Abstract
Telomere length is maintained in most eukaryotic cells by telomerase. The core components of this ribonucleoprotein (RNP) enzyme include a protein catalytic subunit, composed of motifs conserved among reverse transcriptases (RT), and an RNA subunit that contains a short template sequence essential for the synthesis of telomeric repeats. We developed an electrophoretic mobility shift assay using active telomerase partially purified from 293 cells and radiolabeled, in vitro-transcribed human telomerase RNA (hTR) to investigate the molecular interactions of the human telomerase RT (hTERT) and telomerase-associated proteins with hTR. A specific hTR–protein complex was identified and shown to contain hTERT and human Staufen by antibody supershift assays. Variants of hTR altered in distinct structural elements were analyzed for their ability to competitively inhibit complex formation. Human telomerase RNAs lacking the CR4-CR5 domain were poor inhibitors of hTR–protein complex formation, suggesting that the CR4-CR5 domain of hTR is a potential protein-binding site. Furthermore, alterations in the telomerase RNA pseudoknot’s P3 helix, the CR7 domain, or the H/ACA box efficiently inhibited formation of the complex, indicating that these domains are dispensable for the assembly of a telomerase RNP in vitro. Potential telomerase-associated proteins that bind hTR were also identified using a UV cross-linking assay.
INTRODUCTION
Telomerase is a ribonucleoprotein (RNP) complex with RNA-dependent DNA polymerase activity. In most eukaryotic cells, this specialized reverse transcriptase mediates the synthesis of guanine-rich DNA repeats onto telomeres, the ends of chromosomes, using a template sequence within its integral RNA subunit (1). In humans, telomeres consist of tandem repeats of the hexanucleotide sequence TTAGGG and are bound by a variety of proteins (2). This DNA–protein complex protects chromosomes from nuclease digestion, end-to-end fusion and other chromosomal rearrangement events (3).
The 451-nt human telomerase RNA (hTR) is most likely expressed in its premature form as a polyadenylated RNA polymerase II transcript (4–6), similarly to the yeast telomerase RNA (7). A secondary structure was recently proposed for vertebrate telomerase RNA (8). This model predicts the evolutionary conservation of four domains: a pseudoknot, the CR4-CR5 domain, the H/ACA box and the CR7 domain (8; see Fig. 2A). The cellular accumulation of human and mouse telomerase RNAs requires folding of their 3′-end into a structure similar to the H/ACA box of a family of small nucleolar RNAs (snoRNAs) (9,10). Subcellular fractionation experiments (9) and microinjection of fluorescent RNAs in Xenopus oocytes (11) also suggest that hTR localizes to the nucleolus.
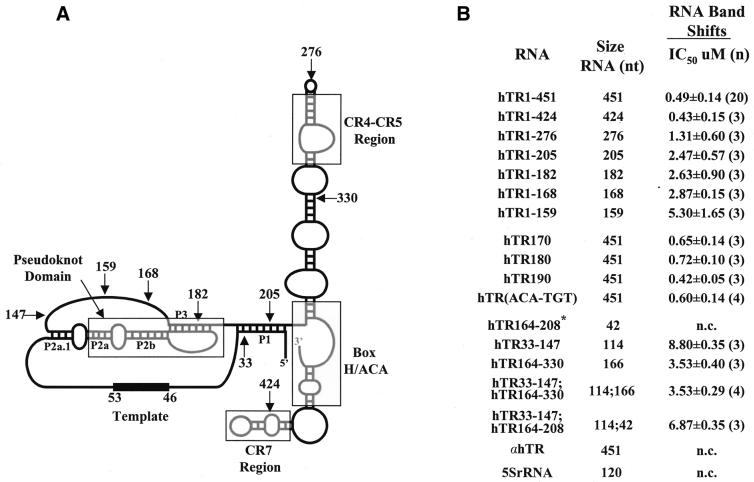
Figure 2.
Summary of hTR mutations analyzed in this study and their inhibition of hTR–protein complex formation. (A) Secondary structure of hTR with its telomeric template sequence (nucleotides 46–53) and its 5′ and 3′ ends [adapted from Chen et al. (8)]. Four universally conserved structural elements among vertebrate telomerase RNAs including the pseudoknot, the CR4-CR5 region, the H/ACA box and the CR7 region are boxed in gray. The P1, P2a.1, P2a, P2b and P3 helices are indicated. Arrows indicate the nucleotide position of the 5′ and 3′ ends of the different hTR truncations used in this study. (B) Summary of hTR variants analyzed for association with hTERT and telomerase-associated proteins in vitro. The ability of wild-type and hTR derivatives to competitively inhibit the binding of radiolabeled hTR to proteins of a partially purified extract from 293 cells was determined using an EMSA. IC50 values (in µM) represent the concentration of competitor RNAs that inhibit complex formation by 50%. The calculated standard deviations (±) and number (n) of times the experiment was repeated are also indicated. n.c., no competition. *Residues 200 and 201 are deleted in hTR164–208.
Human telomerase activity can be reconstituted by the addition of in vitro-transcribed hTR to: (i) rabbit reticulocyte lysates (RRL) expressing the human telomerase reverse transcriptase (hTERT) (12,13); (ii) hTERT immunoprecipitated from yeast extracts (14); or (iii) recombinant hTERT purified from baculovirus-infected insect cells (15). These results suggest direct interactions between hTR and the telomerase catalytic subunit, hTERT. The presence of two distinct hTERT-binding sites within hTR was recently reported (16,17). The integrity of the evolutionary conserved CR4-CR5 domain, in both human and mouse telomerase RNAs, is critical for the reconstitution of telomerase activity in vitro (17) and in vivo (10,16).
Though evidence suggest that hTERT and hTR are sufficient for the reconstitution of human telomerase activity in vitro, hTR appears to interact with a number of cellular proteins in vivo. The telomerase-associated protein-1 (TEP1) was shown to specifically interact with the mammalian telomerase RNA in the yeast three-hybrid system (18). A genetic screen for open reading frames expressing proteins that associate with hTR identified the human homolog of the Drosophila Staufen protein and the ribosomal-associated protein L22 (19). Furthermore, protein components from heterogeneous nuclear RNPs have been reported to bind human telomerase (20–22). Antisera specific for dyskerin and human Gar1, two proteins associated with the maturation and processing of H/ACA box snoRNAs, coimmunoprecipitate hTR from cellular extracts (23,24). However, the relationship and significance of these different interactions with the human telomerase RNP in vivo is not clearly understood.
We developed an electrophoretic mobility shift assay (EMSA) to investigate the interactions between hTR, hTERT and telomerase-associated proteins. We used active telomerase partially purified from transformed human embryonic kidney (293) whole cell extracts and radiolabeled, in vitro-transcribed, wild-type hTR. A specific protein-dependent complex was identified that could be competitively inhibited by unlabeled wild-type hTR, but not heterologous non-specific RNAs. Addition of antibodies specific for hTERT and human Staufen promoted the formation of a supershifted complex, indicating that these two proteins are part of the identified hTR–protein complex. Alterations in hTR that modified specific structural elements, identified potential protein binding sites within the hTR. hTR-binding proteins of 40, 42, 58 and 125 kDa were identified using a UV cross-linking assay.
MATERIALS AND METHODS
Preparation of human telomerase extracts
The preparation of human telomerase extracts from 293 cells has been described previously (25). Briefly, total proteins from an S100 cytoplasmic extract of 293 cells were precipitated using 40% ammonium sulfate and collected by centrifugation. Following dialysis, the extract was subjected to two rounds of purification using anion exchange (Toyopearl Q 650M column from TosoHass) and size exclusion (Toyopearl HW-65F column from TosoHass) chromatography. Pooled fractions (1.7 mg/ml total protein) were purified ∼120-fold with respect to the specific telomerase activity of the starting S100 cytoplasmic extracts (25).
hTERT and hTR plasmid constructs
Cloning of nucleotides 1–451 of the hTR into the pUC119 plasmid (phTR+1) was described previously (25). The mutagenic substitution of hTR nucleotides 170–179 (hTR170), 180–189 (hTR180) and 190–199 (hTR190) was also reported (25). The plasmids expressing hTRs hTR33–147, hTR164–330, hTR164–208 and hTR ACA-TGT were constructed as described by Bachand and Autexier (17). To generate phTRα, the hTR was amplified by PCR from the pGRN33 (4) vector using the 5′ primer 5′-CGCGGATCCCGGCAGCGCACCGGGTTGCGG-3′ and the 3′ primer 5′-CGCGGATCCGCATGTGTGAGCCGAGTCCTGGGT-3′, both containing BamHI restriction sites. The BamHI-digested PCR fragment was cloned into the pBluescript SK vector (Stratagene). Following transformation into Escherichia coli, clones were screened by restriction digests for hTR inserts in the antisense orientation (αhTR).
Preparation of gel-purified human telomerase RNAs
The RNAs used in the EMSAs were transcribed in vitro using T7 RNA polymerase (New England Biolabs) as described previously (17). The hTR probe used in the EMSA and UV cross-linking assays was radiolabeled during the transcription of 1 µg of FspI-digested phTR+1 plasmid with T7 RNA polymerase (25 U) and 100 µCi [α-32P]UTP (800 Ci/mmol; NEN) as recommended by the manufacturer (New England Biolabs). Following a 1–2 h incubation at 37°C, the RNA was treated with RNase-free DNase I (Amersham Pharmacia Biotech) and purified on a denaturing 4% acrylamide gel as described previously (26). 5S E.coli rRNA was purchased from Boehringer Mannheim.
Electrophoretic mobility shift assays
In the standard binding reactions, partially purified telomerase extract (∼4 µg total protein) was adjusted to 5 mM EDTA in a final volume of 10 µl containing 20 mM HEPES pH 7.9, 1 mM DTT, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, 100 mM NaCl, 0.1% NP-40, 0.1 µg/µl yeast tRNA (Sigma), 3.8 U/µl RNAguard (Amersham Pharmacia Biotech) and 0.25 pmol 32P-labeled hTR riboprobe. Following a 10 min incubation at 30°C, reactions were placed on ice and adjusted to 10 mM MgCl2. Competitor RNAs were added either before or in conjunction with the labeled hTR probe without any difference in the results. For the proteinase K treatment experiment, partially purified telomerase fractions were treated with 0.8 µg/µl proteinase K for 10 min at 30°C. The supershift assays were similar to the standard EMSA binding reactions described above, but were subsequently supplemented with different antibodies for 15 min at 30°C. Kep1 antiserum was a gift from Dr Stéphane Richard (McGill University) (27). hTERT antibody (K370) was a gift from Dr Maria Blasco (Centro Nacional de Biotecnología–CSIC) (28). Staufen antisera were donated by Dr Luc Desgroseillers (Université de Montréal) (29,30). Antibodies against TEP1 were generously donated by Dr Lea Harrington (University of Toronto) (18). GST and T7 antibodies were purchased from Amersham Pharmacia Biotech and Novagen, respectively. Binding reactions were analyzed on a non-denaturing composite gel system modified from Nelson and Green (31) and consisted of 2.5–3.0% acrylamide, 0.1% piperazine di-acrylamide (Bio-Rad), 0.5% agarose, 10% glycerol, 0.5–1.0× TBE (1× TBE: 90 mM Tris-borate, 2 mM EDTA). Gels were run at 150–200 V (20 mA) for 5–6 h at 4°C in 0.5–1.0× TBE, dried, and exposed either to PhosphorImager screens (Molecular Dynamics) or X-ray films.
The amount of competitor RNA resulting in a percentage inhibition of binding was calculated as previously described (26). Briefly, the amount of bound hTR versus the total amount of radiolabeled hTR in each lane represented the percentage of hTR bound to the complex. A non-linear curve fit was applied to the percentage inhibition–concentration data and 50% effective concentration (IC50) was calculated using Microsoft Excel. The IC50 values for each mutant were determined from a number of experiments (three to four) and are expressed with the calculated standard deviations (±SD).
UV cross-linking assays
Binding reactions were prepared as for the EMSA except that more hTR riboprobe (0.5–0.75 pmol; 2 × 105 c.p.m.) and larger amounts of partially purified telomerase extracts (3.4–8.5 µg total protein) were used. Following the adjustment of the binding reactions to 10 mM MgCl2, they were transferred onto 96-well microtiter plates previously cooled at –20°C and irradiated with 500 mJ in a GS gene linker UV chamber (Bio-Rad). Samples were then treated with 30 µg of RNase A for 30 min at 37°C. UV-treated protein extracts were boiled and subjected to electrophoresis on 10% SDS–PAGE gels.
RESULTS
Identification and characterization of a specific human telomerase RNA–protein complex
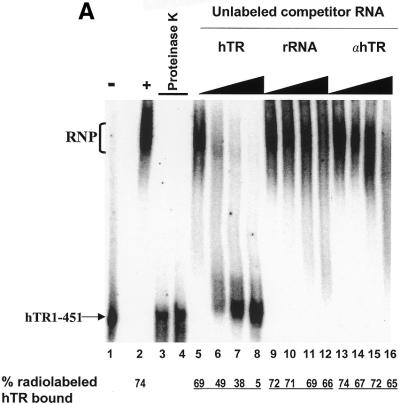
We developed an EMSA to investigate the interaction of hTR with hTERT and telomerase-associated proteins. We used active telomerase partially purified from 293 whole cell extracts and radiolabeled, in vitro-transcribed wild-type hTR. The size of the active telomerase complex purified by anion exchange and size exclusion chromatography is between 250 and 600 kDa (data not shown). This partially purified extract contains hTERT as it is positive for telomerase activity, and may contain telomerase-associated proteins such as TEP1 (18,32), dyskerin (23), chaperone proteins (33), human Staufen (19), hnRNP proteins (20–22), and other as yet unidentified proteins. Incubation of this extract, under conditions that stimulate reversible RNA binding (26,34,35), with radiolabeled hTR resulted in the formation of a complex that retarded the migration of radiolabeled, full-length hTR in a non-denaturing gel (Fig. 1A, compare lanes 1 and 2). Pre-treatment of the extract with proteinase K completely abolished complex formation (lanes 3 and 4), indicating that protein component(s) are required for the formation of this complex. The EMSA was performed with excess unlabeled hTR or non-specific RNAs to determine whether binding was specific for the hTR. A previously described quantification of this method (26; see Materials and Methods) was used to calculate the percentage of complex-bound hTR. Increasing amounts of either unlabeled E.coli 5S rRNA (lanes 9–12) or antisense hTR (lanes 13–16) did not inhibit the formation of the hTR–protein complex. As much as 66% (lane 12) and 65% (lane 16) of complex-bound hTR was observed in the presence of a 64-fold molar excess of these non-specific competitor RNAs and compared favorably to the 74% bound in the absence of competitor (lane 2). However, the hTR–protein complex reconstituted in the presence of a 64-fold molar excess of specific unlabeled hTR resulted in only 5% of the labeled hTR remaining bound by the complex (lane 8). These results are consistent with the identification of a specific hTR–protein complex in vitro.
Figure 1.
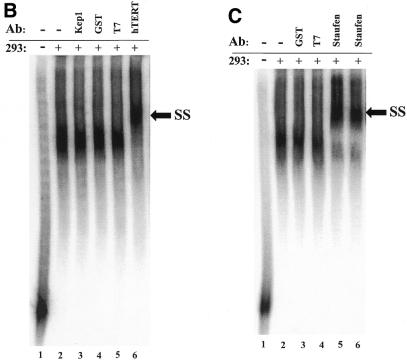
Identification of a specific hTR–protein complex in vitro containing hTERT and human Staufen. (A) 32P-labeled hTR (0.25 pmol) was incubated in the absence (lane 1) or presence (lanes 2–16) of 3.4 µg partially purified telomerase extract from 293 cells that were (lanes 3 and 4) or were not (lanes 2 and 5–16) pretreated with proteinase K. The arrow points to the position of radiolabeled wild-type hTR, and the bracket indicates the RNP complex. In lanes 5–16, the binding reactions were performed with increasing concentrations (0.05, 0.4, 0.8 and 1.6 µM) of unlabeled specific hTR (hTR, lanes 5–8), non-specific E.coli 5S rRNA (rRNA, lanes 9–12) or antisense hTR (αhTR, lanes 13–16). The percentage (%) of radiolabeled complex-bound hTR is indicated at the bottom for each lane. (B and C) Identification of the catalytic subunit of human telomerase (hTERT) and the human Staufen protein in the specific hTR–protein complex using antibody supershift assays. Specific antibodies for hTERT and human Staufen, or control antibodies specific for the Drosophila RNA-binding protein Kep1, for the glutathione S-transferase (GST), and for the T7 epitope (T7) were added following the binding reactions as described in the Materials and Methods. The arrows indicate the respective supershifted (SS) complexes.
EMSAs were then performed to probe for specific proteins that are predicted to form a complex in the human telomerase RNP. A specific complex between proteins of the partially purified extract and radiolabeled hTR (Fig. 1B, lane 2) was resolved and clearly supershifted by the addition of specific hTERT antibodies (lane 6). Control antibodies (lanes 3–5) that recognize heterologous proteins unrelated to telomerase did not affect complex mobility. Supershifts were also detected with two different polyclonal antibodies raised against recombinant human Staufen (Fig. 1C, lanes 5 and 6), a protein that was previously shown to associate with hTR (19). The addition of antibodies specific for the human telomerase-associated protein TEP1 (18,32) did not result in a detectable supershift (data not shown). Results of the EMSA indicate that the hTR-specific complex contains hTERT and the human Staufen protein.
Sequences or structures between nucleotides 276 and 424 of hTR are necessary to competitively inhibit the formation of the hTR–protein complex
Using micrococcal nuclease-treated, partially purified telomerase extract from 293 cells, terminal deletions in hTR were previously tested for their ability to reconstitute human telomerase activity in vitro (25). The minimal region of hTR required for the reconstitution of human telomerase activity in the micrococcal nuclease assay consists of nucleotides 44–205 (25). In order to identify protein-binding domains in hTR, hTR variants (Fig. 2) were tested for their ability to competitively inhibit the formation of the hTR–protein complex characterized in Figure 1.
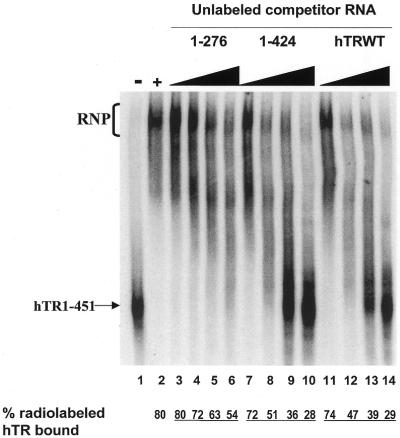
hTR 3′ truncations were generated by in vitro run-off transcription (see Materials and Methods and Fig. 2B) and used as competitor RNAs in the EMSA (Fig. 3 and data not shown). hTR1–424, in which the conserved ACA box and CR7 domain are perturbed (Fig. 2A), inhibited complex formation as efficiently as wild-type hTR (Fig. 3, compare lanes 7–10 with 11–14). The calculated IC50 values (concentration of competitor RNA resulting in 50% inhibition of binding) for hTR1–424 and hTR1–451 were similar (Fig. 2B). A deletion of 175 nt at the 3′ end of the hTR (hTR1–276), removing part of the conserved CR4-CR5 domain (Fig. 2A), altered the ability of competitor hTR to inhibit the formation of the hTR–protein complex (Fig. 3, lanes 3–6; Fig. 2B). The percentages of complex-bound hTR in the presence of maximal concentration of the competitor RNAs hTR1–451, hTR1–424 and hTR1–276 were 29, 28 and 54%, respectively (Fig. 3, compare lanes 14, 10 and 6). The calculated IC50 values for the hTR1–205, hTR1–182 and hTR1–168 RNAs (data not shown) ranged between 2.5 and 2.9 µM (Fig. 2B), indicating that these truncated hTRs were ∼5-fold less effective than wild-type hTR at inhibiting the binding of full-length radiolabeled hTR to hTERT and telomerase-associated proteins in vitro. hTR spanning nucleotides 1–159 was 10-fold less efficient than hTR1–451 at competitively inhibiting the formation of the hTR–protein complex (IC50 values of 5.3 and 0.5 µM, respectively; Fig. 2B).
Figure 3.
Inhibition of the hTR–protein telomerase complex formation by hTR1–424 and hTR1–276. 32P-labeled hTR (0.25 pmol) was incubated in the absence (lane 1) or presence (lanes 2–14) of 3.4 µg of partially purified telomerase extracts from 293 cells. Standard binding reactions were performed in the absence (lane 2) or presence (lanes 3–14) of increasing concentrations (0.05, 0.4, 0.8 and 1.6 µM) of unlabeled hTR1–276 (lanes 3–6), hTR1–424 (lanes 7–10) and hTR1–451 (lanes 11–14). The arrow points to free radiolabeled wild-type hTR, and the bracket indicates the RNP complex. The percentage (%) of radiolabeled complex-bound hTR is indicated at the bottom for each lane.
Mammalian telomerase RNA contains a conserved structural element, the H/ACA box (8; Fig. 2A), which is critical for RNA accumulation and telomerase activity reconstitution in vivo (9,10,23), but not in vitro (13,17,25,36). We substituted TGT for the ACA trinucleotides within the conserved ACA box of hTR and tested the ability of this modified RNA to competitively inhibit complex formation. hTR(ACA-TGT) inhibited the formation of the hTR–protein complex as efficiently as hTR1–451 (data not shown; IC50 values of 0.6 and 0.5 µM, respectively; Fig. 2B). These results indicate that nucleotides 1–424 of hTR are sufficient for the formation of a stable telomerase RNP and that sequences or structures between nucleotides 276 and 424 of hTR are important for the formation of a human telomerase complex in vitro.
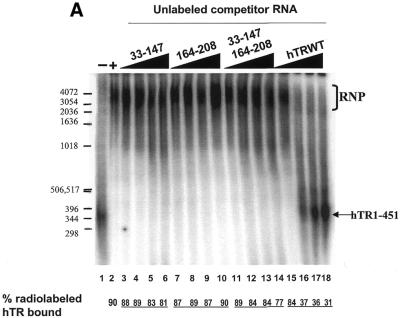
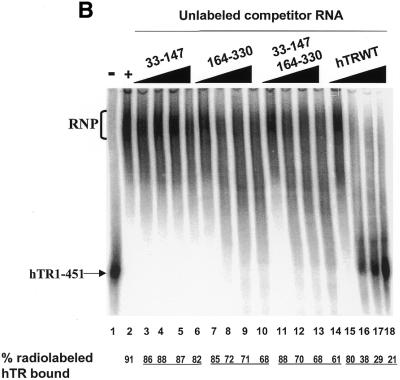
hTR domains 33–147 and 164–330 do not cooperate to inhibit hTR–protein complex formation as efficiently as full-length hTR
Several studies have previously determined that sequences or structures between nucleotides 160 and 210 of hTR are functionally important for reconstituting human telomerase activity in vitro (17,25,36). Two inactive fragments of hTR (33–147 and 164–325) complement one another to reconstitute human telomerase activity when incubated with hTERT previously synthesized in RRL (36). Interestingly, hTR fragments spanning nucleotides 33–147 and 164–330 can independently associate with hTERT (17), suggesting the presence of two hTERT-binding sites within hTR.
The ability of hTR sequences 33–147, 164–208 and 164–330 (alone or in combination) to competitively inhibit the formation of the hTR–protein complex was tested to determine whether these RNAs constitute distinct protein-binding domains of the human telomerase RNP. The addition of excess unlabeled hTR33–147 (Fig. 4A, lanes 3–6) and hTR164–208 (Fig. 4A, lanes 7–10) did not significantly inhibit the binding of radiolabeled hTR to hTERT and telomerase-associated proteins in vitro. In the presence of a 64-fold molar excess of hTR33–147 and hTR164–208, the percentage of complex-bound hTR was 81% (lane 6) and 90% (lane 10), respectively, which was not significantly different from the 90% bound in the absence of competitor RNA (lane 2). hTR164–330 was more effective than either hTR33–147 or hTR164–208 at inhibiting complex formation (68% complex-bound hTR at the highest concentration of competitor RNA; Fig. 4B, lane 10; and a calculated IC50 value of 3.5 µM; Fig. 2B). These results suggest that the individual hTR fragments hTR33–147, hTR164–208 or hTR164–330 are not as efficient as full-length hTR in forming stable associations with hTERT and telomerase-associated proteins.
Figure 4.
hTR33–147 and hTR164–330 do not cooperate to inhibit the formation of the hTR–protein telomerase complex as efficiently as wild-type hTR. (A) Effect of hTR33–147 and hTR164–208 on hTR–protein telomerase complex formation in vitro. 32P-labeled hTR (0.25 pmol) was incubated in the absence (lane 1) or presence (lanes 2–18) of 3.4 µg partially purified telomerase extracts from 293 cells. Standard binding reactions were performed in the absence (lane 2) or presence (lanes 3–18) of increasing concentrations (0.05, 0.4, 0.8 and 1.6 µM) of unlabeled hTR33–147 (lanes 3–6), hTR164–208 (lanes 7–10), hTR33–147 with hTR164–208 (lanes 11–14), and wild-type hTR (lanes 15–18). The arrow points to free radiolabeled wild-type hTR, and the bracket indicates the RNP complex. The percentage (%) of radiolabeled complex-bound hTR is indicated at the bottom for each lane. DNA molecular weight standards are indicated on the left (in bp). (B) Effect of hTR33–147 and hTR164–330 on hTR–protein telomerase complex formation in vitro. 32P-labeled hTR (0.25 pmol) was incubated in the absence (lane 1) or presence (lanes 2–18) of 3.4 µg partially purified telomerase extracts from 293 cells. Standard binding reactions were performed in the absence (lane 2) or presence (lanes 3–18) of increasing concentrations (0.05, 0.4, 0.8 and 1.6 µM) of unlabeled hTR33–147 (lanes 3–6), hTR164–330 (lanes 7–10), hTR33–147 with hTR164–330 (lanes 11–14), and wild-type hTR (lanes 15–18). The arrow points to free radiolabeled wild-type hTR and the bracket indicates the RNP complex. The percentage of radiolabeled complex-bound hTR is indicated at the bottom for each lane.
In order to determine whether subgenomic hTR regions can cooperate to mediate protein binding, we tested the ability of hTR33–147 to competitively inhibit complex formation in combination with either hTR164–208 or hTR164–330. The combination of hTR33–147 and hTR164–208 slightly increased the ability of these two RNAs to inhibit complex formation (Fig. 4A, lanes 11–14; Fig. 2B). In contrast, the calculated IC50 values for the hTR33–147/hTR164–330 combination and for hTR164–330 were identical, 3.53 ± 0.29 µM and 3.53 ± 0.40 µM, respectively (Figs 2B and 4B, compare lanes 11–14 with 7–10). The results suggest that hTR segments spanning nucleotides 33–147 and 164–330 complement one another to reconstitute a catalytically active enzyme in vitro (17,36), but in combination do not form an hTR–protein complex as efficiently as full-length hTR.
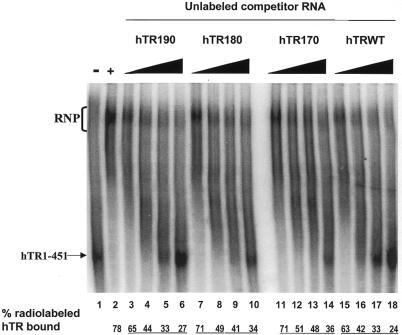
hTR170 and hTR180 are catalytically inactive in vitro but efficiently inhibit the formation of the hTR–protein complex
The secondary structure model of vertebrate telomerase RNA predicts the presence of a pseudoknot, as previously proposed for ciliate telomerase RNA (8). The helices P2a, P2b and P3 establish the structural elements of the pseudoknot domain of hTR (Fig. 2A). The 10-nt substitutions encoded by hTR170 and hTR180 (see Materials and Methods) are predicted to disrupt the P3 helix within the pseudoknot, whereas the hTR190 substitution is predicted to disrupt the P1 helix (Fig. 2A). hTR170 and hTR180 poorly reconstitute human telomerase activity, whereas hTR190 reconstitutes some activity in vitro (17,25).
We performed our competitive EMSA using excess unlabeled hTR170, hTR180 and hTR190 RNAs to determine whether these mutated hTRs are defective in binding to hTERT and telomerase-associated proteins in vitro. All three substitutions inhibited complex formation as efficiently as wild-type hTR (Fig. 5). The calculated IC50 values for hTR170, hTR180 and hTR190 (0.65, 0.72 and 0.42 µM, respectively) are comparable to the IC50 value for hTR1–451 (0.49 µM). These results suggest that sequences or structures affected by these substitutions are critical for the catalytic action of the telomerase RNP, but not for the association of hTR with hTERT and telomerase-associated proteins in vitro.
Figure 5.
The catalytically inactive hTR substitutions hTR170, hTR180 and hTR190 efficiently inhibit the formation of the hTR–protein telomerase complex. 32P-labeled hTR (0.25 pmol) was incubated in the absence (lane 1) or presence (lanes 2–18) of 3.4 µg partially purified telomerase extracts from 293 cells. Standard binding reactions were performed in the absence (lane 2) or presence (lanes 3–18) of increasing concentrations (0.05, 0.4, 0.8 and 1.6 µM) of unlabeled hTR190 (lanes 3–6), hTR180 (lanes 7–10), hTR170 (lanes 11–14) and wild-type hTR (lanes 15–18). The arrow points to free radiolabeled wild-type hTR, and the bracket indicates the RNP complex. The percentage (%) of radiolabeled complex-bound hTR is indicated at the bottom for each lane.
Identification of hTR-binding proteins in the partially purified human telomerase extract
We used UV cross-linking as a first step toward the identification of cellular proteins that interact with the hTR. Partially purified human telomerase extract was pre-incubated with radiolabeled hTR prior to irradiation with UV light. Following digestion with RNase A, proteins covalently cross-linked to radiolabeled hTR oligomers were analyzed by SDS–PAGE. Four bands of apparent molecular weight 40, 42, 58 and 125 kDa were identified (Fig. 6, lanes 2 and 3). These bands were not observed when the protein extract was omitted from the reaction or following proteinase K treatment (data not shown). In addition, these different hTR-binding proteins were undetectable in the absence of UV irradiation (Fig. 6, lane 1). These results indicate that at least four proteins in the partially purified telomerase extract bind the hTR in vitro.
Figure 6.
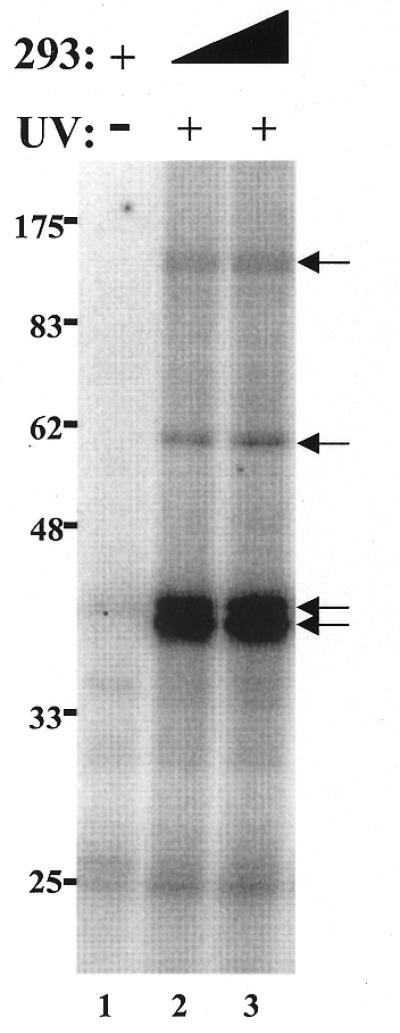
UV Cross-linking of four proteins to the hTR in vitro. Radiolabeled hTR was incubated in the presence of 3.4 µg (lane 2) or 8.5 µg (lanes 1 and 3) partially purified telomerase extracts from 293 cells. Following standard binding reactions, the samples were (lanes 2–3) or were not (lane 1) submitted to UV irradiation, digested with RNase A, boiled, and analyzed on a 10% SDS–PAGE. The position of the protein markers are indicated on the left (in kDa). The arrows indicate the position of the four main hTR-binding proteins.
DISCUSSION
The in vitro reconstitution of human telomerase suggests that hTERT and hTR are minimally required for activity (12–15). However, purification of the telomerase holoenzyme indicates that the approximate molecular mass of the human telomerase complex is 500–1500 kDa (32,37), thus suggesting that other components are associated with hTERT and hTR as part of a multi-subunit telomerase complex. In this study, active telomerase partially purified from 293 whole cell extract was used to investigate hTR–protein interactions. Using an EMSA, we identified an hTR–protein complex. This complex highlighted a specific interaction with the hTR through competitive studies where wild-type hTR, but not heterologous RNAs, inhibited its formation. Addition of antibodies that specifically recognize hTERT and human Staufen promoted a supershift, indicating that these proteins are components of the identified hTR–protein complex. One of our concerns was that the exogenous radiolabeled hTR only bound hTERT and human Staufen proteins that were not associated with endogenous hTR. Telomerase proteins may exist exclusively in a complex with hTR, and since the extracts were not pretreated to remove endogenous telomerase RNA, the association of labeled hTR with telomerase-associated proteins may not be detectable. However, we have used conditions that favor exchanges between the endogenously telomerase-associated hTR and the exogenously added recombinant hTR to allow the detection of hTR associations with telomerase proteins in the complex. Telomerase extracts were pre-treated with EDTA to favor exchanges between endogenous and radiolabeled hTR. The integrity of many RNPs is dependent on divalent ions and chelating agents such as EDTA have been previously used to stimulate reversible RNA binding of several RNPs (26,34,35). Nonetheless, the possibility that the in vitro complex we observe is lacking certain proteins that interact with hTR in vivo cannot be excluded.
The strong conservation of elements predicted by the secondary structure of hTR suggests important functional roles and may be indicative of protein-binding domains within the telomerase RNA. Secondary structure motifs such as hairpins, internal loops, bulges and helices commonly define the recognition sites for RNA-binding proteins (38,39). Using the EMSA, we analyzed a variety of hTR deletions and mutations altered in different secondary structure domains for their ability to bind hTERT and telomerase-associated proteins in vitro. A recent study also used a gel mobility shift assay to investigate the in vitro assembly of H/ACA box snoRNPs, including hTR (24). We are confident that the hTR–protein complex observed in our RNA band shift assay is specific for telomerase components rather than for H/ACA box binding proteins. First, the 293 cell extract used in this study was partially purified for catalytically active human telomerase (25). Secondly, electrophoretic mobility supershift assays demonstrated that hTERT, the telomerase catalytic subunit, is present in the complex. Thirdly, using the EMSA we demonstrated that the deletion of the ACA box (hTR1–424) and the substitution of the conserved ACA trinucleotide did not interfere with the ability of these RNAs to act as specific competitors in the formation of the hTR–protein complex. In contrast, a similar mutation in the ACA box of hTR affected its ability to competitively inhibit the assembly of radiolabeled hTR206–451 with H/ACA binding proteins in vitro (24). The observation that hTR1–424 and hTR(ACA-TGT) were as effective as wild-type hTR in inhibiting the formation of the telomerase complex suggests that the CR7 domain and the H/ACA box are not required for binding telomerase proteins in vitro. This is also supported by the reconstitution of human telomerase activity in vitro with hTRs containing alterations or deletions in these two structural elements (13,17,25,36).
We and others have recently identified the CR4-CR5 domain of hTR as a binding site for hTERT (16,17). Results presented in this work further support this structural element as an important RNA-binding site within the human telomerase RNP complex. hTR1–276, in which the CR4-CR5 domain of hTR lacks the evolutionary conserved CR5 sequence (8), was ∼3-fold less efficient than wild-type hTR at inhibiting complex formation (Figs 3 and 2B). Furthermore, hTR164–330, which contains the complete CR4-CR5 domain (see Fig. 2A), partially inhibited complex formation, in contrast to hTR164–208, which does not contain the CR4-CR5 domain and failed to inhibit complex formation (Fig. 2B). The functional requirement for the CR4-CR5 domain to reconstitute mammalian telomerase activity (10,16,17) is likely due to direct interactions between mammalian TERT and the CR4-CR5 domain of the telomerase RNA; however, binding of TERT to the telomerase RNA could be mediated via a telomerase-associated protein or catalytic cofactor. The availability of recombinant human telomerase protein components will be essential to characterize the domains required for the direct molecular interactions between the different subunits.
Results obtained in this study support the conclusions of recent analyses of in vitro reconstitution of telomerase activity and hTERT–hTR interactions in RRL (16,17,36). The combination of hTR fragments 33–147 and 164–330 does not reconstitute human telomerase activity at levels as robust as full-length hTR (17,36). Furthermore, immunoprecipitation of hTERT expressed in RRL showed that hTR164–330 binds hTERT more efficiently than hTR33–147 and that the binding of either RNA to hTERT is not increased when the two RNAs are combined (17). These results support our current observations that: (i) hTR164–330 competitively inhibits complex formation more efficiently than hTR33–147, and (ii) that the efficiency in competing with full-length hTR for complex formation does not increase in the presence of both RNA domains. The complementation of the two independent domains to reconstitute human telomerase activity in vitro may be due to the contribution of a template domain by hTR33–147 and an hTERT-binding site (between nucleotides 208 and 330) by hTR164–330, both required for activity.
The 10-nt substitutions in the hTR variants hTR170 and hTR180 are predicted to partially disrupt the P3 helix within the pseudoknot domain of hTR and were used to study the role of the pseudoknot in telomerase protein-binding in vitro. Addition of these hTR variants to micrococcal nuclease-treated, partially purified human telomerase extracts or to RRL expressing recombinant hTERT restores low levels of telomerase activity (17,25). Sequence alterations predicted to destabilize the pseudoknot of the mouse telomerase RNA also demonstrated a critical role for this domain in the reconstitution of telomerase activity (10). hTR170, hTR180 and hTR190 efficiently inhibit the formation of the hTR–protein complex (Figs 2B and 5) and indicate that substitutions in the P3 helix of hTR do not affect RNA association with hTERT and/or telomerase-associated proteins in vitro. The ability of these mutated hTRs to associate with hTERT is also supported by in vitro coimmunoprecipitation results (17). The role of the pseudoknot domain of the telomerase RNA in telomerase catalysis remains uncharacterized. Contrary to the effect of mammalian telomerase RNA pseudoknot alterations, nucleotide substitutions within the pseudoknot of the Tetrahymena telomerase RNA do not significantly affect in vitro telomerase activity (40,41). However, in vivo expression of mutated Tetrahymena telomerase RNA that affects the base-pairing within the pseudoknot domain prevented the assembly of a catalytically active telomerase RNP (42). Thus, further studies will be required to define the precise role of the pseudoknot domain of telomerase RNAs from both ciliates and mammals.
A UV cross-linking approach was also used to identify proteins that can associate with hTR. Four proteins of apparent molecular weights 40, 42, 58 and 125 kDa were detected upon irradiation of the binding reactions with UV light. The molecular weights of the 125 and 58 kDa proteins correspond to the predicted masses of hTERT and human Staufen, respectively. Moreover, their cross-linking to hTR is supported by results from the supershift assays (Fig. 2B). The identities of the prevalent 40 and 42 kDa proteins observed in our cross-linking experiments are unknown. The absence of cross-linked proteins with molecular weights that correspond to previously reported or to novel hTR-associated proteins may be explained by either the loss of specific proteins during the partial purification of the 293 cell extract or to the inefficient cross-linking of these proteins to hTR.
The EMSA and UV cross-linking results that we describe have provided a characterization of hTR domains involved in protein binding. These methods will be useful in supporting the identification of telomerase-associated proteins that may bind the hTR. A detailed understanding of the different interactions occurring between the hTR, hTERT and telomerase-associated proteins is an essential step toward the elucidation of the mechanism of assembly and function of this critical ribonucleoprotein complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Carol Greider, in whose laboratory many of the reagents used in these experiments were generated, R. Pruzan for helpful discussions, S. Richard, L. Desgroseillers, M. Blasco and L. Harrington for antibodies, G. Kukolj, M. Esmail, T. Moriarty, S. Huard and S. Dupuis for comments on the manuscript, and T. Moriarty for help in the preparation of some of the figures. F.B. is the recipient of a Medical Research Council of Canada (MRCC) Studentship Award. This work was supported by a grant from the MRCC (MT-14026) to C.A.
References
- 1.Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- 2.Collins K. (2000) Mammalian telomeres and telomerase. Curr. Opin. Cell Biol., 12, 378–383. [DOI] [PubMed] [Google Scholar]
- 3.Lundblad V. (2000) DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res., 451, 227–240. [DOI] [PubMed] [Google Scholar]
- 4.Feng J., Funk,W.D., Wang,S.-S., Weinrich,S.L., Avilion,A.A., Chiu,C.-P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J., Le,S., West,M.D., Harley,C.B., Andrews,W.H., Greider,C.W. and Villeponteau,B. (1995) The human telomerase RNA component. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 5.Hinkley C., Blasco,M., Funk,W., Feng,J., Villeponteau,B., Greider,C. and Herr,W. (1998) The mouse telomerase RNA 5′-end lies just upstream of the telomerase template sequence. Nucleic Acids Res., 26, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaug A.J., Lingner,J. and Cech,T.R. (1996) Method for determining RNA 3′ ends and application to human telomerase RNA. Nucleic Acids Res., 24, 532–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon C., Cech,T. and Zaug,A. (1997) Polyadenylation of telomerase RNA in budding yeast. RNA, 3, 1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J.-L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell J., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Rivera L. and Blasco,M. (2001) Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J. Biol. Chem., 276, 5856–5865. [DOI] [PubMed] [Google Scholar]
- 11.Narayanan A., Lukowiak,A., Jady,B., Dragon,F., Kiss,T., Terns,R. and Terns,M. (1999) Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J., 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 13.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 14.Bachand F. and Autexier,C. (1999) Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem., 274, 38027–38031. [DOI] [PubMed] [Google Scholar]
- 15.Masutomi K., Kaneko,S., Hayashi,N., Yamashita,T., Shirota,Y., Kobayashi,K. and Murakami,S. (2000) Telomerase activity reconstituted in vitro with purified human telomerase reverse transcriptase and human telomerase RNA component. J. Biol. Chem., 275, 22568–22573. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell J. and Collins,K. (2000) Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell, 6, 361–371. [DOI] [PubMed] [Google Scholar]
- 17.Bachand F. and Autexier,C. (2001) Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell. Biol., 21, 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington L., McPhail,T., Mar,V., Zhou,W., Oulton,R., Bass,M.B., Arruda,I. and Robinson,M.O. (1997) A mammalian telomerase-associated protein. Science, 275, 973–977. [DOI] [PubMed] [Google Scholar]
- 19.Le S., Sternglanz,R. and Greider,C.W. (2000) Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell, 11, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallaire F., Dupuis,S., Fiset,S. and Chabot,B. (2000) Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem., 275, 14509–14516. [DOI] [PubMed] [Google Scholar]
- 21.Ford L., Suh,J., Wright,W. and Shay,J. (2000) Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol., 20, 9084–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaBranche H., Dupuis,S., Ben-David,Y., Bani,M., Wellinger,R. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell J., Wood,E. and Collins,K. (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- 24.Dragon F., Pogacic,V. and Filipowicz,W. (2000) In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol., 20, 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Autexier C., Pruzan,R., Funk,W.D. and Greider,C.W. (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J., 15, 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 26.Autexier C. and Triki,I. (1999) Tetrahymena telomerase ribonucleoprotein RNA–protein interactions. Nucleic Acids Res., 27, 2227–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Fruscio M., Chen,T., Bonyadi,S., Lasko,P. and Richard,S. (1998) The identification of two Drosophila K homology domain proteins. Kep1 and SAM are members of the Sam68 family of GSG domain proteins. J. Biol. Chem., 273, 30122–30130. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Rivera L., Herrera,E., Albar,J.P. and Blasco,M.A. (1998) Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl Acad. Sci. USA, 95, 10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duchaine T., Wang,H., Luo,M., Steinberg,S., Nabi,I. and DesGroseillers,L. (2000) A novel murine Staufen isoform modulates the RNA content of Staufen complexes. Mol. Cell. Biol., 20, 5592–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham L., Duchaine,T., Luo,M., Nabi,I. and DesGroseillers,L. (1999) Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol., 19, 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson K.K. and Green,M.R. (1988) Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev., 2, 319–329. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama J., Saito,M., Nakamura,H., Matsuura,A. and Ishikawa,F. (1997) TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell, 88, 875–884. [DOI] [PubMed] [Google Scholar]
- 33.Holt S.E., Aisner,D.L., Baur,J., Tesmer,V.M., Dy,M., Ouellette,M., Trager,J.B., Morin,G.M., Toft,D.O., Shay,J.W., Wright,W.E. and White,M.A. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter P. and Blobel,G. (1983) Disassembly and reconstitution of signal recognition particle. Cell, 34, 525–533. [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee A.K. and Sarkar,S. (1981) The translational inhibitor 10S cytoplasmic ribonucleoprotein of chick embryonic muscle. J. Biol. Chem., 256, 11301–11306. [PubMed] [Google Scholar]
- 36.Tesmer V.M., Ford,L.P., Holt,S.E., Frank,B.C., Yi,X., Aisner,D.L., Ouellette,M., Shay,J.W. and Wright,W.E. (1999) Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol., 19, 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnapp G., Rodi,H., Rettig,W., Schnapp,A. and Damm,K. (1998) One-step affinity purification protocol for human telomerase. Nucleic Acids Res., 26, 3311–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermann T. and Westhof,E. (1999) Non-Watson–Crick base pairs in RNA–protein recognition. Chem. Biol., 6, 335–343. [DOI] [PubMed] [Google Scholar]
- 39.Cusack S. (1999) RNA–protein complexes. Curr. Opin. Struct. Biol., 9, 66–73. [DOI] [PubMed] [Google Scholar]
- 40.Autexier C. and Greider,C.W. (1998) Mutational analysis of the Tetrahymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res., 26, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Licht J.D. and Collins,K. (1999) Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev., 13, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilley D. and Blackburn,E.H. (1999) The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl Acad. Sci. USA, 96, 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]