CTX/NOR/LEV | S83 L | D87N | S80I | + | − | − |
| SBSF1604173 | KF/KZ/MA/CRO/CTX/NOR/LEV | S83 L | D87N | S80I | + | − | − |
| GBSF1611290 | KF/KZ/MA/CTX/CFP/NOR/ENR | S83 L | D87N | S80I | + | − | − |
| GBSF1501026 | KF/KZ/MA/CTX/CFP/NOR/ENR | S83 L | D87N | S80I | + | + | − |
| GBSF1512425 | KF/KZ/MA/CRO/CTX/NOR/ENR/LEV | S83 L | D87N | S80I | + | + | − |
| GBSF1602098 | KF/KZ/MA/CRO/CTX/CFP/NOR/ENR/CIP | S83 L | D87Y | S80I | + | − | + |
| GBSF1512433 | KF/KZ/MA/FOX/CRO/CTX/CFP/NOR/ENR/LEV | S83 L | D87N | S80I | + | + | − |
+: Presence corresponding genes
-: Absence corresponding genes
PFGE pattern analysis
PFGE was performed to determine the genetic relatedness among the isolates and to study the molecular epidemiology in specific geographical regions [25]. The PFGE patterns of the 54 NotI-digested S. flexneri isolates were heterogeneous, and multiple PFGE patterns were present among the strains. Thus, diverse factors such as geography and environment may affect PFGE patterns. At an 80% similarity level, S. flexneri isolates generated 31 reproducible and unique PFGE patterns, including 11 common types (CT) and 20 single types (ST) (Fig. 3).
Among all isolates, the majority of S. flexneri 2a (26/54, 48.15%) isolates were classified into 11 PFGE patterns (4 CT and 7 ST). These PFGE patterns were closely related to each other, except the Tibet (TYSF1412001) and Qinghai (QYSF1511395) isolates, suggesting the strains isolated from different geographical locations exhibit diverse PFGE patterns and a capricious genetic diversity.
Discussion
ARGs are widespread and cause problems when present in pathogens [26]. Over the past decade, MDR Shigella has been reported in many countries [27]. However, only a few studies have described the prevalence of Shigella in animals worldwide. In the present study, we investigated the epidemiology of S. flexneri in cows in northwest China. During a 2-year survey, 54 S. flexneri isolates were obtained. Unfortunately, 16S rRNA gene sequence analysis does not effectively distinguish between closely related strains in a superfamily, such as Shigella and E. coli [28], and conventional biochemical and serological techniques are also insufficient. Therefore, PFGE was utilized to analyze the molecular characteristics of these isolates, to determine the relatedness among isolates and to study the molecular epidemiology in specific geographical regions. The clustering results allowed us to analyze the epidemiological trends of S. flexneri. Characterization of these isolates will be helpful for clinical diagnosis, treatment, prevention and the control of shigellosis [15].
Antimicrobial resistance has emerged as a serious problem [29], particularly for conventional, older-generation antibiotics such as P, AMP, TE, and E. According to the results of our antimicrobial susceptibility tests, cephalosporin and fluoroquinolone resistance rates in our isolates were higher than those in human isolates [19, 26]. Notably, the predominant S. flexneri 2a isolates were all resistant to cephalosporins, fluoroquinolones and multiple antibiotics. Two isolates (GBSF1505314 and GBSF1602098) were also resistant to ciprofloxacin, which is the first-line antibiotic treatment for shigellosis. The universal emergence of resistant and MDR strains in animals may be attributable to the unrestricted and excessive use of antibiotics in veterinary clinics. The widespread presence of MDR strains has reduced the selectivity of clinical medications to treat shigellosis [30]. Notably, our PFGE dendrogram showed various genetic patterns for S. flexneri, and there were diverse resistance profiles associated with each pattern. Based on these results, S. flexneri has the ability to adapt to the selective pressures of different antibiotics.
The high levels of resistance of S. flexneri 2a to cephalosporin/fluorquinolones, which are the most effective treatments for severe gastrointestinal infections caused by pathogenic bacteria, prompted us to study potential molecular resistance mechanisms. The emergence of ESBL-producing Shigella spp. has been observed in many countries [31]. In the current study, only 3 ARG genotypes (bla OXA-1, bla TEM-1 and bla CTX-M-14) were detected. Among them, the bla TEM-1 gene was detected in all 19 cephalosporin-resistant isolates. In total, 174 bla TEM variants resistant to penicillin and other ß-lactamase antibiotics have been recorded. TEM-1 confers resistance to ampicillin and cephalothin [32]. bla OXA-type ARGs are class D β-lactamases, which were named for their ability to hydrolyze oxacillin [32]. Initially, bla OXA-beta-lactamases were reported in P. aeruginosa, although now the bla OXA gene has been detected in plasmids and integrons in many Gram-negative organisms [32, 33]. According to some studies, the probable host preference for bla OXA-type β-lactamase is S. flexneri [34]. In the present study, 15/19 (78.95%) isolates harbored bla OXA- type genes, and sequencing results indicated that all the bla OXA genes were bla OXA-1. Additionally, bla CTX-M has become one of the most prevalent extended-spectrum-β-lactamases (ESBLs) [35]. This gene was widely harbored by S. flexneri 2a isolated from beef cattle. Interestingly, all S. flexneri 2a isolated from yaks were negative for bla CTX-M type ARGs.
Fluoroquinolones are highly effective for the treatment of shigellosis worldwide [36]. The primary mechanism of quinolone resistance involves the accumulation of sequential mutations in QRDRs that encode DNA gyrase and topoisomerase IV [37]. The most prevalent mutations in Shigella spp. are the point mutations in gyrA codons 83, 87 and 211, and parC codon 80 [38, 39]. Novel mutations in QRDRs are also being discovered [39]. In the present study, three mutations in gyrA codon 83 (S → L) and/or 87 (D → N or Y) and parC codon 80 (S → I) were detected in each fluoroquinolone-resistant isolate. All substitutions are responsible for reduced affinity. In addition, the amino acid diversity at the same position may lead to different levels of quinolone resistance [40, 41]. GyrA D87Y mutations were detected in only two ciprofloxacin-resistant isolates. However, the role of this mutation in ciprofloxacin resistance is unclear and requires further investigation.
Over the past few years, PMQR determinants have been deemed the most common ARGs in Enterobacteriaceae worldwide [42]. PMQR determinants mediate only low-level quinolone resistance. However, these resistance genes are usually associated with mobile or transposable elements that allow for dissemination among Enterobacteriaceae. In addition, the presence of PMQR genes may facilitate the selection of QRDR mutations that result in higher levels of quinolone resistance [37, 43, 44]. The aac(6′)-Ib-cr gene encodes an acetyltransferase that is known to reduce quinolone activity. In the present study, all 14 isolates resistant to fluoroquinolones were positive for aac(6′)-Ib-cr, indicating the aac(6′)-Ib-cr gene is widespread in S. flexneri 2a. Compared with the aac(6′)-Ib-cr gene, the transmembrane segment efflux pump qepA gene was scarcely detected in Shigella, and we found only one ciprofloxacin-resistant isolate that was qepA-positive. The qnr family (which includes the first PMQR genes) contains a variety of subtypes, including qnrA, qnrB, qnrC, qnrD and qnrS and several qnr family genes that have been reported in Shigella [39, 45]. The Qnr proteins protect DNA gyrase against quinolones and facilitate the selection of QRDR mutations that improve resistance to these antimicrobials.
Conclusion
In conclusion, cephalosporin and/or fluoroquinolone resistance in Shigella has been widely reported. To increase our understanding of Shigella in cattle, we investigated Shigella in calves with diarrhea and analyzed the genetic relatedness, antimicrobial susceptibility, QRDR mutations, and prevalence of PMQR and ß-lactamase in S. flexneri 2a isolates from five provinces in northwest China. However, this study also had limitations, including the lack of a systematic surveillance system to prospectively or retrospectively detect and analyze shigellosis in veterinary clinics. Furthermore, we are unable to effectively monitor and control antibiotic abuse and the resulting spread of ARGs. Therefore, it is essential to continually monitor rates of shigellosis and the development of resistance patterns.
Acknowledgements
The authors thank all the farms for providing the samples in this study.
Funding
This work was supported by funds from the National Natural Science Foundation of China (31,272,603, 31,101,836).
Availability of data and materials
The data supporting the findings of this study are contained within the manuscript.
Authors’ contributions
Conceived and designed the experiments: JYZ and ZZ. Performed the experiments: ZZ and MZC. Analyzed the data: ZZ, BL, and XZZ. Contributed reagents/materials/analysis tools: MZC and FSC. Wrote the paper: ZZ. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Permission to work in specific locations, information regarding the number of samples harvested, and an associated permit number for calves were not required, and no endangered or protected species were involved or harmed during this study.
Consent for publication
All authors agreed on the publication of the paper.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhen Zhu, Email: zhuzhen234@yeah.net.
Mingze Cao, Email: caomignze123@163.com.
Xuzheng Zhou, Email: xuzhengzhou1@163.com.
Bing Li, Email: libingaa1@163.com.
Jiyu Zhang, Email: infzjy@sina.com.
References
- 1.Nandre R, Matsuda K, Lee JH. Efficacy for a new live attenuated Salmonella Enteritidis vaccine candidate to reduce internal egg contamination. Zoonoses Public Health. 2014;61:55–63. doi: 10.1111/zph.12042. [DOI] [PubMed] [Google Scholar]
- 2.Nandre RM, Ruan X, Duan Q, Sack DA, Zhang W. Antibodies derived from an enterotoxigenic Escherichia coli (ETEC) adhesin tip MEFA (multiepitope fusion antigen) against adherence of nine ETEC adhesins: CFA/I, CS1, CS2, CS3, CS4, CS5, CS6, CS21 and EtpA. Vaccine. 2016;34:3620–3625. doi: 10.1016/j.vaccine.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rausch D, Ruan X, Nandre R, Duan Q, Hashish E, Casey TA, et al. Antibodies derived from a toxoid MEFA (multiepitope fusion antigen) show neutralizing activities against heat-labile toxin (LT), heat-stable toxins (STa, STb), and Shiga toxin 2e (Stx2e) of porcine enterotoxigenic Escherichia coli (ETEC) Vet Microbiol. 2017;202:79–89. doi: 10.1016/j.vetmic.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, Marohn ME, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis. 2014;59:933–941. doi: 10.1093/cid/ciu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linton DS. “War dysentery” and the limitations of German military hygiene during world war I. Bull Hist Med. 2010;84:607–639. doi: 10.1353/bhm.2010.0036. [DOI] [PubMed] [Google Scholar]
- 7.Baker KS, Mather AE, McGregor H, Coupland P, Langridge GC, Day M, et al. The extant world war 1 dysentery bacillus NCTC1: a genomic analysis. Lancet. 2014;384:1691–1697. doi: 10.1016/S0140-6736(14)61789-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Q, Lan R, Wang J, Xia S, Wang Y, Wang Y, et al. Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS One. 2013;8:e70238. doi: 10.1371/journal.pone.0070238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu S, Xu X, Yang C, Wang J, Liang B, Li P, et al. Shift in serotype distribution of Shigella species in China, 2003–2013. Clin Microbiol Infect. 2015;21:252. doi: 10.1016/j.cmi.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Priamukhina NS, Kilesso VA, Tikhomirov ED. Animal carriers of Shigella and their possible epidemiological importance. Zh Mikrobiol Epidemiol Immunobiol. 1984;11:20–24. [PubMed] [Google Scholar]
- 11.Fang H, Wang H, Cai L, Yu Y. Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomic survey. Environ Sci Technol. 2015;49:1095–1104. doi: 10.1021/es504157v. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Qian L, Jiang S, Cai C, Ma D, Gao P, et al. Safety evaluation of neo transgenic pigs by studying changes in gut microbiota using high-throughput sequencing technology. PLoS One. 2016;11:e0150937. doi: 10.1371/journal.pone.0150937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed AN, Abdel-Latef GK, Abdel-Azeem NM, El-Dakhly KM. Ecological study on antimicrobial-resistant zoonotic bacteria transmitted by flies in cattle farms. Parasitol Res. 2016;115:3889–3896. doi: 10.1007/s00436-016-5154-7. [DOI] [PubMed] [Google Scholar]
- 14.Qu F, Bao C, Chen S, Cui E, Guo T, Wang H, et al. Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn Microbiol Infect Dis. 2012;74:166–170. doi: 10.1016/j.diagmicrobio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Sun Q, Wei X, Klena JD, Wang J, Liu Y, et al. Genetic characterization of Shigella flexneri isolates in Guizhou Province, China. PLoS One. 2015;10:e0116708. doi: 10.1371/journal.pone.0116708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui X, Wang J, Yang C, Liang B, Ma Q, Yi S, et al. Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front Microbiol. 2015;6:435. doi: 10.3389/fmicb.2015.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q, Lan R, Wang Y, Zhao A, Zhang S, Wang J, et al. Development of a multiplex PCR assay targeting O-antigen modification genes for molecular serotyping of Shigella flexneri. J Clin Microbiol. 2011;49:3766–3770. doi: 10.1128/JCM.01259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical Laboratory Standards Institute . Performance standards for antimicrobial disk susceptibility testing; twenty-second informational supplement. Wayne: Clinical and Laboratory Standards Institute; 2013. pp. m100–ms23. [Google Scholar]
- 19.Cui X, Yang C, Wang J, Liang B, Yi S, Li H, et al. Antimicrobial resistance of Shigella flexneri serotype 1b isolates in China. PLoS One. 2015;10:e0129009. doi: 10.1371/journal.pone.0129009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong Z, Partridge SR, Thomas L, Iredell JR. Dominance of blaCTX-M within an Australian extended-spectrum beta-lactamase gene pool. Antimicrob Agents Chemother. 2008;52:4198–4202. doi: 10.1128/AAC.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Zeng T, Su G, Lin LY, Zhao Y, Yang WQ, et al. The dissemination mode of drug-resistant genes in Enterobacter Cloacae. Indian J Med Microbiol. 2015;33:87–92. doi: 10.4103/0255-0857.150967. [DOI] [PubMed] [Google Scholar]
- 22.Colobatiu L, Tabaran A, Flonta M, Oniga O, Mirel S, Mihaiu M. First description of plasmid-mediated quinolone resistance determinants and β-lactamase encoding genes in non-typhoidal Salmonella isolated from humans, one companion animal and food in Romania. Gut Pathog. 2015;7:16. doi: 10.1186/s13099-015-0063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu LF, Li JB, Ye Y, Li X. Mutations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in clinical strains of fluoroquinolone-resistant Shigella in Anhui, China. J Microbiol. 2007;45:168–170. [PubMed] [Google Scholar]
- 24.Komp Lindgren PD, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003;47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta S, Jain P, Nandy S, Matsushita S, Yoshida S. Molecular characterization of serologically atypical provisional serovars of Shigella isolates from Kolkata, India. J Med Microbiol. 2014;63:1696–1703. doi: 10.1099/jmm.0.081307-0. [DOI] [PubMed] [Google Scholar]
- 26.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4:54. doi: 10.1186/s40168-016-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed AM, Furuta K, Shimomura K, Kasama Y, Shimamoto T. Genetic characterization of multidrug resistance in Shigella spp. from japan. J Med Microbiol. 2006;55:1685–1691. doi: 10.1099/jmm.0.46725-0. [DOI] [PubMed] [Google Scholar]
- 28.Demarta A, De Respinis S, Dolina M, Peduzzi R. Molecular typing of Yersinia frederiksenii strains by means of 16s rDNA and gyrB genes sequence analyses. FEMS Microbiol Lett. 2004;238:423–428. doi: 10.1016/j.femsle.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Ud-Din AI, Wahid SU, Latif HA, Shahnaij M, Akter M, Azmi IJ, et al. Changing trends in the prevalence of Shigella species: emergence of multi-drug resistant Shigella sonnei biotype g in Bangladesh. PLoS One. 2013;8:e82601. doi: 10.1371/journal.pone.0082601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright DJ, Drasar BS. Dysentery in world war 1: Shigella a century on. Lancet. 2014;384:1651–1653. doi: 10.1016/S0140-6736(14)61940-1. [DOI] [PubMed] [Google Scholar]
- 31.Tajbakhsh M, García Migura LM, Rahbar M, Svendsen CA, Mohammadzadeh M, Zali MR, et al. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? J Antimicrob Chemother. 2012;67:1128–1133. doi: 10.1093/jac/dks023. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Sugumar M, Kumar KM, Manoharan A, Anbarasu A, Ramaiah S. Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS One. 2014;9:e91800. doi: 10.1371/journal.pone.0091800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navia MM, Capitano L, Ruiz J, Vargas M, Urassa H, Schellemberg D, et al. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999;37:3113–3117. doi: 10.1128/jcm.37.10.3113-3117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robin F, Beyrouthy R, Bonacorsi S, Aissa N, Bret L, Brieu N, et al. Inventory of extended-spectrum-beta-lactamase-producing enterobacteriaceae in France as assessed by a multicenter study. Antimicrob Agents Chemother. 2017;61 [DOI] [PMC free article] [PubMed]
- 36.Taneja N, Kumar A, Appannanavar S, Verma G, Sharma M. Plasmid-mediated quinolone resistance in Shigella isolates over a decade in India. J Glob Antimicrob Resist. 2014;2:59–60. doi: 10.1016/j.jgar.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Yang H, Duan G, Zhu J, Zhang W, Xi Y, Fan Q. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int J Antimicrob Agents. 2013;42:173–177. doi: 10.1016/j.ijantimicag.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Kamonporn P, Ladaporn B, Sivhour C, Wilawan O, Sirigade R, Panida N, et al. Multidrug-resistant infections in patients with diarrhea, Cambodia, 2014–2015. Emerg Infect Dis. 2016;22:1640–1643. doi: 10.3201/eid2209.152058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu B, Qin TT, Fan WT, Bi RR, Chen Y, Li Y, et al. Novel mutations in gyrA and parC among Shigella sonnei strains from Jiangsu Province of China, 2002–2011. Int J Infect Dis. 2017;59:44–49. doi: 10.1016/j.ijid.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 41.Tamanna RJ, Ramana J. Structural insights into the fluoroquinolone resistance mechanism of Shigella flexneri DNA gyrase and topoisomerase IV. Microb Drug Resist. 2016;22:404–411. doi: 10.1089/mdr.2015.0018. [DOI] [PubMed] [Google Scholar]
- 42.Pu XY, Pan JC, Gu YM, Zheng W, Li J, Yu H. Complete sequences and characterization of two novel plasmids carrying aac(6′)-Ib-cr and qnrS gene in Shigella flexneri. Microb Drug Resist. 2016;22:115–122. doi: 10.1089/mdr.2015.0082. [DOI] [PubMed] [Google Scholar]
- 43.Folster JP, Pecic G, Bowen A, Rickert R, Carattoli A, Whichard JM. Decreased susceptibility to ciprofloxacin among Shigella isolates in the United States, 2006 to 2009. Antimicrob Agents Chemother. 2011;55:1758–1760. doi: 10.1128/AAC.01463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez-Martínez JM, Machuca J, Cano ME, Calvo J, Martínez-Martínez L, Pascual A. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat. 2016;29:13–29. doi: 10.1016/j.drup.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Zaidi MB, Estrada-García T, Campos FD, Chim R, Arjona F, Leon M, et al. Incidence, clinical presentation, and antimicrobial resistance trends in Salmonella and Shigella infections from children in Yucatan, Mexico. Front Microbiol. 2013;4:288. doi: 10.3389/fmicb.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are contained within the manuscript.


