Abstract
Robust diagnostics for many human genetic disorders are much needed in the pursuit of global personalized medicine. Next-generation sequencing now offers new promise for biomarker and diagnostic discovery, in developed as well as resource-limited countries. In this broader global health context, X-linked intellectual disability (XLID) is an inherited genetic disorder that is associated with a range of phenotypes impacting societies in both developed and developing countries. Although intellectual disability arises due to diverse causes, a substantial proportion is caused by genomic alterations. Studies have identified causal XLID genomic alterations in more than 100 protein-coding genes located on the X-chromosome. However, the causes for a substantial number of intellectual disability and associated phenotypes still remain unknown. Identification of causative genes and novel mutations will help in early diagnosis as well as genetic counseling of families. Advent of next-generation sequencing methods has accelerated the discovery of new genes involved in mental health disorders. In this study, we analyzed the exomes of three families from India with nonsyndromic XLID comprising seven affected individuals. The affected individuals had varying degrees of intellectual disability, microcephaly, and delayed motor and language milestones. We identified potential causal variants in three XLID genes, including PAK3 (V294M), CASK (complex structural variant), and MECP2 (P354T). Our findings reported in this study extend the spectrum of mutations and phenotypes associated with XLID, and calls for further studies of intellectual disability and mental health disorders with use of next-generation sequencing technologies.
Keywords: : diagnostic medicine, genotype–phenotype association, mental retardation, neurodevelopmental disorders, next-generation sequencing
Introduction
Intellectual disability (ID) is a neurodevelopmental disorder characterized by significant deficits in intellectual functions and limitations in adaptive behaviors with onset before the age of 18. Studies estimate ID to occur at 1–3% frequency in the general population (Leonard and Wen, 2002; Roeleveld et al., 1997). X-linked intellectual disability (XLID) accounts for 5–10% of the total ID cases, which occurs predominantly in males (Lubs et al., 2012). It is observed that 30% more males were affected than females (McLaren and Bryson, 1987). XLID is genetically heterogeneous and is also accompanied by a spectrum of phenotypic variability.
Until recently, identification of genetic cause of inherited disorders involved linkage analysis, candidate gene analysis, cytogenetic studies, fluorescence in situ hybridization, or array-based comparative genomic hybridization. Advances in next-generation sequencing technologies have enabled unbiased analysis of whole genomes and exomes for the presence of causal alterations in common (Black and Wang, 2015) and rare diseases (Hekim et al., 2016). Next-generation omics technologies have found applications beyond medicine such as in ecology and environmental health as well (Kumar et al., 2015).
Next-generation sequencing has created a paradigm shift in clinical diagnosis of several diseases. High-depth whole-genome sequencing enables genome-wide sampling of genomic variations such as single nucleotide variants (SNVs), indels, structural variants, and copy number variants. However, exome sequencing is the most widely used method focused only on the protein-coding regions for identification of SNVs and indels. Exome sequencing does not reveal genomic variants that occur outside the protein-coding regions such as the gene regulatory regions.
However, exome sequencing is currently preferred over whole-genome sequencing as the initial approach for identification of genetic cause of inherited genetic disorders because of following advantages: (1) low cost of sequencing, (2) shorter turnaround time, (3) avoids nonspecific or incidental findings, (4) computationally easy to handle raw sequencing data, (5) more specific to identify molecular targets, (6) easy to interpret data, (7) allows deep sampling for reliable identification of variants, and (8) circumvents the problems caused by repetitive sequences. Whole-genome sequencing can be considered as an alternative approach second pass search when whole-exome sequencing results do not lead to identification of causative variants. As costs decrease and our ability to handle whole-genome data improves, we anticipate people to employ whole-genome sequencing over exome sequencing.
Next-generation sequencing has led to a dramatic increase in the identification of disease variants in many new and previously unresolved cases of familial and sporadic genetic disorders, including XLID (Grozeva et al., 2015; Hu et al., 2016). A curated list of 746 genes is reported to be associated with ID (Kochinke et al., 2016). Of these, growing literature on XLID has led to the identification of more than 100 genes on the X-chromosome (Lubs et al., 2012). However, several loci identified through linkage analysis still remain uncharacterized (Lubs et al., 2012) with the cause for ID remaining unknown for several cases.
In this study, we employed whole-exome sequencing to analyze seven affected individuals from three independent Indian families with evidence for mild-to-moderate ID. Based on the X-linked inheritance pattern observed in these families, we identified disease-relevant variants in three genes, including p21 (RAC1)-activated kinase 3 (PAK3), calcium-/calmodulin-dependent serine protein kinase (CASK), and methyl-CpG binding protein 2 (MECP2).
Materials and Methods
Patients
The inclusion criteria for the recruitment of families were as follows: (1) male subjects with mild-to-severe ID with intelligence quotient (IQ) <70, and (2) two or more intellectually disabled male subjects in the same family with an identifiable pattern of X-linked inheritance.
The study was approved by the institutional ethics committee at the National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore, India. Informed consent was obtained from the study participants, and the parents or legal guardians of the affected individuals provided consent for use of samples in the study. Diagnosis of ID (otherwise known as mental retardation) was based on International Classification of Diseases (ICD-10) (WHO) criteria for mental retardation for all the affected participants.
Clinical and phenotype descriptions
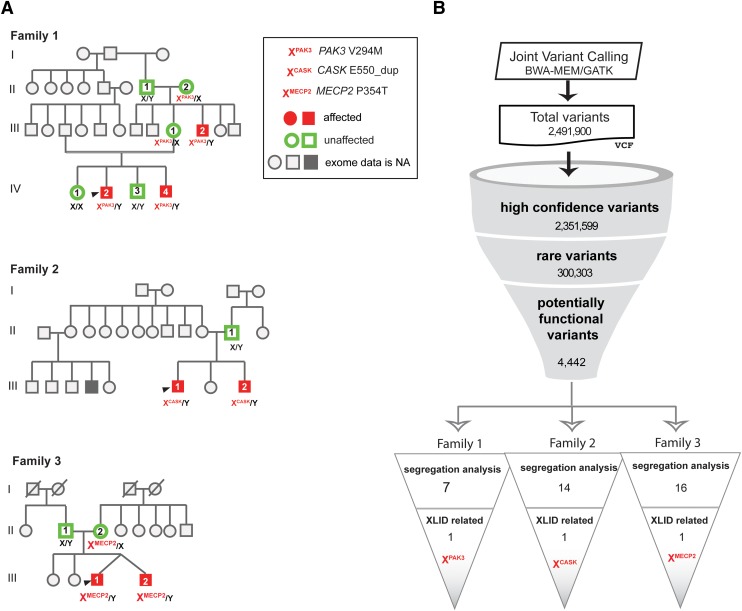
Family 1 was a nonsyndromic XLID family with three members, comprising two male siblings (Family1.IV.2 and Family1.IV.4) and a maternal uncle (Family1.III.2) affected with moderate ID (Fig. 1A). The geographical origin of this family is Bangalore from the state Karnataka, India. Parents are related and healthy. At the time of clinical presentation, the siblings were 7 and 3 years old and the maternal uncle was 25 years old. Motor and language milestones were delayed in all three.
FIG. 1.
XLID families. (A) Pedigrees of three XLID families from India. Solid red or green circles or boxes indicate sequenced samples. Genes with causal variants identified in each family are shown in the pedigree chart. (B) Flowchart depicting the joint variant calling and analysis performed on all the 15 samples studied. Causal variants identified following segregation analysis involving the family pedigree information is reported. NA, not available; XLID, X-linked intellectual disability.
The following phenotypic features were common to all the three: varying degrees of microcephaly, elongated face, bushy eyebrows, synophrys, long and/or prominent low set ears, short neck, and pes planus. Notably, brother of the proband (Family1.IV.4) had hypogenitalism as evidenced by micropenis and hypoplastic testes. Behaviorally, both siblings had attention-deficit hyperactivity disorder (ADHD) and the maternal uncle had clinically significant aggression (Supplementary Table S1). Blood samples were also available from unaffected members in the family, including grandfather (age = 60 years) (Family1.II.1), grandmother (age = 45 years) (Family1.II.2), mother (age = 24 years) (Family1.III.1), sister (age = 10 years) (Family1.IV.1), and an unaffected brother (age = 7 years) (Family1.IV.3).
Family 2 comprised two male siblings (Family2.III.1 and Family2.III.2) with nonsyndromic XLID (aged 14 and 7 years at their presentation to the clinic) (Fig. 1A). They were moderately affected and both had gross delay in motor and language milestones. Their parents are unrelated and healthy. The other shared features were microcephaly, short stature, straight eyebrows, low anterior hairline, low set and posterior ears, and clinodactyly. Behaviorally, both had ADHD (Supplementary Table S2). The geographical origin of this family is Bangalore in the state of Karnataka, India. Blood sample was also available from unaffected father (age = 35) (Family2.II.1).
Family 3 comprised monozygotic twins (age = 7) with the proband (Family3.III.1) affected with moderate ID (IQ = 42) and his brother (Family3.III.2) with borderline intelligence (IQ = 72) (Fig. 1A). They were born preterm at 7 months. Their parents (Family3.II.1 and Family3.II.2) are unrelated and healthy. The first born of the twins (Family3.III.2) was small for gestational age (1.25 kg) and received incubator care for 1 month. The younger twin (Family3.III.1) had fetal distress, had a birth weight of 2 kg, and also received incubator care for 1 month. Language milestones were delayed in both, much more so in the younger one (Family3.III.1).
Both were assessed to have bilateral sensorineural hearing impairment and they use hearing aids. A few facial dysmorphic features and minor congenital anomalies were noted in both. The younger boy (Family3.III.1) also was diagnosed to have autism spectrum disorder and attention-deficit hyperactive disorder (Supplementary Table S3). The geographical origin of this family is from Gaya district, Bihar, India. Blood samples were also available from the unaffected mother (age = 25) (Family3.II.2) and unaffected father (age = 42) (Family3.II.1) to carryout segregation analysis.
Cytogenetic analysis
Blood samples from all the seven affected individuals were processed for cytogenetics and genomic DNA isolation. Peripheral blood lymphocytes were cultured using routine cytogenetics protocol (Moore et al., 2002). Chromosomes were stained by G banding, according to Seabright (1971). Chromosomes were GTG banded.
Array-based comparative genomic hybridization
Genomic DNA was isolated from whole blood using the QIAamp DNA Blood Mini Kit (Cat. No. 51104; Qiagen) according to manufacturer's protocol. Copy number variations in patients' DNA were measured in relation to reference DNA (Promega, USA) by two-color array comparative genomic hybridization. The experiment was carried out as per manufacturer's instructions. Briefly, patient genomic DNA and reference genomic DNA were digested using AluI and RsaI restriction enzymes to generate DNA fragments ∼100–500 bp long, which were confirmed by gel electrophoresis. Patient DNA (Human Genomic DNA, Male, Cat. No. G1471; Promega, USA) was labeled with Cy3 and reference DNA was labeled with Cy5 by random priming. Equal amounts of denatured Cy3- and Cy5-labeled patients and reference DNA samples were hybridized on to Agilent human whole genome 8x44K and 8x60K comparative genomic hybridization microarrays. Microarray slides were washed and scanned using Agilent microarray 3 μm scanner system and TIFF images were generated. Agilent's Feature Extraction software was used to extract raw intensities and quantify the fluorescence data.
The raw data were analyzed using Agilent DNA analytics software version 6.0. ADM-2 aberration algorithm with default threshold 6.0 was used to call copy number variants, and Fuzzy zero algorithm was applied to correct false positives. The aberration filter was set at 3 as single- and double-probe alterations are more prone to be false calls.
Whole exome and X-chromosome panel sequencing
Three micrograms of genomic DNA was sheared to produce 150–200 bp fragments for library preparation. Following size selection, end-repair, phosphorylation, and A-tailing adapters were ligated to the DNA fragments according to the manufacturer's protocol (SureselectXT Human all exon V5 kit/SureselectXT custom 6–11.9 Mb for X-chromosome). Hybridization was carried out using 700 ng of library and 5 μL of biotin-labeled RNA probe sets (Agilent), designed specifically for the desired targets (whole exome or X-chromosome custom panel).
The library and probe sets were incubated at 65°C for 16 h. Capture of resulting DNA-RNA duplexes was performed by the addition of Myone streptavidin T1 beads (Invitrogen, USA). Multiple washes at high stringency were used to remove off-target material and any nonhybridized fragments. Indexed primers and Herculase II Fusion DNA Polymerase (Agilent, USA) were used for the amplification of specific libraries. Exome Library QC was checked on a Bioanalyzer (Agilent, USA) and quantified using Qubit (Invitrogen, USA). The libraries were diluted to 10 nM concentration in the TE buffer.
The libraries were denatured and further diluted to 8.5 pM concentrations using the HT buffer to be loaded for cluster generation in the cBOT (Illumina, USA). Cluster amplification of denatured templates was performed according to manufacturer's protocol (Illumina, USA) using V3 Chemistry and V3 Flow cells. Paired-end sequencing was performed on Hi-Seq2500 to obtain 2 × 75 bp reads for whole exome or 2 × 100 bp reads for X-chromosome panel sequencing, using V3 Illumina sequencing by synthesis chemistry (Illumina, USA).
Joint variant calling
X-chromosome panel and whole-exome sequencing data obtained from seven affected and eight unaffected individuals were combined and then processed using the Genome Analysis Toolkit (GATK) best practices recommendations (Van der Auwera et al., 2013) to call SNVs and small insertions/deletions. Briefly, the raw sequencing reads were aligned to the human reference genome version hg19 (GRCh37) using BWA-MEM (version 0.7.10) (Li and Durbin, 2010). Picard tools (version 1.126) were used for removing duplicate reads. Indel realignment and base quality score recalibration were performed using GATK (version v3.5-0-g36282e4) to improve alignment accuracy and recalibrate base quality (DePristo et al., 2011). HaplotypeCaller (in reference confidence model) was applied for each sample to generate genomic variant call format (gVCF) files. The resulting gVCF files from all samples were used for joint variant calling using GenotypeGVCFs walker. Finally, Variant Quality Score Recalibration (VQSR) was carried out to estimate the confidence of called variants.
Detection of structural variation
Detection of small structural variants from the exome samples was performed using SoftSV software (version 1.4) with default parameters (Bartenhagen and Dugas, 2016). Structural variants detected in XLID-relevant genes were manually reviewed using integrative genomics viewer (IGV) (Robinson et al., 2011). The structural variant identified in CASK gene was confirmed using Sanger sequencing.
Identification of causal variants
All variants resulting from the joint variant calling were annotated using SnpEff (version 4.2) (Cingolani et al., 2012) and analyzed following the steps shown in Figure 1B. To identify potentially causative variants, we applied a number of filtering steps. Briefly, only the high-confident variants that passed GATK VQSR and had a GATK call quality of at least 20 in any sample were retained. We retained variants that were hemizygous in all affected individuals and were heterozygous in the obligate carriers. The variants were discarded if they were observed in unaffected males in hemizygous state or were observed in homozygous state in unaffected females.
We filtered out common variants with alternate allele frequency ≥ 1%, if it is present, in either the 1000 Genome Project (Genomes Project et al., 2012), National Heart, Lung, and Blood Institute-exome variant server (NHLBI-EVS) Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) or Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/). Sorting tolerant from intolerant (SIFT) (Kumar et al., 2009), Polyphen (Adzhubei et al., 2010), and American College of Medical Genetics and Genomics (ACMG) criteria (Richards et al., 2015) were then used to define a set of potentially functional variants.
The genes and corresponding mutations that qualified these filtering criteria were further investigated to determine their significance and relevance in XLID. Genes that are previously associated with XLID were qualified for investigation irrespective of predictions from SIFT and PolyPhen. An additional variant interpretation was done using Qiagen's Ingenuity Variant Analysis software (www.qiagen.com/ingenuity). The final list of variants was manually reviewed using IGV (Robinson et al., 2011).
Results
Cytogenetics and copy number alterations
We first performed cytogenetic analysis of the seven patient samples for the presence of chromosomal abnormalities. We did not observe karyotype alterations in the affected individuals. We then applied array-comparative genomic hybridization to assess copy number alterations in the patients. We did not observe any significant copy number alterations in all the three families.
Exome sequencing and data analyses
To identify variants that are potentially responsible for XLID in affected individuals, we performed whole-exome sequencing of all 15 study subjects that included seven affected individuals and eight unaffected individuals from three unrelated Indian families (Fig. 1A). Given the X-linked inheritance observed in all the three families, we carried out targeted X-chromosome exome sequencing on all the 15 samples to provide additional coverage and orthogonal validation for variant calls. We obtained 40–60x mean coverage for the whole exome and 199–468x mean coverage for the targeted X-chromosome exome panel (X-panel) sequencing data. On average, 85% and 99% of the bases sequenced had at least 20x coverage for the whole exome and X-panel, respectively (Supplementary Table S4).
Following this, we combined both the exome and X-panel data to improve the coverage before joint genotype variant calling. We performed kinship analysis using the variants with the KING software (Manichaikul et al., 2010) and confirmed the identity and relatedness of the samples within families (Supplementary Fig. S1).
A PAK3 causal variant identified in family 1
In the proband from family 1 (Family1.IV.2), we identified a nonsynonymous variant V294M in PAK3 (Table 1). The pattern of inheritance of the mutant allele showed that the proband's unaffected maternal grandmother (Family1.II.2) and mother (Family1.III.1) to be obligate carriers. This is consistent with the manifestation of intellectual disability in the proband, his brother (Family1.IV.4), and his maternal uncle (Family1.III.2) as indicated in the pedigree chart (Fig. 1A). Also, as expected, one of proband's unaffected brothers (Family1.IV.3) did not carry the mutant alleles. As, the unaffected sister (Family1.IV.1) did not carry the mutant allele, she was found not to be a carrier (Supplementary Fig. S2).
Table 1.
Potential Causal Variants Identified from Three Unrelated Indian Families
| Sample name | Phenotype | Mutation | Genotype | Annotation | Transcript ID |
|---|---|---|---|---|---|
| Family1.II.2 | Unaffected | PAK3:c.880G > A (p.V294M) | Heterozygous | Missense and Splice region | NM_002578 |
| Family1.III.1 | Unaffected | PAK3:c.880G > A (p.V294M) | Heterozygous | Missense and Splice region | NM_002578 |
| Family1.III.2 | Affected | PAK3:c.880G > A (p.V294M) | Hemizygous | Missense and Splice region | NM_002578 |
| Family1.IV.2 | Affected | PAK3:c.880G > A (p.V294M) | Hemizygous | Missense and Splice region | NM_002578 |
| Family1.IV.4 | Affected | PAK3:c.880G > A (p.V294M) | Hemizygous | Missense and Splice region | NM_002578 |
| Family2.III.1 | Affected | CASK:(E550_dup) | Hemizygous | Stop gain and in-frame insertion | NM_003688 |
| Family2.III.2 | Affected | CASK:(E550_dup) | Hemizygous | Stop gain and in-frame insertion | NM_003688 |
| Family3.II.2 | Unaffected | MECP2:c.1060C > A (p.P354T) | Heterozygous | Missense variant | NM_001110792 |
| Family3.III.1 | Affected | MECP2:c.1060C > A (p.P354T) | Hemizygous | Missense variant | NM_001110792 |
| Family3.III.2 | Affected | MECP2:c.1060C > A (p.P354T) | Hemizygous | Missense variant | NM_001110792 |
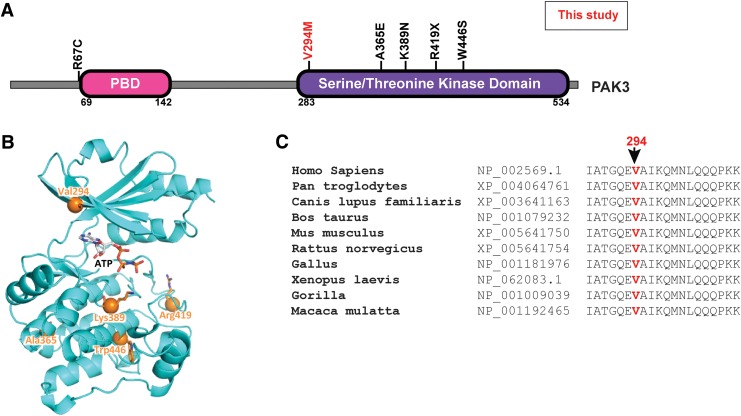
The PAK3 variant V294M identified in this study maps to the kinase domain (Fig. 2A) and is located very close to the ATP binding site in the kinase domain (Fig. 2B). The valine at position 294 in PAK3 is evolutionarily conserved (Fig. 2C), suggesting that a critical functional role for this residue and nonsynonymous substitution at this position are not likely to be tolerated. The PAK3V294M variant has not been reported previously and was not found in the 1000 genomes or ExAC databases. These observations suggest that the V294M substitution likely alters the kinase activity of PAK3. The impaired PAK3 function is likely to be responsible for the observed XLID phenotype, given its known role in synaptic plasticity and deficiency in memory and learning (Boda et al., 2004; Meng et al., 2005).
FIG. 2.
PAK3 mutation. (A) Graphical representation of PAK3 protein showing the conserved protein domains. Mutations in PAK3 that have been previously reported to cause XLID are shown in black and the mutation identified in this study is shown in red. (B) PAK3 mutations shown in the context of structure of PAK3 kinase domain. Homology model was generated using the closely related PAK1 structure as a template. An ATP molecule model is shown in sticks. V294 is on the “roof” of the ATP binding site. (C) Multiple sequence alignment of PAK3 homologs showing the residues and conservation of the mutated valine 294 site (red) in PAK3.
CASK alteration identified in family 2
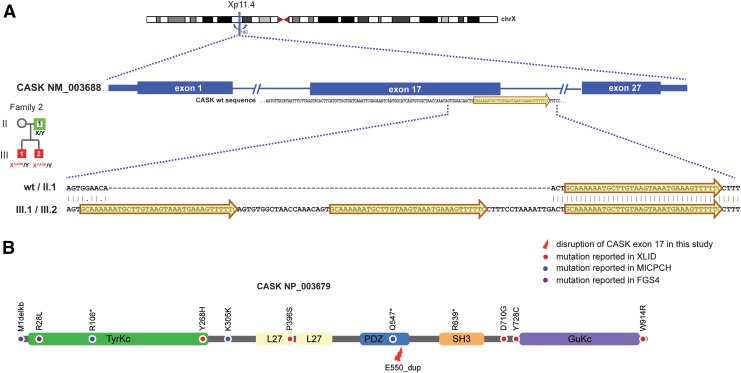
Analysis of exome from two affected (Family2.III.1 and Family2.III.2) and an unaffected parent (Family2.II.1) in family 2 identified evidence for a short tandem duplication, spanning an exon–intron junction in calcium-/calmodulin-dependent serine protein kinase (CASK) in both the affected probands (Family2.III.1 and Family2.III.2) and not in their father (Family2.II.1) (Supplementary Fig. S3A). Sanger sequencing for a 457 bp region (chrX:41,420,622–41,421,078) that covers the tandem duplication confirmed the complex genomic alteration in the two affected probands (Family2.III.1 and Family2.III.2) (Fig. S3B). The duplication involves two copies of a 34 bp sequence (GCAAAAAATGCTTGTAAGTAAATGAAAGTTTTTC) that overlaps the splice donor site in CASK (NM_003688) exon 17 (Fig. 3A). The identified variant along with the previously reported variants in published literature are depicted in Figure 3B.
FIG. 3.
Complex alteration in CASK. (A) Local alignment using Smith–Waterman algorithm of CASK sequences obtained from Sanger sequencing for the region that covers the complex alteration. (B) Known mutations depicted on a cartoon of CASK protein and its protein domains.
A variant in MECP2 in family 3
In the monozygotic twins from family 3 (Family3.III.1 and Family3.III.2), we identified a missense variant (P354T) in methyl-CpG binding protein 2 (MECP2) (OMIM: 300005) located at Xq28 (Table 1). The mother (Family3.II.2) is unaffected and carries the variant (Fig. 1A). The variant P354T was found to be hemizygous in both the affected individuals (Family3.III.1 and Family3.III.2), heterozygous in the mother (Family3.II.2), and not present in the father (Family3.II.1), showing segregation of the variant in the family (Supplementary Fig. S4). The mutated residue is highly conserved in MECP2 (Supplementary Fig. S5) and it is not previously reported to cause ID. SIFT, Polyphen-2, and Mutation Taster predicted the effect of the mutation as deleterious.
Discussion
In this study, we identified novel mutations in three genes PAK3, CASK, and MECP2 that have been previously implicated in XLID. These three genes have been previously shown to be expressed in the brain, involved in synaptic plasticity, and known to play a role in dendritic spine morphogenesis. These novel mutations were inferred to be potentially pathogenic based on the domain that is affected and conservation of the affected residue in addition to segregation of inheritance pattern of the mutation. For example, in family 1, among seven variants that passed segregation analysis (hemizygous in affected males, heterozygous in carriers, and not present in unaffected individuals), V294M in PAK3 was the only variant located on the X-chromosome.
PAK3 is a serine/threonine kinase that is involved in several processes, including actin organization and migration (Arias-Romero and Chernoff, 2008; Holderness Parker et al., 2013), embryonic development of the brain and neuron (Jaffer and Chernoff, 2002), synaptic transmission (Thevenot et al., 2011), dendrite spine formation, and synaptic plasticity (Kreis et al., 2007; Node-Langlois et al., 2006). PAK3 shows significant expression in the brain, more specifically in the hippocampus, cerebral cortex, and neurons (Boda et al., 2004).
Knockout mice and genetically altered PAK3 mice have been previously reported to show reduction in dendritic spine density, abnormalities in synaptic plasticity, and deficiency in memory and learning (Boda et al., 2004; Meng et al., 2005). Mutations in PAK3 kinase domain that reduce or abolish its kinase activity have been found in XLID patients (Kreis et al., 2007; Magini et al., 2014). The novel mutation identified in this study lies in kinase domain of the protein and is conserved in mammals. These observations suggest that this variant is likely to be pathogenic.
Similarly, the genomic duplication event identified in CASK gene may affect the PDZ domain. CASK is a synaptic scaffolding protein in neurons and belongs to MAGUK (membrane-associated guanylate kinase) protein family (Dimitratos et al., 1997). CASK is expressed in brain and is found in various subcellular locations of neurons (Hsueh et al., 1998, 2000). Mutations in CASK have been previously reported in FG syndrome 4 (FGS4) (OMIM: 300422) as well as in intellectual disability and microcephaly with pontine and cerebellar hypoplasia (MICPCH) (OMIM: 300749).
The complex genomic alteration identified in this study affects the PDZ domain of CASK. PDZ domain containing proteins are involved in the regulation of receptor and channel protein localization to the synapses and tight junctions. They are also involved in scaffolding intracellular signaling protein complexes (Dunn and Ferguson, 2015). CASK is expressed in dendritic spine tip where it interacts with syndecan-2 through its PDZ domain and is involved in spine morphogenesis (Chao et al., 2008; Hu and Hsueh, 2014). Hence, the duplication observed in PDZ domain is likely pathogenic, which could alter the function of CASK and thereby affect cognition-related functions. Our study reports the first mutation in PDZ domain in CASK gene associated with XLID.
Mutations in MECP2 have been reported to cause Rett syndrome predominantly in females and intellectual disability in males (Moog et al., 2006). Also, MECP2 mutations contribute to encephalopathy (Imessaoudene et al., 2001) and autism spectrum-associated disorders (Lam et al., 2000; Yu et al., 2013). MECP2 mutations have been identified in other ID-related cases in high-throughput sequencing studies (Grozeva et al., 2015). MECP2 is expressed predominantly in the brain, including hippocampus, the granule cell layer of the dentate gyrus, as well as in the thalamus, striatum, and neocortex (Kishi and Macklis, 2004). MECP2 contains a methyl binding domain (MBD), transcriptional repression domain (TRD), and a C-terminal domain (CTD) along with two nuclear localization signals.
The mutation identified in this study is located in CTD. Mutant mice lacking the fourth exon of MECP2, which contains partial MBD, TRD, and CTD, showed changes in dendritic spine morphology and spine density (Belichenko et al., 2009; Smrt et al., 2007). The mutation observed in MECP2 is predicted to be pathogenic and likely contributed to the intellectual disability observed in family 3.
One of the limitations of our study is the lack of in vitro or in vivo functional analyses of the identified variants to determine the pathogenicity of the identified variants. In addition, we have not carried out extensive validation in unrelated nonfamilial sporadic low IQ cases. However, we have carried out careful analysis to establish potential pathogenicity of the reported variants. Considering other genetic variations in these genes that have been previously implicated in XLID, it is likely that the reported variants are causative. Although we have filtered out common variants found in 1000 genome project, NHLBI-EVS Exome Sequencing Project, and ExAC database, we realize that the representation of Indian population is lower in these cohorts (Sengupta et al., 2016). Further validation in a larger Indian cohort representing the ancestry of the patients should be performed to preclude the possibility that the identified variants are not polymorphisms.
Conclusions
We report clinical and genetic characteristics of three Indian families with at least two individuals affected with a varied degree of intellectual disability and a spectrum of clinical phenotypes. We have identified mutations in PAK3, CASK, and MECP2 that likely contribute to intellectual disability. These genes have previously been implicated in XLID and their roles in cognition-related functions have been reported in animal models (Belichenko et al., 2009; Boda et al., 2004; Chao et al., 2008; Meng et al., 2005; Node-Langlois et al., 2006; Smrt et al., 2007; Zhao et al., 2008, 2009).
Consequently, identification of these potentially causative variants extends the spectrum of mutations observed in XLID. Molecular assays involving these genes will help understand the cause of intellectual disability and enable screening for carriers. Proper counseling is likely of help to carrier parents. Our findings demonstrate the power of next-generation sequencing for identifying candidate causal variants in familial disease when appropriate samples from both affected and unaffected families are sequenced.
Supplementary Material
Abbreviations Used
- ACMG
American College of Medical Genetics and Genomics
- ADHD
attention-deficit hyperactivity disorder
- ATP
adenosine triphosphate
- BWA
Burrows-Wheeler aligner
- CASK
calcium-/calmodulin-dependent serine protein kinase
- CGH
comparative genomic hybridization
- CTD
C-terminal domain
- ExAC
Exome Aggregation Consortium
- FGS4
FG syndrome 4
- GATK
Genome Analysis Toolkit
- GTG Banding
Giemsa banding
- gVCF
genomic variant call format
- ID
intellectual disability
- IGV
integrative genomics viewer
- IQ
intelligence quotient
- MAGUK
membrane-associated guanylate kinase
- MBD
methyl binding domain
- MECP2
methyl-CpG binding protein 2
- MICPCH
mental retardation and microcephaly with pontine and cerebellar hypoplasia
- NHLBI-EVS
National Heart, Lung, and Blood Institute-exome variant server
- PAK3
p21 (RAC1)-activated kinase 3
- QC
quality control
- RAC1
Ras-related C3 botulinum toxin substrate 1
- SIFT
sorting tolerant from intolerant
- TRD
transcriptional repression domain
- VQSR
variant quality score recalibration
- XLID
X-linked intellectual disability
Acknowledgments
We are grateful to all family members who participated in this study. We thank the Department of Biotechnology (DBT), Government of India, for research support to the Institute of Bioinformatics (IOB). A part of this work was funded by DBT through a grant on X-linked intellectual disability (BT/PR10345/Med/30/79/2007). L.D.N.S. is a recipient of research associateship from DBT. IOB is supported by DBT Program Support on Neuroproteomics and infrastructure for proteomic data analysis (BT/01/COE/08/05). We thank the Infosys Foundation for research support to IOB.
Author Disclosure Statement
Genentech authors hold Roche shares. The other authors declare no conflicts of interest.
References
- 1000 Genomes Project Consortium; Abecasis GR, Auton A, et al. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. (2010). A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Romero LE, and Chernoff J. (2008). A tale of two Paks. European Cell Biol Org 100, 97–108 [DOI] [PubMed] [Google Scholar]
- Bartenhagen C, and Dugas M. (2016). Robust and exact structural variation detection with paired-end and soft-clipped alignments: SoftSV compared with eight algorithms. Brief Bioinform 17, 51–62 [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, et al. (2009). Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: Evidence for disruption of neuronal networks. J Comp Neurol 514, 240–258 [DOI] [PubMed] [Google Scholar]
- Black M, and Wang W. (2015). Ischemic Stroke: From Next Generation Sequencing and GWAS to Community Genomics? OMICS 19, 451–460 [DOI] [PubMed] [Google Scholar]
- Boda B, Alberi S, Nikonenko I, et al. (2004). The mental retardation protein PAK3 contributes to synapse formation and plasticity in hippocampus. J Neuroscience 24, 10816–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HW, Hong CJ, Huang TN, Lin YL, and Hsueh YP. (2008). SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol 182, 141–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitratos SD, Woods DF, and Bryant PJ. (1997). Camguk, Lin-2, and CASK: Novel membrane-associated guanylate kinase homologs that also contain CaM kinase domains. Mech Dev 63, 127–130 [DOI] [PubMed] [Google Scholar]
- Dunn HA, and Ferguson SS. (2015). PDZ Protein Regulation of G Protein-Coupled Receptor Trafficking and Signaling Pathways. Mol Pharmacol 88, 624–639 [DOI] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O, et al. (2015). Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutation 36, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekim N, Batyraliev T, Trujillano D, et al. (2016). Whole exome sequencing in a rare disease: A patient with anomalous left coronary artery from the pulmonary artery (Bland-White-Garland Syndrome). OMICS 20, 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderness Parker N, Donninger H, Birrer MJ, and Leaner VD. (2013). p21-activated kinase 3 (PAK3) is an AP-1 regulated gene contributing to actin organisation and migration of transformed fibroblasts. PLoS One 8, e66892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Wang TF, Yang FC, and Sheng M. (2000). Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature 404, 298–302 [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Yang FC, Kharazia V, et al. (1998). Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J Cell Biol 142, 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Haas SA, Chelly J, et al. (2016). X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 21, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HT, and Hsueh YP. (2014). Calcium influx and postsynaptic proteins coordinate the dendritic filopodium-spine transition. Dev Neurobiol 74, 1011–1029 [DOI] [PubMed] [Google Scholar]
- Imessaoudene B, Bonnefont JP, Royer G, et al. (2001). MECP2 mutation in non-fatal, non-progressive encephalopathy in a male. J Med Genet 38, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffer ZM, and Chernoff J. (2002). p21-activated kinases: Three more join the Pak. Int J Biochem Cell Biol 34, 713–717 [DOI] [PubMed] [Google Scholar]
- Kishi N, and Macklis JD. (2004). MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci 27, 306–321 [DOI] [PubMed] [Google Scholar]
- Kochinke K, Zweier C, Nijhof B, et al. (2016). Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 98, 149–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P, Thevenot E, Rousseau V, Boda B, Muller D, and Barnier JV. (2007). The p21-activated kinase 3 implicated in mental retardation regulates spine morphogenesis through a Cdc42-dependent pathway. J Biol Chem 282, 21497–21506 [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, and Ng PC. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols 4, 1073–1081 [DOI] [PubMed] [Google Scholar]
- Kumar RR, Goswami S, Sharma SK, et al. (2015). Harnessing next generation sequencing in climate change: RNA-Seq analysis of heat stress-responsive genes in wheat (Triticum aestivum L.). OMICS 19, 632–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CW, Yeung WL, Ko CH, et al. (2000). Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet 37, E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard H, and Wen X. (2002). The epidemiology of mental retardation: Challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 8, 117–134 [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs HA, Stevenson RE, and Schwartz CE. (2012). Fragile X and X-linked intellectual disability: Four decades of discovery. Am J Hum Genet 90, 579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magini P, Pippucci T, Tsai IC, et al. (2014). A mutation in PAK3 with a dual molecular effect deregulates the RAS/MAPK pathway and drives an X-linked syndromic phenotype. Hum Mol Genet 23, 3607–3617 [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, and Chen WM. (2010). Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren J, and Bryson SE. (1987). Review of recent epidemiological studies of mental retardation: Prevalence, associated disorders, and etiology. Am J Ment Retard 92, 243–254 [PubMed] [Google Scholar]
- Meng J, Meng Y, Hanna A, Janus C, and Jia Z. (2005). Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J Neurosci 25, 6641–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog U, Van Roozendaal K, Smeets E, et al. (2006). MECP2 mutations are an infrequent cause of mental retardation associated with neurological problems in male patients. Brain Dev 28, 305–310 [DOI] [PubMed] [Google Scholar]
- Moore CB, Siopes TD, Steele CT, and Underwood H. (2002). Pineal melatonin secretion, but not ocular melatonin secretion, is sufficient to maintain normal immune responses in Japanese quail (Coturnix coturnix japonica). Gen Comparative Endocrinol 126, 352–358 [DOI] [PubMed] [Google Scholar]
- Node-Langlois R, Muller D, and Boda B. (2006). Sequential implication of the mental retardation proteins ARHGEF6 and PAK3 in spine morphogenesis. J Cell Science 119, 4986–4993 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, et al. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. (2011). Integrative genomics viewer. Nature Biotech 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis GA, and Gabreels F. (1997). The prevalence of mental retardation: A critical review of recent literature. Dev Med Child Neurol 39, 125–132 [DOI] [PubMed] [Google Scholar]
- Seabright M. (1971). A rapid banding technique for human chromosomes. Lancet 2, 971–972 [DOI] [PubMed] [Google Scholar]
- Smrt RD, Eaves-Egenes J, Barkho BZ, et al. (2007). Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis 27, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenot E, Moreau AW, Rousseau V, et al. (2011). p21-Activated kinase 3 (PAK3) protein regulates synaptic transmission through its interaction with the Nck2/Grb4 protein adaptor. J Biol Chem 286, 40044–40059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, et al. (2013). From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protocols Bioinformatics 11, 1110–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). The ICD-10 classification of mental and behavioural disorders: Clinical descriptions and diagnostic guidelines. Geneva, 1992. http://www.who.int/classifications/icd/en/bluebook.pdf Accessed March16, 2017 [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, et al. (2013). Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, DA D, Lim WK, et al. (2009). The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell 17, 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, et al. (2008). The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol 10, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.