Abstract
Proprotein convertases are serine proteases responsible for the cleavage and subsequent activation of protein substrates, many of them relevant for the development of an ample variety of diseases. Seven of the PCs, including furin and PACE4, recognize and hydrolyze the C-terminal end of the general sequence RXRR/KXR, whereas PCSK-9 recognizes a series of non-basic amino acids. In some systems, PC-mediated substrate activation results in the development of pathological processes, such as cancer, endocrinopathies, and cardiovascular and infectious diseases.
After establishing PCs as relevant contributors to disease processes, research efforts were directed towards the development of inhibition strategies, including small and large molecules, anti-sense therapies, and antibody-based therapies. Most of these inhibitors mimic the consensus sequence of PCs, blocking the active site in a competitive manner. The most promising inhibitors were designed as bioengineered proteins; however, some non-protein and peptidomimetic agents has also proved to be effective.
These efforts led to the design of pre-clinical studies and clinical trials utilizing inhibitors to PCs. Although the initial studies were performed using non-selective PCs inhibitors, such as CMK, the search for more specific, and compartmentalized selective inhibitors resulted in specific activities ascribed to some, but not all of the PCs. For instance, PACE4 inhibitors were effective in decreasing prostate cancer cell proliferation, and neovascularization. Decreased metastatic ovarian cancer utilizing furin inhibitors represents one of the major endeavors, currently in a phase II trial stage. Antibodies targeting PCSK-9 decreased significantly the levels of HDL-cholesterol, in a phase III trial.
The study of Proprotein convertases has reached a stage of maturity. New strategies based on the alteration of their activity at the cellular and clinical level represent a promising experimental pharmacology field. The development of allosteric inhibitors, or specific agents directed against individual PCs is one of the challenges to be unraveled in the future.
Keywords: Proprotein convertases, tumor progression, molecular therapy
Chemical compounds cited in this article: Dec-RVKR-CMK (PubChem CID 9962075)
Graphical abstract
1. Introduction
Proprotein convertases subtilisin and kexin type (PCSK) constitute a group of nine Calcium-dependent serine proteases; PC1/2. Furin, PC4, PC5/6, PC7, and PACE4 [1], SKI-1[2], and PCSK9 [3]. All, but the last two PCs listed, recognize and cleave at the C-terminal end of the basic sequence RXR/KR. Several inactive protein precursors gain full activity after a specific PC-mediated proteolysis. These protein substrates include growth factors and growth factor receptors, metalloproteinases, clotting factors, viral proteins, and others. Increased levels of active proteins lead to the promotion of pathophysiological processes as diverse as cell proliferation, degradation of extracellular matrix, activation of the immune response, and activation of viral proteins that allow for efficient entry into the host cells, to mention just a few [4–6]. Table 1 summarizes the main characteristics, subcellular localization, and main pathophysiological processes associated to each of the PCs. The activation of these proteins may result in the development of disease, pointing to PCs as targets for therapeutic interventions. Several strategies aiming at inhibiting the activity of PCs, including the design of competitive inhibitors and interference with mRNA transcription, have been extensively investigated to abolish or minimize the proteolytic activation of these substrates [4, 7–9].
Table 1.
Proprotein convertases. The table depicts the main characteristics of the PCs, including the recognition and cleavage sequence, localization, and cognate pathophysiological processes.
| PC | Cleavage sequence | Subcellular localization | Tissue distribution | Proteins activated after PC cleavage | Pathophysiological processes | Ref. |
|---|---|---|---|---|---|---|
| PCSK-1 | (K/R)-(X)-(K/R) | Vesicles in neuroendocrine tissue | Neuroendocrine tissue, brain Pancreas, heart | Proenkephalin, Proopiomelanocortin | Dwarfism Obesity | [102] [103–107] |
| PCSK-2 | (K/R)-(X)-(K/R) | Vesicles | Neuroendocrine tissue, brain | Proopiomelanocortin Prodynorphin | Susceptibility to diabetes | [108] |
|
PCSK-3 Furin |
(K/R)-(X)-(K/R) | TGN/PM | Ubiquitous | Membrane-type metalloproteinases, IGF-1R | Cancer | |
| PCSK-4 | (K/R)-(X)-(K/R) | (acrosomal) PM | Epididymis Spermatocyte | ADAM-1, IGF-2 (?) | Infertility | [109, 110] |
| PCSK-5 | (K/R)-(X)-(K/R) | PCSK-5A: extracellular PCSK-5B: dense secretory granules | Heart, great blood vessels Kidneys, small intestine |
Pro-renin Integrin α subunit MT1-MMP | Cardiovascular disease | [111] |
| PCSK-6 | (K/R)-(X)-(K/R) | Extracellular | Ubiquitous | Similar to furin | Prostate cancer | [16, 17] |
| PCSK-7 | (K/R)-(X)-(K/R) | TGN/PM | Ubiquitous | Pro-EGF | Learning and memory impairments | [112, 113] |
|
PCSK-8 (SKI-1) |
(R/K)X(hydrophobic)Z | TGN/PM | Ubiquitous | Pro-neurotrophic derived factor SREBP-1 and 2 | Viral glycoproteins | [2, 114, 115] |
| PCSK-9 | LVFAQSIP | ER/endosomes | Hepatocytes | PCSK-9 targets LDLR to lysosomal degradation | Atherosclerosis | [32, 116] |
On the other hand, PCs seem to have minor differences in the cleavage preferences, and most of them can be equally efficient in the activations of their cognate substrates. Subtle differences among these proteases may be attributed to different tissue distribution, or subcellular locations [7, 10, 11]. In this context, PC-1 and PC-3 are restricted to neuroendocrine tissues, whereas PC4 is expressed almost exclusively in testes. However, the rest of the PCs, furin, PACE4, PC-5 and PC-7 demonstrated expression in virtually every tissue. Although PACE4 is fundamentally an extracellular proteins, furin, PC-5 and PC7 cycle between the endoplasmic reticulum and Golgi pointing to similar specificity and activity in analogous cellular and subcellular environments [9]. Although this statement may be valid in many cases, some substrates show a unique sensitivity to a specific PC. For instance, furin appears to be the only PCs that cleaves and activates TGF-β in vivo, supporting a non-redundant role for the PCs [12]. In addition, some PCs may show a differential expression in cancer or normal tissue. In fact, PACE4 is expressed predominantly in cells derived from the relatively normal ovarian surface epithelium, and silenced in ovarian neoplasia, whereas furin seems to follow the opposite pattern; highly expressed in tumors and tumor-derived cell lines, and relatively much less expressed in normal tissue and cell lines derived from them [13, 14]. Furthermore, PACE4 overexpression promotes the proliferation of prostate cancer cell lines in vitro and in vivo, whereas overexpression of other PCs do not seem to be similarly effective [15, 16].
Furin has emerged as the prototype of the family; however, several reports point to significant functions of PACE4 in prostate cancer [15, 16] and in the development of skin squamous cell carcinoma [17, 18] and musculoskeletal diseases [19] [20]. Hence, furin has been more extensively studied pointing to associations with several diseases, including cancer [6], inflammation [21, 22], cardiovascular disease [23–25] and viral infections [26], among others.
Furin is also essential for embryonic development. Knockdown of furin results in a lethal phenotype [1, 27–29]. Moreover, mouse embryos with the furin gene knockdown, specifically in endothelial cells, are born with several cardiovascular defects, including septal and valvular defects, attributable to impaired processing of TGF-β, adrenomedulin, endothelin, and bone morphogenetic protein 4; all of them furin substrates [30, 31]. On the other hand, furin knockouts in liver of adult mice are viable, pointing to certain degree of redundancy among PCs during adult life [11].
PCSK9 represents a unique case among the PCs since it does not cleave at the typical basic motif characteristic of the PCs in atherosclerosis. This PC cleaves and inactivates the low density lipoprotein receptor, LDLR, resulting in elevated blood LDL and VLDL [32]. Elevated levels of these lipoproteins are associated with hyperlipidemia, and the devastating effects of atherosclerosis. Inhibition of this PC resulted in increased levels of LDLR, with the concomitant decrease of plasma LDL and VLDL [33] [34]. These inhibitors have been the focus of successful clinical trials. At this stage, the first two PCSK9 inhibitors, alirocumab and evolocumab, have been in clinical use for lowering LDL-particle concentrations for cases in which classical statins and other drugs were not effective or badly tolerated [35, 36]. Recently, several excellent reviews have been published regarding the spectacular effects of PCSK9 inhibition in cardiovascular therapeutics. The reader is directed to these articles [32, 37–45]
2. Paralyzing the master switches: Inhibition of PCs
Several strategies have been studied to inhibit PCs in vitro and in vivo, from small molecule inhibitors to proteins and antibodies directed against these proteases. Furthermore, subtle differences in the amino acid sequences surrounding the active site have been exploited to increase specificity of an inhibitor towards a PC.
2.1 Small molecule inhibitors
Most of the small molecule inhibitors for the PCs block the accessibility of substrates to the catalytic pocket in a competitive manner. The structure of these inhibitors contain a tetrapeptide containing the furin cleavage site, that is, RXR/KR, or modifications thereof, conserving the crucial cationic nature of the PCs recognition site [46]. These short peptides may not offer specificity towards a particular PCs; however, they proved to bind the catalytic cleft with high affinity, with Ki in the micromolar or nanomolar range. Upon binding these inhibitors, in an induced-fit manner, both PC and inhibitor structure change their conformation, favoring the formation of an antiparallel beta sheets, characteristic of furin, PC5, PC7, and PACE4, pointing some degree of redundancy among these PCs [47]. However, differences in the amino acids surrounding the pocket where the substrate binds may facilitate the design of specific PC inhibitors.
2.1.1 Chloromethylketones
Chloromethylketone derivatives of the consensus sequence RVKR provided the first and strongest evidences of the tremendous potential of inhibiting PCs in terms of the PC mechanism of action, substrate processing, and potential clinical application [48]. The compound contains the recognition sequence for the PCs, an N-terminal decanoyl extension that increases its cell permeability, and a C-terminal chloromethyketone that blocks the possible in vivo protease-mediated degradation (figure 1A). The main caveat associated to these agents is their lack of specificity for any particular PC, and its toxicity at higher doses.
Figure 1.
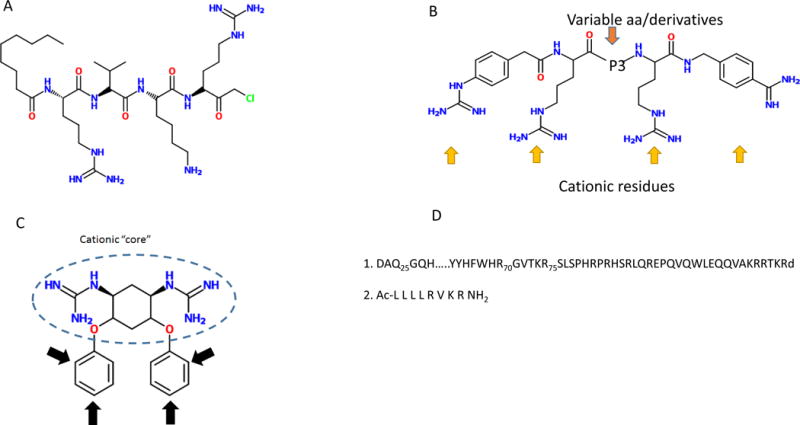
Structure of main furin inhibitors. (A) Decanolyl R-V-K-R chloromethyl ketone, (B) General structure of polyarginines (C) Guanidilated streptamine. (D) 1. Prodomains of furin, 2. Inhibitory peptide directed against PACE4
2.1.2 Poly-arginine derivatives
Poly-arginines represent an extension of one of the basic motif recognized by PCs; RRRR. One of the first inhibitors synthesized was the hexa-D-arginine, an effective inhibitor of inflammation in vitro and in vivo [49]. More than a decade ago, a slightly longer peptide, poly-D-nonaarginine (D9R) proved to be an efficient furin inhibitor (Ki in the order in the nanomolar range), and administration of this peptide protected cells against the anthrax toxin with an EC50 of 3.6 μM [50] [51]. This highly cationic peptide not only provides the sequence that binds to the active site of furin, but to the highly acidic motifs surrounding the active site. Although the delivery of this nonapeptide presents difficulties, it has provided the bases for potential modifications that led to promising newly formulated inhibitors [52] (figure 1B). More recently, cyclic peptides constructed using arginines and variable side chains demonstrated inhibitory activity against furin, combined with an enhanced ability for cell penetration [53].
2.1.3 Streptamine derivatives
As already stated, most of the PCs inhibitors are peptides or proteins. Although they are in general excellent inhibitors of PCs, the low turnover of peptides and proteins, their potential cytotoxicity, and the considerable molecular weight, (especially proteins), may limit their clinical use. Non-peptide inhibitors may circumvent these drawbacks, while still exerting a strong inhibitory effect on PCs. The elucidation of the crystal structure of furin has facilitated the modeling of these non-peptide inhibitors [54, 55]. Guanidinilated derivatives of 2-dideoxystreptamine, modeled using docking experiments and the crystal structure of furin as template, proved to be efficient (Ki in the nanomolar range) inhibitors of furin in vitro [56]. These derivatives mimic the cationic character of the PCs recognition site, and bind the active site of furin, hence, acting in a competitive manner (figure 1). These inhibitors seem to inhibit furin (and PC6B) with a ten-fold higher efficiency than PACE4 or PC7. In fact, guanidilated streptamine derivatives bind to PACE4 and PC7 with ten-fold and 100-fold lower efficiency, respectively(figure 1B) [55].
After the development of these derivatives to dideoxystreptamine, other groups developed the bisguanidinephenyl ethers derivatives of 2–5 dideoxystreptamine containing two guanidine residues [57]. These two positively charged guanidine group are attached to a phenyl group, respectively, and the guanidine phenyl moieties are linked by a three carbon bridge. This positive charge-bridge-positive charge structure resembles the minimal recognition site for the PCs-RXXR. In addition, the phenyl group increases the molecule’s hydrophobicity resulting in an enhanced penetration into the cell. The residues, bond by ether groups, confer extra chemical and biochemical stability (figure 1C).
Some of the bisguanidylated derivatives exhibit poor cell penetration, making them ideal for diseases that require a membrane-bound furin, which mainly catalyzes the cleavage of extracellular substrates, such as the anthrax toxin protective antigen. Variations in the positioning of the guanydil substituents in the aromatic group are localized to different intracellular compartments, such as endosomes and Golgi. As different substrates are putatively processed in different subcellular compartments, the selection of derivatives with a particular substitution pattern may affect the activation of different substrates, depending on the final destination of the substituted compound administered. In the future, these compounds may represent a breakthrough in PC.s, especially furin- inhibition, and may stimulate research in non-peptide PC inhibitors to increase the repertoire of drugs at our disposal.
2.2 Peptidomimetics
Small peptidomimetics combine the best of both worlds; small molecule and full-protein inhibitors. As small molecules, they exhibit better pharmacokinetic properties, better formulation, and delivery. As these compounds usually contain the PC recognition site embedded in a peptide moiety, they allow for specific interactions outside the binding pocket that are present in some, but not all of the PCs. These extra interactions strengthen the specific binding of the peptidomimetic to PC in a selective manner [58] (figure 1D).
Levesque et al (2012) [59] have synthesized a peptide containing the recognition sequence for PCs (RVKR) with a four Leucine residues extension at the N terminal end of this sequence (figure 1D). Although the binding site for furin and PACE4 are virtually identical, these investigators showed that some regions, specifically alpha helices 3 and 4, surrounding its catalytic site, contain a different array of amino acids. Furin exhibit in a highly anionic pocket between alpha helices 3 and 4 with an overall charge of −7, whereas this region is devoid of charge in PACE4. This lack of overall charge can be exploited to strengthen the inhibitor-PACE4 interaction. The four leucine residues extension provides a hydrophobic environment resulting in increased binding forces to PACE4. On the other hand, the higher hydrophobicity prohibits binding to furin. In fact, this inhibitor has proved to bind PACE4 with Ki in the nM range and about twenty times lower than furin, and is an efficient inhibitor of cancer cell proliferation [60, 61].
3. Work in progress: Peptides and proteins as models for the development of new therapeutic strategies
3.1 Pro-domain
PCs follow the secretory biosynthetic pathways, transiting from the endoplasmic reticulum to vesicles to the Golgi apparatus to the trans-Golgi compartment. PCs then cycle between the trans-Golgi network and the plasma membrane, transported back and forth through endocytic vesicles [62]. Hence, PCs exert their catalytic activity in these three compartments. PCs do not become activated until they reach the trans-Golgi network since the catalytic site is masked by the prosegment of the protein. After cleavage from the protein, the prosegment remains attached to the catalytic site, precluding substrate binding. In the trans-Golgi network, changes in pH favors the dissociation of the prosegment to the PC core, enabling the enzyme to exert its catalytic activities [62] (figure 1D). This approach has been used in several research studies [63] Furin prosegment, for instance decreased the activity of MMP-9 in breast cancer cells [64], inhibition of growth and invasiveness in head and neck cancer cell lines [65], prohepcidin maturation [66], and inhibition of the maturation of brain-derived neurotrophic factor [67]. Despite these promising studies, prosegments have not been evaluated in vivo, probably because full proteins display a similar or better inhibitory activity.
3.2 Alpha-1-antitrypsin and derivatives
Proteins provide not only the tetrapeptide domain that specifically binds PCs, but a scaffold that may provide specificity, increased affinity, and improved kinetic properties. Alpha1- antitrypsin, var Portland (PDX), is a bioengineered variant of antitrypsin containing the PCs recognition sequence. The original sequence of α1-antitrypsin contains the sequence AIPM r 358 and selectively inhibits the activity of elastase. The change AIPM to AIPR, named α1-antitrypsin Pittsburgh, not only lacks affinity to elastase, but originates a motif that serves as thrombin inhibitor. In fact, this variant has been discovered in a patient suffering from uncontrolled hemorrhages [68]. A bioengineered change in the sequence resulted in the motif RIPR 358 creating the minimal consensus sequence for PC binding [69]. This consensus sequence has minimal effect on the overall structure of protein, since it is in a loop that projects away from the core structure of the protein, a fact that has been exploited to inhibit extracellular substrates. This antitrypsin variant forms a stable, SDS and heat-resistant complex with PCs, especially furin, and PC6B, and to a lesser extent with PACE4, [70, 71]. Several in vitro and in vivo systems have employed PDX in order to decrease the activation of substrates relevant to cancer progression [72–74] viral infection and inflammation [75, 76].
PDX, a 50kDa protein, may establish multiple interactions with PCs besides those in the active site. In this context, some variants contain a recognition sequence that depart from the minimal consensus for PCs, and still binds efficiently to PCs. In this context, Hada et al, [77] demonstrated that the variant AVNR (instead of RIPR) binds furin with great selectivity, albeit with less efficiency. The selectivity displayed by these variants, that do not contain the minimal consensus for PCs, suggests that other important domains in the protein may contribute to the efficiency and selectivity of protein-based inhibitors. PDX has been a source of inspiration for many protein-based approaches.
4. Nanobodies: beyond competitive inhibition
PCs efficient cleavage relies not only on the integrity of the active site, but on the P-domain, located towards the C-terminal end of the protein, as shown in mutagenesis experiments [78]. Nanobodies, the variable region of the heavy chain of antibodies, VHH fragments, have been produced to target the P-domain of furin. The resulting antibody, Nb14, binds to the P-domain, without interfering with the activity of the catalytic domain, measured as binding and processing of small substrates [79]. Interestingly, binding of the nanobody Nb14 to furin precludes activation of full-length proteins, such as factor X. These somewhat puzzling results suggest that Nb14 may cover partially the catalytic site, preventing molecules with large molecular weights such as proteins to reach the binding site and be cleaved. In contrast, Nb14 binding to the P domain does not seem to disrupt catalytic activity, since small molecules, such as the fluorescent substrate 7-amino-4 methyl-coumarin, can be cleaved with both, furin or NB14-bound furin. Furthermore, Dec-RVKR-CMK binding to the catalytic site occurs regardless of the presence or not of Nb14.
The design and usage of a nanobody represents a novel finding in the field of PC inhibition. Nb14 binds to a regulatory region (P-region) within the molecule of furin rather than the catalytic site as most of the inhibitors developed so far. This non-competitive mode of inhibition represents a promising approach, and may stimulate the design of similar allosteric inhibitors with potential applications in the clinic. Furthermore, the mode of non-competitive action of Nb14 differs markedly from the mechanism of actions of other antibodies used to block the activity of other proteases, where these nanobodies bock the catalytic site, or induce a conformational change in the active site, preventing the binding of any substrate-large or small.
Nb14 is specifically recognizes furin P-domain; other PCs, such as PACE-4, PC1 and PC2 do not bind this nanobody. Cleavage of substrates that are specific for furin, such as hemagglutinin, or substrates that are cleaved with poor efficiency by other PCs, can be effectively blocked using nanobodies directed against furin. Further research may evaluate the effectiveness of this approach to treat pathologies associated to furin activity as well as to develop nanobodies specific for other PCs. Figure 2 provides a scheme of this therapeutic approach.
Figure 2.
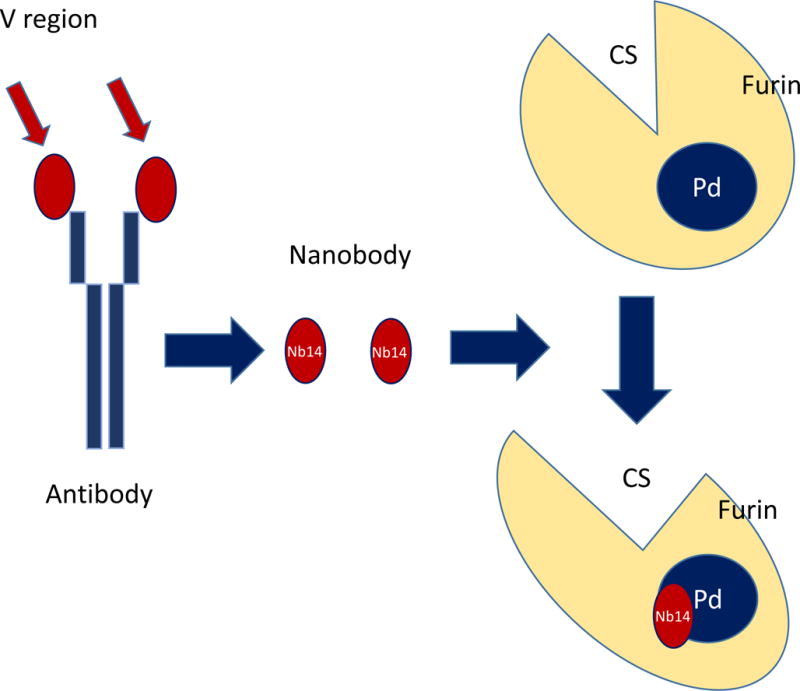
Furin nanobodies. Furin nanobodies consisting of the variable region of an antibody specific for the P-domain of furin, binds this regulatory region, inhibiting the binding of furin to physiological substrates. Note the change in shape in the catalytic site upon Nb14 binding to the P-site.
5. Furin silencing: strengthening the immune response
Carcinogenesis is characterized in part by the expression of certain genes and the consequent production of new proteins that enable tumor cells to successfully compete with normal cells, leading to their survival. One of the properties that tumor cells acquire is the ability to escape the normal immune mechanisms mounting strong immunosuppressive responses. Although tumor cells exhibit a repertoire of antigens not present in normal cells and tissues, these potentially immunogenic markers are not recognized by the immune system. One of the culprits for immunosuppression is the transforming growth factor beta- TGFβ, both variants 1 and 2. Many tumor cell lines overexpress this extracellular factor, resulting in an extra advantage for survival, especially since TGF-β inhibits the secretion of CD8+T cell-dependent Granulocyte-Macrophage Colony Stimulating Factor (GMCSF), and, consequently, the maturation of dendritic cells in the bone marrow [80]. Low levels of dendritic cells translate into a deficient antigen presentation, thwarting the possibility of immune-mediated elimination of cancer cells.
Furin has been identified as the main activator of TGF-β. This growth factor is synthesized in the rER, and after separation of the signal peptide, it transits through the endomembrane system where furin cleaves this protein at the C-terminal end of the basic sequence RHRR A279 [81]. Although the other PCs can process TGF-β, it has been shown that furin is the most efficient among the PCs as demonstrated by restoration of normal levels of TGF-β after incorporating the furin cDNA into the furin-deficient cells LoVo, and PDX-mediated inhibition studies [12]. Furin processing is indispensable for activity since engineered mutants in the PC cleavage sequence (RHRR) produced secreted proteins, albeit inactive, which can act as dominant negative [82]. Hence, blocking furin activity may impair the activation of TGF-β and lead to decreased immune responses and successful elimination of tumor cells.
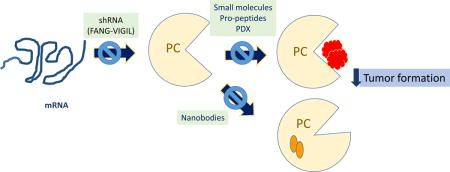
This approach has been investigated and seems to be promising in the various malignancies. The cDNA from GMCSF and a bidirectional small hairpin RNA interference targeting furin [83]were placed under the control of the strong cytomegalovirus promoter. This construct express elevated GMCSF stimulating the immune response and shRNAi for furin that silence this PC. When transfected to autologous cancer cells, this, these cells produced GMCSF, and efficiently silenced furin. Lack of furin expression led to impairment of TGF-β activation. These transfected, patient-derived tumor cells, were used as cellular vaccines (formerly named FANG, and now called VIGIL in the clinical trials) and reinjected in patients. These tumor cells were unable to effect immunosuppression, hence, becoming sensitive to GMCSF-derived responses. Elevated levels of GMCSF also resulted in increased activation of bone marrow-derived dendritic cells, facilitating the elimination of tumor cells. In a phase I trial, Nemunaitis et al [84–86] produced vaccines from a wide variety of tumor sources, including, small cell lung cancer, breast cancer, colon cancer, liposarcoma, and ovarian cancer [84] This approach is still underway, but anticipates positive outcomes in decreasing the relapse in patients diagnosed with hepatocellular carcinoma [84], metastatic advanced Ewing’s sarcoma [87, 88], advanced stage ovarian cancer [89].
These vaccines represent a safe therapy. Overall these vaccines point to effective treatments of advanced tumors, which frequently relapse, with poorer outcomes with successive relapses. The phase II clinical trial for advanced ovarian cancer showed a significant (P= 0.033) increase in relapse free survival from 481 days (control) to 826 days (VIGIL treatment). Few toxic effects have been reported, most of them at the injection site, including erythema, induration, pain, pruritus, swelling, and tenderness. Other effects might be associated to the therapy itself, such as joint function, back pain, and fatigue; however no definite association between treatment and these effects can be drawn with certainty.
6. Activating prodrugs at the right target: Taking advantage of the PCs cleavage sequence
Specific targeting of tumor cells poses a problem that has required creative approaches. Most drugs, small synthetic drugs or biologicals, are delivered systemically. Drugs may be toxic to normal cells and display unacceptable toxic effects. Prodrugs have provided a partial response to this (and other) problems however, the activation of the prodrug may not be achieved in the target cells. In response to this difficulty, researchers have focused on furin as a potential prodrug activating enzyme [90]. An early approach consisted to deliver the Tumor necrosis factor related apoptosis induced ligand (TRAIL) specific to cancer cells, avoiding TRAIL’s toxic effects on normal hepatocytes [91]. The soluble N-terminal domain of TRAIL was fused to the extracellular domain of a Fms-like tyrosine kinase (hFlex), a highly immunogenic peptide. Between these two genes, a DNA fragment containing the sequence for PCs cleavage was inserted in phase. Transfection of several cancer cell lines with this fusion plasmid (hFlex-furin sequence-soluble TRAIL) resulted in induction of apoptosis and cytolysis. Similar results were observed in vivo, after intratumoral treatment of subcutaneous tumors with this plasmid.
Other approaches to specifically deliver toxins to tumor cells have employed fusion proteins that include a peptide containing the PCs cleavage sequence acting as a linker between two structurally and functionally different proteins [92, 93] One of the most popular therapies using fusion proteins consisted to a fragment of the variable chain of the HER-2 antibody (e23sFv) fused to an internalization sequence, such as the Pseudomonas endotoxin A(PEA), linked to a proapoptotic or toxic proteins though a peptide containing the PCs cleavage sequence [93]. Several tumors, including up to 30% of breast cancers, colon, and lung cancers express the protein HER-2 [94–97] which is localized to the plasma membrane. The easy access to this protein facilitates the binding of the variable chain of HER-2 antibody tethering the fusion protein specifically to cancer cells expressing HER-2. When fused to the translocation domain of the Pseudomonas endotoxin A (PEA), the fusion protein containing the e23sFv fragment internalizes to the endomembrane system [98–100]. Once in the endomembrane system, the fusion protein is cleaved, releasing the proapoptotic or toxic protein. One of the pro-apopototic proteins fused to the HER-2, tBID (truncate BH3 interacting domain death agonist) was used in many cells and animal systems. This protein is transported to the mitochondria and stimulates the release of proapototic proteins such as Smac/DIABLO and AIF [101, 102], specifically eliminating cancer cells.
7. Conclusion
PCs play an important role in several physiological and pathological processes from embryonic development to cancer, viral infections to inflammation, apoptosis to angiogenesis. Although some PCs exhibit a restrictive tissue distribution, PC1 and PC2 in neuroendocrine tissues, and PC4 in testis, others, like furin, PACE4, PC5, and PC7 are ubiquitously expressed. The latter PCs are overexpressed in a series of malignancies. In addition, PCs are crucial for some virus to infect host cells, bacterial pathogenicity, and cardiovascular diseases. The development of PC inhibitors has shown great potential to cure or improve certain pathological conditions. Newer approaches are being continuously developed. Several clinical trials are currently being conducted, or initiated (table 2). It is hoped that future inhibitors may surpass the effectiveness of the ones already at hand. The possibility to mitigate and eliminate some of the most common lethal diseases of mankind, such as cancer, with the use of PC inhibitors and other anticancer approaches seems quite realistic at this time.
Table 2.
Ongoing clinical trials designed to evaluate therapies based on furin inhibition.
| Clinical trial | PC targeted | Target | Phase | NCT number |
|---|---|---|---|---|
| Bi: shRNA-Furin GMCSF | Furin (PCSK-3) | Ewin Sarcoma Non small lung cancer Liver cancer | I | 01061840 |
| FANG™ vaccine + Carboplatinum | Furin (PCSK-3) | Stage III and stage IV Ovarian cancer | II | 01867086 |
| FANG™ vaccine + Bevacizumab | Furin (PCSK-3) | Stage III and stage IV Ovarian cancer | II | 0155145 |
| FANG™ vaccine | Furin (PCSK-3) | Advanced melanoma | II | 01453361 |
| VIGIL + atezolizumab | Furin (PCSK-3) | Gynecological cancers | II | 03073525 |
Abbreviations
- AIF
Apoptosis-inducing factor
- cDNA
complementary DNA
- DIABLO
Direct IAP binding protein with low pI
- EGF
Epidermal growth factor
- GMCSF
Granulocyte Macrophage colony-stimulating factor
- HER-2
Human epidermal growth factor receptor 2
- IAP
Inhibitor of apoptosis proteins
- LDL
Low density lipoprotein
- LDLRL
Low-density lipoprotein receptor
- MMP-9
Matrix metalloproteinase 9
- MT1-MMP
Membrane Type 1 metalloproteinase
- PEA
Pseudomonas endotoxin A
- PC
Proprotein convertases
- PCSK
Proprotein convertases subtilisin and kexin type
- PDX
Alpha1- antitrypsin, var Portland
- PM
Plasma membrane
- shRNAi
Short hairpin interference ribonucleic acid
- Smac
Second mitochondria-derived activator of caspases
- SREBP-1
Sterol regulatory element binding protein 1
- TGN
Trans Golgi network
- TGF-β
Transforming growth factor beta
- tBID
Truncated BH3 interacting domain death agonist
- TRAIL
Tumor necrosis factor related apoptosis induced ligand
- VLDL
Very low density lipoproteins
- VHH
Variable region of the heavy chain of antibodies
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nature reviews. Mol Cell Biol. 2002;3(10):753–66. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidah NG, Mowla SJ, Hamelin J, Mamarbachi AM, Benjannet S, Toure BB, et al. Mammalian subtilisin/kexin isozyme SKI-1: A widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. PNAS. 1999;96(4):1321–6. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. PNAS. 2003;100(3):928–33. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–83. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 5.Seidah NG, Sadr MS, Chretien M, Mbikay M. The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J Biol Chem. 2013;288(30):21473–81. doi: 10.1074/jbc.R113.481549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassi DE, Fu J, Lopez de Cicco R, Klein-Szanto AJ. Proprotein convertases: “master switches” in the regulation of tumor growth and progression. Mol Carcinog. 2005;44(3):151–61. doi: 10.1002/mc.20134. [DOI] [PubMed] [Google Scholar]
- 7.Taylor NA, Van De Ven WJ, Creemers JW. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 2003;7(10):1215–27. doi: 10.1096/fj.02-0831rev. [DOI] [PubMed] [Google Scholar]
- 8.Basak A. Inhibitors of proprotein convertases. J Mol Med (Berlin, Germany) 2005;83(11):844–55. doi: 10.1007/s00109-005-0710-0. [DOI] [PubMed] [Google Scholar]
- 9.Couture F, D’Anjou F, Day R. On the cutting edge of proprotein convertase pharmacology: from molecular concepts to clinical applications. Biomolecular concepts. 2011;2(5):421–438. doi: 10.1515/bmc.2011.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creemers JW, Khatib AM. Knock-out mouse models of proprotein convertases: unique functions or redundancy? Frontiers in bioscience: a journal and virtual library. 2008;13:4960–71. doi: 10.2741/3055. [DOI] [PubMed] [Google Scholar]
- 11.Roebroek AJ, Taylor NA, Louagie E, Pauli I, Smeijers L, Snellinx A, et al. Limited redundancy of the proprotein convertase furin in mouse liver. J Biol Chem. 2004;279(51):53442–50. doi: 10.1074/jbc.M407152200. [DOI] [PubMed] [Google Scholar]
- 12.Dubois CM, Blanchette F, Laprise MH, Leduc R, Grondin F, Seidah NG. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am J Pathol. 2001;158(1):305–16. doi: 10.1016/s0002-9440(10)63970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Campbell EJ, Shepherd TG, Nachtigal MW. Epigenetic regulation of proprotein convertase PACE4 gene expression in human ovarian cancer cells. Mol cancer Res: MCR. 2003;1(8):569–76. [PubMed] [Google Scholar]
- 14.Page RE, Klein-Szanto AJ, Litwin S, Nicolas E, Al-Jumaily R, Alexander P, et al. Increased expression of the pro-protein convertase furin predicts decreased survival in ovarian cancer. Cell Oncol. 2007;29(4):289–99. doi: 10.1155/2007/930321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Anjou F, Routhier S, Perreault JP, Latil A, Bonnel D, Fournier I, et al. Molecular Validation of PACE4 as a Target in Prostate Cancer. Translational oncology. 2011;4(3):157–72. doi: 10.1593/tlo.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couture F, D’Anjou F, Desjardins R, Boudreau F, Day R. Role of proprotein convertases in prostate cancer progression. Neoplasia (New York, NY) 2012;14(11):1032–42. doi: 10.1593/neo.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassi DE, Cenna J, Zhang J, Cukierman E, Klein-Szanto AJ. Enhanced aggressiveness of benzopyrene-induced squamous carcinomas in transgenic mice overexpressing the proprotein convertase PACE4 (PCSK6) Mol Carcinog. 2015;54(10):1122–31. doi: 10.1002/mc.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassi DE, Zhang J, Cenna J, Litwin S, Cukierman E, Klein-Szanto AJ. Proprotein convertase inhibition results in decreased skin cell proliferation, tumorigenesis, and metastasis. Neoplasia (New York, NY) 2010;12(7):516–26. doi: 10.1593/neo.92030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malfait AM, Arner EC, Song RH, Alston JT, Markosyan S, Staten N, et al. Proprotein convertase activation of aggrecanases in cartilage in situ. Arch Biochem Biophys. 2008;478(1):43–51. doi: 10.1016/j.abb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Malfait AM, Seymour AB, Gao F, Tortorella MD, Le Graverand-Gastineau MP, Wood LS, et al. A role for PACE4 in osteoarthritis pain: evidence from human genetic association and null mutant phenotype. Ann Rheum Dis. 2012;71(6):1042–8. doi: 10.1136/annrheumdis-2011-200300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordova ZM, Gronholm A, Kytola V, Taverniti V, Hamalainen S, Aittomaki S, et al. Myeloid cell expressed proprotein convertase FURIN attenuates inflammation. Oncotarget. 2016;7(34):54392–54404. doi: 10.18632/oncotarget.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz J, Broder C, Helmstetter A, Schmidt S, Yan I, Muller M, et al. Short-term TNFalpha shedding is independent of cytoplasmic phosphorylation or furin cleavage of ADAM17. BBA. 2013;1833(12):3355–67. doi: 10.1016/j.bbamcr.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Ichiki T, Burnett JC., Jr Post-transcriptional modification of pro-BNP in heart failure: is glycosylation and circulating furin key for cardiovascular homeostasis? Eur Heart J. 2014;35(43):3001–3. doi: 10.1093/eurheartj/ehu381. [DOI] [PubMed] [Google Scholar]
- 24.Lei X, Basu D, Li Z, Zhang M, Rudic RD, Jiang XC, et al. Hepatic overexpression of the prodomain of furin lessens progression of atherosclerosis and reduces vascular remodeling in response to injury. Atherosclerosis. 2014;236(1):121–30. doi: 10.1016/j.atherosclerosis.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turpeinen H, Seppala I, Lyytikainen LP, Raitoharju E, Hutri-Kahonen N, Levula M, et al. A genome-wide expression quantitative trait loci analysis of proprotein convertase subtilisin/kexin enzymes identifies a novel regulatory gene variant for FURIN expression and blood pressure. 2015;134(6):627–36. doi: 10.1007/s00439-015-1546-5. [DOI] [PubMed] [Google Scholar]
- 26.Day PM, Schiller JT. The role of furin in papillomavirus infection. Future microbiology. 2009;4(10):1255–62. doi: 10.2217/fmb.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, et al. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development (Cambridge, England) 1998;125(24):4863–76. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 28.Reichhart JM. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15(12):659–65. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.van Tetering G, Vooijs M. Proteolytic cleavage of Notch: “HIT and RUN”. Curr Mol Med. 2011;11(4):255–69. doi: 10.2174/156652411795677972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W, Essalmani R, Szumska D, Creemers JW, Roebroek AJ, D’Orleans-Juste P, et al. Loss of endothelial furin leads to cardiac malformation and early postnatal death. Mol Cell Biol. 2012;32(17):3382–91. doi: 10.1128/MCB.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez de Cicco R, Watson JC, Bassi DE, Litwin S, Klein-Szanto AJ. Simultaneous expression of furin and vascular endothelial growth factor in human oral tongue squamous cell carcinoma progression. Clin Cancer Res. 2004;10(13):4480–8. doi: 10.1158/1078-0432.CCR-03-0670. [DOI] [PubMed] [Google Scholar]
- 32.Seidah NG, Abifadel M, Prost S, Boileau C, Prat A. The Proprotein Convertases in Hypercholesterolemia and Cardiovascular Diseases: Emphasis on Proprotein Convertase Subtilisin/Kexin 9. Pharmacol Rev. 2017;69(1):33–52. doi: 10.1124/pr.116.012989. [DOI] [PubMed] [Google Scholar]
- 33.Elbitar S, Khoury PE, Ghaleb Y, Rabes JP, Varret M, Seidah NG, et al. Proprotein convertase subtilisin / kexin 9 (PCSK9) inhibitors and the future of dyslipidemia therapy: an updated patent review (2011–2015) Expert Opin Ther Pat. 2016;26(12):1377–1392. doi: 10.1080/13543776.2016.1206080. [DOI] [PubMed] [Google Scholar]
- 34.Mazhar F, Haider N. Proprotein convertase subtilisin/kexin type 9 enzyme inhibitors: An emerging new therapeutic option for the treatment of dyslipidemia. J Pharmacol Pharmacother. 2016;7(4):190–193. doi: 10.4103/0976-500X.195906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito MK, Santos RD. PCSK9 Inhibition With Monoclonal Antibodies: Modern Management of Hypercholesterolemia. J Clin Pharmacol. 2017;57(1):7–32. doi: 10.1002/jcph.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natarajan P, Kathiresan S. PCSK9 Inhibitors. Cell. 2016;165(5):1037. doi: 10.1016/j.cell.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Alkindi M, Siminovitch KA, Gupta M, Genest J. Monoclonal Antibodies for the Treatment of Hypercholesterolemia: Targeting PCSK9. Can J Cardiol. 2016;32(12):1552–1560. doi: 10.1016/j.cjca.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Blom DJ, Dent R, Castro RC, Toth PP. PCSK9 inhibition in the management of hyperlipidemia: focus on evolocumab. Vasc Health Risk Manag. 2016;12:185–97. doi: 10.2147/VHRM.S102564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon DL, Trankle C, Buckley L, Parod E, Carbone S, Van Tassell BW, et al. A review of PCSK9 inhibition and its effects beyond LDL receptors. J Clin Lipidol. 2016;10(5):1073–80. doi: 10.1016/j.jacl.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Page MM, Watts GF. PCSK9 inhibitors – mechanisms of action. Australian prescriber. 2016;39(5):164–167. doi: 10.18773/austprescr.2016.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng W, Qiang F, Peng W, Qian Z, Ke Z, Yi L, et al. Therapeutic efficacy of PCSK9 monoclonal antibodies in statin-nonresponsive patients with hypercholesterolemia and dyslipidemia: A systematic review and meta-analysis. Int J Cardiol. 2016;222:119–29. doi: 10.1016/j.ijcard.2016.07.239. [DOI] [PubMed] [Google Scholar]
- 42.Rallidis LS, Lekakis J. PCSK9 inhibition as an emerging lipid lowering therapy: Unanswered questions. Hellenic J Cardiol. 2016;57(2):86–91. doi: 10.1016/j.hjc.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Robinson JG. Nonstatins and Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Inhibitors: Role in Non-Familial Hypercholesterolemia. Prog Cardiovasc Dis. 2016;59(2):165–171. doi: 10.1016/j.pcad.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Schmidli R. PCSK9 inhibitors – clinical applications. Australian prescriber. 2016;39(5):168–170. doi: 10.18773/austprescr.2016.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White K, Mohan C, Rocco M. PCSK9 inhibition: A promise fulfilled? Cleve Clin J Med. 2016;83(11 Suppl 2):S36–s44. doi: 10.3949/ccjm.83.s2.05. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Vallejo F, Martinez-Mayorga K. Furin inhibitors: importance of the positive formal charge and beyond. Bioorg Med Chem Lett. 2012;20(14):4462–71. doi: 10.1016/j.bmc.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 47.Tian S, Jianhua W. Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors. Int J Biol Sci. 2010;6(1):89–95. doi: 10.7150/ijbs.6.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angliker H. Synthesis of tight binding inhibitors and their action on the proprotein-processing enzyme furin. J Med Chem. 1995;38(20):4014–8. doi: 10.1021/jm00020a016. [DOI] [PubMed] [Google Scholar]
- 49.Sarac MS, Cameron A, Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect Immun. 2002;70(12):7136–9. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kacprzak MM, Peinado JR, Than ME, Appel J, Henrich S, Lipkind G, Houghten RA, Bode W, Lindberg I. Inhibition of furin by polyarginine-containing peptides: nanomolar inhibition by nona-D-arginine. J Biol Chem. 2004;279(35):36788–94. doi: 10.1074/jbc.M400484200. [DOI] [PubMed] [Google Scholar]
- 51.Cameron A, Appel J, Houghten RA, Lindberg I. Polyarginines are potent furin inhibitors. J Biol Chem. 2000;275(47):36741–9. doi: 10.1074/jbc.M003848200. [DOI] [PubMed] [Google Scholar]
- 52.Fugere M, Appel J, Houghten RA, Lindberg I, Day R. Short polybasic peptide sequences are potent inhibitors of PC5/6 and PC7: Use of positional scanning-synthetic peptide combinatorial libraries as a tool for the optimization of inhibitory sequences. Mol Pharmacol. 2007;71(1):323–32. doi: 10.1124/mol.106.027946. [DOI] [PubMed] [Google Scholar]
- 53.Ramos-Molina B, Lick AN, Nasrolahi Shirazi A, Oh D, Tiwari R, El-Sayed NS, et al. Cationic Cell-Penetrating Peptides Are Potent Furin Inhibitors. PloS one. 2015;10(6):e0130417. doi: 10.1371/journal.pone.0130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahms SO, Arciniega M, Steinmetzer T, Huber R, Than ME. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. PNAS. 2016;113(40):11196–11201. doi: 10.1073/pnas.1613630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J Mol Biol. 2005;345(2):211–27. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 56.Jiao GS, Cregar L, Wang J, Millis SZ, Tang C, O’Malley S, et al. Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. PNAS. 2006;103(52):19707–12. doi: 10.1073/pnas.0606555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramos-Molina B, Lick AN, Blanco EH, Posada-Salgado JA, Martinez-Mayorga K, Johnson AT, et al. Identification of potent and compartment-selective small molecule furin inhibitors using cell-based assays. Biochem Pharmacol. 2015;96(2):107–18. doi: 10.1016/j.bcp.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker GL, Lu Y, Hardes K, Strehlow B, Levesque C, Lindberg I, et al. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J Biol Chem. 2012;287(26):21992–2003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levesque C, Fugere M, Kwiatkowska A, Couture F, Desjardins R, Routhier SP, et al. The Multi-Leu peptide inhibitor discriminates between PACE4 and furin and exhibits antiproliferative effects on prostate cancer cells. J Med Chem. 2012;55(23):10501–11. doi: 10.1021/jm3011178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwiatkowska A, Couture F, Levesque C, Ly K, Beauchemin S, Desjardins R, Neugebauer W, et al. Novel Insights into Structure-Activity Relationships of N-Terminally Modified PACE4 Inhibitors. ChemMedChem. 2016;11(3):289–301. doi: 10.1002/cmdc.201500532. [DOI] [PubMed] [Google Scholar]
- 61.Levesque C, Couture F, Kwiatkowska A, Desjardins R, Guerin B, Neugebauer WA, et al. PACE4 inhibitors and their peptidomimetic analogs block prostate cancer tumor progression through quiescence induction, increased apoptosis and impaired neovascularisation. Oncotarget. 2015;6(6):3680–93. doi: 10.18632/oncotarget.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molloy SS, Thomas L, Kamibayashi C, Mumby MC, Thomas G. Regulation of endosome sorting by a specific PP2A isoform. J Cell Biol. 1998;142(6):1399–411. doi: 10.1083/jcb.142.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong M, Munzer JS, Basak A, Benjannet S, Mowla SJ, Decroly E, et al. The prosegments of furin and PC7 as potent inhibitors of proprotein convertases. In vitro and ex vivo assessment of their efficacy and selectivity. J Biol Chem. 1999;274(48):33913–20. doi: 10.1074/jbc.274.48.33913. [DOI] [PubMed] [Google Scholar]
- 64.Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, et al. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells’ malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res. 2007;67(19):9030–4. doi: 10.1158/0008-5472.CAN-07-0807. [DOI] [PubMed] [Google Scholar]
- 65.Lopez de Cicco R, Bassi DE, Zucker S, Seidah NG, Klein-Szanto AJ. Human carcinoma cell growth and invasiveness is impaired by the propeptide of the ubiquitous proprotein convertase furin. Cancer Res. 2005;65(10):4162–71. doi: 10.1158/0008-5472.CAN-04-2820. [DOI] [PubMed] [Google Scholar]
- 66.Scamuffa N, Basak A, Lalou C, Wargnier A, Marcinkiewicz J, Siegfried G, et al. Regulation of prohepcidin processing and activity by the subtilisin-like proprotein convertases Furin, PC5, PACE4 and PC7. Gut. 2008;57(11):1573–82. doi: 10.1136/gut.2007.141812. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Zhang J, Deng M. Furin mediates brain-derived neurotrophic factor upregulation in cultured rat astrocytes exposed to oxygen-glucose deprivation. J Neurosci research. 2015;93(1):189–94. doi: 10.1002/jnr.23455. [DOI] [PubMed] [Google Scholar]
- 68.Owen MC, Brennan SO, Lewis JH, Carrell RW. Mutation of antitrypsin to antithrombin. alpha 1-antitrypsin Pittsburgh (358 Met leads to Arg), a fatal bleeding disorder. NEJM. 1983;309(12):694–8. doi: 10.1056/NEJM198309223091203. [DOI] [PubMed] [Google Scholar]
- 69.Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason AJ, et al. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. PNAS. 1998;95(13):7293–8. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dufour EK, Denault JB, Hopkins PC, Leduc R. Serpin-like properties of alpha1-antitrypsin Portland towards furin convertase. FEBS letters. 1998;426(1):41–6. doi: 10.1016/s0014-5793(98)00307-x. [DOI] [PubMed] [Google Scholar]
- 71.Tsuji A, Hashimoto E, Ikoma T, Taniguchi T, Mori K, Nagahama M, et al. Inactivation of proprotein convertase, PACE4, by alpha1-antitrypsin Portland (alpha1-PDX), a blocker of proteolytic activation of bone morphogenetic protein during embryogenesis: evidence that PACE4 is able to form an SDS-stable acyl intermediate with alpha1-PDX. J Biochem. 1999;126(3):591–603. doi: 10.1093/oxfordjournals.jbchem.a022491. [DOI] [PubMed] [Google Scholar]
- 72.Bassi DE, Lopez De Cicco R, Mahloogi H, Zucker S, Thomas G, Klein-Szanto AJ. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. PNAS. 2001;98(18):10326–31. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Cicco RL, Bassi DE, Benavides F, Conti CJ, Klein-Szanto AJ. Inhibition of proprotein convertases: approaches to block squamous carcinoma development and progression. Mol Carcinogen. 2007;46(8):654–9. doi: 10.1002/mc.20331. [DOI] [PubMed] [Google Scholar]
- 74.Scamuffa N, Siegfried G, Bontemps Y, Ma L, Basak A, Cherel G, et al. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest. 2008;118(1):352–63. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson ED, Thomas L, Hayflick JS, Thomas G. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J Biol Chem. 1993;268(33):24887–91. [PubMed] [Google Scholar]
- 76.Jean F, Thomas L, Molloy SS, Liu G, Jarvis MA, Nelson JA, et al. A protein-based therapeutic for human cytomegalovirus infection. PNAS. 2000;97(6):2864–9. doi: 10.1073/pnas.050504297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hada K, Isshiki K, Matsuda S, Yuasa K, Tsuji A. Engineering of alpha1-antitrypsin variants with improved specificity for the proprotein convertase furin using site-directed random mutagenesis. Protein Eng Des Sel. 2013;26(2):123–31. doi: 10.1093/protein/gzs091. [DOI] [PubMed] [Google Scholar]
- 78.Dahms SO, Creemers JW, Schaub Y, Bourenkov GP, Zogg T, Brandstetter H, et al. The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers. Sci Rep. 2016;6:34303. doi: 10.1038/srep34303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu J, Declercq J, Roucourt B, Ghassabeh GH, Meulemans S, Kinne J, et al. Generation and characterization of non-competitive furin-inhibiting nanobodies. Biochem J. 2012;448(1):73–82. doi: 10.1042/BJ20120537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamaguchi Y, Tsumura H, Miwa M, Inaba K. Contrasting effects of TGF-beta 1 and TNF-alpha on the development of dendritic cells from progenitors in mouse bone marrow. Stem Cells. 1997;15(2):144–53. doi: 10.1002/stem.150144. [DOI] [PubMed] [Google Scholar]
- 81.Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor beta to the mature polypeptide. Mol Cell Biol. 1988;8(10):4162–8. doi: 10.1128/mcb.8.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osada SI, Wright CV. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development (Cambridge, England) 1999;126(14):3229–40. doi: 10.1242/dev.126.14.3229. [DOI] [PubMed] [Google Scholar]
- 83.Rao DD, Maples PB, Senzer N, Kumar P, Wang Z, Pappen BO, et al. Enhanced target gene knockdown by a bifunctional shRNA: a novel approach of RNA interference. Cancer Gene Ther. 2010;17(11):780–91. doi: 10.1038/cgt.2010.35. [DOI] [PubMed] [Google Scholar]
- 84.Nemunaitis J, Barve M, Orr D, Kuhn J, Magee M, Lamont J, et al. Summary of bi-shRNA/GM-CSF augmented autologous tumor cell immunotherapy (FANG) in advanced cancer of the liver. Oncology. 2014;87(1):21–9. doi: 10.1159/000360993. [DOI] [PubMed] [Google Scholar]
- 85.Senzer N, Barve M, Kuhn J, Melnyk A, Beitsch P, Lazar M, et al. Phase I Trial of “bi-shRNAifurin/GMCSF DNA/Autologous Tumor Cell” Vaccine (FANG) in Advanced Cancer. Mol Ther. 2012;20(3):679–686. doi: 10.1038/mt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghisoli M, Barve M, Mennel R, Lenarsky C, Horvath S, Wallraven G, et al. Three-year Follow up of GMCSF/bi-shRNA(furin) DNA-transfected Autologous Tumor Immunotherapy (Vigil) in Metastatic Advanced Ewing’s Sarcoma. Mol Ther. 2016;24(8):1478–83. doi: 10.1038/mt.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghisoli M, Barve M, Schneider R, Mennel R, Lenarsky C, Wallraven G, et al. Pilot Trial of FANG Immunotherapy in Ewing’s Sarcoma. Mol Ther. 2015;23(6):1103–9. doi: 10.1038/mt.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu X, Hui KM. Induction of potent TRAIL-mediated tumoricidal activity by hFLEX/Furin/TRAIL recombinant DNA construct. Mol Ther. 2004;9(5):674–81. doi: 10.1016/j.ymthe.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 91.Armeanu S, Lauer UM, Smirnow I, Schenk M, Weiss TS, Gregor M, et al. Adenoviral gene transfer of tumor necrosis factor-related apoptosis-inducing ligand overcomes an impaired response of hepatoma cells but causes severe apoptosis in primary human hepatocytes. Cancer Res. 2003;63(10):2369–72. [PubMed] [Google Scholar]
- 92.Shan LQ, Ma S, Qiu XC, Wang T, Yu SB, Ma BA, et al. A novel recombinant immuno-tBid with a furin site effectively suppresses the growth of HER2-positive osteosarcoma cells in vitro. Oncology Rep. 2011;25(2):325–31. doi: 10.3892/or.2010.1074. [DOI] [PubMed] [Google Scholar]
- 93.Wang T, Zhao J, Ren JL, Zhang L, Wen WH, Zhang R, et al. Recombinant immunoproapoptotic proteins with furin site can translocate and kill HER2-positive cancer cells. Cancer Res. 2007;67(24):11830–9. doi: 10.1158/0008-5472.CAN-07-1160. [DOI] [PubMed] [Google Scholar]
- 94.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science (New York, NY) 1989;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 95.Kapitanovic S, Radosevic S, Kapitanovic M, Andelinovic S, Ferencic Z, Tavassoli M, et al. The expression of p185(HER-2/neu) correlates with the stage of disease and survival in colorectal cancer. Gastroenterology. 1997;112(4):1103–13. doi: 10.1016/s0016-5085(97)70120-3. [DOI] [PubMed] [Google Scholar]
- 96.Osman I, Mikhail M, Shuch B, Clute M, Cheli CD, Ghani F, et al. Serum levels of shed Her2/neu protein in men with prostate cancer correlate with disease progression. J Urol. 2005;174(6):2174–7. doi: 10.1097/01.ju.0000181205.23233.65. [DOI] [PubMed] [Google Scholar]
- 97.Turken O, Kunter E, Cermik H, Isitmangil T, Kandemir G, Yaylaci M, et al. Prevalence and prognostic value of c-erbB2 expression in non-small cell lung cancer (NSCLC) Neoplasma. 2003;50(4):257–61. [PubMed] [Google Scholar]
- 98.Jia LT, Zhang LH, Yu CJ, Zhao J, Xu YM, Gui JH, et al. Specific tumoricidal activity of a secreted proapoptotic protein consisting of HER2 antibody and constitutively active caspase-3. Cancer Res. 2003;63(12):3257–62. [PubMed] [Google Scholar]
- 99.Wang SY, Chen B, Zhan YQ, Xu WX, Li CY, Yang RF, et al. SU5416 is a potent inhibitor of hepatocyte growth factor receptor (c-Met) and blocks HGF-induced invasiveness of human HepG2 hepatoma cells. J Hepatol. 2004;41(2):267–73. doi: 10.1016/j.jhep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 100.Weldon JE, Skarzynski M, Therres JA, Ostovitz JR, Zhou H, Kreitman RJ, et al. Designing the furin-cleavable linker in recombinant immunotoxins based on Pseudomonas exotoxin A. Bioconjugate Chem. 2015;26(6):1120–8. doi: 10.1021/acs.bioconjchem.5b00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis, Nature reviews. Cancer. 2002;2(4):277–88. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 102.Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol. 2002;71(6):907–20. [PubMed] [Google Scholar]
- 103.Zhu X, Zhou A, Dey A, Norrbom C, Carroll R, Zhang C, et al. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. PNAS. 2002;99(16):10293–8. doi: 10.1073/pnas.162352599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nature Genet. 2008;40(8):943–5. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 105.Kilpelainen TO, Bingham SA, Khaw KT, Wareham NJ, Loos RJ. Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Hum Mol Gen. 2009;18(18):3496–501. doi: 10.1093/hmg/ddp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loffler D, Behrendt S, Creemers JW, Klammt J, Aust G, Stanik J, et al. Functional and clinical relevance of novel and known PCSK1 variants for childhood obesity and glucose metabolism. Mol Metab. 2017;6(3):295–305. doi: 10.1016/j.molmet.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nordang GB, Busk OL, Tveten K, Hanevik HI, Fell AK, Hjelmesaeth J, et al. Next-generation sequencing of the monogenic obesity genes LEP, LEPR, MC4R, PCSK1 and POMC in a Norwegian cohort of patients with morbid obesity and normal weight controls. Mol Genet Metab. 2017 doi: 10.1016/j.ymgme.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 108.Wang L, Sui L, Panigrahi SK, Meece K, Xin Y, Kim J, et al. PC1/3 Deficiency Impacts Pro-opiomelanocortin Processing in Human Embryonic Stem Cell-Derived Hypothalamic Neurons. Stem Cell Reports. 2017;8(2):264–277. doi: 10.1016/j.stemcr.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leak TS, Keene KL, Langefeld CD, Gallagher CJ, Mychaleckyj JC, Freedman BI, et al. Association of the proprotein convertase subtilisin/kexin-type 2 (PCSK2) gene with type 2 diabetes in an African American population. Mol Genet Metab. 2007;92(1–2):145–50. doi: 10.1016/j.ymgme.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iamsaard S, Vanichviriyakit R, Hommalai G, Saewu A, Srakaew N, Withyachumnarnkul B, et al. Enzymatic activity of sperm proprotein convertase is important for mammalian fertilization. J Cell Physio. 2011;226(11):2817–26. doi: 10.1002/jcp.22626. [DOI] [PubMed] [Google Scholar]
- 111.Mbikay M, Tadros H, Ishida N, Lerner CP, De Lamirande E, Chen A, et al. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. PNAS. 1997;94(13):6842–6. doi: 10.1073/pnas.94.13.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marchesi C, Essalmani R, Lemarie CA, Leibovitz E, Ebrahimian T, Paradis P, et al. Inactivation of endothelial proprotein convertase 5/6 decreases collagen deposition in the cardiovascular system: role of fibroblast autophagy. J Mol Med (Berlin, Germany) 2011;89(11):1103–11. doi: 10.1007/s00109-011-0776-9. [DOI] [PubMed] [Google Scholar]
- 113.Rousselet E, Benjannet S, Marcinkiewicz E, Asselin MC, Lazure C, Seidah NG. Proprotein convertase PC7 enhances the activation of the EGF receptor pathway through processing of the EGF precursor. J Biol Chem. 2011;286(11):9185–95. doi: 10.1074/jbc.M110.189936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wetsel WC, Rodriguiz RM, Guillemot J, Rousselet E, Essalmani R, Kim IH, et al. Disruption of the expression of the proprotein convertase PC7 reduces BDNF production and affects learning and memory in mice. PNAS. 2013;110(43):17362–7. doi: 10.1073/pnas.1314698110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bergeron E, Vincent MJ, Nichol ST. Crimean-Congo hemorrhagic fever virus glycoprotein processing by the endoprotease SKI-1/S1P is critical for virus infectivity. J Virol. 2007;81(23):13271–6. doi: 10.1128/JVI.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J Biol Chem. 2002;277(13):11265–75. doi: 10.1074/jbc.M109011200. [DOI] [PubMed] [Google Scholar]
- 117.Basak A, Palmer-Smith H, Mishra P. Proprotein convertase subtilisin kexin9 (PCSK9): a novel target for cholesterol regulation. Protine Pept Lett. 2012;19(6):575–85. doi: 10.2174/092986612800494020. [DOI] [PubMed] [Google Scholar]


