Abstract
Background
Zinc finger homeodomain proteins (ZHD) constitute a plant-specific transcription factor family with a conserved DNA binding homeodomain and a zinc finger motif. Members of the ZHD protein family play important roles in plant growth, development, and stress responses. Genome-wide characterization of ZHD genes has been carried out in several model plants, including Arabidopsis thaliana and Oryza sativa, but not yet in tomato (Solanum lycopersicum).
Results
In this study, we performed the first comprehensive genome-wide characterization and expression profiling of the ZHD gene family in tomato (Solanum lycopersicum). We identified 22 SlZHD genes and classified them into six subfamilies based on phylogeny. The SlZHD genes were generally conserved in each subfamily, with minor variations in gene structure and motif distribution. The 22 SlZHD genes were distributed on six of the 12 tomato chromosomes, with segmental duplication detected in four genes. Analysis of Ka/Ks ratios revealed that the duplicated genes are under negative or purifying selection. Comprehensive expression analysis revealed that the SlZHD genes are widely expressed in various tissues, with most genes preferentially expressed in flower buds compared to other tissues. Moreover, many of the genes are responsive to abiotic stress and phytohormone treatment.
Conclusion
Systematic analysis revealed structural diversity among tomato ZHD proteins, which indicates the possibility for diverse roles of SlZHD genes in different developmental stages as well as in response to abiotic stresses. Our expression analysis of SlZHD genes in various tissues/organs and under various abiotic stress and phytohormone treatments sheds light on their functional divergence. Our findings represent a valuable resource for further analysis to explore the biological functions of tomato ZHD genes.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4082-y) contains supplementary material, which is available to authorized users.
Keywords: ZF-HD, Solanum lycopersicum, Phytohormone, Abiotic stress, Organ-specific expression, Fruit development
Background
Various regulatory proteins systematically controlled the many developmental processes in plants; among them, transcription factors (TFs) are major regulatory proteins with sequence-specific DNA or nucleotide binding activity [1–4]. TFs control a range of biological processes in plants, such as growth, development, metabolism, cell cycle progression, and responses to environmental stimuli. For example NF-Y, MYB, AP2, TCP, WRKY, NAC, GRF, and SPL TFs play important role in stress tolerances, whereas NAC, SPL, and GRF TFs are involved in root growth, flower, seed development, and plant transition [5]. Zinc finger homeodomain (ZHD) TFs, harboring a homeodomain (HD) and a C2H2-type zinc finger motif (ZF), are involved in plant development and stress responses [3]. ZHD TFs have distinct sequence characteristics compared to other plant HD-containing proteins identified to date [3, 6].
The HD is a DNA-binding domain containing approximately 60 amino acids that is present in numerous transcription factors in all eukaryotic organisms [7, 8]. HD proteins participate widely in development by regulating the expression patterns of target genes in both plants and animals [7]. Most HD proteins are associated with additional
domain(s) or motifs for protein–protein interactions and/or other regulatory functions [9]. HD-containing proteins are classified into six distinct families based on the presence of different motifs: leucine zipper-associated HD (HD-Zip), zinc finger motif-associated HD (ZF-HD), WUSCHEL-related homeobox (WOX), Bell type HD, finger domain associated to a HD (PHD finger), and Knotted-related homeobox (KNOX) proteins [10].
The zinc finger, one of the most important structural motifs, consists of a zinc ion in the core surrounded by several amino acid residues (cysteines or histidines in most cases) [11]. Zinc finger domains are widely present in many regulatory proteins and are actively involved in sequence-specific binding to DNA/RNA and in protein–protein interaction [11–13]. Zinc finger motifs are classified into different categories based on the presence of Cys and His residues, for instance, C3H, C2H2, and C2C2 [11]. Zinc finger TFs, especially C2H2-type TFs, play crucial roles in many metabolic pathways in plants, including stress responses and defense activation [14].
A ZHD protein was identified in Flaveria trinervia as a potential regulator of the gene encoding C4 phosphoenolpyruvate carboxylase (PEPCase) [15]. ZHD protein family members were subsequently identified in several model plants including Arabidopsis thaliana with 17 members and rice (Oryza sativa), with 15 members [8, 15]. In Arabidopsis, ZHD proteins act as transcriptional regulators with unique biochemical properties that function in the regulation of floral development [16]. AtZHD1 is a transcriptional regulator that binds to the promoter region of ERD1 (EARLY RESPONSE TO DEHYDRATION STRESS 1), and its expression is induced by drought, salinity, and abscisic acid (ABA) [17]. The overexpression of NAC and AtZHD1 increases drought tolerance in Arabidopsis [20]. Soybean ZHD1 and 2 (GmZF-HD1 and GmZF-HD2) are upregulated upon pathogen inoculation, and GmZF-HD1 and GmZF-HD2 bind to the promoter region of a gene encoding calmodulin isoform 4 (GmCaM4) [18]. Three Arabidopsis MINI ZINC FINGER (AtMIF) proteins and their homologs share high levels of sequence similarity with the ZF domain of Arabidopsis ZHD proteins [19]. The presence of only a zinc finger motif without a HD in MIF genes suggests that MIF proteins might interfere with the functions of ZHD proteins via their ZF domains [8, 19]. Phylogenetic and sequence analyses of ZHD and MIF genes demonstrated that both are land plant-specific and that ZHDs and MIFs belong to two different groups of the ZHD protein family [8]. However, the origin, evolutionary history, and relationship of those two groups remain unclear [8].
As ZHD protein family members function as transcriptional regulators of floral development and stress responses in Arabidopsis, it is possible that they play similar roles in tomato (Solanum lycopersicum). Although ZHD genes have been investigated in Arabidopsis and several other species, no systematic, comprehensive investigation of the ZHD subfamily has been reported for any solanaceous crop. Therefore, in this study, we performed comprehensive genome-wide analysis of the ZHD gene family in tomato to explore their potential roles in organ development and responses to a wide range of stresses. We also analyzed the predicted gene structures, chromosomal locations, duplication events, and evolutionary divergence of the tomato ZHD genes and classified them based on phylogenetic analysis. Finally, we predicted the functions of ZHD genes based on their expression profiles and the presence of putative cis-elements in their upstream promoter regions. Our results lay the foundation for further studies aimed at uncovering the important biological functions of ZHD proteins in plants.
Results
Identification of ZHD family genes in tomato
We identified a total of 22 non-redundant putative ZHD genes, which we designated SlZHD1–SlZHD22 (Sl for Solanum lycopersicum, Z for zinc finger, HD for homeodomain) according to their physical locations on the chromosomes (Table 1). The lengths of the open reading frame (ORF) of tomato ZHD genes range from 252 bp (SlZHD14) to 2418 bp (SlZHD22). The deduced encoded proteins of tomato ZHDs range in size from 83 aa (SlZHD14) to 805 aa (SlZHD22). In addition, their isoelectric points (pIs) range from 5.76 (SlZHD12) to 9.75 (SlZHD21) and their molecular weights (MWs) range from 9037.14 kDa (SlZHD14) to 90,496.49 kDa (SlZHD22). Information about these genes, including the chromosome locations and introns, is provided in Table 1.
Table 1.
Detailed information about the SlZHD genes and corresponding proteins in tomato
| Gene name | Locus name | ORF (bp) | Location | Length (aa) | Domain (start-end) | Mol. wt (kDa) | pI | Exons | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| SlZHD1 | Solyc01g014970 | 690 | SL2.50ch01:16,338,200…16,339,565 (−strand) | 229 | 20–71 | 25,789.04 | 8.12 | 2 | Extracellular |
| SlZHD2 | Solyc01g102980 | 873 | SL2.50ch01:91,639,500…91,641,389 (+ strand) | 290 | 64–118 | 32,600.98 | 7.76 | 2 | Nuclear |
| SlZHD3 | Solyc01g103810 | 351 | SL2.50ch01:92,343,498…92,343,848 (− strand) | 116 | 14–68 | 13,144.06 | 9.46 | 1 | Cytoplasmic |
| SlZHD4 | Solyc01g103820 | 285 | SL2.50ch01:92,348,106…92,348,390 (− strand) | 94 | 14–68 | 10,361.66 | 8.27 | 1 | Cytoplasmic |
| SlZHD5 | Solyc01g103830 | 633 | SL2.50ch01:92,352,642…92,353,696 (− strand) | 210 | 10–64 | 24,492.28 | 9.68 | 2 | Extracellular |
| SlZHD6 | Solyc01g103840 | 255 | SL2.50ch01:92,360,326…92,360,580 (− strand) | 84 | 10–64 | 9529.72 | 6.27 | 1 | Extracellular |
| SlZHD7 | Solyc02g067310 | 879 | SL2.50ch02:37,494,131…37,495,541 (+ strand) | 292 | 62–118 | 32,086.75 | 8.20 | 2 | Nuclear |
| SlZHD8 | Solyc02g067320 | 996 | SL2.50ch02:37,520,623…37,521,618 (− strand) | 331 | 53–109 | 36,516.54 | 7.75 | 1 | Nuclear |
| SlZHD9 | Solyc02g067330 | 309 | SL2.50ch02:37,527,458…37,527,766 (− strand) | 102 | 29–83 | 11,208.36 | 6.18 | 1 | Cytoplasmic |
| SlZHD10 | Solyc02g085160 | 882 | SL2.50ch02:48,141,575…48,142,456 (− strand) | 293 | 89–143 | 33,312.95 | 8.14 | 1 | Nuclear |
| SlZHD11 | Solyc02g087970 | 273 | SL2.50ch02:50,205,827…50,206,099 (+ strand) | 90 | 23–75 | 10,104.24 | 9.07 | 1 | Cytoplasmic |
| SlZHD12 | Solyc03g061620 | 267 | SL2.50ch03:31,305,510…31,305,776 (− strand) | 88 | 23–77 | 9648.53 | 5.76 | 1 | Cytoplasmic |
| SlZHD13 | Solyc03g098060 | 540 | SL2.50ch03:60,406,311…60,406,850 (− strand) | 179 | 7–64 | 19,951.89 | 8.47 | 1 | Nuclear |
| SlZHD14 | Solyc03g116070 | 252 | SL2.50ch03:65,585,458…65,585,709 (− strand) | 83 | 21–74 | 9037.14 | 8.75 | 1 | Cytoplasmic |
| SlZHD15 | Solyc04g014260 | 744 | SL2.50ch04:4,560,854...4561970 (− strand) | 247 | 54–107 | 27,722.19 | 7.16 | 2 | Nuclear |
| SlZHD16 | Solyc04g074990 | 432 | SL2.50ch04:60,885,156...60886298 (+ strand) | 143 | 45–99 | 16,096.89 | 5.97 | 3 | Cytoplasmic |
| SlZHD17 | Solyc04g080490 | 873 | SL2.50ch04:64,650,886…64,652,793 (− strand) | 290 | 57–111 | 31,547.28 | 8.72 | 2 | Nuclear |
| SlZHD18 | Solyc05g007580 | 894 | SL2.50ch05:2,120,151…2,121,044 (− strand) | 297 | 52–108 | 32,814.23 | 7.26 | 1 | Nuclear |
| SlZHD19 | Solyc05g018740 | 360 | SL2.50ch05:23,129,391...23129750 (− strand) | 119 | 14–68 | 13,674.54 | 9.30 | 1 | Cytoplasmic |
| SlZHD20 | Solyc05g020000 | 360 | SL2.50ch05:25,503,046...25503405 (− strand) | 119 | 14–68 | 13,702.57 | 9.44 | 1 | Cytoplasmic |
| SlZHD21 | Solyc05g051420 | 504 | SL2.50ch05:61,721,814...61724132 (− strand) | 167 | 1–50 | 19,300.40 | 9.75 | 2 | Nuclear |
| SlZHD22 | Solyc09g089550 | 2418 | SL2.50ch09:69,252,813...69257262 (− strand) | 805 | 554–608 | 90,496.49 | 6.97 | 4 | Extracellular |
ORF Open reading frame, bp base pair, aa amino acid, pI isoelectric point, kDa kilodaltons
Phylogenetic and gene structure analysis of the tomato ZHD gene family
To obtain insights into the evolutionary relationships among tomato ZHD family proteins, a phylogenetic tree was constructed from 150 amino acid sequences of tomato (22), potato (38), tobacco (20), Arabidopsis (17), rice (15), Chinese cabbage (31) and Selaginella moellendorffii (7) (Fig. 1). The ZHD protein family was divided into six well-conserved clades (I–VI), with significant bootstrap support [8, 20]. Among these, clade V contained the previously described MIF proteins from Arabidopsis, rice and Chinese cabbage together with four tomato ZHD proteins. Therefore, our analysis separated the MIF proteins from the other ZHD proteins. Clade IV contained the fewest ZHD members, with no SlZHD protein found in this clade. The largest number of SlZHD proteins was in Clade VI.
Fig. 1.
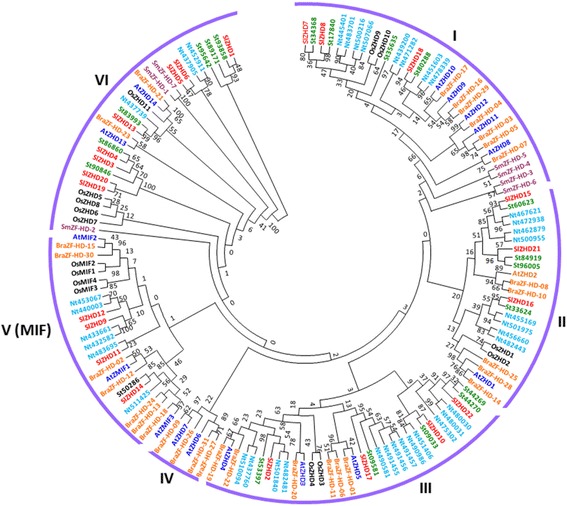
Phylogenetic relationship of Arabidopsis(AtZHD), rice(OsZHD), potato (St, Solanum tuberosum is used instead of PGSC0003DMT4000), tobacco (Nt, Nicotiana tabacum is used instead of XP_0164), Chinese cabbage (BraZF-HD), Selaginella moellendorffii, (SmZF-HD) and tomato (SlZHD) ZHD genes. The conserved ZF-HD_ dimer domain sequences of Arabidopsis, rice, potato, tobacco, Chinese cabbage, Selaginella moellendorffii, and tomato genes were aligned using ClustalX, and the tree were constructed by the neighbor-joining (NJ) method with MEGA 6.0. The numbers on the branches indicate bootstrap support values from 1000 replications. The protein sequences used in the phylogenetic analysis are listed in Additional file 4, along with their accession numbers. The tree was divided into six subfamilies according to bootstrap support values and evolutionary distances
To further investigate the diversity of the tomato ZHD genes, we analyzed SlZHD protein motifs using the MEME online server. Ten conserved motifs were identified, i.e., motif 1 to 10 (Fig. 2, Additional file 1: Fig. S1). An overview of these protein motifs is presented in Additional file 1: Fig. S1. Motif 1 and 2, the most common motifs, comprise the ZF-HD dimer domain. Among the 22 gene products, motif 1 was absent in SlZHD1 and motif 2 was absent in SlZHD21. Motif 10 was mainly found in subfamily III proteins (Figs. 1 and 2). Motif 9 was mainly present in subfamily VI, except for SlZHD10, a subfamily IV protein (Figs. 1 and 2). Motif 5 was mainly found in subfamily I and II proteins (Figs. 1 and 2). The subfamily-specific distribution of conserved motifs may have contributed to the functional divergence of ZHD genes in tomato.
Fig. 2.
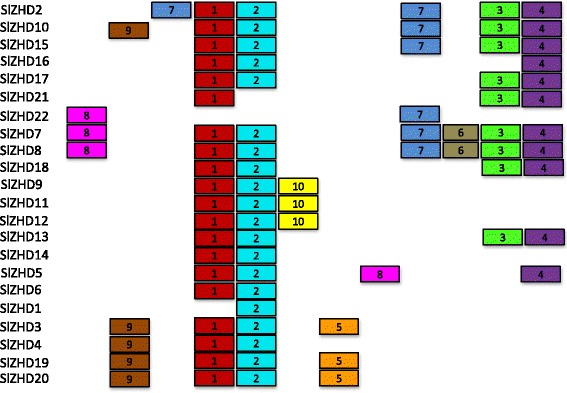
Schematic representation of the 10 conserved motifs in SlZHD proteins. SlZHD protein motifs were identified using the online MEME program. Members of same group are arranged sequentially according to phylogenetic classification. Different colored boxes represent different motifs, where the number in center of each boxes indicates their name (Motif 1 to 10). The colored boxes were drawn and ordered manually according to the results of MEME analysis. The length of each box in the figure does not represent the actual motif size in the proteins
To gain further insights into the structural diversity of ZHD genes in tomato, we constructed a phylogenetic tree based on the 22 SlZHD proteins using their full-length protein sequences (Fig. 3a). The SlZHD proteins were also classified into six subfamilies in this phylogenetic tree, which is in an agreement with the results shown in Fig. 1 based on phylogenetic analysis of the seven plant species (Figs. 1 and 3a). Analyzing the genetic structural diversity among the proteins of a multigene family is a useful way to perform evolutionary analysis. We therefore deduced the exon-intron organization of individual SlZHD genes to examine their structural diversity (Fig. 3b). Most SlZHD genes (13 out of 22) lack introns, whereas the remaining nine have one to three introns. Most closely related members in the same subfamily share almost identical exon-intron organization (Fig. 3a and b). For example, SlZHD genes in subfamily I lack introns. However, the exon-intron organization was not always conserved for most sister gene pairs. For example, SlZHD5/−6 and SlZHD7/−8 have different numbers of exons and introns (Fig. 3a and b).
Fig. 3.
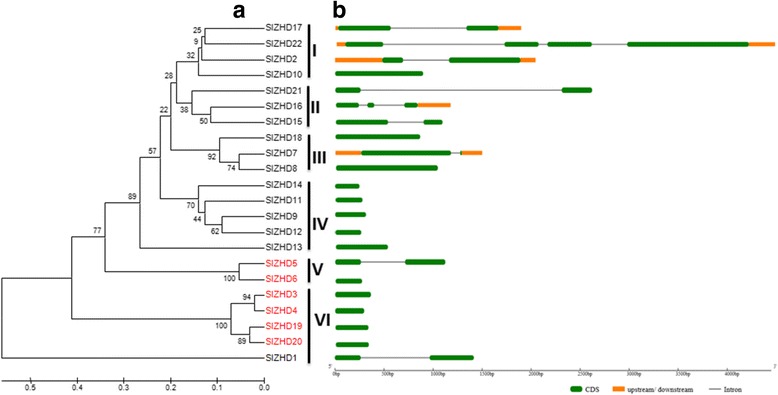
Phylogenetic relationships and gene structures of SlZHD genes. a. Phylogenetic tree constructed among the 22 SlZHD genes using full-length amino acid sequences with MEGA 6.0 following the UPGMA method with 1000 bootstrap replicates. b. Exon-intron organization of SlZHD genes. Exon and introns are represented by green boxes and gray lines, respectively. Untranslated regions are indicated by orange boxes
Chromosomal location, gene duplication, and microsynteny analysis
We mapped the 22 SlZHD genes onto the 12 tomato chromosomes (Additional file 1: Fig. S2), finding that they are unevenly distributed on six of the 12 chromosomes. Chromosome 1 contains the highest number of ZHD family genes (six genes), followed by chromosome 2 and 5 (five and four genes, respectively). Both chromosome 3 and 4 contain three genes, while chromosome 9 possesses only one gene, and no ZHD genes are present on the six remaining tomato chromosomes. To determine the segmental duplication events between the genes, we used the criteria [21]; when the query coverage percentage and identity of the candidate genes was ≥80% they were considered to be duplicated genes. Therefore, segmental duplication analysis showed that four pairs of SlZHD genes, SlZHD3-SlZHD4, SlZHD5-SlZHD6, SlZHD19-SlZHD20, and SlZHD4-SlZHD19, originated through segmental duplication (Additional file 2: Table S2). According to the criterion of tandem duplication (when two genes were separated by five or fewer genes within a 100-kb region on a chromosome), no pair of SlZHD genes originated by tandem duplication (Additional file 1: Fig. S2). To determine the selection constraints on the duplicated SlZHD genes, we estimated the Ka/Ks ratio of each pair of paralogous genes using the method of Nei & Gojobori [22] and found that the ratios for seven paralogous pairs <1 (Additional file 2: Table S2). This result suggests that these genes experienced strong purifying/negative selection pressure, with little variation taking place after duplication. The duplication of paralogous gene pairs is estimated to have occurred 4.3 to 10.13 million years ago (Mya) (Additional file 2: Table S2). Based on a comparative microsyntenic map of Arabidopsis versus tomato and potato (S. tuberosum), 15 pairs of ZHD orthologous genes between S. lycopersicum and S. tuberosum, 16 pairs between A. thaliana and S. lycopersicum, whereas, 11 pairs of orthologous gene pairs were found between A. thaliana and S. tuberosum (Fig. 4). These results confer that during species divergence 16 tomato ZHD and 11 potato ZHD genes are derived from Arabidopsis.
Fig. 4.
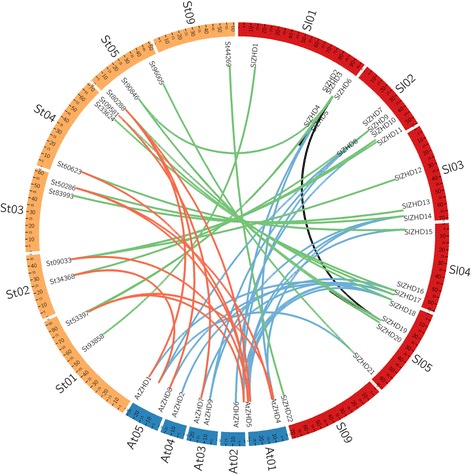
Microsynteny analyses of ZHD genes among S. lycopersicum, S. tuberosum, and A. thaliana. The chromosomes from the three species are indicated in different colors: red, yellow, and blue represent the S. lycopersicum, S. tuberosum, and A. thaliana chromosomes, respectively. Black lines represent duplicated SlZHD genes on tomato chromosomes
Expression profiling of tomato ZHD genes in various organs
To obtain a first glance at the roles of SlZHD genes during various developmental process of tomato, the transcript accumulation levels were investigated in 13 tomato organs (using root, stem, leave, flower bud, full blooming flower, and six fruit developmental stages) by qRT-PCR. Most ZHD genes, except for SlZHD3, SlZHD4, SlZHD5, and SlZHD9, exhibited tissue-specific expression patterns (Fig. 5). Four genes (SlZHD3, SlZHD4, SlZHD5, and SlZHD9) were not expressed in any of the organs examined. These genes exhibited little or no expression in the RNA-Seq data set (Additional file 3). Alternatively, we may have failed to detect their expression in the organs/under the conditions examined, or these four genes might be expressed in other organs or could be pseudogenes. Five genes (SlZHD2, SlZHD10, SlZHD15, SlZHD16, and SlZHD17) and two genes (SlZHD12 and SlZHD21) were highly expressed in flower buds and fully open flowers, respectively, compared to vegetative tissue and developing fruits (Fig. 5). By contrast, three genes (SlZHD7, SlZHD8, and SlZHD19) were strongly expressed in leaves, one gene (SlZHD13) was highly expressed in stems, and one gene (SlZHD14) was highly expressed in roots and stems compared to reproductive tissue and developing fruits. In addition, eight genes (SlZHD1, SlZHD6, SlZHD10, SlZHD11, SlZHD12, SlZHD19, SlZHD20, and SlZHD22) exhibited differential expression profiles in fruits at six developmental stages (Fig. 5). Among these, three genes (SlZHD1, SlZHD20, and SlZHD22) were highly expressed at B3 (breaker stage [after 3 days]), two (SlZHD6 and SlZHD11) were highly expressed at MG (mature green stage), and one (SlZHD19) was highly expressed at the ripening stages (B, B3, and B7) of fruit development (Fig. 5). Similar expression patterns were found in different organs for some paralogous gene pairs, e.g., SlZHD7 and SlZHD8 were strongly expressed in leaves, followed by flower buds (Figs. 3 and 5). However, some gene pairs exhibited differential expression patterns in different organs. For example, SlZHD16 was highly expressed in flower buds, whereas its paralog, SlZHD21, was highly expressed in flowers (Figs. 3a and 5).
Fig. 5.
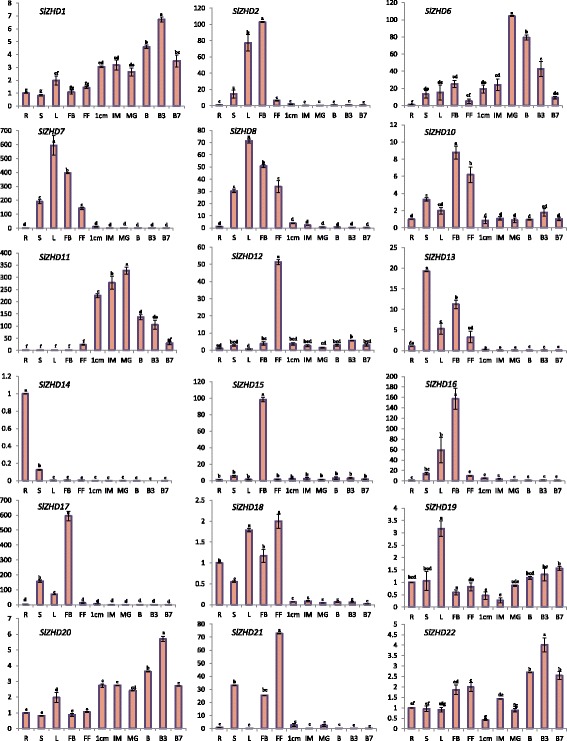
Expression profiles of SlZHD genes in various tomato tissues. Root (R), stem (St), meristem (M), leaves (L), flower bud (FB), full blooming flower (FF), and fruits at six developmental stages (1 cm: 1 cm-sized fruit, IM: immature fruit, MG: mature green fruit, B: breaker, B3: 3 days after breaker, B7: 7 days after breaker) analyzed by qRT-PCR. Relative gene expression levels were normalized to EF1a expression levels. Error bars represent standard deviations of the means of three independent replicates. Statistically significant variations in expression and mean values at different sampling points (ANOVA, p < 0.01 for all 12 genes) are indicated with different letters. Y axis indicates the relative expressions of the genes
Expression profiling of tomato ZHD genes in response to abiotic stress and phytohormone treatment
We analyzed the responses of the SlZHD genes to abiotic stress (drought, NaCl, heat, and cold) and phytohormone (ABA) treatment (Fig. 6). The expression of SlZHD11 and SlZHD21 did not differ between control and abiotic stress- or phytohormone-treated plants. However, many genes were up- or downregulated by these treatments at various time points.
Fig. 6.
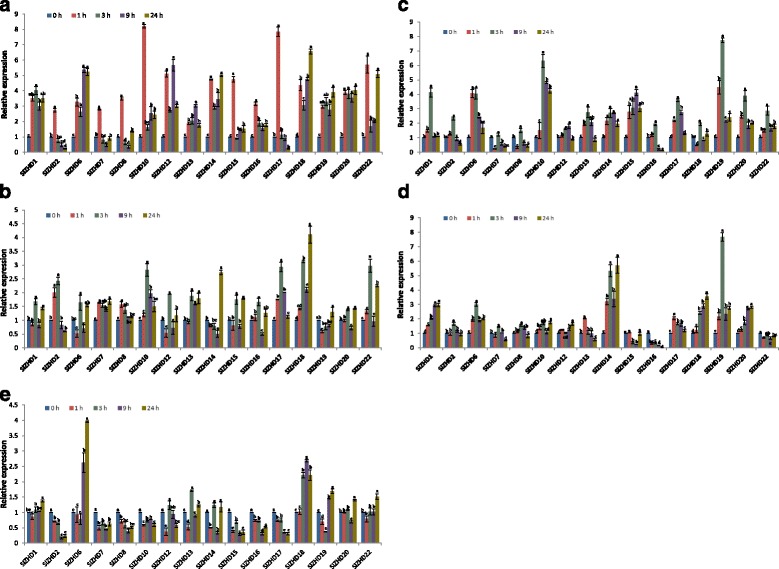
Expression analysis of 16 SlZHD genes by qRT-PCR: the relative expression levels of SlZHD genes under different abiotic and phytohormone treatments: a drought, b NaCl, c heat, d cold, e ABA; error bars indicate the standard error among three replicates. Different letters associated with each treatment indicate statistically significant differences at the 5% level, where the same letter indicates that the values did not differ significantly at P < 0.05 according to Tukey’s pairwise comparison tests
Drought treatment caused a marked change in the transcription levels of 16 SlZHD genes at different time points (Fig. 6a). Eleven of the 16 genes (except SlZHD2, SlZHD7, SlZHD8, SlZHD15, and SlZHD17) were significantly upregulated (2- to 8-fold) within 24 h after treatment compared to the control. More importantly, the expression levels of most of these genes peaked during early stages of treatment. For instance, SlZHD2, SlZHD7, SlZHD8, SlZHD10, SlZHD15, SlZHD16, and SlZHD17 exhibited maximum expression (more than 2.5- to 8-fold vs. the control) at 1 h after treatment. SlZHD12 and SlZHD14 were the most highly expressed at 1 h and at 9 h after treatment, and SlZHD6 expressed peaked at 9 h and 24 h after treatment. The expression of SlZHD22 peaked at both early and last time points (1 h and 24 h after treatment). By contrast, the expression of SlZHD18 and SlZHD19 peaked at 24 h after drought treatment.
The expression levels of 16 SlZHD genes changed in response to NaCl treatment (Fig. 6b). Seven of the 16 SlZHD genes (SlZHD1, SlZHD6, SlZHD12, SlZHD15, SlZHD16, SlZHD20, and SlZHD22) exhibited maximum expression (more than 1.5- to 4-fold vs. the control) at 3 h and 24 h of treatment, whereas two genes (SlZHD14 and SlZHD19) were the most highly upregulated only at last time point (24 h) compared to the control. SlZHD7 and SlZHD8 were slightly upregulated (0.5- to 1.5-fold) from 1 to 24 h, whereas SlZHD18 expression peaked at 24 h (relative expression ~4-fold higher) compared to the control. SlZHD10 and SlZHD13 were upregulated (1- to 2-fold) from 3 h to 24 h of treatment compared to the control. SlZHD2 was strongly induced at 1 h and 3 h, where SlZHD17 was significantly upregulated at 1 h to 9 h after treatment as compared to the control.
Under heat treatment, SlZHD6, SlZHD14, SlZHD15, SlZHD19, SlZHD20, and SlZHD22 were significantly upregulated at various time points compared to the control (Fig. 6c). The expression of SlZHD7, SlZHD8, and SlZHD12 was relatively low (<0.5-fold control levels) under heat treatment, whereas SlZHD10 was strongly upregulated (>4- to 6.5-fold) at 1 h to 24 h after treatment. SlZHD13 and SlZHD17 genes were upregulated by >1- to 3-fold from 3 h to 9 h of heat treatment compared to the control. Four genes (SlZHD1, SlZHD2, SlZHD16, and SlZHD18) were also expressed at higher levels (>1- to 3.5-fold) at 1 h of treatment compared to the control.
The expression of SlZHD2, SlZHD7, SlZHD8, SlZHD10, SlZHD12, SlZHD15, SlZHD16, and SlZHD22 was not significantly altered (<0.5-fold) by cold treatment (Fig. 6d). By contrast, SlZHD1, SlZHD18, and SlZHD20 were upregulated by this treatment, whereas SlZHD16 was downregulated from 1 h to 24 h after treatment vs. the control. SlZHD6, SlZHD14, and SlZHD19 were significantly upregulated (>1- to 7-fold) from 1 h to 24 h after cold treatment compared to the control, and SlZHD13 was induced at 1 h after treatment compared to the control.
Under ABA treatment, seven of the 16 genes (SlZHD2, SlZHD7, SlZHD8, SlZHD10, SlZHD15, SlZHD16, and SlZHD17) were downregulated compared to the control (Fig. 6e). SlZHD6 and SlZHD19 were significantly induced at 9 and 24 h after ABA treatment compared to the control, and three genes (SlZHD1, SlZHD20, and SlZHD22) were slightly upregulated at 24 h after treatment compared to the control. SlZHD18 was highly expressed (> 2.5- to 3-fold) at 3 h to 24 h after treatment compared to the control, while SlZHD12, SlZHD13, and SlZHD14 exhibited constitutively weak expression levels under ABA treatment. The expression of SlZHD12 and SlZHD13 declined at 1 h, but these genes were slightly upregulated at the remaining time points compared to the control. Finally, SlZHD14 was upregulated at 3 and 24 h after treatment compared to the control.
Putative stress- and hormone-responsive cis-elements in the promoter regions of SlZHD genes
Cis-acting elements in the promoter region of genes have vital roles in determining the tissue-specific or stress-responsive expression patterns of genes under variable environmental conditions [23]. Significant positive correlations have been reported between the density of cis-elements and multi-stimulus response genes in upstream regions [24]. A web search was performed using PlantCare database to identify possible stress and hormone-responsive cis-acting elements in the promoter regions of tomato SlZHD genes. Cis-elements responsive to developmental cue, tissue specific expression, and stress responses are found in the promoter of SlZHD family members. In addition, we found five abiotic-stress responsive cis-elements: MBS (MYB binding site) present in 11 different SlZHD genes responsive to drought, ABRE (ABA-responsive element) in 11 genes, HSE (heat stress responsiveness) in 12 genes, LTR (low-temperature responsiveness) in 3 genes, and DRE (dehydration, low-temp, salt stresses) in one SlZHD gene (Additional file 2: Table S3). Among those genes, we found higher expression of SlZHD10, SlZHD12, SlZHD18 and, SlZHD22, which also bear the cis-element MBS for drought stress responsiveness. In addition, ABA-induced upregulation was found in case of SlZHD18 and SlZHD22, coinciding with the presence of ABRE cis-element. The LTR is found in SlZHD1, in agreement with the cold-induced higher expression of that gene. The heat responsive cis-element HSE was found in the genes SlZHD1, SlZHD6, SlZHD10, SlZHD19, SlZHD20, and SlZHD22, consistent with their heat stress-induced upregulation (Fig. 6a-e and Additional file 2: Table S3).
Discussion
The plant-specific ZHD gene family has been found in major groups of land plants, but not in algae [8], suggesting that these genes may have evolved before the divergence of land plants but after the separation of land plant lineages from single-celled algae. In the current study, we identified and characterized 22 ZHD genes from tomato via genome-wide analyses. The number of ZHD genes in tomato was somewhat higher than that of Arabidopsis (17), rice (15), and Selaginella moellendorffii (7) but lower than that of Brassica rapa (31). However, compared with the differences in overall genome size between tomato (950 Mb) and smaller-genomes plants such as Arabidopsis (164 Mb), rice (441 Mb), B. rapa (283.8 Mb), and Selaginella moellendorffii (212.5 Mb), the number of ZHD genes in tomato was relatively small, suggesting that genome duplication events might have contributed to the expansion of the ZHD gene families in plants with smaller genome sizes. For instance, four different large-scale duplication events occurred in the Arabidopsis genome, and more than half of the AtZHD genes likely arose as a result of genome duplication [8, 25, 26]. The deduced protein parameters and conserved ZF-HD_dimer domains of SlZHD family genes are consistent with those of other plant species [3, 8], indicating that ZHD proteins are structurally similar. The 22 SlZHD proteins were classified into six subfamilies. Among these, two subfamilies (I and VI) including proteins found in seedless vascular plants, eudicots, and/or monocots, suggesting that these proteins might have been generated during the early evolution of land plants, considerably before the divergence of major groups of angiosperms (Fig. 1). MIF proteins from various crops form a phylogenetically distinct clade from ZHD proteins, suggesting structural divergence among these proteins and that MIF genes might be derived from ZHD genes after losing the HD [8]. Analysis using the MEME server identified various conserved motifs in SlZHD proteins, with similar motifs found in the most closely related members in the phylogenetic tree, revealing the functional similarity among the same subfamily proteins (Figs. 1 and 2). Gene structure analysis confirmed that 13 of the 22 SlZHD genes lack introns, whereas the nine remaining genes contain one to three introns. The majority of plant ZHD genes were previously found to be intronless [3, 8, 20], but our data do not support this finding for tomato. Three main mechanisms, including exon/intron gain/loss, exonization/pseudo-exonization, and insertion/deletion, are responsible for the variation in exon-intron structures of a gene, with each mechanism contributing to the structural divergence of genes alone or in combination [27, 28]. The variable exon-intron structures of ZHD genes observed in tomato compared to other plants suggests that there is structural divergence in this gene family in S. lycopersicum. Moreover, the similar exon-intron organization in different subfamilies suggests that these genes were highly conserved during evolution (Fig. 3).
Gene duplication mechanisms, including segmental duplication, tandem duplication, and transposition (retro and replicate transposition), are important contributors to biological evolution [29]. Among these, segmental duplication is a principal contributor to the amplification of many gene families [30]. In the current study, we found that four pairs of paralogous genes developed through segmental duplication, whereas no evidence of tandem duplication was detected for any gene pair, indicating that segmental duplication rather than tandem duplication has played a prominent role in the expansion of the tomato ZHD gene family. The Ka/Ks ratios of the duplicated gene pairs indicate that they have undergone purifying selection during the process of evolution. Furthermore, our calculation of the duplication times of the paralogous gene pairs predicted that the segmental duplication event in the SlZHD gene family was occurred 4.3 to 10.13 Mya ago (Additional file 2: Table S2).
The expression of four SlZHD genes (SlZHD3, SlZHD4, SlZHD5, and SlZHD9) was not detected in any tissue examined, indicating that they might be pseudogenes or might be expressed only at specific developmental stages or under specific conditions not included in our study. ZHD transcription factors are involved in regulating various biological processes in plants, including development and responses to abiotic stress and phytohormones [3, 8, 20]. Our expression analysis indicates that most SlZHD genes have tissue-preferential expression patterns. In fact, five of the SlZHD genes were predominately expressed in flower buds, suggesting they play important roles in flower bud development (Fig. 5). Three SlZHD genes (SlZHD7, SlZHD8, and SlZHD19) were highly expressed in leaves, and two genes (SlZHD12 and SlZHD21) were highly expressed only in flowers, suggesting they are involved in leaf and flower development. ZHD gene family members in other plants (e.g. Arabidopsis) are preferentially expressed in floral tissues, revealing their vital regulatory role in floral tissue development [6]. The root is the main organ responsible for water and nutrient acquisition in plants. The predominant expression of SlZHD14 in roots suggests that it might be involved in root development and/or water and nutrient uptake.
Fruit development and subsequent growth is a multifaceted biological process that ultimately leads to the production of crops for harvest. Fruit development is controlled by various transcriptional regulatory networks involving transcription factors such as members of the NAC, MADS-box, and EIN3/EIL families [31]. However, to date, potential roles of ZHD family proteins in fruit development have been characterized only in grape (Vitis vinifera) [20]. In the current study, the transcript levels of five SlZHD genes (SlZHD1, SlZHD19, SlZHD20, and SlZHD22) were generally higher in ripening tomato fruit compared to other stages (B, B3), suggesting they might function in tomato fruit growth and development, especially at the ripening stage. SlZHD11 was upregulated during the early and later phases of green fruit development (1 cm, IM and MG), indicating its potential involvement in cell division and elongation, perhaps to increase fruit size. SlZHD6 expression was highest at the MG stage and then gradually declined thereafter, suggesting it might be involved in increasing fruit size and shape (Fig. 5). Similarly, tissue-specific expression was detected for ZHD family genes in Arabidopsis, Chinese cabbage, maize, and grape across a variety of tissues [3, 8, 20, 32]. The similar expression patterns of duplicated paralogous gene pairs (i.e., SlZHD19 and SlZHD20) suggests that their functions might have been conserved even after duplication and that they might play redundant roles in regulating tissue development. However, the diverse expression patterns of several pairs of paralogs suggest that they likely play different roles in tomato development.
Transcription factors having specific DNA binding motifs (such as zinc fingers, MYB motif) are induced by various signals during specific developmental processes and under various stress conditions [33, 34]. ZHD family proteins contain a DNA binding motif and a zinc finger type motif, suggesting that when they are induced by various signals under different environmental conditions they could play an important role in mediating adaptive responses to various stresses. ZHD genes from Arabidopsis were recently shown to be induced by various abiotic stresses including drought and salt stress, as well as ABA treatment [35]. Therefore, we investigated the expression profiles of ZHD genes in tomato after stress treatment, finding that the expression patterns of many genes differed in response to stress treatments (Fig. 6). SlZHD2, SlZHD7, SlZHD8, and SlZHD15 were upregulated within 1 h of drought and NaCl treatment, but their expression was not markedly altered by cold stress. SlZHD18 was markedly induced by drought, NaCl, and cold treatment, but not by heat stress. Therefore, SlZHD genes might play important roles in various abiotic stress responses. Particularly, the transcript levels of two segmentally duplicated genes, SlZHD19 and SlZHD20, markedly increased from 1 h to 24 h of drought, heat, and cold stress treatment, whereas we detected only minor changes in their expression in response to NaCl stress, supporting the involvement of these genes in the responses to drought, heat, and cold stress rather than salinity stress.
Transcription factors are involved in the regulation of stress signaling and stress-responsive gene expression through various mechanisms that rely on a combination of cis-acting elements present in numerous stress-related genes [2, 36]. Many stress-associated cis-acting elements have been identified in plants [37]. In the current study, several stress-responsive cis-elements were widely found in the promoter regions of most abiotic stress-induced ZHD genes in tomato (Additional file 2: Table S3). Overall 16 SlZHD genes were differentially regulated (up−/downregulated) in response to at least one stress condition, and the presence of stress responsive cis-elements suggests that these genes play important roles in regulating gene expression in response to abiotic stress and in environmental adaptation. Further studies on the putative cis-elements in tomato ZHD genes are needed to unravel the complex regulatory mechanisms involving the cis-elements and stress tolerance in tomato. AtZHD1 is related to some NAC (NAM/ATAF1/2/CUC2) genes, and the overexpression of AtZHD1 and NAC increases drought tolerance in Arabidopsis [38]. In addition, 31 Chinese cabbage ZHD genes were found to be regulated by abiotic and hormonal stress [3].
ABA is a key regulator in the adaptation of plants to unfavorable environmental conditions such as high salinity, drought, and cold [39]. Almost all 16 SlZHD genes were differentially regulated by ABA, with many (SlZHD2, SlZHD7, SlZHD8, SlZHD10, SlZHD15, SlZHD16, and SlZHD17) downregulated by this treatment. Two genes, SlZHD6 and SlZHD18, were markedly upregulated from 9 h to 24 h and 3 h to 24 h after ABA treatment, respectively. The five remaining genes had different expression patterns at different time points, suggesting that ZHD genes play regulatory roles in stress responses by modulating ABA signaling. Indeed, ZHD genes are regulated by ABA stress in several plant species, including AtZHD1 in Arabidopsis, VvZHD4 and VvZHD13 in grape, and TaZHD1 in wheat [33, 40–42].
Most SlZHD genes within the same subfamily in the phylogenetic tree had different expression patterns. For example, SlZHD15 and SlZHD16 in subfamily II showed diverse expression patterns in response to all abiotic stress treatment, indicating that the regulatory sequences in these genes that respond to stress conditions have diverged significantly during gene evolution. However, some genes in the same subfamily showed similar expression patterns under all treatments, such as SlZHD7 and SlZHD8, indicating that the regulatory sequences in these genes that respond to stress conditions share high sequence similarity and were conserved during the course of evolution.
Conclusions
ZHD genes have been comprehensively characterized in several model plant species. However, to the best of our knowledge, no systematic study of this gene family has been performed in any Solanaceous species. In this study, we identified 22 ZHD genes in Solanum lycopersicum. Our systematic analysis revealed some structural diversity among tomato ZHD proteins, suggesting they play diverse roles in plant adaptation to environmental stress during particular stages of development. Our expression profiling analysis of SlZHD genes in various tissues/organs and under different abiotic stress and phytohormone (ABA) treatments should facilitate the identification of appropriate candidate genes for further functional characterization. The information obtained in this study lays the foundation for further analysis of the biological functions of ZHD proteins in tomato.
Methods
Identification of ZHD genes and encoded proteins in tomato
To conduct genome-wide characterization of ZHD family gene (Z for Zinc finger, HD for homeodomain) in tomato, the keyword ZF-HD was used as a query identify ZHD genes in the tomato genome using the Sol Genomics Network (SGN) (https://www.solgenomics.net//) [43]. After removing redundant sequences using blast search of conserved domain in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) 22 ZHD genes were identified and confirmed through comparisons with the iTAK (Plant Transcription factor and Protein Kinase Identifier and Classifier) database (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) [44] and the Tomato Genomic Resources Database (TGRD) (http://webcache.googleusercontent.com/search?q=cache:UgWJjBHR92EJ:59.163.192.91/tomato2/contact.html+&cd=2&hl=en&ct=clnk&gl=kr = zf-HD) [45]. All of the predicted ZHD proteins had typical “ZF-HD” dimer domains (Pfam accession number PF04770), which were confirmed by comparing these sequences with previously identified Arabidopsis ZHD domain sequences using the web tool from EMBL (http://smart.embl-heidelberg.de/) [46]. Protein sequences and CDS of the identified tomato ZHD genes were obtained from SGN (https://solgenomics.net/) [43]. TGRD (http://59.163.192.91/tomato2/getTF_family.php?trans_fac_family = zf-HD) [45] and iTAK (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) [44] were used to verify the identified sequences.
Sequence analysis of tomato ZHD genes
The primary structures including the length, molecular weight (Mw), and isoelectric point (pI) of the deduced tomato ZHD proteins were analyzed using ProtParam (http://web.expasy.org/protparam/) [47] (Table 1). The tomato ZHD CDS and their corresponding genomic sequences were aligned with the GSDS (http://gsds.cbi.pku.edu.cn/) [48] web tool to analyze the exon/intron structures of the tomato ZHD genes. MEME software (http://meme-suite.org/) [49] was employed to analyze the tomato ZHD protein motifs. The distinctive motifs were identified using the following parameters: (1) width of optimum motif ≥6 and ≤500 nt; (2) maximum number of motifs 10. The similarity among the 22 tomato ZHD proteins was analyzed using the NCBI protein BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) [50]. The subcellular locations of the ZHD proteins in S. lycopersicum were predicted using ProtComp 9.0 from SoftBerry (http://linux1.softberry.com/berry.phtml) [51].
Phylogenetic analysis of tomato ZHD proteins
The deduced amino acid sequences of Arabidopsis, rice, and Chinese cabbage ZHD proteins containing ZF-HD dimer domain were obtained from TAIR (https://www.arabidopsis.org/) [52], TIGR-Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) [53], and the BRAD Brassica database (http://brassicadb.org/brad/) [54], respectively. The amino acid sequences from potato, tobacco and Selaginella moellendorffii were obtained from iTAK ((http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) [44] and the plant transcription factor database (http://planttfdb.cbi.pku.edu.cn/) [55]. The deduced amino acid sequences from tomato, Arabidopsis, rice, Chinese cabbage, potato, tobacco and Selaginella moellendorffii were aligned using the multiple alignment tool ClustalX (http://www.clustal.org/clustal2/) [56]. A phylogenetic tree was constructed using the most conserved ZF-HD dimer domains with MEGA6.0 software following the neighbor-joining (NJ) algorithm [57] with 1000 bootstrap replicates. A second phylogenetic tree of the full-length amino acid sequences of 22 tomato ZHD proteins was constructed following the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method using complete deletion of amino acids with 1000 bootstrap replicates.
Analysis of putative cis-element in the promoter regions of SlZHD genes
Approximately 5 to 10 bp putative cis-elements in tomato ZHD genes were detected using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [58] web-based tool. The region from the start codon of each gene to the 2000 bp upstream sequence [59] was used to identify cis-regulatory elements, because cis-elements bound by transcription factors are present in these upstream regions that regulate target genes [60].
Chromosomal locations, gene duplication, and microsynteny analysis of tomato ZHD genes
The start and end positions of each tomato ZHD gene, including subgenome information, were obtained from SGN (https://solgenomics.net/) [43]. The position of each gene on the tomato chromosomes was analyzed using the MapGene2Chromosome2 ((http://mg2c.iask.in/) [61] web tool. A NCBI BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) [50] of the tomato ZHD genes against each other was conducted to identify duplicated genes based on the query coverage percentage and identity of each gene. When the query coverage percentage and identity of the candidate genes was ≥80% [21], they were considered to be segmentally duplicated genes. Duplication lines among the segmentally duplicated genes were drawn manually on the chromosomes. Paralogous genes were considered to be tandemly duplicated when two genes were separated by five or fewer genes in a 100-kb region on a chromosome [22]. A BLAST search against the whole genomes of Arabidopsis, tomato, and potato was conducted to determine the microsyntenic relationships of ZHD genes among these species; the results were displayed using Circos software (http://circos.ca/) [62].
Calculation of Ka/Ks ratios
The synonymous (KS) and non-synonymous (Ka) nucleotide substitution rates of SlZHD genes were calculated based on their coding sequence alignments following the Nei and Gojobori model with Mega 6.0 [56] software. The Ka/Ks ratios between duplicated genes were analyzed to identify the mode of selection. In general, Ka/Ks ratio > 1, <1, and =1 indicates accelerated evolution with positive selection, functional constraint with purifying selection, and neutral selection of genes, respectively [63]. Divergence time (T) of each duplicated gene pair was calculated using the formula T = Ks/2r Mya (Millions of years) where, Ks is the synonymous substitutions per site and r is considered 1.5 × 10−8 substitutions per site per year for dicot plants [64].
Plant material preparation and collection
Tomato seeds of the cultivar ‘Ailsa Craig’ were germinated in potted soil in a growth room. Seed germination and seedling growth performed in a growth room with a controlled environment at 25 °C day/20 °C night temperatures, with a 16/8 h (light dark) photoperiod, 55–70% relative humidity, and 300 μmol m−2s−1 light intensity. Leaf, stem, and root tissues were collected from 28-day-old seedlings for organ-specific expression analysis, and the remaining seedlings were transferred to a greenhouse with at 18 ± 2 °C and 65–80% relative humidity. Flower samples were collected during full bloom at the anthesis stage. Six developmental stages (based on approximate fruit size and color) of fruits were collected: i) 1 cm, 2 weeks after pollination; ii) immature (IM), 2 cm in diameter and 20 days after pollination; iii) mature green (MG); iv) breaker stage (B), when the fruit color turned from green to yellowing-orange; v) (B + 5), 5 days after the breaker stage of fruit; and vi) (B + 10), 10 days after the breaker stage [65, 66]. Samples were collected from three biological replicates, immersed in liquid nitrogen, and stored at −80 °C for further analysis.
Potted seedlings (28 days old) were subjected to various stress treatments. Three fresh seedlings per treatment were incubated at 40 °C and 4 °C for 24 h to confer heat and cold stress, respectively. For drought stress treatment, the seedlings were removed from the soil and transferred to a dry paper towel for 24 h. NaCl stress treatment involved submerging the roots of seedlings in 200 mM NaCl for 24 h. For ABA treatment, the leaves of seedlings were sprayed with 100 μM ABA solution. Plants growing in pots under normal conditions (25 °C) were used as the 0 h controls for heat and cold, drought, NaCl, and ABA treatment. After applying different treatments, leaf samples from three biological replicates were collected at different time points (0 h, 1 h, 3 h, 9 h, and 24 h), immediately immersed in liquid nitrogen, and stored at −80 °C for RNA extraction and cDNA synthesis.
Expression analysis of tomato ZHD genes
Total RNA was isolated from the plant samples using an RNeasy mini kit (Qiagen, Valencia, USA) and purified with a Qiagen RNase free DNase1 kit. RNA concentrations were measured using NanoDrop® 1000 Spectrophotometer (Wilmington, DE, USA). First-strand cDNA was synthesized using 6 ng total RNA per sample with a Superscript® III First-Strand cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA). Gene-specific primers for the candidate tomato ZHD genes were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm) [67] (Additional file 2: Table S1). The primers for EF1a (F: TCAGGTAAGGAACTTGAGAAGGAGCCT, R: AGTTCACTTCCCCTTCTTCTGGGCAG) from S. lycopersicum were used as an internal control [68]. The reaction mixture for qRT-PCR (10 μL total volume) contained 75−80 ng/μL of cDNA, 2 μL forward and reverse primers, 2 μL double distilled water, and 5 μL iTaq™ from the SYBR® Green PCR kit (California, USA). A Light cycler® 96SW 1.1 (Roche, Germany) was used for amplification and detection using the following PCR parameters: pre-denaturation at 95 °C for 5 min followed by 40 cycles of 94 °C for 10 s, annealing at 58 °C for 10 s, and extension at 72 °C for 15 s. The 2−∆∆Ct method was used for data analysis [69]. Relative gene expression levels were normalized against the expression of the housekeeping gene EF1a. The significance of differences among relative expression levels of the genes for different samples and time points was analyzed using one-way analysis of variance (ANOVA) with MINITAB statistical software 17 (Minitab Inc., State College, PA, USA). Tukey’s pairwise comparison test was employed to determine the mean separation of expression values. The RNAseq data of SlZHD genes were downloaded from the Solgenomics database (https://solgenomics.net/) [47] and Tomato functional Genomic Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi) [70].
Additional files
Fig. S1. Logos of 10 conserved motif identified by MEME software. Fig. S2. Chromosomal locations of SlZHD genes. The 22 genes are widely distributed on six of the 12 tomato chromosomes. The chromosomes number is indicated at the top of each vertical bar. The duplicated genes are connected with pink dotted line. The colored box in front of each gene indicates the subfamily according to phylogenetic tree. The scale indicates the length of the chromosome (PPTX 304 kb)
Table S1. List of primers used for qRT-PCR analysis and their sequence, product size, primer length, primer-designing site, GC%, and melting temperature. Table S2. Pairwise identities and divergence between paralogous pairs of ZHD genes from tomato, and details about the segmental duplication of these genes. Table S3. Putative cis-elements >5 bp identified in 22 SlZHD genes from Solanum lycopersicum using the PlantCARE database (XLSX 43 kb)
Table S1. Information of ZHD genes of Putative tomato, Arabidopsis, potato, tobacco, chinese cabbage, rice and Selaginella moellendorffii used in in silico analysis. Table S2. Online RNA sequencing data (PKRM) downloaded from Solgenomics database (https://solgenomics.net/) and Tomato functional Genomic Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi) (XLSX 35 kb)
Fig. S1. Multiple sequence alignment of the conserved domain of ZHD protein family of tomato, potato, tobacco, Arabidopsis, chinese cabbage, rice and Selaginella moellendorffii, in where black and grey shading indicating 100% and 60% identity, respectively. Fig. S2. Phylogenetic relationship of Arabidopsis(AtZHD), rice(OsZHD), potato (St, Solanum tuberosum is used instead of PGSC0003DMT4000), tobacco (Nt, Nicotiana tabacum is used instead of XP_0164), Chinese cabbage (BraZF-HD), Selaginella moellendorffii, (SmZF-HD) and tomato (SlZHD) ZHD proteins. The conserved ZF-HD_ dimer domain sequences of Arabidopsis, rice, potato, tobacco, Chinese cabbage, Selaginella moellendorffii, and tomato proteins were aligned using ClustalX, and the tree were constructed by the Maximum likelihood method with MEGA 6.0. The numbers on the branches indicate bootstrap support values from 1000 replications. The protein sequences used in the phylogenetic analysis are listed in Additional file 1, along with their accession numbers. The tree was divided into six subfamilies according to bootstrap support values and evolutionary distances (PPTX 818 kb)
Acknowledgments
Special thanks to Dr. Manosh Kumar Biswas, Department of Horticulture, Sunchon National University, Suncheon, Korea Republic for helping to do microsynteny analysis.
Funding
This research was supported by Golden Seed Project (Center for Horticultural Seed Development, No. 213007–05-1-SBG10) Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS), South Korea. The funding bodies were not involved in the design of the study or in any aspect of the collection, analysis and interpretation of the data or paper writing.
Availability of data and materials
We declare that the dataset(s) supporting the conclusions of this article are included within the article (and its additional file(s) will be available in journal web page.
Abbreviations
- aa
amino acid
- ABA
Abscisic acid
- At
Arabidopsis thaliana
- bp
base pair
- BRAD
Brassica database
- Ka
Nonsynonymous substitutions per nonsynonymous site
- kDa
Kilo Dalton
- Ks
Synonymous substitutions per synonymous site
- MW
Molecular weight
- MYA
Million year
- ORF
Open reading frame
- pI
isoelectric points
- qRT-PCR
Quantitative polymerase chain reaction
- Sl
Solanum lycopersicum
- Sm
Selaginella moellendorffii
- TIGR
Rice Genome Annotation Project
- ZHD
Zinc finger homeodomain
Authors’ contributions
The work presented here was carried out in collaboration among all authors. MYC designed and supervised the work. KK designed the work and conducted the in silico analysis, grew the plants, isolated RNA and carried out the expression analysis, and prepared a draft of the manuscript. UKN analyzed the data and extensively revised the manuscript. JIP, DJL, MBK, CKK, KBL and ISN supervised and monitored the experimental work. AHKR analyzed part of the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The seed materials of the cultivar ‘Ailsa Craig’ used in this study was provided by Jim Giovannoni, Adjunct Professor, Section of Plant Biology, School of Integrative Plant Science, Boyce Thomson Institute, Cornell University, USA. The corresponding author of this manuscript was a postDoc research fellow of that lab and still has bilateral collaboration at personal level. The plant materials are maintained following the institutional guidelines of department of Agricultural industry economy and education, Sunchon National University, South Korea.
Consent for publication
All of the authors of this manuscript give their consent to publish the findings in BMC Genomics.
Competing interests
The authors declare that there are no conflicts of interest to publish in this journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4082-y) contains supplementary material, which is available to authorized users.
Contributor Information
Khadiza Khatun, Email: kkr@pstu.ac.bd.
Ujjal Kumar Nath, Email: ujjalnath@gmail.com.
Arif Hasan Khan Robin, Email: gpb21bau@bau.edu.bd.
Jong-In Park, Email: jipark@sunchon.ac.kr.
Do-Jin Lee, Email: djlee@sunchon.ac.kr.
Min-Bae Kim, Email: salt@sunchon.ac.kr.
Chang Kil Kim, Email: ckkim@knu.ac.kr.
Ki-Byung Lim, Email: kblim@knu.ac.kr.
Ill Sup Nou, Email: nis@sunchon.ac.kr.
Mi-Young Chung, Phone: +82-61-750-0587, Email: queen@sunchon.ac.kr.
References
- 1.Yanagisawa S. Transcription factors in plants: physiological functions and regulation of expression. J Plant Res. 1998;111:363–371. doi: 10.1007/BF02507800. [DOI] [Google Scholar]
- 2.Lata C, Yadav A, Prasad M. Role of plant transcription factors in abiotic stress tolerance. Abiotic Stress Response in Plants, INTECH Open Access Publishers. 2011;10:269–296. [Google Scholar]
- 3.Wang W, Wu P, Li Y, Hou X. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol Gen Genomics. 2016;291:1451–1464. doi: 10.1007/s00438-015-1136-1. [DOI] [PubMed] [Google Scholar]
- 4.Rohrmann J, McQuinn R, Giovannoni JJ, Fernie AR, Tohge T. Tissue specificity and differential expression of transcription factors in tomato provide hints of unique regulatory networks during fruit ripening. Plant Signal Behav. 2012;7:1639–1647. doi: 10.4161/psb.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samad AF, Sajad M, Nazaruddin N, Fauzi IA, Murad AM, Zainal Z, Ismail I. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan QKG, Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006;140:1095–1108. doi: 10.1104/pp.105.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bürglin TR. A comprehensive classification of homeobox genes. Guidebook to the homeobox genes. 1994:25–71.
- 8.Hu W, DePamphilis CW, Ma H. Phylogenetic analysis of the plant specific zinc finger homeobox and mini zinc finger gene families. J Integr Plant Biol. 2008;50:1031–1045. doi: 10.1111/j.1744-7909.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 9.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-zip family. Trends Plant Sci. 2007;12:419–426.17. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Kawagashira N, Ohtomo Y, Murakami K, Matsubara K, Kawai J, Carninci P, et al. Multiple zinc finger motifs with comparison of plant and insect. Genome Inform. 2001;12:368–369. [Google Scholar]
- 11.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers SURVEY AND SUMMARY. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39:1073–1078. doi: 10.1023/A:1006184519697. [DOI] [PubMed] [Google Scholar]
- 13.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/S0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 14.Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants. Cell Mol Life Sci. 2008;65:1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M, Tyagi AK, Khurana JP. Genome wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008;275:2845–2861. doi: 10.1111/j.1742-4658.2008.06424.x. [DOI] [PubMed] [Google Scholar]
- 16.Windhovel A, Hein I, Dabrowa R, Stockhaus J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria Trinervia. Plant Mol Biol. 2001;45:201–214. doi: 10.1023/A:1006450005648. [DOI] [PubMed] [Google Scholar]
- 17.Tran LS, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, et al. Coexpression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 18.Park HC, Kim ML, Lee SM, Bahk JD, Yun DJ, Lim CO, et al. Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res. 2007;35:3612–3623. doi: 10.1093/nar/gkm273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W, Ma H. Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 2006;45:399–422. doi: 10.1111/j.1365-313X.2005.02626.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Yin X, Li X, Wang L, Zheng Y, Xu X, Wang X. Genome-wide identification, evolution and expression analysis of the grape (Vitis Vinifera L.) zinc finger-homeodomain gene family. Int J Mol Sci. 2014;15:5730–5748.43.57. doi: 10.3390/ijms15045730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013;14:433. doi: 10.1186/1471-2164-14-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Walther D, Brunnemann R, Selbig J. The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 26.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filiz E, Tombuloğlu H. Genome-wide distribution of superoxide dismutase (SOD) gene families in Sorghum Bicolor. Turk J Biol. 2014;39:49–59. doi: 10.3906/biy-1403-9. [DOI] [Google Scholar]
- 29.Kong H, Landherr LL, Frohlich MW, LeebensMack J, Ma H, DePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 30.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of seg-mental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng BH, Han YC, Xiao YY, Kuang JF, Fan ZQ, Chen JY, et al. The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J Exp Bot. 2016;67:2263–2275. doi: 10.1093/jxb/erw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abu-Romman S. Molecular cloning and expression analysis of zinc finger-homeodomain transcription factor TaZFHD1 in wheat. S Afr J Bot. 2014;91:32–36. doi: 10.1016/j.sajb.2013.11.014. [DOI] [Google Scholar]
- 33.Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. doi: 10.1016/S1360-1385(97)82562-9. [DOI] [Google Scholar]
- 34.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and crosstalk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. doi: 10.1016/S1369-5266(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 35.Nakashima K, Yamaguchi-Shinozaki K. Regulations involved in osmotic stress responsive and cold stress responsive gene expression in plants. Physiol Plant. 2006;126:62–71. doi: 10.1111/j.1399-3054.2005.00592.x. [DOI] [Google Scholar]
- 36.Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol Plant. 2010;54:201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 37.Saeediazar S, Zarrini HN, Ranjbar G, Heidari P (2014) Identification and study of cis regulatory elements and phylogenetic relationship of TaSRG and other salt response genes. JBES. 5:1-5].
- 38.Tran LSP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, et al. Co-expression of the stress inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 39.Davies WJ, Jones HG. Abscisic acid physiology and biochemistry. Oxford: BIOS Scientific Publishers; 1991. [Google Scholar]
- 40.Huang WZ, Ma XR, Wang QL, Gao YF, Xue Y, Niu XL, Yu GR, Liu YS. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays) Plant Mol Biol. 2008;68:451–463. doi: 10.1007/s11103-008-9382-9. [DOI] [PubMed] [Google Scholar]
- 41.Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis Thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006;29:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- 42.Rodrigues SM, Andrade MO, Gomes APS, DaMatta FM, Baracat-Pereira MC, Fontes EPB. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J Exp Bot. 2006;57:1909–1918. doi: 10.1093/jxb/erj132. [DOI] [PubMed] [Google Scholar]
- 43.Mueller LA, Solow TH, Taylor N, Skwarecki B, Buel R, Binn J, Herbst EV. The SOL genomics network. A comparative resource for Solanaceae biology and beyond Plant physiol. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Fei Z. iTAK–identification and classification of plant transcription factors and protein kinases. Plant and Animal Genome: In Plant and Animal Genome XXII Conference; 2014. [Google Scholar]
- 45.Suresh BV, Roy R, Sahu K, Misra G, Chattopadhyay D. Tomato genomic resources database: an integrated repository of useful tomato genomic information for basic and applied research. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. Humana press:571-607).
- 48.Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Zhongguo yi chuan xue hui bian ji. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 49.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic acids res. gkp335. [DOI] [PMC free article] [PubMed]
- 50.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer E, Freeman T. Investigation into the use of C-and N-terminal GFP fusion proteins for subcellular localization studies using reverse transfection microarrays. Comp Funct Genomics. 2004;5:342–353. doi: 10.1002/cfg.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Radenbaugh A. The Arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Buell CR. The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 2003;31:229–233. doi: 10.1093/nar/gkg059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Wang X (2011) BRAD, the genetics and genomics database for brassica plants. BMC Plant Boil.11: 136. [DOI] [PMC free article] [PubMed]
- 55.Perez-Rodriguez P, Riano-Pachon DM, Correa LGG, Rensing SA, Kersten B, Mueller-Roeber B (2009) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic acids res. gkp805. [DOI] [PMC free article] [PubMed]
- 56.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Peng Z, Liu S, He Y, Cheng L, Kong F, Lu G. Genome-wide analysis of aux/IAA gene family in Solanaceae species using tomato as a model. Mol Gen Genomics. 2012;287:295–311. doi: 10.1007/s00438-012-0675-y. [DOI] [PubMed] [Google Scholar]
- 60.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Gen Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 61.Jiangtao C, Yingzhen K, Qian W, Yuhe S, Daping G, Jing L, Guanshan L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Zhongguo yi chuan xue hui bian ji. 2015;37:91–97. doi: 10.16288/j.yczz.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 62.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nekrutenko A, Makova KD, Li WH. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12:198–202.51. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17(10):1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 65.Khatun K, Robin AHK, Park JI, Kim CK, Lim KB, Kim MB, et al. Genome-wide identification, characterization and expression profiling of ADF family genes in Solanum lycopersicum L. Genes. 2016;7:79. doi: 10.3390/genes7100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khatun K, Robin AHK, Park JI, Ahmed NU, Kim CK, Lim, et al (2016) Genome-wide identification, characterization and expression profiling of LIM family genes in Solanum lycopersicum L. Plant Physiol Biochem 108: 177–190. [DOI] [PubMed]
- 67.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115-e115. [DOI] [PMC free article] [PubMed]
- 68.Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K, Ooga K. Large-scale analysis of full length cDNAs from the tomato (Solanum lycopersicum) cultivar micro-tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 70.Fei Z, Joung JG, Tang X, Zheng Y, Huang M, Lee JM, Giovannoni JJ. Tomato functional genomics database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011;39:D1156–D1163. doi: 10.1093/nar/gkq991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Logos of 10 conserved motif identified by MEME software. Fig. S2. Chromosomal locations of SlZHD genes. The 22 genes are widely distributed on six of the 12 tomato chromosomes. The chromosomes number is indicated at the top of each vertical bar. The duplicated genes are connected with pink dotted line. The colored box in front of each gene indicates the subfamily according to phylogenetic tree. The scale indicates the length of the chromosome (PPTX 304 kb)
Table S1. List of primers used for qRT-PCR analysis and their sequence, product size, primer length, primer-designing site, GC%, and melting temperature. Table S2. Pairwise identities and divergence between paralogous pairs of ZHD genes from tomato, and details about the segmental duplication of these genes. Table S3. Putative cis-elements >5 bp identified in 22 SlZHD genes from Solanum lycopersicum using the PlantCARE database (XLSX 43 kb)
Table S1. Information of ZHD genes of Putative tomato, Arabidopsis, potato, tobacco, chinese cabbage, rice and Selaginella moellendorffii used in in silico analysis. Table S2. Online RNA sequencing data (PKRM) downloaded from Solgenomics database (https://solgenomics.net/) and Tomato functional Genomic Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi) (XLSX 35 kb)
Fig. S1. Multiple sequence alignment of the conserved domain of ZHD protein family of tomato, potato, tobacco, Arabidopsis, chinese cabbage, rice and Selaginella moellendorffii, in where black and grey shading indicating 100% and 60% identity, respectively. Fig. S2. Phylogenetic relationship of Arabidopsis(AtZHD), rice(OsZHD), potato (St, Solanum tuberosum is used instead of PGSC0003DMT4000), tobacco (Nt, Nicotiana tabacum is used instead of XP_0164), Chinese cabbage (BraZF-HD), Selaginella moellendorffii, (SmZF-HD) and tomato (SlZHD) ZHD proteins. The conserved ZF-HD_ dimer domain sequences of Arabidopsis, rice, potato, tobacco, Chinese cabbage, Selaginella moellendorffii, and tomato proteins were aligned using ClustalX, and the tree were constructed by the Maximum likelihood method with MEGA 6.0. The numbers on the branches indicate bootstrap support values from 1000 replications. The protein sequences used in the phylogenetic analysis are listed in Additional file 1, along with their accession numbers. The tree was divided into six subfamilies according to bootstrap support values and evolutionary distances (PPTX 818 kb)
Data Availability Statement
We declare that the dataset(s) supporting the conclusions of this article are included within the article (and its additional file(s) will be available in journal web page.


