Abstract
Cocirculation of subtype B and CRF01_AE in Southeast Asia has led to the establishment of new recombinant forms. In our previous study, we found five samples suspected of being recombinants between subtype B and CRF01_AE, and here, we analyzed near full-length sequences of two samples and compared them to known CRFs_01B, subtype B, and CRF01_AE. Five overlapped segments were amplified with nested PCR from PBMC DNA, sequenced, and analyzed for genome mosaicism. The two Indonesian samples, 07IDJKT189 and 07IDJKT194, showed genome-mosaic patterns similar to CRF33_01B references from Malaysia, with one short segment in the 3′ end of the p31 integrase-coding region, which was rather more similar to subtype B than CRF01_AE, consisting of unclassified sequences. These results suggest gene-specific continuous diversification and spread of the CRF33_01B genomes in Southeast Asia.
As a retrovirus, HIV-1 is prone to natural error of viral reverse transcriptase. In combination with a high rate of virion production in infected individuals and genome recombination during genome synthesis, HIV-1 exhibits an extensive genetic diversity in nature.1,2 Cocirculation of two or more different subtypes and/or circulating recombinant forms (CRFs) in HIV-1 high-risk populations can lead to HIV-1 intersubtype recombination.3–5 To date, 9 different subtypes and 43 CRFs have been reported.6 In addition to the reported CRFs, numerous unique recombinant forms (URFs) were also identified worldwide, including Southeast Asia,5,7,8 where cocirculation of subtype B and CRF01_AE as the main circulating subtypes resulted in the generation of CRFs, such as CRF15_01B,9 CRF33_01B,10 and CRF34_01B.11 These CRFs originated and mostly circulated in their countries of origin.9–11
Our previous study showed that CRF01_AE was the main circulating subtype in Indonesia, followed by small number of subtype B. Furthermore, we also found five samples suspected of being CRF_01B URFs.12 In this current study we analyzed near full-length sequences of two samples showing recombination patterns similar to CRF33_01B previously reported in Malaysia. These samples were obtained during a cross-sectional study of molecular epidemiology in 2007, from the Sulianti Saroso Infectious Diseases Hospital in Northern Jakarta, Indonesia. 07IDJKT189 is a 25-year-old male with numerous risk factors; the subject was an injecting drug user (IDU) who also had a history of sexual contact with commercial sex workers whose HIV status was unknown. The patient presented with opportunistic infections such as lung tuberculosis, oral candidiasis, and cytomegalovirus (CMV) retinitis, which led to the diagnosis of HIV. The CD4 count was 17 cells/μl at admission. 07IDJKT194 is a 21-year-old female with heterosexual risk factors. She was screened for HIV infection due to her spouse's HIV status. Her CD4 count was 360 cells/μl at admission. Both subjects were antiretroviral (ARV) naive, without any known relationship to each other (Table 1).
Table 1.
Subjects' Clinical Characteristics
| Characteristic | 07IDJKT189 | 07IDJKT194 |
|---|---|---|
| Gender | Male | Female |
| Age (years) | 25 | 21 |
| Risk factor | IDU and heterosexa | Heterosex |
| Initiation of risky behavior | 2004 | 2006 |
| Ethnicity | Javanese | Batavian |
| Education | High School | High School |
| Occupation | Employee | Housewife |
| Marital status | Not married | Married |
| Sex partner HIV status | Unknown | Positive |
| Treatment status | Naive | Naive |
| CD4 count (cells/μl) | 17 | 360 |
| Disease stage | AIDS | HIV |
| Opportunistic infection | Presentb | Absent |
| Hepatitis B infection | Positivec | Unknown |
| Hepatitis C infection | Negative | Unknown |
IDU, injecting drug use; heterosex, heterosexual transmission.
Opportunistic infections: lung tuberculosis, oropharyngeal candidiasis, and cytomegalovirus retinitis.
Based on positive HBsAg result.
Blood samples were collected during enrollment; peripheral blood mononuclear cells (PBMCs) were separated, and DNA was extracted as described previously.12 First round amplifications of the 9.0-kbp genome cDNAs were performed by long range polymerase chain reaction (PCR) amplification using the Takara ExTaq Hot Start Version (Takara Bio Inc, Otsu, Japan) with the following primer sets: 172A (sense, 5′-ATCTCTAGCAGTGGCGCCCGAACAG-3′) and 9KU5B (antisense, 5′-GGTCTGAGGGATCTCTAGTTACCAG-3′), followed by nested PCR of five overlapping genome segments from 1 kbp to 2.3 kbp using the primers listed in Table 2. The nested PCR products were purified and directly sequenced as reported previously,12 using primers designed to sequence overlapping regions. Five-nucleotide sequence segments were assembled using Sequencer v.4.8 to form a near full-length genome sequence, and aligned with reported genome sequences of various HIV-1 major group M in the world by MEGA v.413 for phylogenetic tree construction and recombination analysis. SimPlot v.3.514 and RDP v.3 software packages were used to analyze possible recombination events.
Table 2.
Primers Used for Nested PCR Amplification of Five Overlapped Segments
| Segment | Primer | Gene | Locationa | Sequence |
|---|---|---|---|---|
| 1 | F: 174A | gag–pol | 683 ⇒ 707 | CTCTCGACGCAGGACTCGGCTTGCT |
| R: RT2-28 | 3012 ⇐ 3031 | TGGAATATTGCTGGTGATCC | ||
| 2 | F: RT3-27 | pol–int | 2798 ⇒ 2818 | AACTCAAGACTTCTGGGAAGT |
| R: Pol-3′ | 4899 ⇐ 4919 | GCTGTCCCTGTAATAAACCCG | ||
| 3 | F: pol/tat-5′ | int–env | 4808 ⇒ 4829 | GTACAGTGCAGGGGAAAGAATA |
| R: env SB-3′ | 6990 ⇐ 7010 | ATTTAACAGCAGTTGAGTTGA | ||
| 4 | F: 106A | env | 6867 ⇒ 6889 | ATACATTATTGTRCTCCRGCTGG |
| R: env (Y,Y)b | 8288 ⇐ 8308 | AAACCTAYYAAGCCTCCTACT | ||
| SK69c | 7911 ⇐ 7937 | CCAGACTGTGAGTTGCAACAG | ||
| 5 | F: env/nef (R,Y)b | env-3′L | 8221 ⇒ 8243 | GGRCAAGTYTGTGGAATTGGTTT |
| SK68c | TR | 7796 ⇒ 7815 | AGCAGCAGGAAGCACTATGG | |
| R: MSR5 | 9606 ⇐ 9627 | CAAGGCAAGCTTTATTGAGGCT | ||
| 2–3 | F: pol/seq2 | pol–vif | 3876 ⇒ 3901 | GATGGGGCAGCTARYAGGGARACTAA |
| R: vif365B | 5782 ⇐ 5805 | GGGTGTCGACATAGCAGAATAGGC |
Location is in reference to HIV HXB2 (K03455).
Primer used for 07IDJKT189.
Primer used for 07IDJKT194.
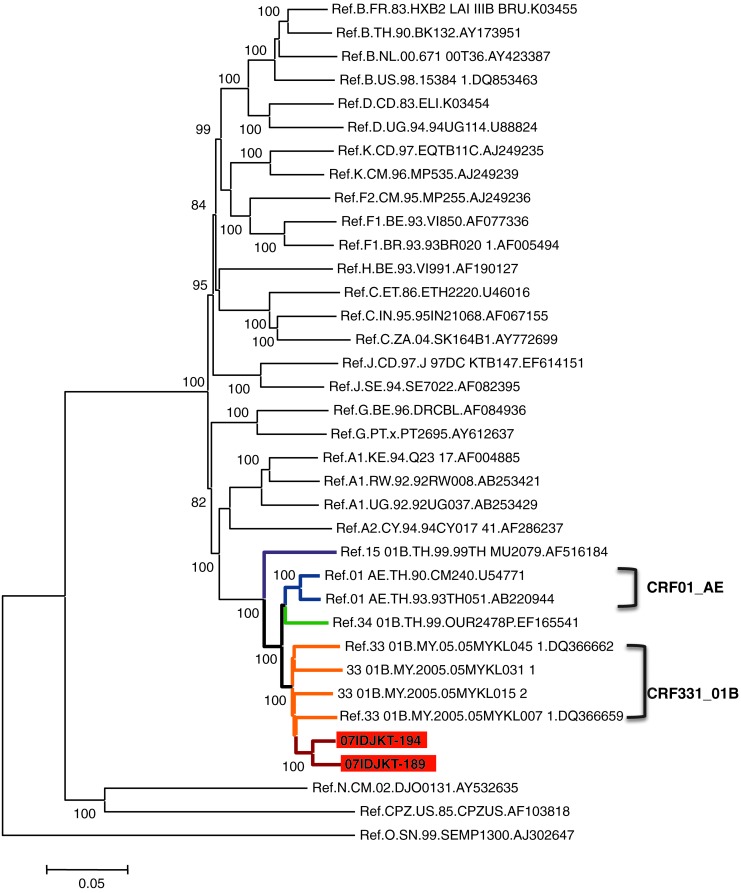
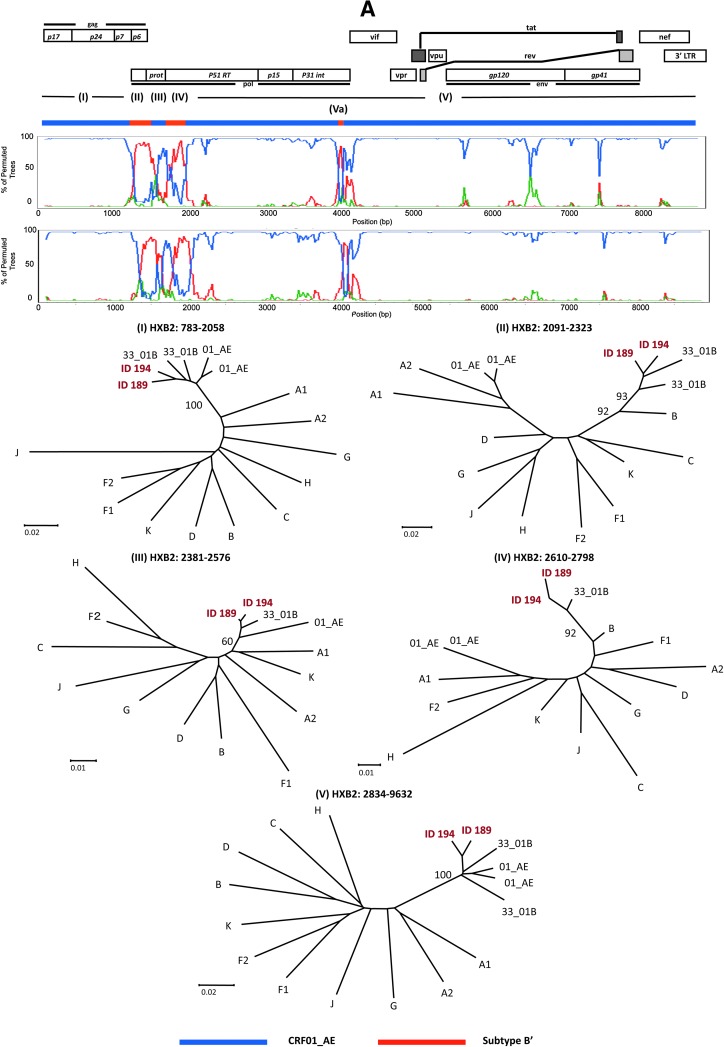
A representative neighbor-joining tree of the near full-length genome sequences showed that both 07IDJKT189 and 07IDJKT194 were clustered closely to each other, and formed a branch node with other CRF33_01B Malaysian sequences with high bootstrap values (Fig. 1). This branch joined the CRF01_AE, CRF34_01B, and CRF15_01B branches and formed a major branch node (Fig. 1). These results raised the possibility that 07IDJKT189 and 07IDJKT194 had mosaic genomes similar to the Malaysian CRF33_01B genomes.10 To assess this possibility, we performed bootscanning plot analysis as described previously.15 The analysis confirmed that 07IDJKT189 and 07IDJKT194 indeed were mosaics of CRF01_AE and subtype B in Southeast Asia (subtype B′) and had a mosaic pattern similar to the CRF33_01B genomes (Fig. 2A, bootscanning plots and phylogenetic trees): the four putative recombination breakpoints located around the 3′ end of the gag gene or the 5′ end of the pol gene constantly resulted in the highest statistical significance with various recombination detection tools in the RDP software package (p < 0.01).
FIG. 1.
Phylogenetic classification of near full-length genome sequences of 07IDJKT189 and 07IDJKT194. The tree was constructed with the neighbor-joining method using MEGA v.4. Reference sequences of HIV-1 major group M were obtained from the Los Alamos National Laboratory HIV Database (http://hiv-web.lanl.gov/). Two more sequences of CRF33_01B from Malaysia (accession numbers DQ366660 and DQ366661) were included in this analysis. The reliability of interior branches in the phylogenetic tree was assessed by the bootstrap method with 500 resamplings.
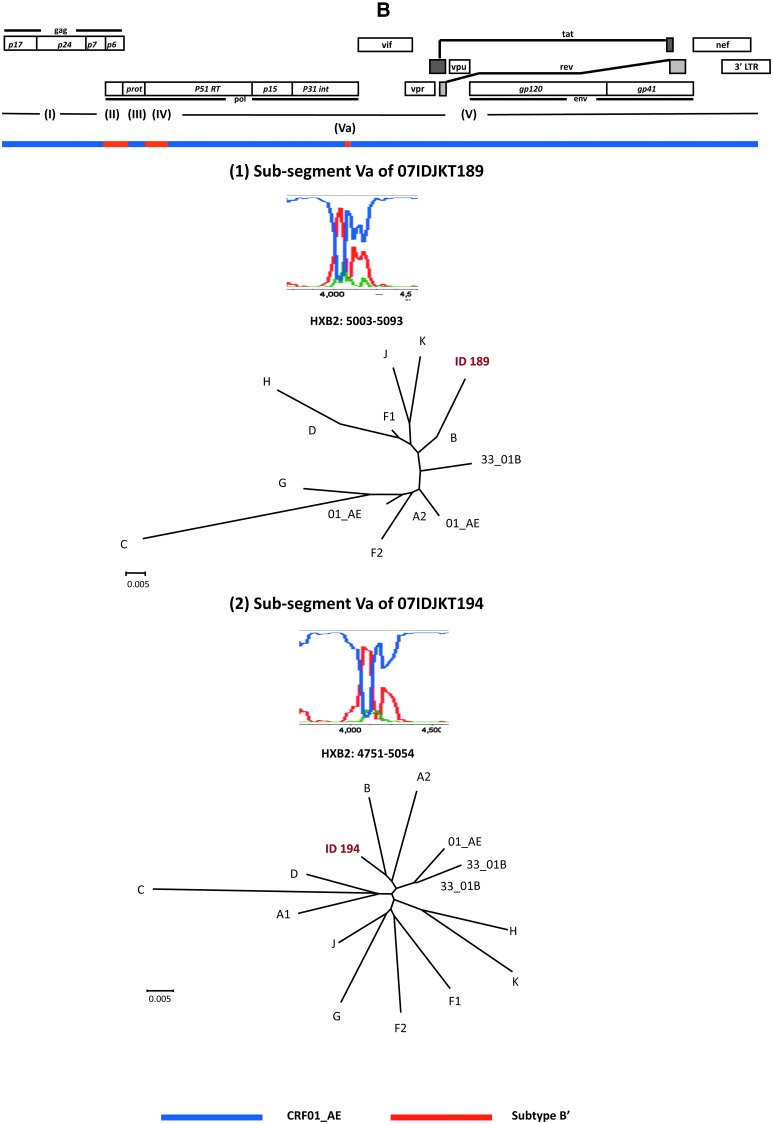
FIG. 2.
Phylogenetic evidence for genome mosaicism of Indonesian HIV-1 strains. (A) Bootscanning plots of nucleotide sequences of near full-length genomes of 07IDJKT189 and 07IDJKT194 from Indonesian blood samples. Each query genome sequence was aligned with three reference sequences with CLUSTAL W software, using subtype B Thai variant (subtype B′; AY173951) and CRF01_AE (U54771) as putative parental strains, with subtype C (AF067155) as the outgroup. Bootstrap values were plotted for a window with 200 bp moving in increments of 20 bp. Neighbor-joining trees were constructed with segment (I), (II), (III), (IV), and (V) sequences and are shown under the bootscanning plots. (B) Phylogenetic relationship of the subgenome segment. The 1.9-kb segment containing portions of the integrase and Vif coding regions of 07IDJKT189 and 07IDJKT194 was amplified, cloned into plasmid, and subjected to bootscanning plot and phylogenetic tree analyses. Trees were constructed with Va region sequences (91 and 304 nt for 07IDJKT189 and 07IDJKT194, respectively). Reference sequences used for phylogenetic tree construction include HIV-1 A1 (AF004885), A2 (AF286238), B (AY173951), C (AF067155), F1 (AF005494), F2 (AY371158), G (U88826), H (AF005496), J (EF614151), K (AJ249239), CRF01_AE (U54771) and (AB220944), and CRF33_01B (DQ3666662) and (DQ366659).
Notably, the short genome segments corresponding to the border of the integrase and Vif coding regions were phylogenetically similar to subtype B and highly diverse among the 07IDJKT189, 07IDJKT194 (subsegment Va in Fig. 2B), and CRF33_01B genomes.16 We suspected that there was a possibility of recombination events in this region. However, we could not define appreciable recombination breakpoints and parental sequences for recombination by using available recombination detection tools in the RDP v.3 software package. This might be because the mosaic segment is too short to accumulate sufficient informative sites for assessing the statistical significance of the recombination events. Therefore it remains unclear at present how and why this region tends to be more diversified than the other regions. Interestingly, bootscanning analysis of this region in Malaysian CRF33_01B sequences obtained from http://hiv-web.lanl.gov/ also shows similar results (data not shown), suggesting this region might be the new breakpoint site for recombination to occur.
To clarify whether the mosaicism and the diversity in the Indonesian CRF33_01B genomes exist at the clonal genome level, we cloned both the gag–pol and pol–vif regions from blood samples. Corresponding segments were amplified with segment 1 and 2–3 primer sets (Table 2) and cloned into plasmids using the TOPO XL PCR Cloning Kit (Invitrogen, Carlsbad, CA). The patterns of the bootscanning plots of the 2.3 kb containing Gag and part of the Pol coding region and the 1.9 kb containing the integrase and Vif coding region were almost identical between the sequences of the uncloned and cloned genome segment (Fig. 2, bootscanning plots). Similarly, results of phylogenetic tree analysis of both regions were consistent with those obtained with uncloned sequences (Fig. 2, phylogenetic trees). Taken together, these results suggest the presence of the mosaicism and diversity at these regions in the single genome segment.
Both Indonesian subjects came from different risk factor groups, and had no connection with each other with no history of traveling to Malaysia or contact with Malaysian people. This suggests that CRF33_01B might have been circulating in Indonesian long before both patients initiated their risky behaviors and have been crossing between risk factor groups. Moreover, the end-of-integrase variations of subtype B′ in Indonesian CRF33_01B sequences suggest an evolutionary process that might have started in Malaysia and further developed in and spread to other countries, including Indonesia. So far several new CRFs in Southeast Asia have been reported, and they tended to circulate locally in the beginning of their endemic period. After a certain period of time, however, they started to spread to other countries.6 The Indonesian CRF33_01B variants not only support this phenomenon, but also suggest that these new CRFs are still evolving in each country and might contribute to the generation of other new CRFs by recombining with local strains of HIV-1.
Continuous study is necessary to see how HIV-1 has spread and evolved in Indonesia and in Southeast Asia in general. Recombination may alter the viral characteristics and thus influence the pathogenesis and/or the ability of the virus to survive, including the ability to generate resistance to antiretroviral drugs, which may affect the regional and global HIV-1 epidemic.
Sequence Data
GenBank accession numbers of nucleotide sequences reported in this article are AB547463 for 07IDJKT189 and AB547464 for 07IDJKT194.
Acknowledgments
We thank Dr. Gunawan Martinus, Dr. Dyah Waluyo, and the staff at Cipto Mangunkusumo National Hospital, Kramat 128 Hospital, and Sulianti Saroso Infectious Disease Hospital for their help during specimen collection. We thank Dr. Wataru Habano for technical advice and Ms. Kumi Furusawa for technical assistance. This work was supported by a grant from the Imai Memorial Trust for AIDS Research, a grant-in-aid for the Strategic Medical Science Research Center of Japan Ministry of Education, Culture, Sports, Science and Technology (2009–2013 period), and a fellowship from the Takeda Science Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Malim MH. Emerman M. HIV-1 sequence variation: Drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 2.Blackard JT. Cohen DE. Mayer KH. Human immunodeficiency virus super infection and recombination: Current state of knowledge and potential clinical consequences. Clin Infect Dis. 2002;34:1108–1114. doi: 10.1086/339547. [DOI] [PubMed] [Google Scholar]
- 3.Tovanabutra S. Beyrer C. Sakkhachornphop S, et al. The changing molecular epidemiology of HIV type 1 among Northern Thai drug users, 1999 to 2002. AIDS Res Hum Retroviruses. 2004;20:465–475. doi: 10.1089/088922204323087705. [DOI] [PubMed] [Google Scholar]
- 4.Tee KK. Saw TL. Pon CK, et al. The evolving molecular epidemiology of HIV type 1 among injecting drug users (IDUs) in Malaysia. AIDS Res Hum Retroviruses. 2005;21:1046–1050. doi: 10.1089/aid.2005.21.1046. [DOI] [PubMed] [Google Scholar]
- 5.Motomura K. Kusagawa S. Kato K. Nohtomi K. Lwin HH. Tun KM, et al. Emergence of new forms of human immunodeficiency virus type 1 intersubtype recombinants in Central Myanmar. AIDS Res Human Retroviruses. 2000;16:1831–1843. doi: 10.1089/08892220050195793. [DOI] [PubMed] [Google Scholar]
- 6.Los Alamos HIV sequence database. http://www.hiv.lanl.gov/ http://www.hiv.lanl.gov/
- 7.Wang B. Lau KA. Ong LY, et al. Complex pattern of the HIV-1 epidemic in Kuala Lumpur, Malaysia: Evidence for expansion of circulating recombinant form CRF33_01B and detection of multiple other recombinants. Virology. 2007;367:288–297. doi: 10.1016/j.virol.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 8.Lau KA. Wang B. Kamarulzaman A. Ng KP. Saksena NK. Near full-length sequence analysis of a unique CRF01_AE/B recombinant from Kuala Lumpur, Malaysia. AIDS Res Hum Retroviruses. 2007;23:1139–1145. doi: 10.1089/aid.2007.0056. [DOI] [PubMed] [Google Scholar]
- 9.Tovanabutra S. Watanaveeradej V. Viputtikul K, et al. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses. 2003;19:561–567. doi: 10.1089/088922203322230923. [DOI] [PubMed] [Google Scholar]
- 10.Tee KK. Li XJ. Nohtomi K, et al. Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr. 2006;43:523–529. doi: 10.1097/01.qai.0000242451.74779.a7. [DOI] [PubMed] [Google Scholar]
- 11.Tovanabutra S. Kijak GH. Betrer C, et al. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in Northern Thailand. AIDS Res Hum Retroviruses. 2007;23:829–833. doi: 10.1089/aid.2006.0300. [DOI] [PubMed] [Google Scholar]
- 12.SahBandar IN. Takahashi K. Djoerban Z, et al. Current HIV Type 1 molecular epidemiology profile, identification of unique recombinant forms Jakarta, Indonesia. AIDS Res Hum Retroviruses. 2009;25:637–646. doi: 10.1089/aid.2008.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. http://www.kumarlab.net/publications. pp. 1596–1599.http://www.kumarlab.net/publications .Publication PDF at. [DOI] [PubMed]
- 14.Lole KS. Bollinger RC. Paranjape RS. Gadkari D. Kulkarni SS. Novak NG. Ingersoll R. Sheppard HW. Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takebe Y. Motomura K. Tatsumi M. Lwin HH. Zaw M. Kusagawa S. High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: Geographical hot spot of extensive recombination. AIDS. 2003;17(14):2077–2087. doi: 10.1097/00002030-200309260-00009. [DOI] [PubMed] [Google Scholar]
- 16.Martin DP. Williamson C. Posada D. RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]