Abstract
Plexiform fibromyxomas are rare neoplasms, being officially recognized as a distinct entity among benign mesenchymal gastric tumors in the 2010 WHO Classification of Tumors of the Digestive System. Characteristically, these tumors have a multinodular/plexiform growth pattern, and histologically contain variably cellular areas of bland myofibroblastic type spindle cells embedded in an abundant myxoid matrix, rich in capillary-type vessels. As of yet, the molecular and/or genetic features of these tumors are unknown. Here, we describe a recurrent translocation t(11;12)(q11;q13) involving the MALAT1 (metastasis associated lung adenocarcinoma transcript 1) long noncoding gene and the GLI1 (glioma-associated oncogene homologue 1) gene in a subgroup of these tumors. The presence of the fusion transcript in our index case was confirmed using polymerase chain reaction on genomic DNA followed by Sanger sequencing. We showed that the truncated GLI1 protein is overexpressed and retains its capacity to transcriptionally activate its target genes. A specific FISH assay was developed to detect the novel MALAT1-GLI1 translocation in formalin-fixed paraffin-embedded material. This resulted in the identification of two additional cases with this fusion, and two cases with polysomy of the GLI1 gene. Finally, immunohistochemistry revealed that the GLI1 protein is exclusively overexpressed in those cases that harbor GLI1/12q13 genomic alterations. In conclusion, overexpression of GLI1 through a recurrent MALAT1-GLI1 translocation or GLI1 upregulation delineates a pathogenically distinct subgroup of plexiform fibromyxomas with activated Sonic Hedgehog signaling pathway.
Keywords: MALAT1, GLI1, oncogenic fusion, upregulation, plexiform fibromyxoma
Introduction
Mesenchymal tumors of the gastrointestinal tract other than gastrointestinal stromal tumors are rare. In 2007, plexiform angiomyxoid myofibroblastic tumors of the stomach were first described by Takahashi and coworkers in two patients [1]. Two years later, Miettinen and colleagues described similar tumors, which they called plexiform fibromyxoma [2]. Although the exact name is still controversial, plexiform fibromyxoma was added to the WHO classification system as diagnostic term [3]. The tumors originate predominantly in the gastric antrum and are characterized microscopically by an irregular multinodular plexiform growth pattern. The nodules extend between layers of the muscularis. Moreover, the bland spindle cells are separated by an abundant myxoid stroma rich in capillary-sized vessels [1].
Based on morphology and immunohistochemistry, plexiform fibromyxomas are composed mainly of myofibroblastic tumor cells. However, subsets of the tumor can contain cells with features of fibroblasts or smooth muscle cells [4].
The low proliferation rate, absence of nuclear atypia and lack of recurrences or metastases suggest a benign nature. Nevertheless, only limited follow-up data are available. To date, excision by partial or distal gastrectomy remains the treatment of choice for plexiform fibromyxoma.
Case studies focusing on the morphologic and immunohistochemical appearance of plexiform fibromyxomas have been published [1, 2]. However, to our knowledge, nothing is known about the underlying genetic changes characterizing these tumors. We therefore set out to study molecular alterations using high-throughput RNA-sequencing of two plexiform fibromyxomas, and extended our study towards other cases which we collected.
Methods
Patients and histopathology
The present study included 16 patients [12 women and 4 men; age range 18–76 (median 43 years)] (Table 1). From the two index patients (case 1 and 2), formalin- fixed paraffin-embedded (FFPE) tissues and surgically removed specimens immediately snap-frozen in −80°C were available for the study. The remaining cases were retrieved from the consultation files of the Departments of Pathology of the KU Leuven, the Brigham and Women’s Hospital, the Memorial Sloan Kettering Cancer Center and the Cochin Hospital. The selection criteria included a typical morphology and immunoprofile and availability of FFPE tissue for fluorescence in situ hybridization (FISH) studies. Clinical details were obtained from the clinical database. Two cases were the subject of a previous case report [5]. The study was approved by the local ethical committees (S52904) and performed in accordance with the Helsinki Declaration.
Table 1.
Clinical and histopathological features of 16 plexiform fibromyxomas under study.
| Case # | Gender | Age | Localization | Size [cm] | MI/10 HPF | Immunohistochemistry | FISH * | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a-SMA | Desmin | S-100 | CD34 | ALK | EMA | CD117 | GLI1 | |||||||
| 1 | M | 19 | G. antrum | max. 5.5 | <5 | pos | neg | neg | neg | neg | nd | neg | pos | GLI1(+), MALAT1(+) |
| 2 | F | 65 | G. antrum | 4.3 ×3.0 × 1.7 | 1 | pos | neg | neg | neg | neg | neg | neg | neg | neg |
| 3 | F | 58 | G. antrum | na | 0 | pos | neg | neg | neg | neg | neg | neg | neg | neg |
| 4 | F | 51 | G. antrum | 9.0 × 8.5 × 5.5 | 1 | pos | focal | neg | nd | nd | nd | nd | nd | GLI1 polysomy |
| 5 | F | 63 | Prox. jejunum | 3.5 × 3.0 × 2.0 | 0 | pos | focal | neg | nd | nd | nd | nd | nd | neg |
| 6 | F | 76 | Stomach | max. 4.0 | 1 | multifoc | pos | nd | nd | nd | nd | nd | nd | neg |
| 7 | F | 62 | Stomach | na | 1 | nd | multifoc | neg | nd | nd | nd | nd | nd | neg |
| 8 | F | 30 | Stomach | max. 2.0 | 2 | pos | pos | nd | nd | nd | nd | nd | nd | neg |
| 9 | M | 28 | G. body | 10.0 × 6.0 × 3.0 | 0 | pos | focal | pos | neg | neg | nd | nd | nd | neg |
| 10 | F | 44 | Stomach | na | 0 | nd | pos | neg | focal | nd | nd | nd | nd | neg |
| 11 | F | 18 | G. antrum | 4.5 × 3.5 × 2.7 | 3 | focal | focal | nd | nd | nd | nd | nd | nd | neg |
| 12 | F | 19 | Stomach | max. 4.5 | 0 | pos | neg | neg | neg | nd | neg | neg | pos | GLI1(+), MALAT1(+) |
| 13 | M | 46 | G. antrum | 3.5 | <1 | pos | neg | neg | neg | nd | nd | nd | neg | neg |
| 14 | M | 29 | G. antrum | 6.5 × 4.5 × 4.0 | 2 | pos | neg | neg | neg | neg | nd | neg | pos | GLI1(+), MALAT1(+) |
| 15 | F | 36 | G. antrum | max. 8.0 | 0 | pos | neg | neg | neg | nd | nd | neg | pos | GLI1 polysomy |
| 16 | F | 47 | G. antrum | 4.5 × 3.7 | 0 | pos | neg | neg | neg | nd | nd | neg | neg | neg |
Abrreviations: M - male; F - female; G. - gastric; Prox. - proximal; max. - maximal diameter; na - not available; MI - mitotic index; HPF - high power fields; pos - diffuse positive; multifoc - multifocal, neg - negative; nd - not done
FISH using GLI1 and MALAT1 split-apart probes
Histopathological examination was performed on FFPE tissues. Five μm sections were used for routine hematoxylin and eosin (H&E) staining, and immunohistochemical (IHC) staining was performed by the avidin-biotin-peroxidase complex method, using the following monoclonal (mc) and polyclonal (pc) antibodies: GLI1 (pc, dilution 1:50, citrate buffer pressure cook) (Santa Cruz Biotechnology, Heidelberg, Germany), desmin (mc, dilution 1:20) (ICN Pharmaceuticals, Aurora, OH, USA), alpha smooth muscle actin (a-SMA, mc, dilution 1:100) (DAKO, Heverlee, Belgium), vimentin (mc, dilution 1:500) (DAKO), CD117 (pc, dilution 1:250) (DAKO), DOG1 (mc, dilution 1:100) (Novacastra, Diegem, Belgium), ALK (pc, dilution 1:500) (R&D Systems, Abingdon, UK), CD35 (mc, dilution 1:10) (Becton Dickinson, Erembodegem, Belgium), S100 (pc, dilution 1:300) (DAKO) and beta-catenin and Epithelial Membrane Antigen (EMA) (mc, dilution 1:200) (DAKO). Appropriate positive and negative controls were used throughout.
Conventional and molecular cytogenetic analysis
Sterile, representative samples of resection specimens from cases 1 and 2 were submitted for cytogenetic analysis, using standard culture and harvest procedures. The karyotypes were described according to the International System for Human Cytogenetic Nomenclature [2011].
Metaphase spreads for FISH analysis were available from the same material that had been analyzed cytogenetically. The DNA bacterial artificial chromosome (BAC) probes, covering and flanking genes that had been identified as potential fusion partners in the RNA-Seq experiment were obtained from the BACPAC Resource of Children’s Hospital of Oakland Research Institute (Supplementary Table 1; http://bacpac.chori.org), and selected based on their location in the UCSC Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway, GRCh37/hg19). DNA isolation, probe labeling and hybridization were performed as previously described [6]. The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes.
Array comparative genomic hybridization (aCGH)
Genomic DNA was extracted from frozen tumor specimens using the Blood & Tissue DNeasy kit with RNase treatment (Qiagen, Venlo, The Netherlands). Array CGH analyses were performed using a 180K oligonucleotide microarray (CytoSure, Oxford Gene Technology, Oxfordshire, UK) according to the manufacturer’s instructions. After washing, the array was analyzed with the Agilent scanner and Feature Extraction software (Agilent Technologies, Diegem, Belgium) and text file outputs from the quantization analysis were imported to the CytoSure Interpret Software (Oxford Gene Technology) for copy number analysis.
KIT/PDGFRA mutational analysis
Mutational analysis of KIT and PDGFRA was performed in cases 1 and 2, based on DNA isolated from frozen tumor biopsies. Mutation of KIT exons 9, 11, 13 and 17 and PDGFRA exons 12, 14 and 18 were identified by Sanger sequencing of PCR products, according to routine protocols previously published [6].
RNA-Sequencing and fusion detection
Total RNA was isolated from frozen tumor specimens (cases 1 and 2) and prepared for RNA-Sequencing in accordance with the standard Illumina mRNA sample preparation protocol. Briefly, after selection of polyA+ RNA, the RNA was converted into cDNA libraries using the TruSeq RNA Sample Preparation kit (Illumina, San Diego, CA, USA). Sequencing paired-end reads of 125 bp with the HiSeq2500 (Illumina) in rapid mode was followed by detection of gene rearrangements using a combination of deFuse (version 0.6.2), Tophat-fusion (version 2.0.13) and FusionCatcher (version 0.99.3e) [7–9]. All software was run using default parameters and with hg19 as reference genome. For the deFuse results we required ≥ 8 reads spanning the fusion and ≥ 5 split reads while excluding fusions between adjacent genes. To increase the rate of true positives, we only focused on those fusions that were detected by ≥ two software programs. The interchromosomal translocation was visualized using the Circos software (version 0.62–1) [10]. RNA-Seq data used in this study have been deposited in the European Nucleotide Archive (ENA) and have study accession number PRJEB12299 (www.ebi.ac.uk/ena/data/view/PRJEB12299).
Western blot
Cell lysis of frozen tumors, SDS-PAGE, and immunoblotting were carried out as previously described [6]. In short, tumor lysate aliquots containing 50 μg of protein were electrophoresed and blotted to PVDF membranes (GE Healthcare, Diegem, Belgium). Membranes were blocked in TBST containing 5% blocking reagent (non-fat milk) and immunoblotted using a rabbit antibody against GLI1 (dilution 1:100, Santa Cruz), diluted in 5% blocking reagent. Total beta-actin (dilution 1:20,000, Sigma Aldrich, Diegem, Belgium) was used as a protein-loading and transfer control. The HRP-conjugated anti-rabbit IgG and anti-mouse IgG (DAKO) were used at a dilution of 1:5,000, and visualized with Enhanced Chemiluminescence (ThermoFisher Scientific, Gent, Belgium).
Fusion validation with Sanger sequencing
Genomic DNA was extracted from frozen tumor specimens. The sequence surrounding the MALAT1-GLI1 fusion was amplified by polymerase chain reaction (PCR), using GoTaq G2 Flexi DNA polymerase (Promega, Leiden, The Netherlands). Genomic sequences were obtained from online databases from the National Center for Biotechnology Information (NCBI), and specific primers for amplified fragments were designed using the NCBI primer-BLAST software [11]. Primer sequences are available upon request. The PCR products were purified using the Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA, USA) followed by direct bi-directional cycle sequencing using the ABI 3730 Genetic Analyzer (Applied Biosystems, ThermoFisher Scientific).
Interphase FISH screening on FFPE
To screen for rearrangements involving the genes of interest in a larger cohort of plexiform fibromyxomas, 4-μm cut FFPE sections from 14 additional cases were retrieved and subjected to interphase FISH with SpectrumOrange (SO) and SpectrumGreen (SG) differentially labeled BAC probes, covering or spanning the GLI1/12q13 and MALAT1/11q12 loci regions. Paraffin-embedded sections were pretreated using the SPoT-Light Tissue Pretreatment kit (Invitrogen, Life Technologies, ThermoFisher Scientific), according to the instructions of the manufacturer. FISH was performed using standard procedures.
FISH images were captured using an epifluorescence microscope equipped with the ISIS digital image analysis system (MetaSystems, Altlussheim, Germany). Tissues were scored by two independent investigators and considered positive if > 25% of at least 200 cells showed split-apart or fused signals. Nuclei with an incomplete set of signals were omitted from the score.
Cell culture and transfection studies
HEK-293T cells were obtained from the American Type Culture Collection and grown at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, ThermoFisher Scientific). This medium was supplemented with 100 μg/ml penicillin, 100 μg/ml streptomycin and 10% HyClone fetal bovine serum (GE Healthcare). For Western blotting, cells were seeded in a 6 well plate, while 96 well plates were used otherwise. Cells were transfected with the TurboFect transfection reagent (ThermoFisher Scientific). To test the transcriptional activity, cells were transfected with a mixture of variable amounts of a luciferase reporter plasmid (preceded by 11 copies of a GLI1 binding site), 100 ng of GLI1 expression vectors and 5 ng of a plasmid with constitutive expression of Renilla luciferase. The latter served as an internal control for transfection efficiency. One day after seeding, the cells were transfected and harvested 48 hours later in Passive Lysis Buffer (Promega). Both luciferases were measured using the Dual-Luciferase reporter assay system (Promega) on a Victor X3 (Perkin Elmer, Zaventem, Belgium). The relative luciferase activities represent the amount of Firefly chemiluminescence corrected for the transfection efficiency by normalizing against the Renilla luciferase activity. The values shown are the averages of at least three independent experiments performed in triplicate. The error bars are the SEM. P < 0.05 was regarded as the threshold value for statistical significance.
Results
Histology and immunohistochemistry
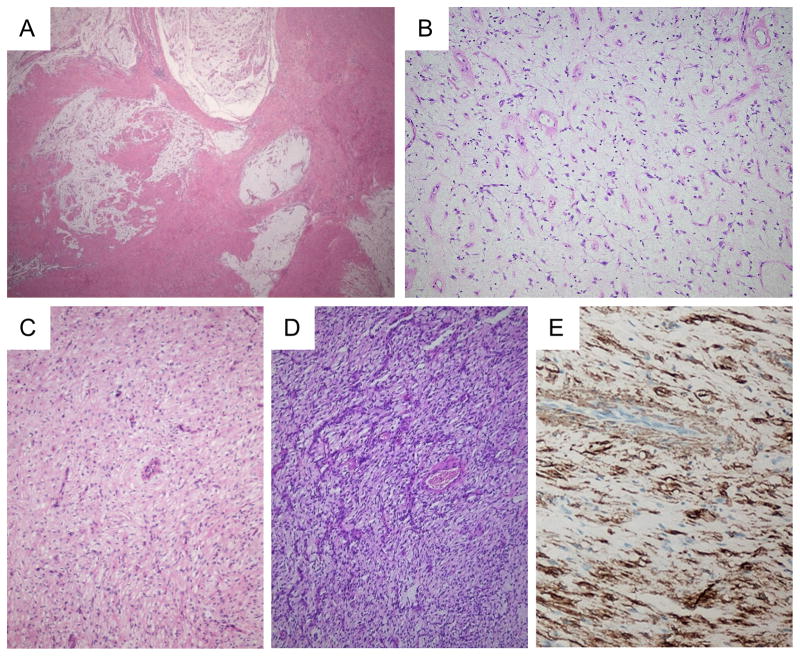
Histologically, the lesions had a lobular architecture and frequently extended from the submucosa to the serosa of the gastric wall, often in a multinodular/plexiform pattern (Figure 1A–B). In some cases a single myxoid nodule was seen. All lesions had a myxoid background, but the cellularity was highly variable (Figure 1B–D). The stellate to spindly tumor cells were bland and mitotic figures were rarely seen. Scattered inflammatory cells were present. A high vascularity was consistently seen, the vessels being relatively small and often branching and curvilinear, sometimes with perivascular hyalinization (Figure 1B–D). Strong expression of alpha smooth muscle actin was present (Figure 1E), but CD117, DOG1, ALK, CD34, S100, and EMA stains were negative. Beta-catenin only stained the cytoplasm of the tumor cells.
Figure 1.
Histology of plexiform fibromyxomas of the stomach. Low power view, showing the lobular and plexiform architecture (A). At high power, the myxoid matrix contains numerous vessels, bland spindle cells and scattered inflammatory cells (B). In some lesions/nodules, the cellularity is somewhat increased. Note the bland appearance of the spindle cells and the prominent vessels (C and D). Tumor cells express alpha smooth muscle actin by IHC (E).
Conventional G-banding and aCGH results
Two cases were cytogenetically analyzed after short-term culturing. In case 1, an abnormal karyotype 46,XY,t(1;12;11)(q21;q13;q11),t(7;18;13;14)(q22;q21;q12;q22) was found (Supplementary figure 1). In case 2, no chromosomal abnormalities were detected (data not shown). Using high-resolution aCGH, we found a simple genomic profile in case 1: arr[hg19] 7q22.1q22.3(99,687,542-104,592,401)x1,11p15.4 (2,905,504-2,907,123)x1,13q12.11(19,675,986-19,684,924)x1 (data not shown). The absence of genomic imbalances at the 1q21, 11q11 and 12q13 breakpoints in case 1 indicated a balanced translocation.
In case 2, the aCGH profile disclosed only minute aberrations in the regions of known polymorphic copy number variations (CNVs), according to the Database of Genomic Variants (http://projects.tcag.ca/variation) and The CNV project at the Children’s Hospital of Philadelphia (CHOP) database (http://cnv.chop.edu).
KIT/PDGFRA mutational analysis
Both analyzed tumor specimens (case 1 and 2) showed a wild-type sequence of the examined exons of KIT and PDGFRA (data not shown).
RNA-Seq analysis identifies the MALAT1-GLI1 gene fusion
To identify the genes involved in the t(1;12;11) translocation, we performed paired-end whole transcriptome RNA-Sequencing with read lengths of 125 bp of case 1 and 2. On average, 13 billion high-quality bases were aligned to the reference genome. We did not find any gene fusions predicted by all three software programs that were used (TopHat, FusionCatcher and deFuse). However, several fusions were predicted by two programs (Supplementary Table 1). We studied gene expression for all the genes involved in the listed fusions, but only GLI1 revealed a change in mRNA expression. The IGFBP5 and KANSL1 genes were recurrently found in the table. However, fusions involving the IGFBP5 gene are all located at the end of the protein coding part, almost eliminating the chance that each fusion has a functional effect. Moreover, the negative results of FISH experiments using split-apart probes for IGFBP5 gene (Supplementary table 2) further gave evidence of the lack of IGFBP5-associated fusions. The KANSL1 gene and its proposed fusion partners are located close together on the same chromosome, indicating that these fusions are most likely read-throughs. Taking this information into account, we decided to focus on the fusion involving the Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and the Glioma-associated oncogene homologue 1 (GLI1) gene as detected in case 1.
Direct sequencing confirmed the presence of the MALAT1-GLI1 fusion
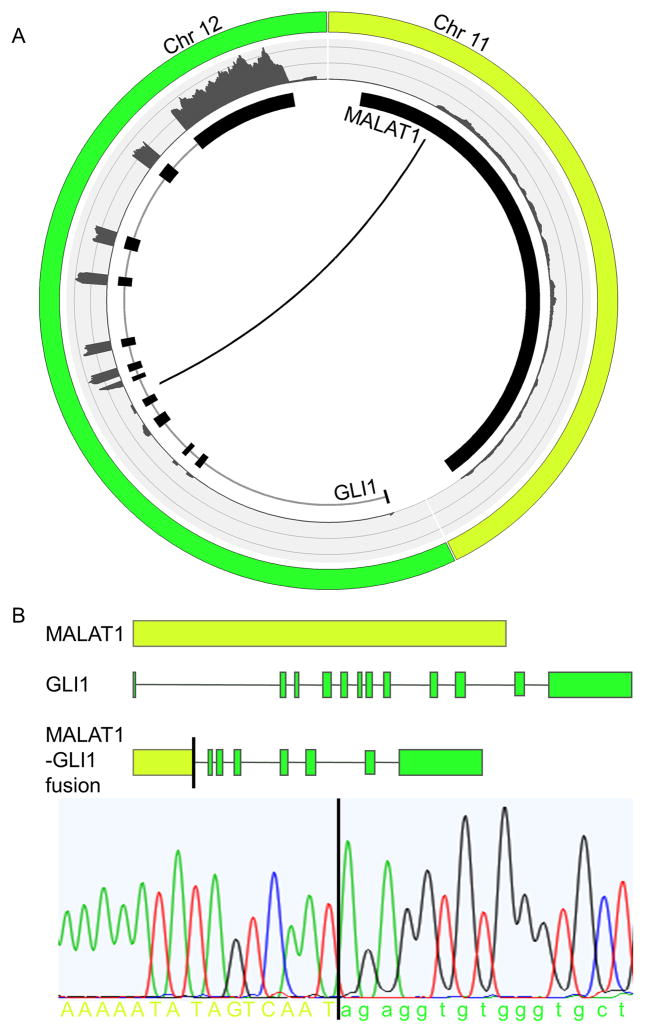
A Circos plot shows the exact breakpoint in intron 5 of the GLI1 gene (Figure 2A). The fusion of the promoter of MALAT1 to the 3′-end of GLI1 results in the increased expression of exons 6 to 12 of GLI1, as shown with the FPKM counts (Figure 2A). The DNA-binding zinc fingers that are encoded by exons 7 to 12 are thus still present in the fusion. To validate the RNA-Sequencing results, specific primers were designed and PCR was performed on genomic DNA of the tumor of case 1 followed by Sanger sequencing. This confirmed the breakpoint in MALAT1 and in intron 5 of GLI1 (Figure 2B).
Figure 2.
Graphical representation of the MALAT1-GLI1 fusion. The Circos plot depicts the interchromosomal translocation and its position relative to the different exons of GLI1 and the non-coding exon of MALAT1. The outer ring shows the FPKM counts representing the expression level of the different exons (A). Schematic representation of the MALAT1-GLI1 gene fusion. Structures for the genes have their basis in the UCSC reference sequences. For validation, a fusion-specific PCR followed by Sanger sequencing was performed on genomic DNA isolated from the tumor of index case 1 (B).
FISH analyses
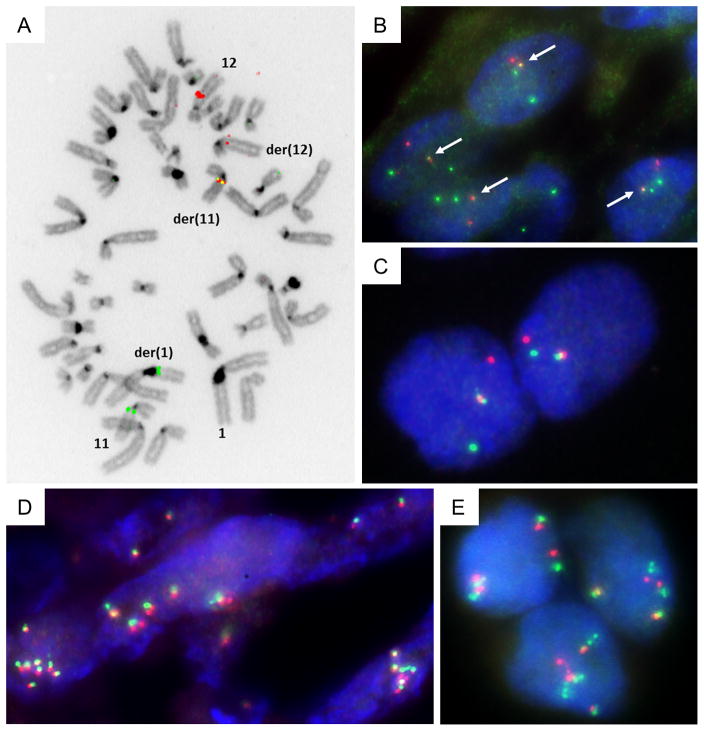
In t(1;12;11)-carrying cells of case 1, metaphase FISH analysis with BAC probes covering GLI1/12q13 and MALAT1/11q12 genes resulted in fused signals (Figure 3A). More specifically, the BAC probes covering the GLI1 and MALAT1 genes resulted in a fused signal over the abnormal der(11) chromosome, the translocated MALAT1 signal over der(1) and the reduced (reductant) GLI1 signal on der(12), while on normal chromosome 11 and 12 homologues the intact signals were observed. The same probes were used on FFPE sections from case 1 to perform interphase FISH (Figure 3B).
Figure 3.
Metaphase dual-color FISH on index case 1 using SG-labeled RP11-472D15 and SO-labeled RP11-181L23 BAC probes that cover respectively the MALAT1 and GLI1 genes, indicate the presence of the MALAT1-GLI1 fusion (A). Interphase dual-color FISH on FFPE tissue from case 1 using SG-labeled RP11-472D15 and SO-labeled RP11-181L23 BAC probes that cover the MALAT1 and GLI1 genes, respectively. Overlapping green and red signals indicate MALAT1-GLI1 fusion (B). Interphase dual-color FISH on case 14 using SG-labeled RP11-1077C21 (centromeric) and SO-labeled RP11-571M06 (telomeric) BAC probes that flank the GLI1 gene. Rearrangement of GLI1 is pointed out by split apart green and red signals (C). Interphase dual-color FISH using probes that flank GLI1 on FFPE tissue from cases 4 and 15, showing polysomy of GLI1 (D and E).
MALAT1-GLI1 fusion is recurrently present in a subgroup of plexiform fibromyxomas
To check the prevalence of the MALAT1-GLI1 fusion, interphase dual-color FISH was performed on tumor sections from 14 additional cases of plexiform fibromyxoma, using break-apart and bring-together approach. Rearrangements of the GLI1 gene were found in two additional plexiform fibromyxomas with classic histology (Table 1; Figure 3C). The presence of the MALAT1-GLI1 fusion in these cases was further confirmed by differentially labeled FISH probes, which map to sequences covering GLI1 and MALAT1 genomic loci. As a result, the frequency of the MALAT1-GLI1 fusion in our group of plexiform fibromyxoma was 18 %. Importantly, polysomy of the chromosome 12q13 region containing the GLI1 gene locus was revealed by interphase FISH in two other tumors (Figure 3D–E).
Overexpression of GLI1 protein as a result of the MALAT1-GLI1 gene fusion
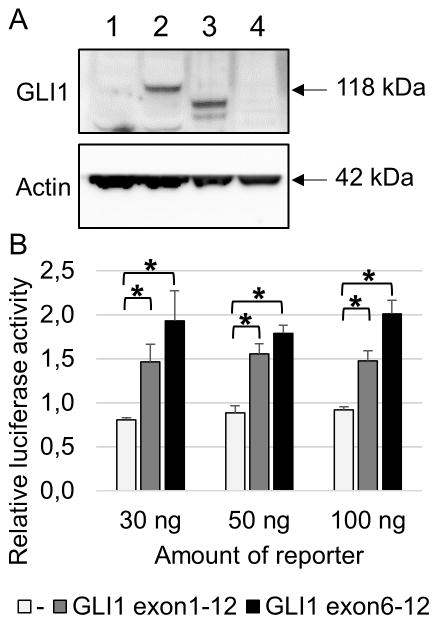
The detected increase in RNA-levels of exons 6 to 12 of GLI1 suggested an overexpression of part of the GLI1 protein. To investigate this, we performed a Western blot for case 1 (with the fusion) and case 2 (without fusion) (Figure 4A). As a control, we used untransfected 293T cells where no endogenous GLI1 protein expression could be observed. We also overexpressed full length GLI1 in 293T cells. As expected, case 1 revealed overexpression of GLI1 protein as a result of the MALAT1-GLI1 gene fusion, while no GLI1 expression could be detected in case 2.
Figure 4.
Western blot showing overexpression of GLI1 protein in case 1 with the MALAT1-GLI1 fusion (lane 3). Case 2 without the fusion does not show GLI1 overexpression (lane 4). As a control, 293T cells were either untransfected (lane 1) or transfected with a full length GLI1 plasmid (lane 2) (A). Transcriptional activity of truncated GLI1 was tested using a dual-luciferase assay. 293T cells were transiently transfected with a luciferase reporter construct containing 11 copies of a GLI1 binding site together with a construct containing either full length GLI1 or only exons 6 to 12 of GLI1. Cells were lysed 48 hours after transfection. Results are represented as means of relative luciferase activity of 3 independent experiments performed in triplicate. Error bars indicate SEM; *p<0.05 (B).
Truncated GLI1 is still transcriptionally active
As a result of the MALAT1-GLI1 gene fusion, exons 6 to 12 of GLI1 are overexpressed. To investigate whether this truncated GLI1 (lacking exons 1 to 5) is still transcriptionally active, 293T cells were transiently transfected with full length GLI1 or with the truncated GLI1 (exons 6–12). The reporter construct contained a luciferase gene under the control of 11 GLI1 binding sites. Both full length GLI1 and truncated GLI1 induced luciferase expression, indicating that the truncated GLI1 retains its full capacity to transcriptionally activate its target genes (Figure 4B).
Overexpression of GLI1 protein using immunohistochemistry
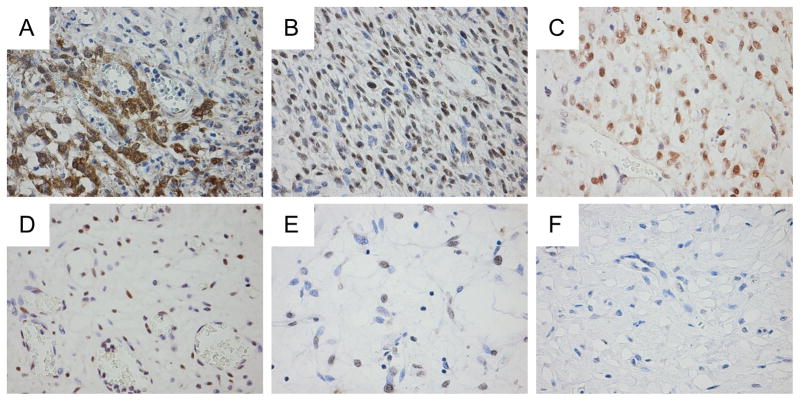
Using the FFPE sections from six patients with plexiform fibromyxomas, we investigated the protein expression level of GLI1. A pericytoma with t(7;12) translocation and hence GLI1 overexpression was used as control (Figure 5A). Indeed, tumors harboring the MALAT1-GLI1 fusion as confirmed by FISH overexpressed GLI1 protein (Figure 5B–C). In addition, tumors polysomic for GLI1/12q13 region also revealed GLI1 overexpression, as would be expected in neoplasms with amplification of the GLI1 gene (Figure 5D). In cases without a GLI1 gene alteration, insubstantial or no expression of GLI1 was detected (Figure 5E–F).
Figure 5.
Immunostaining for GLI1 protein expression. As a positive control with known GLI1 overexpression, a pericytoma carrying t(7;12)(p22;q13) and ACTB-GLI1 fusion showed diffuse cytoplasmic and nuclear immunopositivity (A). Plexiform fibromyxomas with MALAT1-GLI1 fusion (case 1 and 14, respectively) exhibit higher cellularity, and diffuse immunopositivity for GLI1 (B and C). Plexiform fibromyxoma with GLI1 polysomy (case 15), showing immunopositivity for GLI1 in a vast majority of tumor cells (D). Tumors without evident GLI1 involvement demonstrate either insubstantial positivity (case 2) or lack immunopositivity for GLI1 (case 13) (E and F).
Discussion
We have identified a novel fusion in a subgroup of plexiform fibromyxomas that involves the MALAT1 (in 11q12) and GLI1 (in 12q13) genes. At the genomic level, both genes are transcribed from centromere to telomere, thus, a simple balanced translocation is sufficient to generate a functional MALAT1-GLI1 chimeric protein. This recurrent fusion was detected in three out of 16 patients. In two additional patients polysomy of GLI1/12q13 was identified. Notably, both genetic alterations led to upregulation of GLI1 protein. In terms of morphology, GLI1-immunopositive tumors seemed to show an increased cellularity, but numbers are too limited to make firm conclusions. No other differences in pathology or clinical parameters were found in this limited cohort of tumors. We did not detect genomic anomalies in GLI1 in the remaining 11 samples of plexiform fibromyxomas. It is possible there were false negatives among the tumor samples we tested by FISH (for example cryptic fusions) or alternative mechanisms of GLI1 upregulation. Alternatively, increased GLI1 signaling may only contribute to a subset of plexiform fibromyxomas. Unfortunately, due to the restricted number of frozen tissue specimens and limited FFPE material from consultation cases, we were not able to investigate this further.
The long noncoding RNA MALAT1, also known as NEAT2, is an mRNA-like RNA polymerase II transcript that consists of more than 8700 nucleotides [12]. It is broadly expressed at a level comparable with or higher than many (housekeeping) protein-coding genes, including beta-actin and GAPDH [13].
Translocations involving MALAT1 have been described in renal cell carcinomas and undifferentiated embryonal sarcomas. In the former, the MALAT1 promoter was fused upstream of the open reading frame of the transcription factor EB (TFEB), resulting in > 30-fold overexpression of TFEB [14]. In the latter, the t(11;19)(q11;q13.4) translocation was studied, revealing a fusion between MALAT1 and the MHLB1 locus [15].
GLI1 is a component of the Sonic Hedgehog signaling pathway, which is activated when the Sonic Hedgehog ligand binds to its receptor Patched. This relieves the repressive activity on Smoothened (SMO), resulting in its accumulation and activation of the GLI1 transcription factor [16]. The GLI1 protein contains five successive DNA-binding zinc finger motifs of which the first three bind the phosphate backbone, while the last two bind target DNA in a sequence-specific way [17]. The C-terminal region has a transactivation function through modulation of chromatin remodeling. Finally, GLI1 acts as the ultimate effector for the control of specific oncogenic target genes.
Deregulation of GLI1 through a translocation has been described in a distinctive subgroup of pericytic neoplasms, pericytomas with t(7;12)(p21;q13) [18]. In these tumors, the 5′-part of ACTB is fused to the 3′-part of GLI1, leading to deregulation of GLI1 expression and its downstream target genes. Similarly, the strong promoter of MALAT1 likely enhances GLI1 expression in plexiform fibromyxomas. The fused GLI1 sequences include the exons encoding the five zinc finger domains (exons 7 to 10) as well as the transcriptional activation domain within exon 12 [18].
Inappropriate reactivation of the Hedgehog signaling pathway is responsible for the formation as well as the progression of several cancers. Amplification and/or overexpression of GLI1 has been described in a variety of tumors, including brain tumors, sarcomas, B-cell lymphomas and benign fetal rhabdomyomas [16, 19]. High protein levels of GLI1 can be the result of amplification of the GLI1 gene, epigenetically driven overexpression, mutations in the SUFU or PTCH1 gene or post-synthetic modifications [17, 20]. Likewise, activation of the Hedgehog signaling pathway may be possible by low-level gains of the 12q13 region containing the GLI1 gene in two plexiform fibromyxomas of our tumor cohort.
In summary, we have found that overexpression of GLI1 through a recurrent MALAT1-GLI1 translocation and GLI1 polysomy characterizes a subset of plexiform fibromyxomas. The specificity, frequency and biological consequences of these phenomena and the dissection of other mechanisms that induce these tumors await further studies.
Supplementary Material
Supplementary table 1. List of gene fusions in two cases detected by two different software programs.
Supplementary table 2. Custom BAC probes used for FISH analysis.
Representative GTG-banded karyogram of case 1 showing abnormal 46,XY,t(1;12;11)(q21;q13;q11),t(7;18;13;14)(q22;q21;q12;q22) karyotype. Arrows indicate 1q21, 12q13 and 11q11 chromosomal breakpoints.
Acknowledgments
This study was supported by KU Leuven Concentrated Action Grant number 2011/010. The authors acknowledge Lut Mekers, Belinda Carleer and Vanessa Vanspauwen for their excellent technical assistance.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Statement of author contributions
Carrying out experiments, manuscript drafting: LS
Concept and study design, writing of the manuscript: LS, MDR, RS
Acquisition, analysis and interpretation of data: LS, CDMF, CRA, AR, JMC, MDR, RS
All authors had participated in the critical revision of the manuscript for important intellectual content and in final approval of the submitted version.
References
- 1.Takahashi Y, Shimizu S, Ishida T, et al. Plexiform angiomyxoid myofibroblastic tumor of the stomach. Am J Surg Pathol. 2007;31:724–728. doi: 10.1097/01.pas.0000213448.54643.2f. [DOI] [PubMed] [Google Scholar]
- 2.Miettinen M, Makhlouf HR, Sobin LH, et al. Plexiform fibromyxoma: a distinctive benign gastric antral neoplasm not to be confused with a myxoid GIST. Am J Surg Pathol. 2009;33:1624–1632. doi: 10.1097/PAS.0b013e3181ae666a. [DOI] [PubMed] [Google Scholar]
- 3.Ikemura M, Maeda E, Hatao F, et al. Plexiform angiomyxoid myofibroblastic tumor (PAMT) of the stomach. A case report focusing on its characteristic growth pattern. Int J Clin Exp Pathol. 2014;7:685–689. [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi Y, Suzuki M, Fukusato T. Plexiform angiomyxoid myofibroblastic tumor of the stomach. World J Gastroenterol. 2010;16:2835–2840. doi: 10.3748/wjg.v16.i23.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida A, Klimstra DS, Antonescu CR. Plexiform angiomyxoid tumor of the stomach. Am J Surg Pathol. 2008;32:1910–1912. doi: 10.1097/PAS.0b013e3181838fd1. author reply 1912–1913. [DOI] [PubMed] [Google Scholar]
- 6.Debiec-Rychter M, Wasag B, Stul M, et al. Gastrointestinal stromal tumours (GISTs) negative for KIT (CD117 antigen) immunoreactivity. J Pathol. 2004;202:430–438. doi: 10.1002/path.1546. [DOI] [PubMed] [Google Scholar]
- 7.McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicorici D, Satalan M, Henrik E, et al. FusionCatcher: a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014:11. [Google Scholar]
- 9.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015;5:e008653. doi: 10.1136/bmjopen-2015-008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis IJ, Hsi BL, Arroyo JD, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajaram V, Knezevich S, Bove KE, et al. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13. 4) translocation. Genes Chromosomes Cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- 16.Gonnissen A, Isebaert S, Haustermans K. Targeting the Hedgehog signaling pathway in cancer: beyond Smoothened. Oncotarget. 2015;6:13899–13913. doi: 10.18632/oncotarget.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Infante P, Mori M, Alfonsi R, et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J. 2015;34:200–217. doi: 10.15252/embj.201489213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlen A, Fletcher CD, Mertens F, et al. Activation of the GLI oncogene through fusion with the beta-actin gene (ACTB) in a group of distinctive pericytic neoplasms: pericytoma with t(7;12) Am J Pathol. 2004;164:1645–1653. doi: 10.1016/s0002-9440(10)63723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hettmer S, Teot LA, van Hummelen P, et al. Mutations in Hedgehog pathway genes in fetal rhabdomyomas. J Pathol. 2013;231:44–52. doi: 10.1002/path.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelczar P, Zibat A, van Dop WA, et al. Inactivation of Patched1 in mice leads to development of gastrointestinal stromal-like tumors that express Pdgfralpha but not kit. Gastroenterology. 2013;144:134–144. e136. doi: 10.1053/j.gastro.2012.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1. List of gene fusions in two cases detected by two different software programs.
Supplementary table 2. Custom BAC probes used for FISH analysis.
Representative GTG-banded karyogram of case 1 showing abnormal 46,XY,t(1;12;11)(q21;q13;q11),t(7;18;13;14)(q22;q21;q12;q22) karyotype. Arrows indicate 1q21, 12q13 and 11q11 chromosomal breakpoints.