Abstract
Background/Objectives
Elevated triglycerides predict insulin resistance and vascular disease in obesity, but how the inert triglyceride molecule is related to development of metabolic disease is unknown. To pursue novel potential mediators of triglyceride-associated metabolic disease, we used a forward genetics approach involving inbred mice and translated our findings to human subjects.
Subjects/Methods
Hemopexin was identified as a differentially expressed gene within a quantitative trait locus associated with serum triglycerides in an F16 advanced intercross between the LG/J and SM/J strains of mice. Hpx expression was evaluated in both reproductive fatpads and livers of mice representing three strains, LG/J (n = 25), SM/J (n = 27) and C57Bl/6J (n = 19), on high- and low-fat diets. The effect of altered Hpx expression on adipogenesis was studied in 3T3-L1 cells. Circulating HPX protein along with HPX expression were characterized in subcutaneous white adipose tissue samples obtained from a cohort of metabolically abnormal (n = 18) and of metabolically normal (n = 24) obese human subjects. We further examined the relationship between HPX and triglycerides in human atherosclerotic plaques (n = 18).
Results
Hemopexin expression in mouse adipose tissue, but not liver, was regulated by dietary fat regardless of genetic background. Hemopexin increased in concert with adipogenesis in 3T3-L1 cells, and disruption of its expression impaired adipocyte differentiation. RNAseq data from the adipose tissue of obese humans showed differential expression of hemopexin based on metabolic disease status (p < 0.05), and circulating hemopexin levels were correlated with serum triglycerides in these subjects (r = 0.33; p = 0.03). Hemopexin was also found in an unbiased proteomic screen of human atherosclerotic plaques, and shown to display differential abundance based on extent of disease and triglyceride content (p < 0.05).
Conclusions
Our findings suggest that hemopexin is associated with triglycerides and provide a framework for understanding mechanisms underlying lipid metabolism and metabolic disease.
Introduction
Obesity is associated with a constellation of metabolic abnormalities and is an important risk factor for nonalcoholic fatty liver disease, diabetes, and atherosclerosis (1). The mechanisms responsible for the link between excess body fat and metabolic dysfunction, particularly vascular disease, are not clear. Since circulating triglycerides are associated with obesity, decrease with even modest weight loss (2), and are associated with stroke and myocardial infarction (3, 4), we focused on triglycerides and used a genetic approach to identify novel potential pathogenic mediators of increased adiposity. Unexpectedly, our findings implicate hemopexin in adipocyte biology and vascular disease.
Hemopexin (HPX) is an extensively glycosylated protein with the highest known binding affinity to heme (5). Heme is released with erythrocyte lysis and has the potential to intercalate into lipid membranes where it may participate in Fenton chemistry to damage tissues (6). HPX is known to sequester plasma heme and decrease its oxidative and inflammatory effects (7, 8). HPX facilitates this process via low-density lipoprotein receptor-related protein-1 (LRP1)-mediated endocytosis, leading to catabolism by heme oxygenase-1, HO-1, and subsequent release of iron to ferritin (9, 10).
Hpx is mainly expressed by hepatocytes (11), but also by cells of the brain parenchyma (12), retina (13), and peripheral nervous system (14) – tissues with high lipid content. HPX is not known to participate in adipocyte biology, but given known interactions between HPX and LRP1 as well as circulating lipoproteins (15), we pursued the notion that HPX may have broad roles in lipid metabolism. Our results show that HPX impacts adipocyte differentiation in cultured cells, is altered in adipose tissue of high-fat fed mice and people with obesity, is correlated with triglycerides in both mice and humans, and is differentially expressed in human atherosclerotic lesions. These observations suggest that HPX could serve as a biomarker linking triglycerides, adipocyte biology and cardiovascular disease.
Materials/Subjects and Methods
Ethics Statement
All human subjects provided written informed consent, and all data generated were approved by the Human Research Protection Office at Washington University School of Medicine. All animal care and handling procedures conformed to IACUC guidelines.
Animal Care and Phenotyping
The mice used in this study were the progeny of LG/J, SM/J and C57BL/6J animals obtained from the Jackson Labs. Pups from each strain were weaned at three weeks of age and then separated into sex-specific cages of no more than five animals per cage. At this time, one-half of the animals from each litter were fed a high-fat diet (42% calories from fat, TD88137, Harlan Teklad, Madison WI) and one-half were fed a relatively low-fat diet (15% calories from fat, D12284, Research Diets, New Brunswick, NJ, or 13% calories from fat, 5001, Laboratory Rodent Diet, St Louis MO). Animals’ assignment to their respective diet was random. The number of animals used in this study was determined by the number of mice required to have reproducible results and robust metabolic assays.
At 20 weeks of age, animals were fasted for 4h and anesthetized with an overdose of Ketamine/Xylazine cocktail. Blood was collected from the retro-orbital sinus and euthanasia was achieved by cardiac perfusion with PBS in fully anesthetized animals. Serum was frozen at −80°C until assayed. Adipose and liver tissues were flash frozen and stored at −80°C until assayed.
Quantitative Real-Time PCR
Total RNA was isolated from mouse adipose tissue and from 3T3-L1 cells using the RNeasy Lipid Tissue kit (QIAgen). Total RNA was isolated from mouse liver using the TRIzol reagent (Invitrogen). Total RNA was isolated from frozen human subcutaneous adipose tissue samples using an RNeasy Mini kit (QIAgen). iScript cDNA Synthesis kit (Bio-Rad) was used for reverse transcription. Quantitative PCR was performed with an ABI Prism 7700 PCR instrument using SYBR Green reagent (Applied Biosystems). Assays were performed in duplicate and repeated. Primer sequences are available in Supplementary Table 1. cDNA products were analyzed using the ΔΔCT method. Mouse results were normalized to L32 mRNA levels and human results were normalized to RPLO.
Analytical Procedures
Western blotting was performed using a rabbit polyclonal anti-HPX antibody from Abcam (ab90947) and anti-AP2 and anti-GAPDH antibodies from Cell Signaling Technology (D25B3; D16H11). Serum triglyceride and cholesterol levels were measured using reagents from Thermo Scientific (TR13421; TR22421). Serum glucose and free-fatty acids were measured using reagents from Wako Life Sciences (439–90901; 995–34693 and 997–34893). Serum HPX and HPX in cell culture media were measured using an Abcam HPX Elisa kit for mouse (ab157716). Plasma HPX and carotid plaque HPX content were measured using an Abcam HPX Elisa kit for human (ab108859). Carotid plaque total and free cholesterol contents were measured using reagents from Wako Life Sciences (NC9138103; NC9506034). Esterified cholesterol was estimated by subtracting free cholesterol values from total cholesterol values. Carotid plaque analyses were all normalized to protein content of the homogenate. All assays were performed in duplicate with the exception of those on cell culture media, which were performed in triplicate.
Cell Culture
3T3-L1 cells were obtained from ZenBio (SP-L1-F) and maintained in DMEM supplemented with 10% NCS, L-glutamine and penicillin/streptomycin (Invitrogen). Adipocyte differentiation was induced after cells were fully confluent for 48 h. Cells were cultured in differentiation media 1 (DMEM, 10% FBS, 1 µg/mL insulin, 0.25 µg/mL dexamethasone, 0.5 mM IBMX) for 72 h. Cells were then cultured in differentiation media 2 (DMEM, 10% FBS, 1 µg/mL insulin) for 48 h, after which they were cultured in DMEM supplemented with 10% FBS for at least 24 h before experiments using fully differentiated adipocytes; otherwise cells were collected for experiments on their respective day after induction. 293T cells were maintained in DMEM supplemented with 10% FBS.
Lentiviral shRNA-mediated Knockdown
Plasmids encoding shRNA for mouse Hpx (TRCN0000105469, TRCN0000105468, and TRCN0000288248) were obtained from Sigma-Aldrich (St Louis, MO). Only one plasmid (TRCN0000105469) resulted in a satisfactory knockdown so all experiments were performed using this plasmid. Packaging vector psPAX2 (12260), envelope vector pMD2.G (12259) and scrambled shRNA plasmid (1864) were obtained from Addgene. 293T cells in 10 cm dishes were transfected with 2.66 µg psPAX2, 0.75 µg pMD2.G, and 3 µg shRNA plasmid. After 48 h, media were collected, filtered and used to treat 3T3L1 fibroblasts. Cells were selected with puromycin for 48 h and differentiation was induced as described above with the addition of puromycin to the media. Media was collected and knockdown was assessed 24 h after finishing the differentiation protocol. HPX in media was measured by Elisa as described above. This experiment was performed in duplicate and repeated 6 times.
Metabolically Normal Obese (MNO), Metabolically Abnormal Obese (MAO) and Lean Subjects
The human MNO and MAO subjects were part of a larger study previously described (16). All subjects completed a comprehensive medical evaluation, and no subject smoked tobacco or took medications that could affect outcomes. Intrahepatic triglyceride (IHTG) content was measured with magnetic resonance spectroscopy and used to classify subjects as MNO (IHTG <5.6%) or MAO (IHTG >10%). Body fat mass was determined using dual-energy x-ray absorptiometry (Lunar iDXA; GE Healthcare). All sera were obtained after a 12 h fast. HOMA-IR was measured from the subjects’ fasting glucose and insulin levels. Subcutaneous adipose tissue biopsies were obtained from the periumbilical area with a liposuction cannula (Tulip Medical Products) from all MNO and MAO subjects. Subcutaneous adipose tissue biopsies were also obtained from 4 lean female subjects of European-American descent (mean age = 35.75 with SEM = 8.18; mean BMI (kg/m2) = 20.93 with SEM = 1.12; mean serum triglycerides (mg/dL) = 78.52 with SEM = 12.89). All samples and data were collected at the Clinical Research Unit of the Washington University School of Medicine.
A subset of these subjects’ total RNA samples extracted from adipose tissue biopsies (10 MNO, 13 MAO, 4 Lean) were submitted to EA (Durham, NC) for RiboZero library construction and RNA sequencing on an Illumina HiSeq2000, producing ~ 25million, 50 bp, paired end reads per sample. Quality was assessed, reads were aligned to the NCBI GRCh37 USCS hg19 reference genome and counts were generated using the Golden Helix (Bozeman, MT) data analysis pipeline (goldenhelix.com). RefSeq annotations were used as the gene model and differential expression was assessed using the EdgeR software package (17). Reads for the candidate genes examined in this study were analyzed using custom python code.
Carotid Endarterectomy Patient Recruitment and Carotid Plaque Analysis
Patients with > 70% stenosis in the extracranial carotid artery bifurcation were offered enrollment in an IRB approved protocol to collect an intraoperative plasma sample, and carotid endarterectomy plaque specimen. On the day of the operation, consenting patients provided a > 6 h fasting plasma sample, and carotid endarterectomy plaque was removed en bloc. The maximally diseased endarterectomy segment is adjacent to the carotid bifurcation, and the minimally diseased endarterectomy segment is at the distal internal carotid artery. Sections of maximally and minimally diseased carotid artery plaque segments were lysed in hypotonic non-detergent based lysis buffer solution, and homogenized with a rotational power tissue homogenizer. Following centrifugation (12,000 RPM for 5 min) homogenate supernatant solutions were then standardized relative to protein concentration for further analysis.
Carotid Proteomic Screen
Following lysis, homogenization and standardization as described above, samples from two patients were then resolved by SDS-PAGE followed by Coomassie blue staining. A noticeable relative difference in band intensities was observed between 50–75 kDa. Highlighted bands were manually cut from the gel and in-gel trypsin digestion was performed before analysis by LC/MS/MS. NanoHPLC-ESI-MS/MS mass spectrometric analysis was performed on an LTQ-Orbitrap (Thermo) instrument. Samples were loaded with an autosampler onto a 15 cm Magic C18 column (5 µm particles, 300 Å pores, Michrom Bioresources) packed into a PicoFrit tip (New Objective) and analyzed with a nanoLC-2D plus HPLC (Eksigent). Analytical gradients were from 0–80% organic phase (95% acetonitrile, 0.1% formic acid) over 60 min. Aqueous phase composition was 2% acetonitrile, 0.1% formic acid. Eluent was routed into a PV-550 nanospray ion source (New Objective). The LTQ-Orbitrap was operated in a data-dependent mode with the precursor scan over the range m/z 350–2000, followed by twenty MS2 scans using parent ions selected from the MS1 scan. The Orbitrap AGC target was set to 1E06, and the MS2 AGC target was 1E04 with maximum injection times of 300 ms and 500 ms, respectively. Raw data were submitted to Mascot Server 2.0 and searched against the SwissProt database. Results were quantified by analyzing the mascot “dat” file and its respective thermo “raw” file with the home-brewed program (VC++) “SILACLTQ” (Supplementary Table 2). Protein rank and peptide queries matched were evaluated for each sample and hemopexin was identified to be among the top identified proteins in the analysis.
Statistical Analyses
The technician performing the Elisa and qPCR assays was blinded to sample cohort membership. All HPX values ± 3 SD from the mean were considered outliers and removed. All other non-normally distributed parameters were log transformed for analysis. Descriptive statistics in the text and figures are expressed as mean ± SEM. The Pearson’s correlation coefficient was calculated to test for correlation between 2 parameters. Comparisons between two groups were performed using an unpaired 2-tailed Student’s t test. Comparisons between maximally and minimally diseased portions of carotid plaques taken from the same individual were performed using a paired Student’s t test. Dunnett contrasts were used to compare expression among MAO, MNO and lean human subjects, using lean expression as the control. All statistical analyses were performed using R (r-project.org). All p-values <0.05 were considered significant.
Results
QTL genetic effects and identification of Hemopexin as a candidate gene
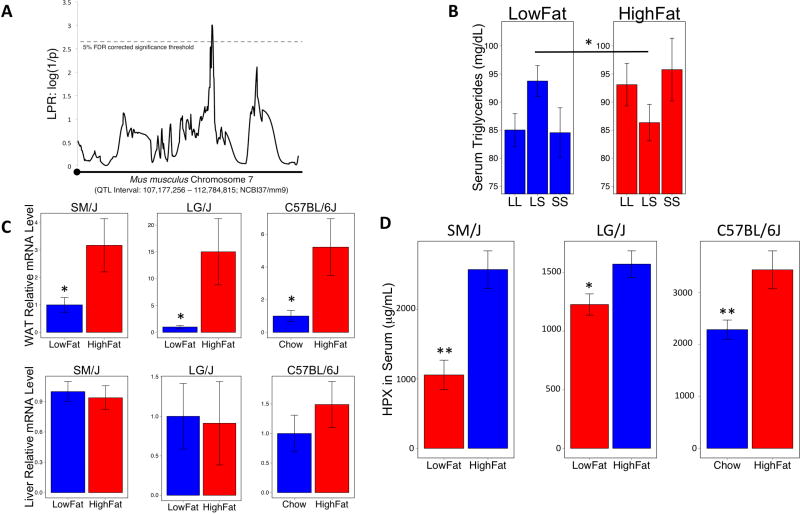
A quantitative trait locus (QTL) associated with variation in serum triglyceride levels was mapped to chromosome 7 (chr7: 100,028,746 – 105,636,301; GRCm38/mm10) in an F16 generation (n=1002) of an advanced intercross between the LG/J and SM/J inbred mouse strains (18), which have proven to be useful for genetic analysis of metabolic traits. The QTL interval overlaps an olfactory receptor cluster and contains 272 genes, of which 85 are protein coding. The genetic effects at the QTL are non-additive and show significant diet and dominance-by-diet interactions (Figure1a and 1b). Previously published microarray data suggest that 2 genes in liver (Inppl1, Smpd1) and 6 genes in white adipose tissue (reproductive fatpad; Arfip2, Frag1, Hpx, Lrrc51, Prkcdbp, Rhog) are differentially expressed on low-fat versus high-fat diets in both the LG/J and SM/J parental strains in this genomic region (19, 20). This pattern of diet-dependent, but not strain-dependent, differential expression is consistent with the diet-dependent genetic effects at the QTL. Of these 8 candidates identified by the microarray, the expression pattern of only 1 gene, Hpx, was validated by qPCR (Supplementary Table 3).
Figure 1. QTL genetic effects and hemopexin.
(A) A QTL associated with serum triglyceride levels mapped to a genomic region on chromosome 7. (B) The QTL shows diet (no difference in genotypic values between homozygotes within diets, but a significant difference between diets) and diet-by-dominance (the heterozygote genotypic value significantly deviates from the mean homozygote values but in different directions depending on diet) genetic effects. (C) Hemopexin was identified as a top candidate gene in the QTL, showing significant differential expression based on diet in white adipose tissue, but not in liver tissue, in three different strains of mice. qPCR normalized to L32; LG/J n= 13 high-fat and 12 low-fat fed animals, SM/J n= 14 high-fat and 13 low-fat fed animals, C57BL/6J n= 10 high-fat and 9 chow fed animals. (D) Serum HPX levels are higher on a high-fat diet, mirroring the expression pattern in white adipose tissue in all three strains. Data shown are from the C57BL/6J strain. * p < 0.05, **p < 0.01
To test the hypothesis that the diet-dependent expression pattern is strain-independent, we examined relative Hpx mRNA levels in liver and white adipose tissues from C57Bl/6J mice fed chow and high-fat diets as well (Figure 1c). We found that eating a high-fat diet increases Hpx mRNA levels in white adipose tissue (reproductive fat pad; p< 0.05), but not in liver, of fasted 20-week old mice representing three different genetic backgrounds. Serum Hpx protein content is also higher in high-fat fed mice relative to low-fat fed controls, regardless of genetic background (Figure 1d).
Hpx is expressed in adipocytes and affects adipogenesis
Because we detected nutritional regulation of Hpx in adipose tissue but not in liver, we tested if Hpx is expressed by the adipocyte itself by examining 3T3-L1 cells, a model of adipocyte differentiation. We found that levels of Hpx mRNA increase after differentiation is induced, with peak levels observed around the time neutral lipid accumulation peaks at 7 days following induction using the standard differentiation protocol described in the Materials/Subjects and Methods section (Supplementary Figure 1a). Since Hpx is known to be secreted (21), we explored whether HPX expression is also altered in the culture media of differentiated 3T3-L1 cells. Hpx protein increases in cell culture media during the differentiation process and is not detected in media that was not exposed to cells (Supplementary Figure 1b). Western blots of these cells during differentiation verified that Hpx protein levels increase in parallel to the increase in message (Supplementary Figure 1c).
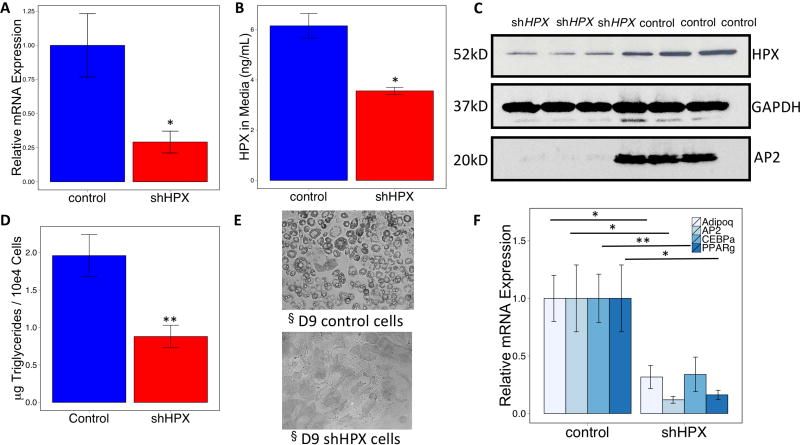
Some genes involved in iron metabolism can affect adipogenesis (22). To test the hypothesis that Hpx affects adipogenesis, we knocked down the expression of this protein in 3T3-L1 fibroblasts and subjected these cells to an adipocyte differentiation protocol. We documented that the Hpx knockdown decreased Hpx message (Figure 2a), Hpx protein in media (Figure 2b) and cells (Figure 2c), and showed that this intervention inhibited adipocyte differentiation. Lipid accumulation was decreased in concert with the lack of development of lipid droplets (Figure 2d,e) and the knockdown reduced expression of adiponectin and of canonical adipogenic genes aP2 (FABP4), CEBPalpha and PPARgamma (Figure 2f).
Figure 2. Knockdown of hemopexin affects adipogenesis.
(A) shRNA-mediated knockdown of Hpx in 3T3-L1 fibroblasts with subsequent puromycin selection and insulin-stimulated induction of differentiation resulted in (B) reduced Hpx in media, (C, D and E) inhibition of adipocyte differentiation, and (F) reduced expression of adipogenic genes. * p < 0.05, **p < 0.01
HPX expression is higher in adipose tissue from metabolically normal obese relative to metabolically abnormal obese human subjects and its circulating levels correlate with plasma triglycerides
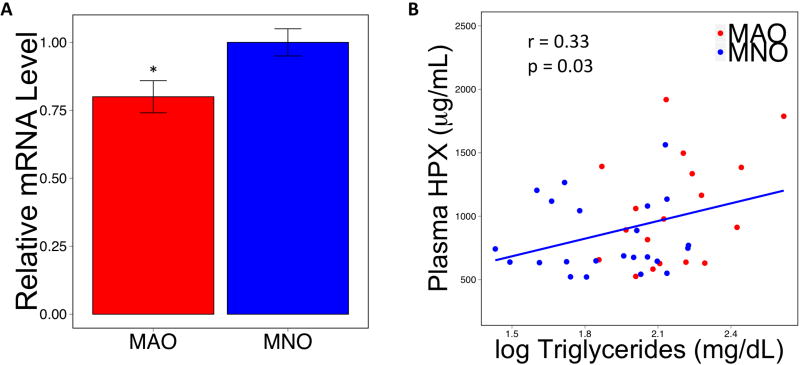
We examined transcriptome data from an RNA sequencing study of subcutaneous white adipose tissue collected from a cohort of human subjects who were categorized as metabolically normal obese (MNO) and metabolically abnormal obese (MAO; Lawson et al., manuscript in preparation). Fasting plasma glucose and insulin concentrations, HOMA-IR, intrahepatic triglyceride content and serum triglyceride concentrations were greater in MAO than MNO subjects (Table 1). None of the human genes that were homologous to the candidates found in the mouse QTL study passed genome-wide significance, but using the mouse results as an a priori hypothesis protects candidates from the extreme burden of multiple tests correction (23). Of the genes represented by high quality reads and without correction for genome-wide multiple tests, only HPX showed a difference between the MNO and MAO groups. HPX message was more than twice as high in the adipose tissue of MNO as compared to MAO subjects (Supplementary Figure 2). Further, HPX message levels in adipose tissue were similar between MNO subjects and a small sample of lean individuals (p = 0.95) but were nearly significantly different between MAO subjects and lean individuals (p = 0.08) (Supplementary Figure 3). The increased expression in MNO relative to MAO adipose tissue was confirmed by qPCR (Figure 3a).
Table 1.
Characteristics of MAO and MNO Subjects
| MAO | MNO | |
|---|---|---|
| Subjects (M/F) | 9/9 | 3/21 |
| Ethnicity (AA/EA)‡ | 2/16 | 11/13 |
| Age (years) | 45.3 ± 2.5 | 42.5 ± 2.2 |
| BMI (kg/m2) | 35.8 ± 0.9 | 33.8 ± 0.6 |
| FM (%) | 43.0 ± 1.4 | 43.8 ± 1.6 |
| IHTG % | 17.7 ± 1.9*** | 2.53 ± 0.2 |
| Glucose (mg/dL) | 99.8 ± 2.3** | 92.0 ± 1.2 |
| Insulin (mU/L) | 22.8 ± 2.5*** | 9.3 ± 4.0 |
| HOMA-IR | 5.7 ± 0.7*** | 2.1 ± 0.2 |
| Total cholesterol (mg/dL) | 186.7 ± 6.6 | 171.3 ± 6.7 |
| HDL-cholesterol (mg/dL) | 39.5 ± 1.8** | 48.3 ± 2.2 |
| LDL-cholesterol (mg/dL) | 115.1 ± 5.2 | 105.3 ± 5.1 |
| Triglycerides (mg/dL) | 161.9 ± 20.1*** | 88 ± 9.07 |
| HPX (µg/mL) | 1044.5 ± 100.6 | 823.7 ± 59.1 |
Values are mean ± SEM;
p=0.05,
p=0.01;
p<0.001
FM – fat mass, IHTG – intrahepatic triglycerides, HPX – hemopexin in plasma
AA- African-American, EA- European-American
Figure 3. Hemopexin is differentially expressed in MNO relative to MAO adipose and correlated with plasma triglycerides.
(A) HPX message is higher in subcutaneous adipose tissue of MNO relative to MAO human subjects. (B) Circulating HPX content in these same subjects has the opposite pattern and (C) is positively correlated with plasma triglyceride levels. * p < 0.05
Circulating HPX levels were positively correlated with log-transformed triglyceride levels in the pooled MNO and MAO subjects (Figure 3b). When the groups are examined separately, the correlation is strongest in the MAO subjects (MAO r = 0.38, p = 0.11; MNO r = 0.22, p = 0.31). The group-specific correlations are not statistically significant, likely because subject sample sizes are small. There was no correlation between circulating HPX and any other circulating parameters measured in these subjects (Table 1; total cholesterol r = 0.07, p = 0.64; HDL-cholesterol r = −0.29, p = 0.06; LDL-cholesterol r = −0.03, p = 0.84; insulin r = 0.17, p = 0.27; glucose r = −0.03, p = 0.80).
HPX levels are increased in maximally diseased segments of carotid artery plaques
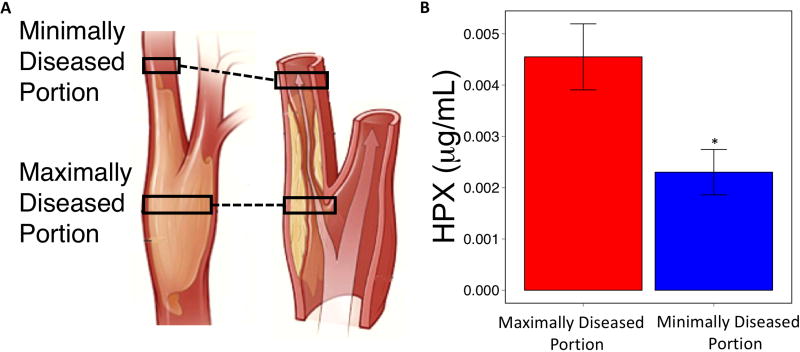
Because vascular disease is the most lethal complication of obesity, we conducted a pilot unbiased proteomics screen comparing maximally and minimally diseased carotid artery plaque segments from two patients undergoing carotid endarterectomy. HPX was identified in maximally diseased segments (Supplementary Table 2). Accordingly, further analysis of HPX expression was evaluated in carotid endarterectomy plaques collected from a cohort of patients with high-grade carotid artery occlusive disease (Table 2). When maximally diseased and minimally diseased segments (Figure 4a) from this cohort were compared, HPX was found to be increased in the maximally diseased samples (Figure 4b). Maximally diseased carotid endarterectomy segments had non-significantly higher levels of esterified cholesterol relative to minimally diseased segments. However, as previously reported (24–26), there was a significant difference in triglyceride content between maximally and minimally diseased carotid plaque segments (Table 2).
Table 2.
Characteristics of Carotid Endarterectomy Patients and of Carotid Artery Plaque Segments
| Patients | ||
|---|---|---|
| Patients (M/F) | 12/6 | |
| Age (years) | 71.3 ± 1.8 | |
| BMI (kg/m2) | 28.9 ± 1.4 | |
| Diabetes status (D/ND) | 11/7 | |
| Hypertension (Y/N) | 14/4 | |
| HPX in plasma (µg/mL) | 1260.5 ± 76.3 | |
| Glucose (mg/dL) | 130.0 ± 11.5 | |
| Triglycerides (mg/dL) | 58.1 ± 6.0 | |
| Total cholesterol (mg/dL) | 67.5 ± 2.7 | |
| Free fatty acids (mmol/L) | 0.44 ± 0.05 | |
|
| ||
| Carotid Plaque Segments | ||
|
| ||
| Maximally Diseased Segment# | Minimally Diseased Segment# | |
|
| ||
| HPX (µg/mL) | 0.0045 ± 0.0006* | 0.0023 ± 0.0004 |
| Esterified cholesterol (µg/mL) | 300.5 ± 53.3 | 182.8 ± 45.1 |
| Triglycerides (mg/dL) | 0.60 ± 0.14* | 0.27 ± 0.04 |
Values are mean ± SEM;
p=0.05
See Figure 4 for illustration of the anatomical distinction between maximally and minimally diseased segments; Value is normalized to weight and protein content of portion assayed
Figure 4. Hemopexin content is higher in maximally diseased carotid artery plaque segments.
(A) Anatomical distinction between maximally and minimally diseased plaque segments at the extracranial carotid artery bifurcation. (B) Difference in hemopexin content between maximally and minimally diseased carotid artery segments. * p < 0.05
When examining the correlation between HPX and triglyceride contents of the maximally diseased and minimally diseased segments, we found a weak positive correlation in the minimally diseased portions (r = 0.42, p = 0.13) and a suggestive negative correlation in the maximally diseased portions (r = −0.51, p = 0.07). We found no correlation between HPX and cholesterol contents of the minimally diseased portions (r = 0.07, p = 0.79), however there is a suggestive negative correlation in the maximally diseased portions (r = −0.49, p = 0.07).
In contrast to our findings in the MNO/MAO subjects, there was no correlation between circulating HPX and serum triglycerides (r = 0.14, p = 0.59) in the carotid endarterectomy patients, a group with less adiposity and lower triglycerides than the MNO/MAO subjects. Similarly, circulating HPX was not correlated with other circulating parameters measured in these patients, although the correlation with cholesterol was suggestive (total cholesterol r = 0.40, p = 0.10; free fatty acids r = −0.12, p = 0.63; glucose r = 0.31, p = 0.21).
Discussion
Triglycerides are a biomarker of human metabolic disease. Their elevation is associated with obesity and insulin resistance (27), as well as increased risk of vascular disease (3, 4). The triglyceride molecule, a neutral lipid, is thought to be metabolically inert so the mechanisms linking triglycerides to disease are not understood. We employed genetic mapping in mice to identify novel potential mediators of the relationship between triglycerides and disease.
In a QTL associated with variation in serum triglycerides, Dserum7b, mapped in the F16 generation of a LG/J × SM/J advanced intercross mouse population (18), Hpx emerged as a promising candidate gene. There are 5 small deletion polymorphisms in the SM/J founder strain relative to the LG/J founder strain in the Hpx genic region (Supplementary Table 4). Four of these polymorphisms fall in intronic sequence while the 5th, a 6bp deletion, falls in an exonic region (28). The expression pattern of Hpx is similar between the two strains, however it is possible that these polymorphisms contribute to the observed difference in expression magnitude (Figure 1c). HPX also falls in two other independently mapped and overlapping QTL associated with variation in serum lipids in mice: Chldq4 identified in a MRL/MpJ × SJL/J cross (29), and Hdlcl1 identified in a C57BL/6J × PERA/EiJ cross (30). HPX is associated with triglycerides (31) in human subjects with diabetes, and with cholesterol in human subjects with coronary artery disease (15). We did not find a significant association between HPX and cholesterol in this study, but our patient numbers are small.
Our observation that HPX is positively associated with serum triglycerides in MAO and MNO people supports our murine findings. We observed increased HPX levels in MAO subjects, who have significantly higher triglyceride levels than the MNO subjects (Table 1). The higher HPX content of maximally diseased segments of carotid plaques relative to minimally diseased segments obtained from endarterectomy patients further reinforces the connection between HPX and triglycerides. There is a significant difference in triglyceride content between minimally and maximally diseased segments in these patients (Table 2), a finding that confirms previous studies (24–26).
A surprising result of this study is the observation that HPX is expressed by the adipocyte. We found that during adipocyte differentiation, Hpx expression increases strikingly with peak levels occurring at the time neutral lipid accumulation peaks in 3T3-L1 cells. Recent research has demonstrated that other genes involved in iron metabolism are expressed by adipocytes (32). We found that knocking HPX down in 3T3-L1 fibroblasts affected differentiation of these cells into adipocytes, which is consistent with other work in 3T3-L1 cells and in human adipose tissue explants showing that disruption of genes involved in iron metabolism antagonizes adipogenesis (22). We find that Hpx expression is higher in the reproductive fat pads of mice fed high-fat diets compared to mice fed low-fat diets, regardless of genetic background. We also find that HPX expression in subcutaneous white adipose tissue is higher in MNO relative to MAO subjects. While the cellular mechanisms responsible for the differences in metabolic function between MNO and MAO individuals are unclear, it is possible that HPX activity in adipose tissue contributes to metabolic benefits in MNO individuals. This may be because HPX is permissive for adipocyte differentiation and affects adiponectin expression, as indicated in cell culture (Figure 2). There is some evidence that MNO people have ‘healthy-fat’ with a greater capacity for storing excess lipids and preventing triglycerides from accumulating in other tissues and increasing disease risk (16). Given that HPX tracks with triglycerides, our results suggest that increased HPX in adipose tissue may be beneficial while in blood and carotid endarterectomy plaque segments increased HPX is a biomarker for elevated triglycerides and associated risk of metabolic disease.
Our study leverages results generated by multiple experimental procedures: mouse QTL mapping, human and mouse gene expression analyses, cell culture, and human and mouse proteomics. In sum, we identified HPX as a top candidate gene in a forward genetics study and found that it is correlated with triglycerides in both mice and human subjects. HPX is expressed in adipose tissue, regulated by nutritional state, participates in lipogenesis and adipogenesis, and affects expression of adiponectin. Increased HPX content localizes to maximally diseased portions of carotid endarterectomy plaque segments, and those segments have higher triglyceride content. Our data reinforces the links among heme, HPX and cardiovascular disease (33, 34) and introduces triglycerides as a new player in this complex. Obesity is associated with serious metabolic complications. Triglycerides are a biomarker for these complications but how they relate to disease pathophysiology is uncertain. Understanding the mechanisms of the triglyceride-HPX axis could provide new insight into how obesity and elevated triglycerides impair vascular health.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Irfan Lodhi for sharing his 3T3-L1 differentiation protocol and Dr Larry Spears for his assistance in trouble shooting the Western blot experiments. This work was supported by NIH-NIDDK K01 DK95003 and P30 DK56341 to HAL, and in part by the EA annual grant contest (2012) co-sponsored by Illumina and Golden Helix to LLG, SK and HAL. The content of this paper is solely the responsibility of the authors.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest
Author Contributions
HAL designed the study, conducted experiments, analyzed data, wrote the manuscript and secured funding and collaborations. MZ performed the carotid endarterectomy operations, prepared the carotid artery plaque homogenates, conducted the pilot proteomics scan, and assisted with manuscript preparation. JPW conducted experiments and assisted with manuscript preparation. EF collected and prepared MNO/MAO subject data. LLG prepared the MNO/MAO RNA samples. SK provided MNO/MAO subject data and samples and assisted with manuscript preparation. CFS assisted in study design, contributed reagents, provided laboratory space and equipment for experiments, and assisted in manuscript preparation.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Pi-Sunyer X, Blackburn G, Brancati FL, Bray GA, Bright R, Clark JM, et al. Reduction in weight and cardiovascular disease risk factors in individuals with type 2 diabetes: one-year results of the look AHEAD trial. Diabetes Care. 2007;30:1374–1383. doi: 10.2337/dc07-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freiberg JJ, Tybjaerg-Hansen A, Jensen JS, Nordestgaard BG. Nonfasting triglycerides and risk of ischemic stroke in the general population. JAMA. 2008;300:2142–2152. doi: 10.1001/jama.2008.621. [DOI] [PubMed] [Google Scholar]
- 4.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 5.Paoli M, Anderson BF, Baker HM, Morgan WT, Smith A, Baker EN. Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains. Nat Struct Biol. 1999;6:926–931. doi: 10.1038/13294. [DOI] [PubMed] [Google Scholar]
- 6.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 7.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 8.Morgan WT, Smith A. Binding and Transport of Iron-Porphyrins by Hemopexin. In: Sykes AG, editor. Advances in Inorganic Chemistry. Vol. 51. Academic Press; San Diego: 2001. [Google Scholar]
- 9.Lin T, Sammy F, Yang H, Thundivalappil S, Hellman J, Tracey KJ, et al. Identification of hemopexin as an anti-inflammatory factor that inhibits synergy of hemoglobin with HMGB1 in sterile and infectious inflammation. J Immunol. 2012;189:2017–2022. doi: 10.4049/jimmunol.1103623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith A, Morgan WT. Hemopexin-mediated heme uptake by liver. Characterization of the interaction of heme-hemopexin with isolated rabbit liver plasma membranes. J Biol Chem. 1984;259:12049–12053. [PubMed] [Google Scholar]
- 11.Baumann H, Jahreis GP, Gaines KC. Synthesis and regulation of acute phase plasma proteins in primary cultures of mouse hepatocytes. J Cell Biol. 1983;97:866–876. doi: 10.1083/jcb.97.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li RC, Saleem S, Zhen G, Cao W, Zhuang H, Lee J, et al. Heme-hemopexin complex attenuates neuronal cell death and stroke damage. J Cereb Blood Flow Metab. 2009;29:953–964. doi: 10.1038/jcbfm.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt RC, Hunt DM, Gaur N, Smith A. Hemopexin in the human retina: protection of the retina against heme-mediated toxicity. J Cell Physiol. 1996;168:71–80. doi: 10.1002/(SICI)1097-4652(199607)168:1<71::AID-JCP9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Swerts JP, Soula C, Sagot Y, Guinaudy MJ, Guillemot JC, Ferrara P, et al. Hemopexin is synthesized in peripheral nerves but not in central nervous system and accumulates after axotomy. J Biol Chem. 1992;267:10596–10600. [PubMed] [Google Scholar]
- 15.Watanabe J, Grijalva V, Hama S, Barbour K, Berger FG, Navab M, et al. Hemoglobin and its scavenger protein haptoglobin associate with apoA-1-containing particles and influence the inflammatory properties and function of high density lipoprotein. J Biol Chem. 2009;284:18292–18301. doi: 10.1074/jbc.M109.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabbrini E, Yoshino J, Yoshino M, Magkos F, Tiemann Luecking C, Samovski D, et al. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Invest. 2015;125:787–795. doi: 10.1172/JCI78425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson HA, Zelle KM, Fawcett GL, Wang B, Pletscher LS, Maxwell TJ, et al. Genetic, epigenetic, and gene-by-diet interaction effects underlie variation in serum lipids in a LG/JxSM/J murine model. J Lipid Res. 2010;51:2976–2984. doi: 10.1194/jlr.M006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson HA, Cady JE, Partridge C, Wolf JB, Semenkovich CF, Cheverud JM. Genetic effects at pleiotropic loci are context-dependent with consequences for the maintenance of genetic variation in populations. PLoS Genet. 2011;7:e1002256. doi: 10.1371/journal.pgen.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge CG, Fawcett GL, Wang B, Semenkovich CF, Cheverud JM. The effect of dietary fat intake on hepatic gene expression in LG/J AND SM/J mice. BMC Genomics. 2014;15:99. doi: 10.1186/1471-2164-15-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Cushman SW, Pannell LK, Hess S. Quantitative proteomic analysis of the secretory proteins from rat adipose cells using a 2D liquid chromatography-MS/MS approach. J Proteome Res. 2005;4:570–577. doi: 10.1021/pr049772a. [DOI] [PubMed] [Google Scholar]
- 22.Moreno-Navarrete JM, Ortega F, Moreno M, Ricart W, Fernandez-Real JM. Fine-tuned iron availability is essential to achieve optimal adipocyte differentiation and mitochondrial biogenesis. Diabetologia. 2014;57:1957–1967. doi: 10.1007/s00125-014-3298-5. [DOI] [PubMed] [Google Scholar]
- 23.Lawson HA. Animal Models of Metabolic Syndrome. In: Conn M, editor. Animal Models for the Study of Human Disease. Elsevier; Cambridge: 2013. pp. 243–264. [Google Scholar]
- 24.Chen Z, Ichetovkin M, Kurtz M, Zycband E, Kawka D, Woods J, et al. Cholesterol in human atherosclerotic plaque is a marker for underlying disease state and plaque vulnerability. Lipids Health Dis. 2010;9:61. doi: 10.1186/1476-511X-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen E, Aviram M, Khatib S, Rabin A, Mannheim D, Karmeli R, et al. Increased Levels of Human Carotid Lesion Linoleic Acid Hydroperoxide in Symptomatic and Asymptomatic Patients Is Inversely Correlated with Serum HDL and Paraoxonase 1 Activity. J Lipids. 2012;2012:762560. doi: 10.1155/2012/762560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mas S, Touboul D, Brunelle A, Aragoncillo P, Egido J, Laprevote O, et al. Lipid cartography of atherosclerotic plaque by cluster-TOF-SIMS imaging. Analyst. 2007;132:24–26. doi: 10.1039/b614619h. [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin T, Abbasi F, Cheal K, Chu J, Lamendola C, Reaven G. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann Intern Med. 2003;139:802–809. doi: 10.7326/0003-4819-139-10-200311180-00007. [DOI] [PubMed] [Google Scholar]
- 28.Nikolskiy I, Conrad DF, Chun S, Fay JC, Cheverud JM, Lawson HA. Using whole-genome sequences of the LG/J and SM/J inbred mouse strains to prioritize quantitative trait genes and nucleotides. BMC Genomics. 2015;16:415. doi: 10.1186/s12864-015-1592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava AK, Mohan S, Masinde GL, Yu H, Baylink DJ. Identification of quantitative trait loci that regulate obesity and serum lipid levels in MRL/MpJ × SJL/J inbred mice. J Lipid Res. 2006;47:123–133. doi: 10.1194/jlr.M500295-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Seidelmann SB, De Luca C, Leibel RL, Breslow JL, Tall AR, Welch CL. Quantitative trait locus mapping of genetic modifiers of metabolic syndrome and atherosclerosis in low-density lipoprotein receptor-deficient mice: identification of a locus for metabolic syndrome and increased atherosclerosis on chromosome 4. Arterioscler Thromb Vasc Biol. 2005;25:204–210. doi: 10.1161/01.ATV.0000149146.32385.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gebre-Medhin M, Ewald U, Tuvemo T. Reduced serum proteins in diabetic children on a twice-daily insulin schedule. Acta Paediatr Scand. 1985;74:961–965. doi: 10.1111/j.1651-2227.1985.tb10065.x. [DOI] [PubMed] [Google Scholar]
- 32.Gabrielsen JS, Gao Y, Simcox JA, Huang J, Thorup D, Jones D, et al. Adipocyte iron regulates adiponectin and insulin sensitivity. J Clin Invest. 2012;122:3529–3540. doi: 10.1172/JCI44421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeney V, Balla G, Balla J. Red blood cell, hemoglobin and heme in the progression of atherosclerosis. Frontiers in Physiology. 2014;5:1–11. doi: 10.3389/fphys.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta NU, Grijalva V, Hama S, Wagner A, Navab M, Fogelman AM, et al. Apolipoprotein E−/− Mice Lacking Hemopexin Develop Increased Atherosclerosis via Mechanisms That Include Oxidatice Stress and Altered Macrophage Function. Arterioscler Thromb Vasc Biol. 2016;36:1152–1163. doi: 10.1161/ATVBAHA.115.306991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.