Abstract
The cotton genus (Gossypium) provides a superior system for the study of diversification, genome evolution, polyploidization, and human-mediated selection. To gain insight into phenotypic diversification in cotton seeds, we conducted coexpression network analysis of developing seeds from diploid and allopolyploid cotton species and explored network properties. Key network modules and functional associations were identified related to seed oil content and seed weight. We compared species-specific networks to reveal topological changes, including rewired edges and differentially coexpressed genes, associated with speciation, polyploidy, and cotton domestication. Network comparisons among species indicate that topologies are altered in addition to gene expression profiles, indicating that changes in transcriptomic coexpression relationships play a role in the developmental architecture of cotton seed development. The global network topology of allopolyploids, especially for domesticated G. hirsutum, resembles the network of the A-genome diploid more than that of the D-genome parent, despite its D-like phenotype in oil content. Expression modifications associated with allopolyploidy include coexpression level dominance and transgressive expression, suggesting that the transcriptomic architecture in polyploids is to some extent a modular combination of that of its progenitor genomes. Among allopolyploids, intermodular relationships are more preserved between two different wild allopolyploid species than they are between wild and domesticated forms of a cultivated cotton, and regulatory connections of oil synthesis-related pathways are denser and more closely clustered in domesticated vs. wild G. hirsutum. These results demonstrate substantial modification of genic coexpression under domestication. Our work demonstrates how network inference informs our understanding of the transcriptomic architecture of phenotypic variation associated with temporal scales ranging from thousands (domestication) to millions (speciation) of years, and by polyploidy.
Keywords: Gossypium, oil seed, polyploid evolution, domestication, gene coexpression network
Introduction
Gossypium is widely known as an important source of textile fibers; however, modern cotton is also a vital oil- and protein-seed crop (Bewley 2006; Liu et al. 2009; Dowd 2015). Although extraction of cotton seed oil for food processing can be traced back to ancient times in the Old World, development of the modern crushing and refinery process dates to late nineteenth century in the nited States. During the Industrial Revolution, American cotton seed oil was initially introduced into the European market to overcome the fats and oils shortage, and subsequently gained popularity both in the Old and New World, dominating the vegetable oil market for almost 100 years until the end of World War II (O’Brien et al. 2005). At present, cotton provides the sixth largest source of vegetable oil in the world. Oilseed yield is usually about 1.5 times higher than fiber yield by weight, and accounts for 10–15% of the total value of cotton crops. In addition to oils, cotton seeds contain about 20% relatively high-quality protein and a low amount of starch (O’Brien et al. 2005; Hu et al. 2011).
Given the importance of cotton seeds as a source of edible oil and protein, the relative dearth of cotton seed research is somewhat surprising, particularly when compared with the extensive research devoted to cotton fibers. Only a few studies have characterized essential cotton seed characteristics. A modest variation in oil content, i.e., 17–23%, was reported within domesticated G. hirsutum lines and hybrids (Shaver and Dilday 1982; Ashokkumar and Ravikesavan 2013; Kouser et al. 2015), whereas approximately double that (10.3–22.9%) was found among 22 diploid cotton species (Gotmare et al. 2004). A more recent and comprehensive genus-wide survey of cotton seed nutritional traits (oil content, protein values, and seed weight) was conducted on five tetraploid and 28 diploid species, examining 2256 accessions from the US National Cotton Germplasm Collection (Hinze et al. 2015). Oil content in these accessions was 8–27%, ranging from the low-content diploid B and E genomes to the high-content tetraploid genomes. Cultivated accessions of both G. barbadense and G. hirsutum had higher oil content and seed weight than did their wild relatives, potentially indicative of a trait altered by domestication. Among allopolyploid cottons, the lowest oil content occurred in G. tomentosum (∼15%), a close relative of G. hirsutum. Despite the importance of natural variability in understanding phenotypic evolution and facilitating crop improvement, molecular characterization of cottonseed development and oil/protein accumulation is scarce and limited to lines of G. hirsutum. In contrast, studies in Arabidopsis (Baud et al. 2002) and major seed oil crops (Weber et al. 2005; Gutierrez et al. 2007) demonstrate that seed developmental processes and phenotypic variation in seed traits reflect a complex network of cellular, biochemical, and metabolic processes that are dynamically regulated, including cell division and differentiation and the accompanying biosynthesis of carbohydrates, amino acids, proteins, lipids, hormones, and secondary metabolites.
While these regulatory networks have not been studied in cotton, transcriptional profiles for seed development have been evaluated to examine the temporal and spatial changes in transcript abundance that correspond with developmental transitions, as in other seed oil plants (Severin et al. 2010; Troncoso-Ponce et al. 2011; Basnet et al. 2013; Jones and Vodkin 2013; Sekhon et al. 2013). Over 17,000 genes are differentially expressed between the cotyledon and embryo axis in 30 days post-anthesis (dpa) Upland cotton seeds (Jiao et al. 2013). In a more recent study, Hovav et al. (2015) analyzed global gene expression profiles at four developmental time points in G. hirsutum, and presented several conclusions. First, differential expression was greatest between 20 and 30 dpa, indicating that most developmental changes take place at the beginning of the seed filling stage. Second, analyses focused on genes of the oil biosynthetic and related pathways revealed preferential usage of certain gene family members; for example, compared to other acyltransferases involved in triacylglycerol synthesis, a predominant role for DGAT3 was found that is unique to cotton seeds. Finally, parental contributions to the allopolyploid seed transcriptome can be unequal, with global biases up to 20% for specific stages.
Here, we expand our understanding of cotton seed development in an evolutionary and phylogenetic context by studying developing seed transcriptomes from four additional Gossypium species/accessions: two model diploid progenitor species, G. arboreum (A2) and G. raimondii (D5); a wild, low-oil tetraploid species, G. tomentosum (AD3); and a wild representative of G. hirsutum (AD1 var. Yucatanense). Allopolyploid cotton species originated within the last 1–2 Myr from a single hybridization event between two divergent diploid parents most similar to modern A- (G. arboreum or G. herbaceum) and D- (G. raimondii) genome species, which differ two-fold in genome size and share a common ancestor 5–10 Ma (Wendel and Grover 2015). Subsequent to polyploid formation, the tetraploid taxa diverged into the six species (“AD1” through “AD6”), including G. hirsutum (AD1) and G. barbadense (AD2) that were domesticated in the New World and which now dominate world cotton production. Together with the domesticated G. hirsutum cultivar (AD1 var. TM1) used in a previous study (Hovav et al. 2015), sequenced seed transcriptomes from these Gossypium species/accessions allow us to conduct a comparative analysis of transcriptional architecture of cotton seed development, in conjunction with an analysis of phenotypic variability and in the context of natural selection/differentiation, allopolyploidization, and artificial selection under domestication. We reveal the network structure that underlies differential expression during cottonseed development and how this is modified in response to polyploidy, and describe the topological changes of the cotton seed developmental network that have accompanied key evolutionary events in Gossypium. We also show that oil and lipid-related pathways in G. hirsutum have become more tightly coregulated as a result of over 5000 years of human domestication and crop improvement.
Materials and Methods
Plant Materials
Seeds from five Gossypium accessions were collected from greenhouse-grown plants (12 h photoperiod; 22 °C /28 °C, night/day). Three accessions of allopolyploid cotton were used: a low-oil, wild species G. tomentosum (AD3) and both a wild (var. yucatanense, Yuc) and cultivated (var. Texas Marker Stock 1, TM1; reported in Hovav et al. 2015) accession of G. hirsutum (AD1). Two diploid species, G. arboreum (A2) and G. raimondii (D5), were used to represent the diploid progenitors of allopolyploid cotton. Three biological replicates were grown for each species/accession. Cotton flowers were tagged on the first day post-anthesis (dpa) and collected at four developmental stages (10, 20, 30, and 40 dpa), which represent, respectively (1) initial seed filling, (2) seed enlargement and oil accumulation, (3) end of seed filling, and (4) maturation. Seeds were extracted from developing fruits at each time point, followed by immediate manual removal of fibers from the seed surface. Delinted seeds were weighed, flash frozen and stored at –80 °C for RNA extraction. The oil content of developing seeds was determined as described (Hovav et al. 2015).
Preprocessing of RNA-seq Data
Total RNA extraction, library construction, and 100 base single-end illumina sequencing were as described in Hovav et al. (2015). Sixty libraries were prepared and sequenced using five lanes of the Illumina HiSeq 2000 sequencer (Illumina, San Diego, CA), and a total of 1.22 billion reads were generated with an average of 20 million reads per library. All Reads have been deposited under the NCBI BioProject PRJNA179447. Following quality filtering and read trimming using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; last accessed November 24, 2016) with parameters described in Hovav et al. (2015), cleaned RNA-seq reads were mapped to the reference cotton genome (D-genome G. raimondii; 37,223 genes) as previously described in (Paterson et al. 2012) using GSNAP (Wu and Nacu 2010) with single-nucleotide polymorphism (SNP) tolerance mapping between the A- and D-genome diploids and their coresident counterparts in allopolyploids (Page and Udall 2015), using mapping parameters as: –nolengths 1 –npaths 1 –nthreads 5 –batch 5 –protein_gen. Corresponding to the gene-level read counts generated for diploid species, the total expression of a homoeolog gene pair in allopolyploids was used for following analyses.
Differential Gene Expression and Coexpression Analysis
Differential gene expression was conducted in R v.3.2.0 (R Foundation for Statistical Computing, Vienna, Austria) with the package DESeq2 (Love et al. 2014). For data visualization and coexpression analysis, rlog (regularized logarithm, built in DESeq2) transformed versions of the count data were used. Differential expression was assessed between the time points and between species. The distribution of P-values was controlled for a false discovery rate (q-value) by the BH method (Benjamini and Hochberg 1995) at α = 0.05.
Differentially coexpressed gene pairs were detected using the R package DiffCorr (Fukushima 2013). Briefly, the Pearson correlation coefficient was calculated for each pair of genes, and Fisher’s z-test was used to identify significant differential correlations between any two Gossypium species/accessions with the local false-discovery rate of <0.05. The percentage of differential correlations among gene pairs was calculated to measure the extent of differential coexpression, or the probability, p, of observed “a differential coexpression gene pair”. For a gene observed in k differential correlation pairs among all possible pairs n, the probability P of a “differential coexpression gene” follows the binomial distribution model as follows:
P was corrected by the BH method (Benjamini and Hochberg 1995) at α = 0.05 for identifying differentially coexpression genes. R scripts for differential analysis and below network analysis are available at https://github.com/Wendellab/SeedDevelopment (last accessed November 24, 2016).
Weighted Gene Coexpression Network Analysis
The WGCNA package in R (Langfelder and Horvath 2008; Horvath 2011) was used to build weighted gene coexpression networks for each Gossypium species/accession and for a global meta-analysis. Network construction was performed using the blockwiseModules function with default parameters, which allows automatic and unsupervised network construction for the input data set. Briefly, this process entails, first, the generation of a matrix of Pearson correlations between all pairs of genes across the measured samples, followed by construction of an adjacency matrix representing the connection strength among genes. The adjacency matrix is calculated by raising the coexpression measure (0.5 + 0.5 × correlation matrix) to a chosen power β = 12 based on the criterion of approximate scale-free topology (Zhang and Horvath 2005), which makes the scale-free topology fit index reach about 0.80. The sum of connection strengths (represented by adjacencies) of a given gene to all other genes is called connectivity, which measures how strongly this gene is coexpressed with all other genes in the network. Genes with high connectivity are termed whole-network hubs. Based on the adjacency matrix, the topological overlap matrix (TOM) is calculated to measure network interconnectedness, i.e., the strength of a coexpression relationship between any two genes with respect to all other genes in the network (Yip and Horvath 2007). Genes with highly similar coexpression relationships were grouped together by performing average linkage hierarchical clustering on the topological overlap dissimilarity measure (1-TOM), and network modules were defined by cutting the clustering tree into branches using a dynamic tree cutting algorithm (Langfelder et al. 2008). Genes belonging to different modules were assigned to different colors for visualization, and genes not assigned to any module were assigned the color gray.
The gene expression pattern in a given module was characterized by the module eigengene, calculated as the first principal component of the scaled (standardized) module expression profiles. The measure of intramodular connectivity is mathematically equivalent to the module membership of a gene, kME, which was estimated by the Pearson correlation between the expression level of that gene and the module eigengene (Horvath and Dong 2008). Module hub genes were identified when kME > 0.9.
For the subnetwork of oil-related genes, topological parameters of density and mean clustering coefficient were calculated to measure network compactness and cohesiveness of neighboring nodes, respectively, using built-in WGCNA functions. A bootstrap approach was used to estimate the P-values for parameter differences between TM1 and Yuc subnetworks, by sampling 1000 random subsets of 433 genes (same number of oil-related genes) for constructing a sampling distribution of parameter differences.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) is a computational method to determine whether a set of genes displays statistically significant differences between two biological samples (e.g., species, tissues, developmental stages, etc.) (Subramanian et al. 2005). Briefly, GSEA examines how members of gene set S are distributed in the ranked gene list L, and calculates an enrichment score (ES) to reflect the degree to which gene set S is overrepresented at the top or bottom of the entire ranked list L. In this context, sets of differentially expressed genes between samples (as S) and genes ranked by module membership kME (as L) were subjected to the GseaPreranked function in GSEA v2.2.0 (http://www.broadinstitute.org/gsea/index.jsp; last accessed November 24, 2016), to compare gene-by-gene differential expression and the organization of coexpression networks. If significant enrichment is detected for a given differential expression pattern (e.g., overexpression in A2 vs. D5) in a module, this pattern becomes highly associated with the module coexpression profile.
Functional annotation of network connectivity and modules were performed using GSEA with gene sets derived from Gossypium Gene Ontology (http://phytozome.jgi.doe.gov/cotton; last accessed November 24, 2016), MapMan pathway (http://mapman.gabipd.org; last accessed November 24, 2016), and published cotton lipid biosynthesis-related gene (Hovav et al. 2015) databases. Cytoscape v3.2.0 (Shannon et al. 2003) with the Enrichment Map Plugin (Merico et al. 2010) was used for interpretation and visualization of the GSEA results. Only gene sets consisting of more than 15 and fewer than 500 genes were taken into account. The enrichment map was generated with enriched gene sets that passed the significance threshold of P < 0.05 and FDR Q < 0.05.
Relating Modules to Seed Traits
To identify modules that are significantly associated with cotton seed traits, including weight and oil content, Pearson correlations were performed between module eigengenes and trait measurements. Significant module-trait associations were considered when BH adjusted P-values (q-value) were < 0.05. The correlations between individual gene expression profiles and traits were defined as Gene Significance (GS), which were used to inspect association of module member genes with traits for each module.
Module Consensus and Module Preservation Analyses
Consensus networks were constructed using the minimum topological overlap similarity measure (TOM) derived from individually constructed and calibrated networks. To assess the preservation of consensus modules between any two genomes, the WGCNA function modulePreservation was performed to provide module preservation statistics. By averaging several preservation statistics generated through 200 permutations of the original data, a Zsummary value was calculated. In general, modules with Zsummary >10 are interpreted as strong preservation, whereas Zsummary between 2 and 10 are weak to moderately preserved and Zsummary <2 indicates no preservation. Pairwise Pearson correlation between module eigengenes was calculated to assess the intermodular relationships, and the resulting network was defined as eigengene network (Langfelder and Horvath 2007). The density D(AEigen) of the eigengene network was defined as an aggregated measure of adjacency preservation of module eigengene; D(AEigen) is close to 1 when the pair of eigengene networks compared are highly conserved (Langfelder and Horvath 2007). To test for differences between two measures of eigengene preservation D(AEigen), paired Student’s t-test was applied for mean connectivity of the adjacency preservation AEigen. To identify overrepresented GO terms (BP—biological process; MF—molecular function; and CC—cellular component) for consensus network modules, Fisher's exact tests were performed in the R package topGO (Alexa and Rahnenfuhrer 2010), followed by multiple testing correction using the procedure by Benjamini and Hochberg (Benjamini and Hochberg 1995). The correspondences between consensus modules and lipid biosynthesis related gene families were tested using Fisher's exact tests with P < 0.05 and associated gene family members >5.
Results
Seed Phenotypes
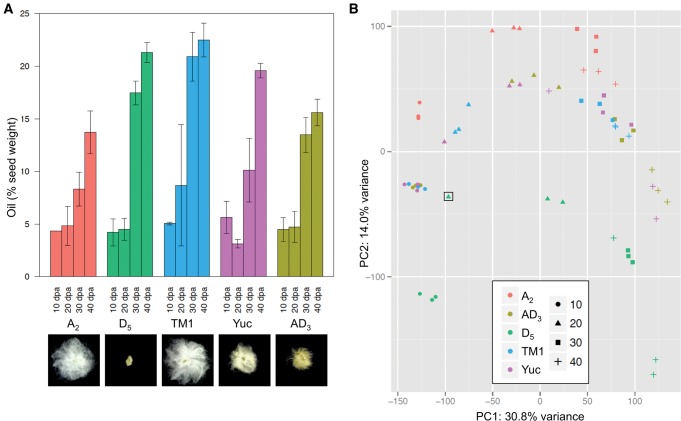
For characterizing phenotypic variation in cotton seed oil accumulation, we measured seed weight and total oil content in three tetraploid and two diploid cotton species at four key stages characterized previously at the physiological level (Hovav et al. 2015): 10 days post-anthesis (dpa), corresponding to the onset of reserve accumulation; 20 dpa, corresponding to peak accumulation of storage proteins and the beginning of oil accumulation; 30 dpa, corresponding to the end of seed filling; and 40 dpa, the stage of physiological maturity and desiccation. As shown in figure 1A, seed oil contents in all cotton species begin to accumulate as early as 20 dpa, followed by a rapid increase until 30 dpa. Different from cultivated G. hirsutum (AD1 var. TM1) and G. tomentosum (AD3), in which oil content peaks around 30 dpa, an extended period of oil accumulation was observed from 30 dpa to 40 dpa in both diploid species, G. arboreum (A2) and G. raimondii (D5), as well as in the wild accession of G. hirsutum (AD1 var. Yuc). The elite cultivar of upland cotton G. hirsutum (AD1 var. TM1) contains the highest oil content at maturity (40 dpa), approximately 15% and 35% more than the wild representative (Yuc) and the low-oil, wild tetraploid G. tomentosum (AD3), respectively. This higher oil content in TM1 is more similar to its diploid progenitor D5 than it is to A2.
Fig. 1.—
Analysis of five cotton representatives at four seed developmental stages. A2— G. arboreum; D5—G. raimondii; AD3—G. tomentosum; TM1—G. hirsutum var. TM1; Yuc—G. hirsutum var. yucatanense. (A) Seed oil content (as % seed weight). (B) Principal component analysis (PCA) of transcript expression profiles. The misidentified D5 20 dpa sample is boxed .
RNA-seq Data Sets
Corresponding to the oil content measurement, RNA-seq datasets were generated for each species at each developmental stage. A total of 60 RNA-seq libraries with an average of 20 million reads per library were acquired (supplementary table S1, Supplementary Material online). RNA-seq reads were mapped onto the reference genome of G. raimondii (D5) while tolerating single nucleotide polymorphisms (SNPs) that distinguish A2 and D5 diploid species, as well as the coresident genomes (homoeo-SNPs) in tetraploid species AD1 and AD3 (Page and Udall 2015). The resulting mapping rates of 71–89% are similar between libraries. One D5 20 dpa library was found to be a misidentified polyploid sample, as half the RNA-seq reads contain A-genome specific SNPs; this sample was therefore excluded from further analyses. The first two PCA components (fig. 1B) of the transcriptional profiles explain 44.8% of sample-to-sample variation, which groups samples according to developmental stages (PC1) prior to species (PC2), suggesting that developmental-stage-specific expression patterns are more similar than are species-specific patterns during cotton seed development. Notably, samples from the three allotetraploid genomes were intermediate between the two model diploid parents; thus, a synthetic midparental representative (Syn), constructed from the average diploid expression values, was included in subsequent analyses to facilitate inference regarding the effects of polyploidization on transcriptomes.
Gene Coexpression Network Analysis
Two different conceptual approaches were explored to help understand gene coexpression network conservation and divergence at various evolutionary timescales and across ploidy levels. First, a multispecies coexpression network was constructed to provide a global view of coexpression network topology among species. Second, individual coexpression networks were constructed and compared for all species included in the study. In conjunction with differential gene pair correlation analysis, this latter analysis permits the diagnosis of altered network edges, i.e., coexpression “rewiring” between species. Given the complexity of large-scale gene networks in our analysis, global and subnetwork properties were examined using both approaches, integrated with functional annotation and oil trait related phenotype and gene family analysis.
Multispecies Coexpression
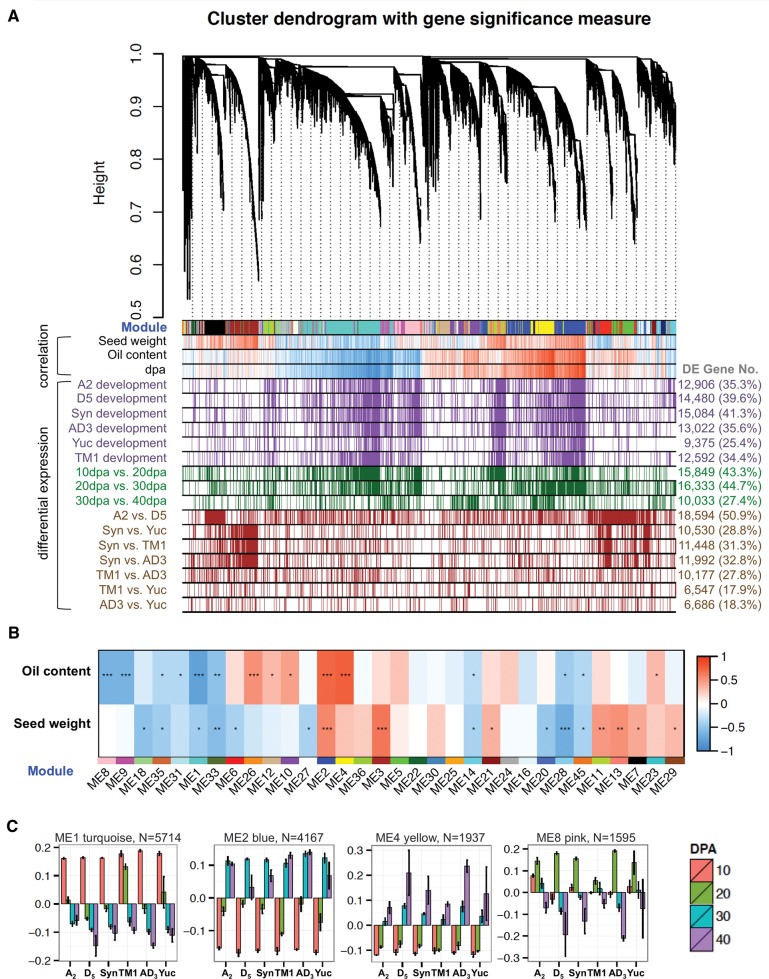
To construct a multispecies coexpression network, weighted gene coexpression network analysis (WGCNA) was applied to the entire RNA-seq dataset containing 70 seed samples derived from six species/accessions (A2, D5, AD3, TM1, Yuc, and Syn; 11 samples from D5 and Syn, and 12 each from others). For the allopolyploids, the total expression of each homoeolog gene pair is referred to as a gene hereafter. Connection strengths among genes were estimated based upon correlation of expression data, and clusters of highly interconnected genes were identified as modules. After removing genes with zero expression or without variance across samples, a multispecies cotton seed development network was constructed with 34,140 genes partitioned into 55 coexpression modules (fig. 2A;supplementary dataset S1, Supplementary Material online). Following a scale-free power law distribution, a few highly connected genes or hubs control a large part of the network while most genes have low connectivity. Functional enrichment for hub genes revealed that seed oil biogenesis and storage, cytoskeleton activity, organ specification, and morphogenesis were enriched (GSEA P < 0.05, FDR Q < 0.05; supplementary dataset S2, Supplementary Material online).
Fig. 2.—
Multi-species co-expression network analysis. (A) Hierarchical cluster tree showing co-expression modules identified using WGCNA. Modules correspond to branches and are labeled by colors as indicated by the first color band underneath the tree. In the “correlation” bars, red and blue color bands indicate genes displaying expression that is highly correlated (red) or anti-correlated (blue) with phenotypic traits (seed weight, percentage oil content) or developmental stage (dpa). In the “differential expression” bars, purple, green, and brown color bands present genes differentially expressed between sampling conditions. Purple = genes differentially expressed between any two adjacent stages for that species; green = genes differentially expressed for all six accessions for the dpa comparison shown; brown = genes differentially expressed between species/accession for any stage. (B) Correlation between co-expressed modules and seed traits. For 32 modules showing significant relationships with developmental and interspecific changes (Anova P < 0.05), module eigengenes (columns labeled by module ID and assigned color) were correlating to trait measurements of oil content and seed weight (rows). Heatmap colors correspond to correlation coefficients and stars to BH corrected P-values (*q < 0.05; **q < 0.001; ***q < 0.0001). (C) Eigengene expression for modules associated with oil content. Bar plots present eigengene values centered by mean across genomes and developmental stages. Error bars represent the standard errors among three biological replicates for each genome at each developmental stage. Module color and number of module member genes are noted above each bar graph.
Patterns of differential gene expression were characterized across species. During cotton seed development, the number of differentially expressed genes peaked between 20 and 30 dpa (fig. 2A, green rows), consistent with the pattern previously reported for upland cotton (Hovav et al. 2015). Similar developmental changes between any two adjacent stages were found in diploid and polyploid cottons (fig. 2A, purple rows); interestingly, a smaller number of differentially expressed genes was found in the wild AD1 accession Yuc (25.4%) than in the other species (34.4–41.3%). Direct interspecific comparisons of gene expression at any stage revealed that the highest amount of differential expression (50.9%) is between the two diploids studied and the lowest amount is between wild and domesticated G. hirsutum (17.9%; fig. 2A, brown rows), consistent with the extent of evolutionary divergence associated with diploid speciation and G. hirsutum domestication.
When network connectivity was examined, developmental changes (in all species) were found to be significantly associated with network hubs (GSEA P < 0.05, FDR Q < 0.05), suggesting that the physiological activities of seed development are controlled through up- and downregulation of high connectivity genes, or network hubs. In contrast, for evolutionary comparisons, significant association with network connectivity was detected only for A2 vs. D5 and TM1 vs. AD3 (GSEA P < 0.05, FDR Q < 0.05), but not for comparisons among other allopolyploids, or between diploids and polyploids. These patterns reflect the differences in network properties between developmental and evolutionary comparisons; that is, differentially expressed genes corresponding to seed development are more connected in the network than are those corresponding to polyploidy and domestication.
Notwithstanding the insights gained from the analysis of whole-network connectivity, it has been shown that when it comes to large and complex coexpression gene networks, subnetwork architecture, summarized by intramodular connectivity, is more relevant to important biological processes (Langfelder et al. 2013). To explore this, within each module expression levels of all member genes were depicted by a heatmap and were summarized by the eigengene value (the first principle component of module expression profiles; supplementary fig. S1, Supplementary Material online). By relating module eigengenes to sample conditions (eigengene ∼ conditions; 24 conditions = 6 genomes × 4 stages), 32 modules were identified with significant genome-specific and/or temporally regulated coexpression patterns (ANOVA P < 0.05). Correspondence tests between intramodular connectivity and differential expression patterns (supplementary table S2, Supplementary Material online; GSEA P < 0.05, FDR Q < 0.05) showed that these modules are enriched with differentially expressed genes, except for three (ME18, 20, and 28). Notably, modules enriched with genes showing differential expression during development (ME1, 2, 7, 8, 9, 11, 13, 23, and 29) collectively contain one third of cotton genes, and exhibit similar developmental expression profiles among diploid and polyploid cotton species.
Phenotypic Association and Functional Annotation of Multispecies Coexpression Modules
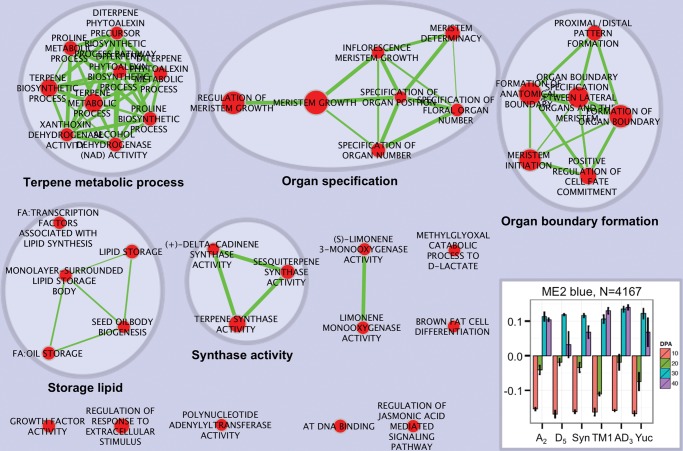
Association analyses between multispecies coexpression modules and seed traits (fig. 2B; supplementary fig. S2, Supplementary Material online) revealed that ME2 and ME4 are the modules most strongly correlated with oil content, thus representing suites of interconnected genes underlying the physiological process of oil synthesis. ME2 was the second largest module detected, with 4,167 genes which as a group are increasingly upregulated from 10 to 30 dpa (figs. 2C and 3); this module includes all seed storage protein genes (legumins, vicilins, and albumin), the majority of oil storage and fatty acid desaturation-related genes, and five key transcription factors associated with fatty acid and triacylglycerol biosynthesis (i.e., ABI3, FUS3, HSL1, HSL2, and WRI1; Hovav et al. 2015). Biological processes and activities enriched with high intramodular connectivity were visualized as interaction networks with Cytoscape and Enrichment Map (fig. 3; see also supplementary dataset S2, Supplementary Material online) (Merico et al. 2010). In addition to lipid storage activities that positively correlate with the seed oil accumulation period, the modular structure of ME2 also associates with numerous biological processes involved in tissue specification (e.g., growth factor activity, organ specification, and organ boundary formation), secondary metabolism (e.g., terpene metabolic process and synthase activity), oxidative homeostasis (e.g., limonene monooxygenase activity), signal transduction, and regulation.
Fig. 3.—
Gene Set Enrichment Analysis (GSEA) delineates biological pathways and processes that characterize ME2. Cytoscape and Enrichment Map were used for visualization of the GSEA results as a network of enriched gene sets (P < 0.05, FDR Q < 0.05). Nodes representing enriched gene sets are grouped and annotated by their similarity according to related gene sets. Node size is proportional to the total number of genes within each gene set. Proportion of shared genes between gene sets is represented as the thickness of the green line between nodes. A bar graph at lower right corner shows the module eigengene. Detailed GSEA results can be found in supplementary dataset 2, Supplementary Material online.
Also positively correlated with oil content, coexpressed genes in ME4 exhibit peak expression levels at 40 dpa (fig. 2B and C). Biological processes of transcription, mRNA spliceosome complex, ribosomal RNA modification, ribosomal organization, and protein peroxisome were enriched with intramodular hubs. Interestingly, many β-oxidation-related genes were found in this module (supplementary fig. S3A, Supplementary Material online), which may suggest an increased level of fatty acid degradation and account for the decreased rate of oil accumulation observed from 30 to 40 dpa (fig. 1A).
The largest module, ME1, with 5714 genes that collectively are downregulated during seed development in each of the five natural genomes and one Syn genome studied, was negatively correlated with oil content (fig. 2B and C). Genes in this module show significant expression change between adjacent developmental stages (fig. 2A, dense purple and green colors corresponding to the turquoise module). Functional enrichment analysis identified cytoskeleton biosynthesis (including microtubule assembly and actin polymerization), basic biosynthetic/metabolic processes, and cell growth signaling, all of which decrease as seeds approach maturity (supplementary dataset S2, Supplementary Material online: GSEA P < 0.05, FDR Q < 0.05). Interestingly, oil-related genes involved in fatty acid elongation, sphingolipid synthesis, and other acyl lipid-related synthesis are interconnected and preferentially present in this module (Fisher’s exact test, P < 0.05; supplementary fig. S3A, Supplementary Material online); the negative correlation between their expression profiles and seed development presumably explains the lack of very-long-chain fatty acids and sphingolipid derivatives in cotton seed oil.
Targeted analysis of the cotton oil- and lipid-related gene families (Hovav et al. 2015) revealed that genes involved in plastid fatty acid synthesis from pyruvate were found to be enriched in ME11, and key transcription factors were enriched in ME13 (supplementary fig. S3A, Supplementary Material online). Because neither ME11 or ME13 is significantly correlated with oil content (fig. 2B), these oil-related genes and their intramodular connections may not play the key role in oil trait variation. Instead, oil-related genes present in the modules that are significantly associated with oil content, such as the oil storage genes enriched in ME2, together with coexpressed transcription factors (i.e., ABI3, FUS3, HSL1, HSL2, and WRI1), are more likely to represent transcriptional connections responsible to phenotypic variation.
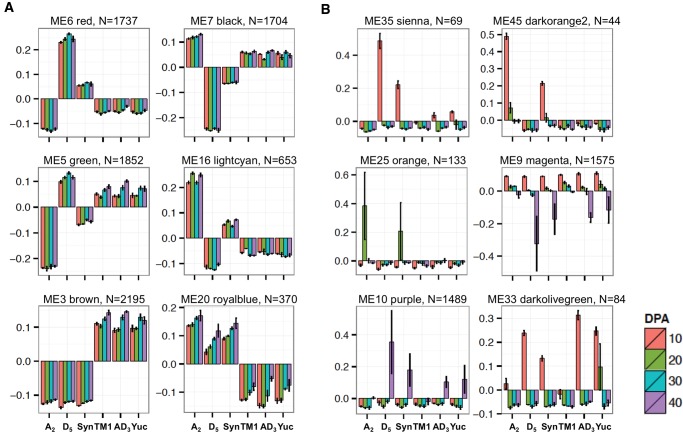
With respect to seed weight, ME3 is the most significantly correlated module (fig. 2B), with coexpressed module genes upregulated in all polyploid species compared with diploids and the synthetic allopolyploid (fig. 4A, left bottom panel). Different from modules related to oil content, the coordinated expression differences between accessions dominate differences among developmental stages, as higher expression levels were seen in tetraploid vs. diploid cottons in ME3. This suggests that the regulatory control of seed weight has been strongly affected by allopolyploidization.
Fig. 4.—
Eigengene expression for modules displaying expression-level dominance and transgression. These co-expression patterns could be consistent between developmental stages (A) or be specific to certain stages and variable between allopolyploid species (B). Bar plots present eigengene values centered by mean across genomes and developmental stages. Error bars represent the standard errors among three biological replicates for each genome at each developmental stage. Module color and number of module member genes are noted above each bar graph.
Modular Characterization of Expression-Level Dominance and Transgression in Allopolyploid Cotton
Parental contributions to total expression patterns in allopolyploids is a topic of broad interest (Grover et al. 2012; Yoo et al. 2014), but there are few examples where gene coexpression has been studied in polyploid plants (Pfeifer et al. 2014). If one assumes that midparent values are expected in allopolyploids, deviations from midparent values relative to progenitor diploids indicate expression modification of progenitor profiles; the application of this logic for coexpressed genes, using their characteristic expression pattern (i.e., eigengenes), offers an opportunity to reveal regulatory modules and hub genes responsible for expression modifications accompanying polyploid evolution.
In an allopolyploid context, expression-level dominance is defined as allopolyploid expression level equivalence to one of the two parents, irrespective of whether it is the higher or lower expressed parent (Grover et al. 2012). This concept has not, to the best of our knowledge, previously been applied to coexpression modules in plants. Here, expression level dominance was observed for ten modules (supplementary fig. S1, Supplementary Material online). Although more modules display expression level dominance for the D-parent (ME5, 16, 25, 31, 33, and 45) than for the A-parent (ME6, 7, 26, and 35), the overall pattern for module member genes was unbalanced towards the A-parent (3638 A-dominant genes) rather than the D-parent (2855 D-dominant genes; Chi-square test, P < 0.05). This unbalance is mainly due to a bigger A-dominant, lower-expression module than a D-dominant, lower-expression module (fig. 4A: A-dominant ME6 with 1737 genes, and D-dominant ME16 with 653 genes). The modular structure of ME16 was found to be associated with translation termination, cellular component disassembly, and positive regulation of cell cycle arrest, which in allopolyploids were expressed at lower levels, as in the D-genome parent. In contrast, the A-dominant ME6 module, associated with tRNA splicing via endonucleolytic cleavage and ligation (supplementary dataset S2, Supplementary Material online), was downregulated in allopolyploids. Interestingly, a D-dominant, higher-expression module, ME5, was also functionally enriched with tRNA splicing via endonucleolytic cleavage and ligation. The module hub genes (kME > 0.9) corresponding to this biological process are several putative RNA 2'-phosphotransferases (Gorai.002G268800 in ME5; Gorai.007G184200, Gorai.008G298600, and Gorai.009G455600 in ME6). These genes likely reflect expression divergence in this gene family that originated subsequent to allopolyploidzation. No functional category was enriched for the A-dominant, higher-expression module ME7.
Cases where expression in the allopolyploids is significantly above or below both parental values are termed transgressive, reflecting either up- or downregulation. Module ME3 and ME20 exemplify this phenomenon, being transgressively up- and downregulated, respectively, in all developmental stages in allopolyploids (fig. 4A), with more member genes transgressively upregulated than downregulated. For ME20, the enriched functional categories were similar to those enriched in ME 16, indicating that these cellular functions were downregulated in allopolyploids relative to one (as in ME16) or both diploid parents (as in ME20). Despite the large number of coexpressed genes and the association with seed weight, no functional categories were enriched for ME3.
Modular expression-level dominance and transgressive expression were also detected that were specific to certain developmental stages (fig. 4B). For example, modules ME35 and ME45 display A-dominant and D-dominant lowered expression at 10 dpa, respectively. D-dominant lower expression was also observed for module ME25 at 20 dpa. Typically, the direction of expression-level dominance was found to be consistent among allopolyploids (fig. 4A), but opposite directions among allopolyploids were observed for modules ME9, ME10, and ME33. Interestingly, in all three of these modules, D-dominant expression in the wild plants AD3 and wild G. hirsutum (Yuc) changed to A-dominant in domesticated G. hirsutum (TM1). Except for kinase activity and transcription regulation enriched in ME9 and ME10, respectively (supplementary dataset S2, Supplementary Material online), no other functional categories were enriched, perhaps due to the small numbers of member genes (N < 150) in these modules.
Individual Coexpression Network Analysis
To investigate topological changes in transcriptional organization during seed development among the six cotton species/accessions (A2, D5, TM1, Yuc, AD3, and Syn), we constructed individual networks for each accession, and applied differential coexpression analysis to identify “rewired edges” between any two individual networks. Using the percentage of rewired edges among all possible edges to measure the extent of network topological changes, we observed 3.4% divergence between diploid networks and less than 0.6% among networks of allopolyploid cottons (table 1). Interestingly, more rewired edges were found between wild and domesticated G. hirsutum than those between the wild polyploid species AD3 and the wild G. hirsutum accession Yuc. When comparing the allopolyploid to both diploid networks, more rewired edges were consistently observed in comparison to the D5 than the A2 network, suggesting that the network topology of allopolyploid cottons resembles A2 more than it does D5.
Table 1.
Differential Co-Expression between Individual Specific Networks
| Network Pairs | Rewired Edges |
Differential Co-Expression Genes (DCGs) |
|||
|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | as DEGs | |
| Between diploids | |||||
| A2 vs. D5 | 15,708,780 | 3.36% | 10,849 | 35.46% | 49.62% |
| Between allopolyploids | |||||
| TM1 vs. Yuc | 1,124,591 | 0.24% | 2,738 | 8.95% | 11.98% |
| TM1 vs. AD3 | 2,744,883 | 0.59% | 4,875 | 15.94% | 24.84% |
| Yuc vs. AD3 | 799,532 | 0.17% | 1,999 | 6.53% | 8.05% |
| Syn vs. TM1 | 5,174,383 | 1.11% | 7,658 | 25.03% | 28.44% |
| Syn vs. Yuc | 962,655 | 0.21% | 2,267 | 7.41% | 22.28% |
| Syn vs. AD3 | 3,286,858 | 0.70% | 5,849 | 19.12% | 30.91% |
| Between allopolyploid and diploid | |||||
| TM1 vs. A2 | 3,676,001 | 0.79% | 6,804 | 22.24% | 38.05% |
| TM1 vs. D5 | 19,240,674 | 4.11% | 10,600 | 34.65% | 44.31% |
| Yuc vs. A2 | 1,982,313 | 0.42% | 4,479 | 14.64% | 34.05% |
| Yuc vs. D5 | 4,843,495 | 1.04% | 6,654 | 21.75% | 36.08% |
| AD3 vs. A2 | 3,636,533 | 0.78% | 6,409 | 20.95% | 41.39% |
| AD3 vs. D5 | 1,1697,128 | 2.50% | 9222 | 30.15% | 38.60% |
| Syn vs. A2 | 568,595 | 0.12% | 1883 | 6.16% | 12.11% |
| Syn vs. D5 | 891,335 | 0.19% | 1998 | 6.53% | 10.81% |
We defined genes enriched with rewired edges as differential coexpression genes (DCGs). Although the percentages of DCGs are comparable to those of differentially expressed genes (DEGs) between species (see fig. 2A, brown rows), the overlap between DEGs and DCGs ranges 8.0–49.6% (table 1). These results suggest that in addition to differential expression of individual genes, changes in gene-to-gene coexpression relationships play a role in the developmental architecture of cotton seed development. In addition, weak to modest correlations (0.15–0.58) of whole-network connectivity (k) were observed between species (supplementary fig. S4, Supplementary Material online), which are strikingly lower than correlations of gene expression profiles (0.89–0.97). This observation confirmed that network topologies were substantially altered between species.
Intra- and Intermodular Topological Analysis of Individual Networks
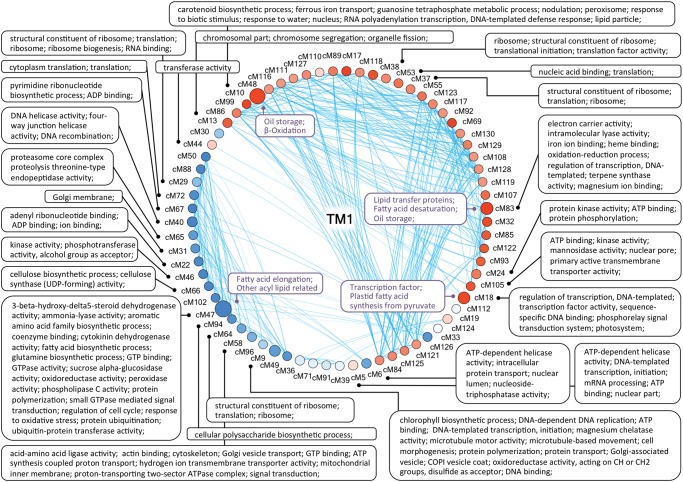
To explore more fully the nature of evolutionary changes in seed coexpression networks, we first identified “consensus modules,” stable clusters of interconnected genes present in all single-species networks. With respect to intramodular topological parameters (e.g., connectivity, density, separability, etc.), these consensus modules, composed of 20,696 genes, are well-preserved among species (all Zsummary above 5; see methods). Next, intermodular relationships among consensus modules were measured by pairwise eigengene correlations and were represented as eigengene networks [see Methods and Langfelder and Horvath (2007)], as exemplified for TM1 (fig. 5). Consensus modules that exhibit similar expression profiles (Pearson’s correlation r > 0.9) were connected; accordingly, modules with increased expression late in development (red nodes, eigengenes positively correlated with developmental stage) appear to be clustered and are separated from those displaying decreased expression as seeds mature (blue nodes, eigengenes negatively correlated with developmental stage). Biological processes and metabolic pathways associated with these consensus modules were identified by GO enrichment analysis (supplementary dataset S3, Supplementary Material online). For oil-related gene families, preserved coexpression patterns were detected for genes involved in “plastid fatty acid synthesis from pyruvate”, “fatty acid elongation”, “other acyl lipid related”, “fatty acid desaturation”, “transcription factors associated with lipid synthesis”, “oil storage”, and “β-oxidation” (supplementary fig. S3B, Supplementary Material online). As exemplified for TM1 (fig. 5), these enriched functional categories and oil-related gene families are shown next to corresponding consensus modules that are conserved across species, whereas the intermodular connections are specific to each species, allowing visual inspection of the functional dependencies within the network.
Fig. 5.—
Eigengene network presenting inter-modular relationships among consensus modules in TM1. Consensus modules represented by nodes are connected by edges when module eigengenes in TM1 are highly correlated (Pearson’s correlation r > 0.9). Node size is proportional to the total number of genes assigned to each consensus module. Node coloring reveals module eigengenes that are highly correlated (red) or anti-correlated (blue) with developmental stage. Significantly enriched GO categories are presented outside the network circle, and inside the circle consensus modules displaying significant correspondences with oil-related gene families are labeled (fisher’s exact test P < 0.05, n > 5).
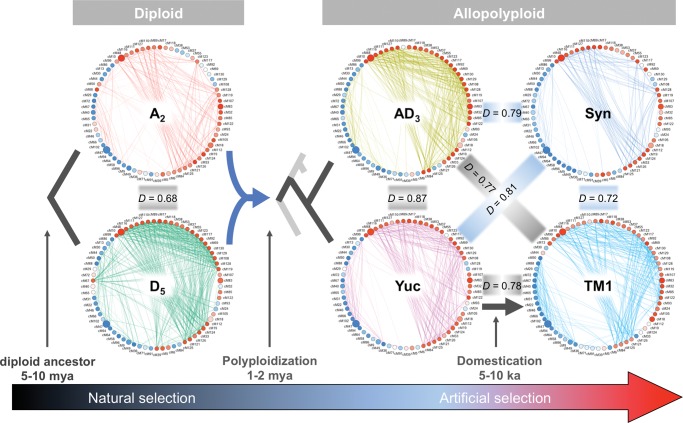
To specify evolutionary modifications in network topology, we compared intermodular relationships among species. Preservation tests of eigengene networks showed that intermodular relationships were least preserved during the 5–10 Myr of divergence at the diploid level (A2 vs. D5; fig. 6, D = 0.68). Notably, there are over twice as many edges between consensus modules in D5 than in A2 (567 vs. 234), suggesting a tighter coordination of the corresponding biological processes relative to those in A2; this appears especially the case for modules displaying higher expression later in seed development (from 11 to 6 o’clock in the circular networks; red node color indicates eigengenes positively correlated with developmental stage). Interestingly, in the D5 network, consensus modules 18, 83, 85, 105, and 107 (3 to 4 o’clock) are disconnected from nearby highly connected, upregulated modules (red nodes) and form a small distinct cluster (supplementary fig. S5, Supplementary Material online), whereas no such separation was evident in the A2 network. This small D5-specific cluster is not only functionally involved in signal transduction and regulation of transcription, but also corresponds to key oil-related gene families, including “plastid fatty acid synthesis from pyruvate”, “transcription factors associated with lipid synthesis”, “fatty acid desaturation”, and “oil storage”. For consensus modules displaying higher expression in early developmental stages (blue nodes indicate eigengenes negatively correlated with developmental stages), key biosynthetic pathways (polysaccharides, amino acids, and fatty acids) and cytoskeleton activities are mostly conserved between the diploid species, whereas translation and proteolysis related modules (1–2 and around 9 o’clock) appear to be controlled by different regulatory systems in A2 and D5 networks.
Fig. 6.—
Preservation of consensus eigengene networks during cotton polyploidization and domestication. The preservation test statistic D is presented for between-genome comparisons. The closer D is to 1, the higher the preservation. Node and edge styles of eigengene network are as described in figure 5.
Notably, the highest preservation of modular structure was found between the wild polyploid species G. tomentosum (AD3) and the wild G. hirsutum accession Yuc (fig. 6, D = 0.87), while intermodular relationships between wild and domesticated forms of the same species, G. hirsutum (Yuc and TM1), were significantly less preserved (D = 0.78; P < 0.05). This unexpected observation, i.e., intraspecific variation is higher than interspecific variation, suggests that transcriptional organization of seed development was substantially altered by domestication. For the remaining comparisons between allopolyploid and diploid networks, the synthetic allopolyploid network (Syn) resembles D5 more than it does A2 (Syn: DA = 0. 80, DD = 0.85; DA < DD, P < 0.05); in Yuc and AD3, the preservation levels of diploid networks decreased and became closer to each other compared to that in Syn (Yuc: DA = 0.76 and DD = 0.78; AD3: DA = 0.75 and DD = 0.76; DA = DD, P > 0.05). Intermodular relationships altered by G. hirsutum domestication (from Yuc to TM1) were further differentiated from those of the diploid progenitors, while skewing the resemblance to become more A-like (TM1: DA = 0. 74, DD = 0.67; DA > DD, P < 0.05), notably opposite to the direction found in the Syn network. Among all consensus modules, the intermodular relationships of modules 33, 86, 13, 44, 24, and 19 were most altered by domestication (supplementary fig. S6, Supplementary Material online), although their corresponding biological processes were not clear.
Oil Synthesis and Lipid Related Gene Network Altered by Domestication
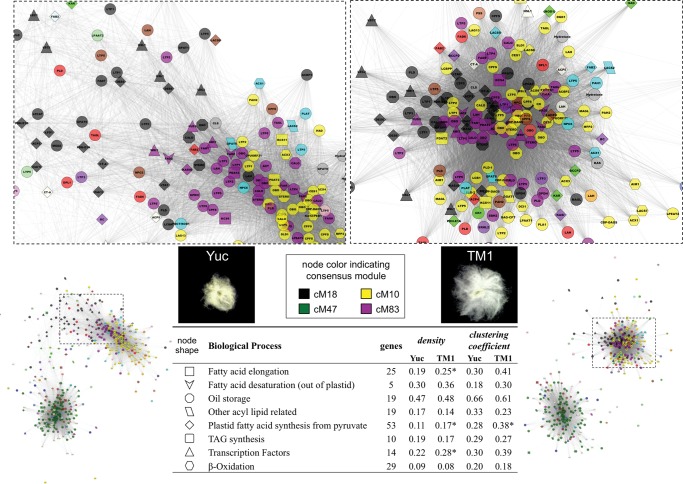
To investigate how domestication reprogrammed cotton seed development and led to an increase in seed oil content from wild to domesticated G. hirsutum (fig. 1), we extracted subnetworks relevant to oil synthesis and lipid-related metabolic processes from the Yuc and TM1 networks. The extracted network of 433 oil-related genes (Hovav et al. 2015) was more densely connected and closely clustered in TM1 than in Yuc (Density = 0.087 and 0.061, mean Clustering Coefficient = 0.33 and 0.26, respectively; bootstrapping P-value < 0.05, see Methods), suggesting an elevated coordination of regulatory control as a consequence of domestication. This increase in network density is also evident at the gene family level for processes “fatty acid elongation”, “plastid fatty acid synthesis from pyruvate”, and “transcription factors associated with lipid synthesis”, while an increased network clustering coefficient was observed only for the process “plastid fatty acid synthesis from pyruvate” (fig. 7, bottom table).
Fig. 7.—
Visualization of the oil related gene co-expression network in wild (Yuc; left panels) and domesticated (TM1; right panels) G. hirsutum developing seeds. The network overview (bottom panels) and a closer view (upper panels) of the “up-regulated” cluster are shown for each genome. The bottom table summarizes the sub-network properties of density and clustering coefficient at the gene family level. A bootstrap approach was used to test differences between TM1 and Yuc parameters, and * denotes high values with significance (P < 0.05). TAG, triacylglycerol.
Visual inspection of oil-related gene networks (fig. 7; supplementary fig. S6, Supplementary Material online) reveals two major coexpression clusters formed between genes upregulated during seed development (mainly composed of black, yellow and dark magenta nodes), and those downregulated (mainly composed of dark green nodes). The downregulated clusters are enriched with gene families of “fatty acid elongation” and “other acyl lipid related”, and the cluster topologies appear similar between wild and domesticated cotton. The upregulated cluster in TM1 (fig. 7; bottom right panel) is more densely connected than in Yuc and with no obvious compartmentation of network space among three major modules as is seen in Yuc (where black, yellow and dark magenta nodes appear to be separated). Oil-related gene families overrepresented in the upregulated clusters, include “plastid fatty acid synthesis from pyruvate” (black diamond), “fatty acid desaturation” (dark magenta V shape), “transcription factors associated with lipid synthesis” (black or dark magenta triangle), “oil storage” (dark magenta or yellow octagon), and “β-oxidation” (yellow hexagon). Among these, “oil storage” genes encoding OBO (oleosin), CALO (caleosin), and STERO (steroleosin) were most connected (Density = 0.47–0.48, mean Clustering Coefficient = 0.61–0.66), whereas “β-oxidation” genes encoding various peroxisome enzymes were loosely scattered in both networks (Density = 0.08–0.09, mean Clustering Coefficient = 0.18–0.20). Among “transcription factors associated with lipid synthesis”, six were clustered with “plastid fatty acid synthesis from pyruvate” genes in the consensus module 18, including LEC1 (LEAFY COTYLEDON 1), LEC2 (LEAFY COTYLEDON 2), HSL1 (HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE-LIKE 1), and WRI1 (WRINKLED 1). In consensus module 83, another three transcriptional factors—ABI3 (ABSCISIC ACID INSENSITIVE 3), FUS3 (FUSCA 3), and WRI1 (WRINKLED 1), were clustered with genes mainly involved in “fatty acid desaturation”, “Lipid Transfer Proteins”, and “oil storage”. Although not enriched in any consensus module, “TAG synthesis” genes (round rectangle) genes such as DGAT (Acyl-CoA: diacylglycerol acyltransferase) and PDAT (Phospholipid:diacylglycerol acyltransferase) were scattered in the upregulated clusters, exhibiting similar subnetwork properties in Yuc and TM1.
Discussion
Here we report on the conservation and divergence of gene expression and coexpression during cotton seed development and evolution. Our comparative and phylogenetic context allowed us to reveal gene expression and coexpression changes accompanying evolutionary stages spanning timescales ranging from millions to thousands of years: at the diploid level subsequent to diversification 5–10 Ma; following genomic merger, polyploidization and speciation at the allopolyploid level 1–2 Ma; and accompanying domestication and crop improvement over the last approximately 5000 years. These analyses provide an overview of seed transcriptional architecture and its conservation vs. evolutionary lability in diploids and polyploids of a single genus, and in response to strong directional human selection.
Coexpression Network Analysis Provides Novel Insight into Phenotypic Evolution
Polyploidy is pervasive among angiosperm species, and is frequently associated with phenotypic changes that are the presumed consequence of the genetic and epigenetic modifications accompanying genomic merger and duplication (Levin 1983; Stebbins 1940; Leitch and Leitch 2008; Jiao et al. 2011; Soltis et al. 2016; Wendel et al. 2016). The massive rewiring of gene expression in allopolyploid species has been evaluated on a genome-wide basis for many polyploid species and their model diploid progenitors (Yoo et al. 2014). Despite the substantial insights gained from these analyses (Doyle et al. 2008; Madlung and Wendel 2013; Yoo et al. 2014; Salmon and Aïnouche 2015), identifying the concerted patterns and causal interactions responsible for phenotypic and large-scale transcriptional changes have remained elusive. This is due not only to the difficulty in distinguishing upstream regulators from the large number of downstream genes that are differentially expressed, but also, more importantly, regulatory processes that are either too subtle to be detected by conventional differential expression analysis or are post-transcriptional (e.g., phosphorylation, ligand binding, formation of “open” euchromatin, etc.). Consequently, phenomena may be overlooked by global expression analyses that focus on gene-by-gene comparisons, which may become apparent in the context of coexpression with other genes in a biological network. For example, an increase of coexpression connectivity was identified for a transcription factor NFYA during soybean domestication, but its expression levels are not differentiated between wild and domesticated accessions (Lu et al. 2016). Other applications of the network approach to plant developmental evolution include studies of the responses to selection accompanying maize domestication (Swanson-Wagner et al. 2012), interacting regulatory mechanisms underlying morphological diversity of Brassica (Basnet et al. 2013) and wheat (Pfeifer et al. 2014) seeds, seasonal regulation and environmental adaptation in pines (Cañas et al. 2015), and for predicting molecular interactions governing tomato leaf complexity (Ichihashi et al. 2014).
Here we show that correlations of whole-network connectivity between species are much lower than are correlations of global gene expression profiles, and also that there is limited overlap between differentially expressed genes and differential coexpression genes. These results confirm that gene-to-gene interconnectivities provide an additional, complementary perspective to traditional differential expression analysis.
In addition to the novel perspectives on differential gene coexpression, the global network analyses provide useful insight into features of cotton seed biology. For example, genes involved in the initial stage of oil synthesis (i.e., de novo plastid fatty acid synthesis from pyruvate) are significantly coexpressed in all species, with peak expression at 20 and 30 dpa (ME11; supplementary fig. S1, Supplementary Material online), while genes involved in fatty acid desaturation and oil storage, which represent the next stages in oil biosynthesis, show similar expression and coexpression among the species studied (ME2; fig. 2C). The temporal expression observed for these late-stage oil synthesis genes is congruent with oil accumulation rates in each species, consistent with our previous results for G. hirsutum, accession TM1 (Hovav et al. 2015) and Arabidopsis (Ruuska et al. 2004). Notably, the higher correlation between ME2 eigengene and oil content suggests that the modular organization of transcriptional regulation for oil storage genes is more relevant to oil content variation between cotton species, than is the set of oil synthesis genes represented by ME11. Another coexpression pattern highly correlated with oil content is the continuously increasing expression, from 10 to 40 dpa, for module ME4 (fig. 2C); the enrichment of β-oxidation genes in this module may partially explain the decreased oil accumulation rate as seed maturation is approached. Indeed, increased expression of β-oxidation genes has also been associated with the later stages of seed development in other plants, including B. napus (Chia et al. 2005), Glycine max (Li et al. 2015), and Arabidopsis (Baud and Lepiniec 2009). This has been proposed as a mechanism to redeposit nutrient reserves to “bridge” between seed maturation and later germination (Angelovici et al. 2010).
Comparisons of species-specific network topologies to each other revealed several new perspectives about seed developmental during evolutionary divergence within Gossypium. First, remarkable differences exist between the two parental diploid networks, with twice the intermodular connectivity found in D5 vs. A2 (567 and 234 intermodular edges, respectively; fig. 6), and a small distinct cluster unique to D5 (supplementary fig. S5, Supplementary Material online). This cluster is composed of consensus modules whose expression generally increases during seed development yet are disconnected from other upregulated modules, and is significantly associated with key oil synthetic processes like “plastid fatty acid synthesis from pyruvate”, “transcription factors associated with lipid synthesis”, “fatty acid desaturation”, and “oil storage”. This distinct cluster was absent from the A2 network, suggesting a difference in regulation for these cellular activities between the two diploid species, which may explain the higher oil content in D5 vs. A2. At the polyploid level, following merger of the A and D genomes, the preservation and recruitment of this D5 cluster was observed only in domesticated G. hirsutum, of the three polyploid accessions studied. It is tempting to speculate that the re-evolution of this cluster, or its “recruitment”, accompanied domestication, which might also help explain the higher oil content in TM1 as in D5. Another notable observation comes from the comparison between wild and domesticated AD1 networks, which summarizes the striking effects of domestication on oil-related gene coexpression networks and oil accumulation; that is, in addition to the elevated level of coordinated control among oil-related genes in domesticated cotton, gene family members of the plastidic fatty acid synthetic genes and related transcription factors are more densely connected with one another, than they are in wild polyploids.
Coexpression Network Analysis Provides Novel Insight into Polyploidy
In addition to providing insight into the genetic underpinnings of phenotypic traits, network analysis can also be used to address longstanding questions regarding the evolutionary genomics of polyploids. In hexaploid bread wheat, for example, Pfeifer et al. (2014) evaluated homoeolog usage for the spatiotemporal control of endosperm development, which suggested expression divergence as a form of subfunctionalization for that developmental series in bread wheat. More interestingly, the authors aggregate homoeolog expression for network construction and relate network organization to the asymmetric contribution of the wheat subgenomes, further demonstrating the power of coexpression network analysis to reveal the evolution of expression divergence and cross-talk within the allopolyploid genome, and ultimately relating these insights to wheat baking quality (Pfeifer et al. 2014). Here, using allopolyploid cotton species, we also aggregated expression for each pair of homoeologs to allow direct comparison between diploid and allopolyploid networks. Although a more sophisticated approach is to construct separate networks for A- and D- homoeologs in contrast to parental A2 and D5 networks, respectively, such an analysis is still hindered by homoeolog gene mapping issues. That is, the ability to distinguish a pair of homoeologs is dependent on the number and distribution of differentiating SNPs, which varies from gene to gene, thereby introducing errors into the estimation of gene-to-gene relationships at the homoeolog level. This direction should become more feasible in future studies, for example, using longer-read technologies that increase the likelihood of unambiguous assignment of reads to homoeolog.
Here, we identified multispecies modules that exhibit expression changes unique to certain developmental stages and species, and comparison of the allopolyploid species with their parental diploids and a synthetic midparental genome permits categorization of allopolyploid coexpression profiles with respect to the contribution of and deviation from parental regulatory patterns (Rapp et al. 2009; Grover et al. 2012; Yoo et al. 2014). Transgressive expression was detected in three modules, two of which were downregulated during seed development, whereas the single upregulated transgressive module, ME3, is strongly correlated with seed weight and contains approximately 5-fold more members than the other two combined. Although no functional categories were enriched for this module, the interconnectivity and other topological features characterized for module member genes provide a resource for the inference of regulatory connections underlying transgressive traits.
We also showed that the recently described phenomenon of expression level dominance (Grover et al. 2012; Yoo et al. 2013) extends beyond the level of individual genes to encompass entire gene modules; that is, in allopolyploids, modules of coexpression may more closely mimic that of one or the other diploid parent, as opposed to simply being an average of the two. This is an entirely unexpected and novel finding. As previously shown for cotton leaves (Yoo et al. 2013), single gene expression-level dominance in allopolyploids in the present study was biased toward the A-genome, but at the coexpression level, more modules exhibit D- than A-dominant expression, suggesting asymmetric regulation such that the coexpression network in the allopolyploid resembles, in this respect, the D-genome network. This is intriguing in providing a partial explanation, perhaps, for the D-like protein storage profiles exhibited by allopolyploid species (Hu et al. 2011), and the D-like oil accumulation in TM1 (fig. 1A).
In general, both cis-regulatory elements and trans-acting factors are known to coordinate the spatiotemporal control of gene expression and thereby present targets for the evolution of gene regulation. As cis elements are spatially aligned with their regulatory targets, differences among genomes may retain parental-like expression in derived allopolyploid genomes more so than do trans-acting elements, thus representing one dimension of the phenomenon of “parental legacy” in allopolyploids (Buggs et al. 2014). Cis regulatory variation often is more responsible for gene expression divergence under natural selection (Emerson and Li 2010), during crop domestication (Lemmon et al. 2014), and accompanying polyploidy (Shi et al. 2012; Xu et al. 2014) than is trans variation, although much remains to be learned before these indications can be considered to be general. The relevance of this to the present study is that for modules exhibiting expression-level dominance or transgression, it may be that differences between species and ploidy levels are caused by trans factors controlling module member genes through shared cis-regulatory elements. This will be a promising direction for future analyses, that is, to identify the cis elements responsible for the observed coexpression dominance or transgressive expression and the variation in the corresponding trans factors, and evaluate these in the context of allopolyploid genome evolution and module function. We see this as an exciting step toward a future systems biology perspective on allopolyploid evolution.
Whereas the multispecies network reveals common gene regulatory patterns in Gossypium, comparing individual species-specific coexpression networks becomes informative with respect to the evolution of network structure, including whole-network connectivity, intramodular topology (i.e., how module member genes are interconnected within each module) and intermodular relationships (i.e., higher-level transcriptional organization among modules). An illustrative example of this is the visualization of divergence of intermodular relationships depicted in figure 6, which provides an informative framework for studying pathway dependencies and regulatory hierarchies that underlie both genome evolution and phenotypic variation, in this case seed nutritional traits. For example, cofactor binding (enriched in consensus module cM83) and transcription factor activity (enriched in cM18) are more connected in D5 and TM1 than they are in A2 and the two wild tetraploids Yuc and AD3, which may reflect the regulatory dependency between seed oil synthesis and subsequent packaging processes, and possibly the recruitment of a D-like topology. Notably, the latter indicates one more perspective revealed by this type of analysis, in that this recruitment did not appear to accompany polyploidy per se, but instead appears to have resulted from domestication. Finally, this example also provides an interesting complement to the observations, discussed above, of the numerically larger number of D- than A-dominant modules. It is an intriguing notion to speculate that the architecture of gene coexpression in the allopolyploid nucleus is an integrated combination of modular structures from both progenitor genomes, each with its individualized contribution to seed development.
Coexpression Analysis and Cotton Domestication
An additional and somewhat surprising observation from comparison of the coexpression networks is that intraspecific divergence in coexpression, i.e., between wild and domesticated G. hirsutum, is greater than that observed between wild G. hirsutum (Yuc) and its sister species, G. tomentosum (AD3). This result underscores the accelerated evolution of intermodular relationships accompanying domestication in cotton. Importantly, this pattern is not apparent using conventional differential expression analysis (fig. 2A, bottom two rows), where similar expression divergence between domesticated cotton and the two wild species is evident. Also, as noted above, domestication appears to have resulted in a tighter, denser coexpression network that strengthens regulatory connections of oil synthesis-related pathways in correspondence to the increased oil content. As discussed earlier, one caveat of our study is that the allopolyploid gene networks were based on the aggregated expression of homoeologous gene pairs. Further analyses dissecting the interaction or distinctness of homoeologous modules and networks in polyploid species will ultimately provide insight into the interaction among genomes, homoeolog usage in the network, and the interdependencies of duplicated gene pathways following polyploidization.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Kara Grupp and Anna Tuchin at Iowa State University for assistance collecting cotton seed tissue. This work was supported by the US-Israel Binational Science Foundation (grant number 2009447) to RH and JW.
Literature Cited
- Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for gene ontology. R package version 2.16.0.
- Angelovici R, Galili G, Fernie AR, Fait A. 2010. Seed desiccation: a bridge between maturation and germination. Trends Plant Sci. 15(4):211–218. [DOI] [PubMed] [Google Scholar]
- Ashokkumar K, Ravikesavan R. 2013. Genetic variation and heterotic effects for seed oil, seed protein and yield attributing traits in upland cotton (Gossypium hirsutum L.). Afr J Biotechnol. 12(33):5183–5191. [Google Scholar]
- Basnet RK, et al. 2013. Genome-wide analysis of coordinated transcript abundance during seed development in different Brassica rapa morphotypes. BMC Genomics 14:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C. 2002. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 40(2):151–160. [Google Scholar]
- Baud S, Lepiniec L. 2009. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem. 47(6):448–455. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300. [Google Scholar]
- Bewley JD. 2006. Cotton - an oilseed In: Bewley JD, Black M, Halmer P, editor. The encyclopedia of seeds: science, technology and uses. London: CAB international; p. 106–109. [Google Scholar]
- Buggs RJ, et al. 2014. The legacy of diploid progenitors in allopolyploid gene expression patterns. Philos Trans R Soc Lond B Biol Sci. 369(1648):20130354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas RA, et al. 2015. Understanding developmental and adaptive cues in pine through metabolite profiling and co-expression network analysis. J Exp Bot. 66(11):3113–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia TYP, Pike MJ, Rawsthorne S. 2005. Storage oil breakdown during embryo development of Brassica napus (L.). J Exp Bot. 56(415):1285–1296. [DOI] [PubMed] [Google Scholar]
- Dowd MK. 2015. Seed In: Fang DD, Percy RG, editor. Cotton. Madison, WI: American Society of Agronomy, Inc., Crop Science Society of America, Inc., and Soil Science Society of America, Inc; p. 745–782. [Google Scholar]
- Doyle JJ, et al. 2008. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 42:443–461. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Li WH. 2010. The genetic basis of evolutionary change in gene expression levels. Philos Trans R Soc Lond B Biol Sci. 365(1552):2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A. 2013. DiffCorr: an R package to analyze and visualize differential correlations in biological networks. Gene 518(1):209. [DOI] [PubMed] [Google Scholar]
- Gotmare V, Singh P, Mayee C, Deshpande V, Bhagat C. 2004. Genetic variability for seed oil content and seed index in some wild species and perennial races of cotton. Plant Breed. 123(2):207–208. [Google Scholar]
- Grover CE, et al. 2012. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196(4):966–971. [DOI] [PubMed] [Google Scholar]
- Grover CE, et al. 2015. Molecular confirmation of species status for the allopolyploid cotton species, Gossypium ekmanianum Wittmack. Genet Resour Crop Evol. 62(1):103–114. [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. 2007. Combined networks regulating seed maturation. Trends Plant Sci. 12(7):294–300. [DOI] [PubMed] [Google Scholar]
- Hinze LL, et al. 2015. Nondestructive measurements of cotton seed nutritional trait diversity in the US national cotton germplasm collection. Crop Sci. 55(2):770–782. [Google Scholar]
- Horvath S, editor. 2011. Weighted network analysis: applications in genomics and systems biology. New York: Springer-Verlag. [Google Scholar]
- Horvath S, Dong J. 2008. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 4(8):e1000117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R, et al. 2015. A transcriptome profile for developing seed of polyploid cotton. Plant Genome 8(1):1–15. [DOI] [PubMed] [Google Scholar]
- Hu G, et al. 2011. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 189(3):1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, et al. 2014. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci U S A. 111(25):E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, et al. 2013. Comparative transcriptomic analysis of developing cotton cotyledons and embryo axis. PLoS One 8(8):e71756.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- Jones SI, Vodkin LO. 2013. Using RNA-Seq to profile soybean seed development from fertilization to maturity. PLoS One 8(3):e59270.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouser S, Mahmood K, A F. 2015. Variations in physicochemical attributes of seed oil among different varieties of cotton (Gossypium hirsutum. L). Pak J Bot. 47(2):723–729. [Google Scholar]
- Langfelder P, Horvath S. 2007. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 1:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Mischel PS, Horvath S. 2013. When Is hub gene selection better than standard meta-analysis? PLoS One 8(4):e61505.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24(5):719–720. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science 320(5875):481–483. [DOI] [PubMed] [Google Scholar]
- Lemmon ZH, Bukowski R, Sun Q, Doebley JF. 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet. 10(11):e1004745.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. Am Nat. 122(1):1–25. [Google Scholar]
- Li L, et al. 2015. A systems biology approach toward understanding seed composition in soybean. BMC Genomics 16 Suppl 3:S9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Singh S, Chapman K, Green A. 2009. Bridging traditional and molecular genetics in modifying cotton seed oil In: Paterson AH, editor. Genetics and Genomics of Cotton. New York: Springer; p. 353–384. [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, et al. 2016. The transcriptomic signature of developing soybean seeds reveals genetic basis of seed trait adaptation during domestication. Plant J. 86(6):530–544. [DOI] [PubMed] [Google Scholar]
- Madlung A, Wendel JF. 2013. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res. 140(2–4):270–285. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5(11):e13984.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien RD, Jones LA, King CC, Wakelyn PJ, Wan PJ. 2005. Cotton seed oil In: Shahidi F, editor. Bailey's Industrial Oil and Fat Products. Hoboken, New Jersey: John Wiley & Sons, Inc; p. 173–279. [Google Scholar]
- Page JT, Udall JA. 2015. Methods for mapping and categorization of DNA sequence reads from allopolyploid organisms. BMC Genet. 16 Suppl 2:S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, et al. 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492(7429):423–427. [DOI] [PubMed] [Google Scholar]
- Pfeifer M, et al. 2014. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345(6194):1250091.. [DOI] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF. 2009. Genomic expression dominance in allopolyploids. BMC Biol. 7:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. 2004.. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136(1):2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A, Aïnouche ML. 2015. Next-generation sequencing and the challenge of deciphering evolution of recent and highly polyploid genomes In: Hörandl E, Appelhans MS, editors. Next generation sequencing in plant systematics. Germany: Koeltz Scientific Books, Chapter3. [Google Scholar]
- Sekhon RS, et al. 2013. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8(4):e61005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, et al. 2010. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver TN, Dilday RH. 1982. Measurement of and correlations among selected seed quality factors for 36 Texas race stocks of cotton. Crop Sci. 22:779–781. [Google Scholar]
- Shi X, et al. 2012. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 3:950. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS. 2016. Polyploidy: Pitfalls and paths to a paradigm. Am J Bot. 103(7):1146–1166 [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1940. The significance of polyploidy in plant evolution. Am Nat 74(750):54–66. [Google Scholar]
- Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner R, et al. 2012. Reshaping of the maize transcriptome by domestication. Proc Natl Acad Sci U S A. 109(29):11878–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, et al. 2011. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68(6):1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. 2005. Molecular physiology of legume seed development. Annu Rev Plant Biol. 56:253–279. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Grover CE. 2015. Taxonomy and evolution of the cotton genus, Gossypium In: Fang DD, Percy RG, editors. Cotton. Madison, WI: American Society of Agronomy; p. 25–42. [Google Scholar]
- Wendel JF, Jackson SA, Meyers BC, Wing RA. 2016. Evolution of plant genome architecture. Genome Biol. 17:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, et al. 2014. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol Biol Evol. 31(5):1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AM, Horvath S. 2007. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE. 2014. Nonadditive gene expression in polyploids. Annu Rev Genet. 48:485–517. [DOI] [PubMed] [Google Scholar]
- Yoo MJ, Szadkowski E, Wendel JF. 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110(2):171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 4:Article17.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.