Abstract
We describe a new assay for in vitro repair of oxidatively induced DNA double-strand breaks (DSBs) by HeLa cell nuclear extracts. The assay employs linear plasmid DNA containing DNA DSBs produced by the radiomimetic drug bleomycin. The bleomycin-induced DSB possesses a complex structure similar to that produced by oxidative processes and ionizing radiation. Bleomycin DSBs are composed of blunt ends or ends containing a single 5′-base overhang. Regardless of the 5′-end structure, all bleomycin-induced DSBs possess 3′-ends blocked by phosphoglycolate. Cellular extraction and initial end joining conditions for our assay were optimized with restriction enzyme-cleaved DNA to maximize ligation activity. Parameters affecting ligation such as temperature, time, ionic strength, ATP utilization and extract protein concentration were examined. Similar reactions were performed with the bleomycin-linearized substrate. In all cases, end-joined molecules ranging from dimers to higher molecular weight forms were produced and observed directly in agarose gels stained with Vistra Green and imaged with a FluorImager 595. This detection method is at least 50-fold more sensitive than ethidium bromide and permits detection of ≤0.25 ng double-stranded DNA per band in post-electrophoretically stained agarose gels. Consequently, our end-joining reaction requires ≤100 ng substrate DNA and ≥50% conversion of substrate to product is achieved with simple substrates such as restriction enzyme-cleaved DNA. Using our assay we have observed a 6-fold lower repair rate and a lag in reaction initiation for bleomycin-induced DSBs as compared to blunt-ended DNA. Also, end joining reaction conditions are DSB end group dependent. In particular, bleomycin-induced DSB repair is considerably more sensitive to inhibition by increased ionic strength than repair of blunt-ended DNA.
INTRODUCTION
DNA double-strand breaks (DSBs) are caused, directly or indirectly, by a variety of DNA-damaging agents, including ionizing irradiation and oxidative metabolism (1–4). These breaks can have serious consequences, including chromosomal aberrations, increased genetic instability, carcinogenesis and cytotoxicity (5). To restore genomic integrity, cells repair DSBs by homologous recombination (HR) or non-homologous end joining (NHEJ) (6,7). The primary mechanism in mammalian cells is NHEJ (8).
Genetic studies, using radiosensitive mammalian cell lines and animals that are deficient in V(D)J recombination during lymphoid development, have been useful in identifying several proteins (Ku70 and Ku86, DNA-PKcs, DNA ligase IV, XRCC4 and the Rad50/Xrs-2/MreII complex) that are involved in the DSB repair process (9–12). However, due to the lack of a quick, simple and versatile in vitro assay, the exact biochemical mechanism of repair remains unknown.
Several DSB repair methods based upon end joining of linear plasmids have been described in the literature (13–16). All of these assays use restriction enzyme cut plasmid DNA as the repair substrate. Although this is an efficient method of linearizing plasmids in vitro, the physical structure of the DSB end does not reflect the structure of DSBs produced naturally in mammalian cells by either endogenous metabolic processes or by many DNA-damaging agents. Most naturally occurring DSBs, particularly the medically relevant DSBs produced by ionizing radiation and some chemotherapeutic agents, result from oxidative processes. Oxidatively induced DSBs are formed by the destructive loss of at least 1 nt in each DNA strand at the break site. In contrast to restriction enzyme cut DNA, the resulting end structure is complex, consisting of nucleotide fragments that render the ends unligatable in the absence of nucleolytic processing (17–19). This is not the case with restriction enzyme cut DNA, in which complementary or blunt ends can be directly ligated. Even in cases where restriction enzyme-produced DSBs consisting of non-complementary abutted overhangs are used as substrates, the overhanging ends can be aligned to permit ligation and/or polymerase-mediated fill-in reactions on the opposite gapped strand (20).
Therefore, to study NHEJ repair using substrates modeled on those expected in vivo, we have developed an in vitro DSB end joining assay that employs a plasmid DNA substrate linearized by the radiomimetic drug bleomycin. Bleomycin-induced strand breaks are formed by the destructive loss of a nucleotide which results in a 1 nt gap flanked by a 5′-PO4 and a 3′-phosphoglycolate (3′-PG) (21). This structure is similar to ionizing radiation-induced strand breaks and strand breaks produced by reactive oxygen species like H2O2 (22–28).
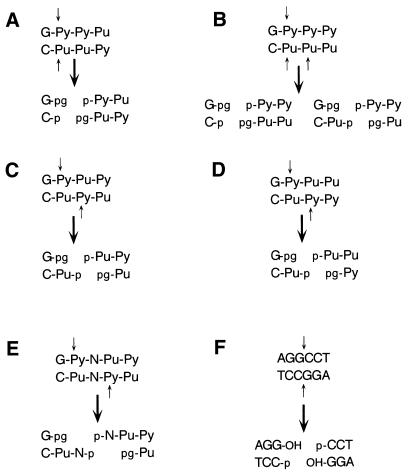
Povirk et al. (29) have presented a comprehensive analysis of bleomycin-induced DSB structures that are summarized in Figure 1A–E. The structures depicted in Figure 1A–D represent nearly 90% of all bleomycin-induced DSB types and are distributed almost equally between 3′-PG blocked blunt ends and ends containing a 3′-PG and a single base 5′-overhang. Therefore, in contrast to ‘non-ligatable’ restriction enzyme generated DSB substrates, which are typically formed by abutting overhangs (3′ or 5′) of four bases per end, bleomycin-induced DSBs resemble blunt-ended DNA. This is noteworthy because eukaryotic cell extracts are reported to be particularly inefficient at end joining blunt-ended substrates (15,30,31). Therefore, bleomycin-induced DSBs present a dual problem for the NHEJ pathway, complex ends, which require processing prior to ligation, and a difficult to ligate blunt-end structure. This dual complexity of the bleomycin DSB suggested that we should optimize our procedure for a simple (i.e. 3′-OH and 5′-P) blunt-ended DNA substrate before attempting repair of complex bleomycin-induced DSBs. Since ligation is the final step of NHEJ and human cell extracts are poor at ligating blunt-ended DNA, we reasoned that unless we could achieve efficient ligation of blunt-ended DNA we were unlikely to achieve efficient end joining with the more complex bleomycin-induced DSB. Consequently, we optimized our method for efficient joining of restriction enzyme-induced blunt ends first and used these conditions to examine repair of the more complex bleomycin-damaged substrate. We show that human HeLa cell nuclear extracts support end joining of these complex DSBs to form multimeric-plasmid products. Interestingly, the optimal repair conditions vary depending upon the chemical structure of the DSB end being rejoined.
Figure 1.
Bleomycin-induced and StuI-induced DSB structures. (A–E) Bleomycin-induced DSB structures as described by Povirk et al. (29). (F) The StuI cut blunt-ended pSP189 linear substrate used in the assay. The top strand is oriented in the 5′→3′ direction. Purines, pyrimidines or either base are indicated by Pu, Py and N, respectively. Small arrows indicate DSB cleavage points. Phosphates are indicated by p, phosphoglycolates are indicated by pg and hydroxyl groups are shown as OH. (A)–(D) represent ∼90% of the possible bleomycin-induced DSB structures, with 3′-pg blocked blunt-ended structures comprising ∼50% of this and 3′-pg blocked DSBs with a 1 base 5′-overhang representing the remainder.
Finally, due to its relatively speedy processing times and lack of radioactive components, this assay can be easily scaled up to levels appropriate for biochemical analysis and protein purification or for high throughput screening methods.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, non-essential amino acids (10 mM), glutamine (100 mM), penicillin/streptomycin (10 000 U/ml), Saccharomyces cerevisiae tRNA and T4 DNA ligase (1 U/µl) were purchased from Life Technologies (Gaithersburg, MD). Bleomycin and phenylmethylsulfonyl fluoride (PMSF) were obtained from Sigma (St Louis, MO). Endonuclease IV (10 U/µl) was purchased from Trevigen (Gaithersburg, MD). StuI (10 000 U/ml) was purchased from New England BioLabs (Beverly, MA). Vistra Green (VG) was obtained from Amersham Pharmacia Biotech (Piscataway, NJ).
Protease inhibitors (leupeptin, bestatin, pepstatin and pefablock) were purchased from Boehringer Mannheim (Indianapolis, IN) and aprotinin was from ICN Biomedicals Inc. (Aurora, OH). Plasmid pSP189 was a generous gift from Dr Michael Seidman (National Institute of Aging, Baltimore, MD).
Cell extraction and fractionation
HeLa S3 cells were grown as monolayers at 37°C in DMEM containing 10% (v/v) fetal bovine serum, 1% (v/v) non-essential amino acids and 1% (v/v) penicillin/streptomycin. Nuclei were isolated from ∼10 g (wet weight) freshly harvested, logarithmically growing cells, by a modification of the method previously described (32). All cellular extraction procedures were performed at 4°C unless stated otherwise. Cells were harvested by scraping, then washed and pelleted twice in ice-cold PBS (800 g). The cell pellet was resuspended in 2.5 vol hypotonic lysis buffer (10 mM Tris–HCl, pH 7.6, 1 mM DTT, 5 mM MgCl2, 1 mM EDTA, 1 mM pefablock, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 µg/ml pepstatin) and swollen on ice for 40 min. The cells were lysed by Dounce homogenization (40 strokes with the ‘loose’ pestle). Lysis was estimated to be ≥90% by Trypan Blue dye exclusion.
The homogenate was brought to 250 mM sucrose and nuclei were recovered by centrifugation at 1000 g for 5 min. The nuclei were resuspended in an equal volume of hypotonic lysis buffer containing 250 mM sucrose and repelleted at 1000 g for 10 min.
Isolated nuclei were resuspended in 4 vol nuclear extraction buffer (20 mM Tris–HCl, pH 7.6, 1 mM DTT, 2 mM EDTA, 20% v/v glycerol, 500 mM NaCl, 1 mM pefablock, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 µg/ml pepstatin). Following incubation on ice for 30 min with occasional gentle mixing, the extract was clarified by centrifugation at 25 000 g for 20 min. The supernatant was dialyzed overnight against buffer A (20 mM Tris–HCl, pH 7.6, 1 mM DTT, 1 mM EDTA, 20% v/v glycerol, 25 mM NaCl, 0.2 mM PMSF), flash frozen in liquid nitrogen and stored at –70°C until needed for chromatography.
Extracts were partially purified by chromatography on HiPrep Sephacryl 200, followed by DEAE–Sephacel chromatography. Nuclear extracts (3 ml) were mixed with 1 mg sheared herring sperm DNA and loaded onto a HiPrep Sephacryl 200 16/60 (Pharmacia, Piscataway, NJ) column pre-equilibrated in buffer A. Fractions (2 ml) containing proteins in excess of 60 kDa, including protein recovered in the void volume, were pooled. Pooled protein from HiPrep Sephacryl 200 chromatography was brought to 200 mM NaCl and loaded onto a MT10 (Bio-Rad, Hercules, CA) DEAE–Sephacel (Amersham/Pharmacia) column equilibrated in buffer B (20 mM Tris–HCl, pH 7.6, 1 mM DTT, 1 mM EDTA, 20% v/v glycerol, 200 mM NaCl). Protein was eluted from the column in two steps. The first step consisted of a 2.5 bed volume wash with buffer B, the second was a 4 bed volume wash with buffer B + 200 mM NaCl. Fractions (4 ml) containing protein eluted at either 200 or 400 mM NaCl were pooled separately and designated A and B, respectively. The recovered protein fractions were dialyzed separately against buffer C (20 mM Tris–HCl, pH 7.6, 1 mM DTT, 1 mM EDTA, 20% v/v glycerol, 1 mM pefablock, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 µg/ml pepstatin), flash frozen and stored at –70°C until needed.
DNA preparation
DNA containing DSBs was prepared from plasmid pSP189 either by restriction enzyme digestion or by treatment with the radiomimetic drug bleomycin. DNA containing DSBs with ligatable (3′-OH and 5′-PO4) blunt ends was produced by complete digestion of pSP189 with StuI restriction endonuclease (Fig. 1F). Linear DNA containing DSBs with unligatable 3′-PG-blocked ends was prepared by treating supercoiled pSP189 plasmid DNA with bleomycin (Fig. 1A–E). Plasmid DNA was treated as described previously under conditions demonstrated to produce strand breaks exclusively (33). Briefly, DNA at 150 µg/ml in 12.5 mM Tris–HCl pH 8.0, 300 mM sucrose, 0.0188% Triton X-100, 1.25 mM EDTA, 5 mM MgCl2, 7.5 mM β-mercaptoethanol (βME) and 250 µg/ml heat-inactivated BSA was treated with bleomycin (0.5 µg/ml) at 37°C for 20 min in the presence of 100 µM ferrous ammonium sulfate. The drug was removed by ethanol precipitation and linearized DNA was band isolated and recovered following 1% agarose gel electrophoresis as described previously (34).
End joining assay
DNA DSB end joining repair reactions were typically conducted in 50 µl total volume. Reactions contained 50 mM Tris–HCl pH 8.2, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 5% polyethyleneglycol (PEG) 8000, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 mM pefablock, 100 ng substrate DNA and partially purified HeLa nuclear extract as indicated in the figures. Repair reactions were stopped by addition of 0.4% SDS and incubation at 65°C for 15 min. DNA was recovered by extraction with phenol:chloroform (1:1) and ethanol precipitation using 0.5 µg tRNA as carrier. Repair products were identified by gel shift following 1% agarose electrophoresis and staining for 1 h with Vista Green (VG) according to the manufacturer’s instructions. Images were digitized with a FluorImager 595 system (Molecular Dynamics, Sunnyvale, CA) and quantified densitometrically using Gel-Pro software (Media Cybernetics, Silver Spring, MD).
Protein assay
Protein determinations were made according to the method of Bradford (35), using bovine plasma γ-globulin as the standard.
RESULTS
Detection method
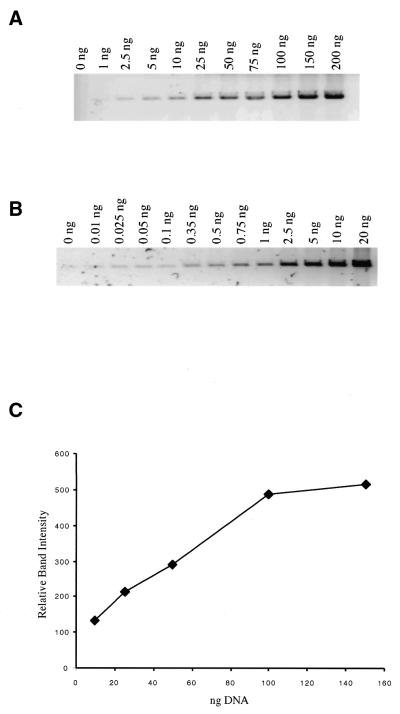
We sought to develop a fast, high throughput assay applicable to protein purification and biochemical analysis. The chief requirement of such an assay is quick, easy and efficient observation and processing of results, preferably by direct observation of repair products in a gel immediately following the repair reaction. We determined that ethidium bromide staining was not sensitive enough for this application. Therefore, we tested VG (which has been reported to be capable of detecting ≤20pg/band duplex DNA; 36,37) for DNA detection sensitivity and linearity of DNA/dye binding with respect to DNA concentration. Measurements were made over the range 0–100 ng using pSP189 plasmid DNA linearized with EcoRI and a FluorImager 595 for detection (Fig. 2). Using scanning conditions optimized for ethidium bromide and VG, respectively, we have found VG staining to be up to 100-fold more sensitive than ethidium bromide (Fig. 2A and B). In practical applications as little as 0.25 ng double-stranded DNA/band can be detected on agarose gels in close proximity to bands with greater fluorescence. In addition, VG dye binding is proportional to DNA concentration from 0.25 to 100 ng (Fig. 2B and C), which encompasses the range of DNA band concentrations observed in our assay.
Figure 2.
DNA staining and detection sensitivity. pSP189 plasmid DNA was linearized with EcoRI and the DNA concentrations indicated in the figure were electrophoresed on a 1% agarose gel. (A) An ethidium bromide stained gel imaged with a Molecular Dynamics FluorImager 595 set for ethidium bromide detection. When imaged with a standard transilluminator, ethidium bromide DNA detection sensitivity is limited to ∼20 ng/band. This is improved by ∼10-fold when ethidium bromide gels are imaged with a FluorImager 595 system (lane 3). (B) A VG stained gel imaged with the FluorImager 595 set to detect VG. VG detection sensitivity is ∼50- to 100-fold greater than ethidium bromide when imaged with the FluorImager system [compare (B) lane 3, and (A) lane 3). (C) VG staining intensity is linear with respect to DNA concentration between 10 and 100 ng and linearity extended to <0.1 ng in additional experiments (data not shown).
HeLa cell extraction and partial purification
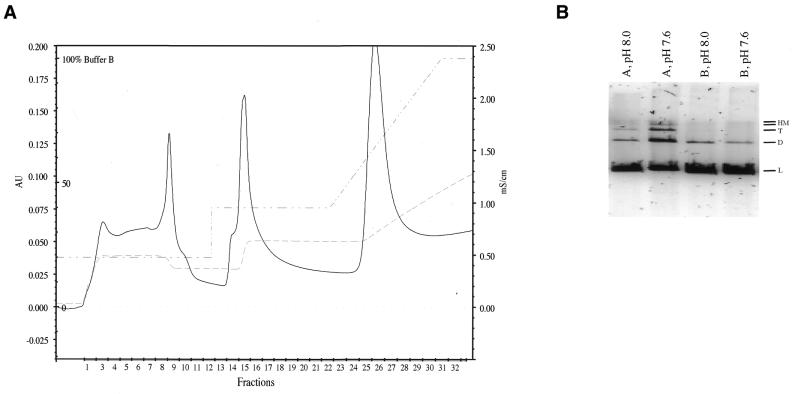
HeLa cell nuclear extracts were produced as described in Materials and Methods. Crude nuclear extracts did not support DSB end joining; therefore, chromatography was performed to produce a protein mixture that would. To enhance for DSB-binding activities, the crude nuclear extracts were mixed with sheared herring sperm DNA, which was added to serve as a substrate for the nucleation of DSB-binding protein complexes. The protein/DNA mixture was then loaded onto a HiPrep Sephacryl 200 16/60 column and proteins which eluted from the column at >60 kDa were pooled. This size range is sufficiently broad to include the known NHEJ proteins. In addition to recovery of potential DNA-bound protein complexes, Sephacryl 200 chromatography helped to reduce small molecular weight nucleases that were not bound to the DNA. The Sephacryl 200 column was followed by a DEAE column, which served to separate the protein component of the mixture from nucleic acids. The added and endogenous nucleic acid fragments act as an inhibitor in our assay and their removal is required to observe end joining. Protein was eluted from DEAE in the 200 (PTN A) and 400 mM NaCl (PTN B) wash steps (Fig. 3A). DNA is retained by the column and does not elute until the gradient reaches a salt concentration >500 mM NaCl.
Figure 3.
(A) DEAE–Sephacel chromatography. Pooled fractions from Sephacryl S-200 chromatography were brought to 200 mM NaCl, applied to DEAE–Sephacel and eluted by washing in two steps with increasing NaCl concentrations. Nucleic acids were retained on the column until the gradient. Protein eluting in fractions 1–11 was pooled and designated PTN A. Fractions 13–19 were pooled and designated PTN B. (B) PTN A and B were assessed for their ability to rejoin StuI cut blunt-ended DNA. Lanes 1 and 2 contain 15 µg PTN A incubated with 100 ng StuI-linearized DNA in 40 mM Bis–Tris–propane, pH 8.0, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 5% PEG 8000, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 mM pefablock or 50 mM Tris–HCl, pH 7.6, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, 5% PEG 8000, 1 µg/ml aprotinin, 0.15 µg/ml leupeptin, 10 µg/ml bestatin, 1 mM pefablock, respectively. Lanes 3 and 4 contain 15 µg PTN B incubated with 100 ng StuI-linearized DNA in the reaction buffers indicated above. DNA substrate and product bands are indicated as follows: L, linear DNA; D, dimer; T, trimer; HM, quatramer and larger high molecular weight products.
Optimization of blunt-end joining
Pooled proteins constituting DEAE PTN A and PTN B were tested for their capacity to rejoin blunt-ended DNA produced by StuI restriction enzyme cleavage of plasmid pSP189. Although both fractions support end joining and produce dimers, only PTN A produces appreciable amounts of trimers and higher molecular weight forms (Fig. 3B, lanes 1 and 2). The yield of end-joined products was buffer dependent, with more product produced in a pH 7.6 Tris buffer containing 1 mM ATP than in a Bis–Tris–propane buffer, pH 8.0, containing 2 mM ATP (Fig. 3B, lanes 1–4). When incubated together, PTN A and B did not complement each other in either buffer (data not shown). The differential end joining activity observed in the two initial test buffers was a function of ATP concentration and not pH. Reactions at 0.5 mM ATP were similar to the 2 mM ATP reactions in that they resulted in a lower yield of repair products than reactions at 1 mM ATP (data not shown). Therefore, 1 mM ATP was used in subsequent end joining reactions. PTN A produced >50% more end-joined product than an equivalent weight of PTN B, therefore PTN A was used in all subsequent end joining optimization procedures.
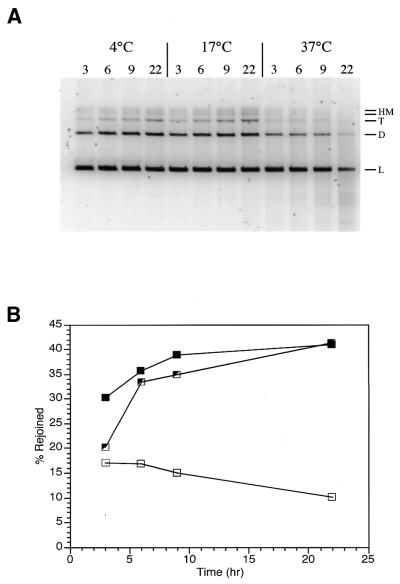
Time and temperature dependence. North et al. have reported that end joining of restriction enzyme-linearized plasmid DNA was a temperature-dependent process with more efficient reactions occurring at lower temperatures (13). We tested this observation with our HeLa cell PTN A extract and found similar results (Fig. 4). Three reaction temperatures were tested: 4°C (Fig. 4A, lanes 1–4), 17°C (Fig. 4A, lanes 5–8) and 37°C (Fig. 4A, lanes 9–12). In addition, the time-dependent yield of end-joined product DNA was determined at all three temperatures. Reactions were conducted with 15 µg PTN A in the standard reaction buffer described in Materials and Methods modified to pH 7.6. Both the 4 and 17°C time course reactions have essentially first order kinetics and both reactions ultimately produced the same relative conversion (∼41%) of substrate DNA to product DNA at 22 h (Fig. 4B). However, the 17°C reactions appeared to have a higher velocity and produced a larger yield of product DNA in a shorter time. Therefore, 17°C was selected as the standard reaction temperature and reactions were typically run for 18–22 h. In the case of the 37°C reactions, the yield of product decreased with increasing time as a result of nucleolytic degradation (Fig. 4A, lanes 9–12). This indicates the presence of nuclease activity in the PTN A fraction that exceeds end joining activity at 37°C.
Figure 4.
Time and temperature dependence for PTN A-mediated blunt-end ligation. (A) Standard repair reactions were conducted at 4, 17 and 37°C with 15 µg PTN A protein/reaction for the times indicated. Linear substrate DNA (L), dimer (D), trimer (T) and high molecular weight multimers (HM) are indicated to the right of the gel. (B) Rejoining kinetics for PTN A-mediated blunt-end ligation. Data from the gel in (A) are plotted as a percentage of total DNA in the lane converted to high molecular weight products. Symbols are as follows: partially open squares, 4°C; closed squares, 17°C; open squares, 37°C.
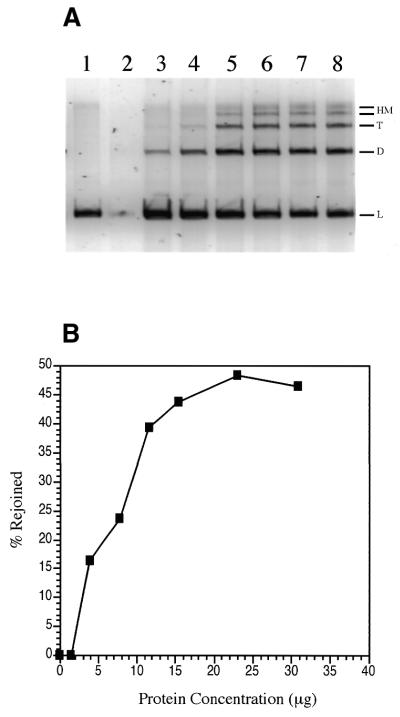
Protein concentration dependence. The amount of partially purified extract needed to maximize the yield of blunt-end DSB joining products was assessed with 100 ng StuI cut pSP189. These reactions were conducted at 17°C for 18 h under standard reaction conditions as described in Materials and Methods. The reaction displayed first order kinetics and maximal blunt-end joining was ∼48% conversion of substrate to product at 23 µg extract protein (Fig. 5A and B). Interestingly, at very low protein concentrations (Fig. 5A, lane 2) not only were no product molecules detected, but the linear substrate DNA itself was degraded; this effect was observed in multiple experiments, including those containing bleomycin-linearized substrate. As observed above, there are nuclease activities in our extract that can degrade the substrate DNA and this reaction is favored at low protein concentrations, even when lower temperatures that favor end joining are used. In contrast, substrate DNA is stabilized at higher protein concentrations and the end joining reaction is favored. It is also important to note that end-joined products are subject to nucleolytic degradation when reactions are conducted in the absence of protease inhibitors. Under these conditions the extract protein itself is unstable and suffers from proteolytic degradation. This probably reflects loss of Ku from the extract in the presence of active proteases.
Figure 5.
Protein concentration dependence for blunt-end ligation. (A) Visualization of linear (L), dimer (D), trimer (T) and high molecular weight multimer (HM) bands. Lane 1, 0 µg PTN A; lane 2, 1.5 µg; lane 3, 3.9 µg; lane 4, 7.7 µg; lane 5, 11.6 µg; lane 6, 15.4 µg; lane 7, 23 µg; lane 8, 30 µg. (B) Data from the gel shown in (A) was quantified as a percentage of DNA in each lane converted to rejoined products. Maximal blunt-end joining was 48% conversion of substrate to product at 23 µg PTN A.
Production and characterization of linear substrate DNA containing bleomycin-induced DSBs
The time, temperature, protein concentration and buffer conditions identified in the previous experiments were established as standard conditions for maximizing end joining of a simple, directly ligatable, blunt-ended substrate. Given these conditions, we were interested in conducting a comparative analysis for end joining of the simple StuI DSB and the complex bleomycin-induced DSB. However, because only a fraction of bleomycin-damaged plasmids will contain DSB, we first sought to maximize the yield of this product and determine its suitability for use as an end joining substrate.
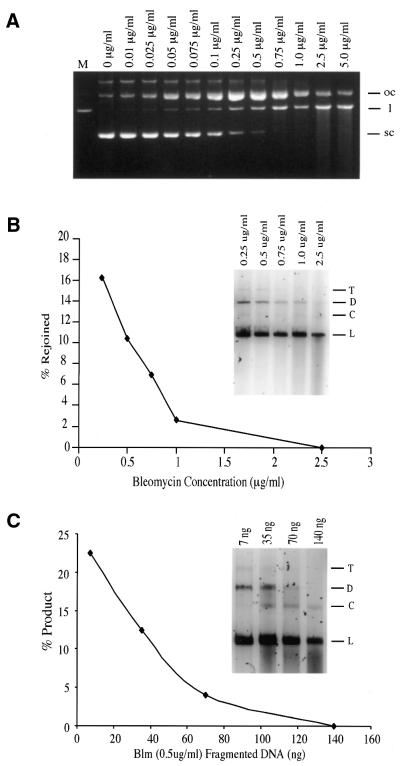
To maximize the yield of linear plasmid containing bleomycin-induced DSBs, a bleomycin dose–response experiment was conducted (Fig. 6A). Bleomycin treatment was performed as described in Materials and Methods and maximum yield of linear DNA was obtained at 0.75 µg bleomycin/ml DNA solution (5.2 × 10–3 µg bleomycin/µg DNA). The linear DNA was isolated and used as substrate in several end joining assays. Although limited amounts of end-joined products were observed with this substrate, the yield (2–5%) was only a fraction of that obtained with the equivalent StuI cut linear DNA substrate (data not shown). This could reflect the inherent difficulty of repairing the more complex bleomycin-induced DSB or it may reflect a problem with the quality of the bleomycin-induced linear DNA substrate. To determine which of these two possibilities were responsible for the low yield of repair products with bleomycin-damaged DNA, we first examined the quality of the purified bleomycin-linearized substrate.
Figure 6.
(A) Bleomycin DNA damage dose–response characteristics. Concentrations are reported as µg bleomycin/ml DNA solution treated (150 µg DNA/ml). Lane M, EcoRI-linearized pSP189 as a marker. Open circular (oc), linear (l) and supercoiled (sc) pSP189 DNA are indicated to the right of the gel. (B) Repair of oxidatively induced DSBs as a function of bleomycin dose used to induce damage. The standard repair assay was conducted with 15 µg PTN A protein at 17°C for 18 h. Linear DNA recovered from the reactions in lanes 8–12 of (A) were used as substrates. The data in the inset gel was plotted as a percentage of substrate converted to high molecular weight products versus the concentration of bleomycin used to produce the linear substrate DNA. (C) Increasing concentrations of DNA fragments recovered from below the linear band in the 0.5 µg/ml bleomycin reaction (A, lane 9) were introduced into a fully functional repair reaction employing PTN A as the repair enzyme source and bleomycin-linearized DNA produced by treatment with 0.5 µg/ml bleomycin (see A, lane 9, and B, inset gel, lane 2). Inhibition of end joining is plotted as a function of added fragmented DNA and reaction results are displayed in the inset gel for comparison of fragment content with the results presented in (A) and (B). Trimers (T), dimers (D), circular (C) and linear (L) forms are indicated to the right of the inset gels.
Substrate quality assessment. Although it had been band purified and was quite clean, the quality of the bleomycin-linearized substrate DNA was questioned because a small amount of the fragmented DNA resulting from bleomycin treatment (Fig. 6A, lane 10) was still detectable in the purified DNA and might act as an inhibitor of the end joining reaction. If so, this phenomenon should be bleomycin dose dependent. To test this, we band purified bleomycin-linearized DNA from bleomycin dose–response curve reactions ranging between 0.25 and 2.5 µg/ml bleomycin (Fig. 6A, lanes 8–12) and subjected it to repair by PTN A protein. The result was a bleomycin dose-dependent decrease in the yield of end-joined product DNA (Fig. 6B). Furthermore, as the repairability of the bleomycin-linearized substrate DNA decreased, the quantity of fragmented DNA in the substrate preparation increased (Fig. 6B, inset). However, based upon the bleomycin dose–response results (Fig. 6A), induction of damage was normally distributed. Therefore, the yield of single-strand breaks proportionate to DSB would also be expected to increase as the bleomycin dose was increased. Consequently, the decrease in end joining observed with DNA prepared by treatment with 0.75 µg/ml bleomycin might reflect a disproportionately large number of single-strand breaks being located immediately proximal to the DSB ends, thus inhibiting processing and end joining. To address this, we recovered bleomycin-fragmented DNA from beneath the linear band in 0.5 µg/ml bleomycin reactions and added it incrementally to otherwise functional end joining reactions using linear DNA recovered from the same reaction (Fig. 6C). Inclusion of modest amounts of fragmented DNA similar to those observed in the reactions in Figure 6B was sufficient to completely inhibit end joining. Furthermore, fragment dose-dependent inhibition of the end joining reaction mirrors the bleomycin dose-dependent inhibition of the reaction almost exactly (Fig. 6B and C). Therefore, to balance yield of linear DNA from bleomycin treatment with the quality and repairability of the resulting purified DSB-containing DNA, we isolated DNA linearized by 0.5 µg/ml bleomycin (3.4 × 10–3 µg bleomycin/µg DNA) for use as substrate DNA.
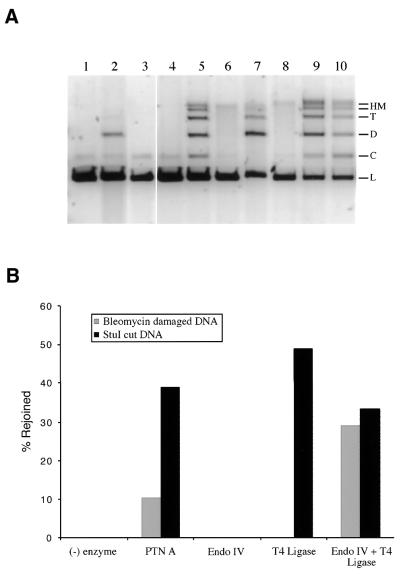
To evaluate the total inherent capacity of this DNA to support end joining in comparison to DNA linearized by StuI, its ability to form end-joined products following processing by recombinant bacterial enzymes or PTN A was examined (Fig. 7). Both the bleomycin-linearized and StuI-linearized DNA were treated with E.coli endonuclease IV and T4 DNA ligase either alone or in combination. Endonuclease IV is a class II AP endonuclease that is also capable of cleaving 3′-nucleotide fragments at DNA strand-break ends and producing a 3′-OH end capable of supporting ligation or polymerization (38). Endonuclease IV treatment alone had no effect on the migration of either bleomycin-linearized substrate DNA or StuI-linearized DNA (Fig. 7A, lanes 3 and 8). In contrast, although treatment with T4 DNA ligase does not affect migration of the bleomycin-linearized substrate, it did produce end-joined products in the presence of StuI-linearized DNA (Fig. 7A, lanes 4 and 9, and Fig. 7B). This result demonstrates the blocked, unligatable nature of the bleomycin-induced DSB. However, when the bleomycin-linearized substrate DNA was reacted with endonuclease IV and T4 DNA ligase in combination, end-joined products were formed with an identical distribution to those produce with StuI-linearized DNA under the same conditions (Fig. 7A, lanes 5, 9 and 10). Most importantly, the yield of end-joined product for both the bleomycin- and StuI-linearized DNA substrates in the presence of endonuclease IV and T4 DNA ligase was essentially the same (Fig. 7B). These results demonstrate that the capacity of the bleomycin-linearized substrate DNA to support end joining repair is equal to that of the blunt-ended StuI-linearized substrate DNA. This observation also confirms the predominately blunt-ended nature of the bleomycin-induced DSB. Finally, the bleomycin- and StuI-linearized substrates were subjected to repair by PTN A and, although high molecular weight end-joined products were formed with both substrates, substantially more product was formed with the StuI-linearized substrate than with the bleomycin-linearized substrate (Fig. 7A, lanes 2 and 7, and Fig. 7B). The difference in the ability of PTN A to catalyze end joining with the two substrates is a function of the extract’s ability to process the blocked ends of the bleomycin-linearized DNA. Therefore, either the bleomycin-linearized DNA is more difficult for PTN A to process due to the complexity of the oxidatively induced DSB or the protein mixture is lacking a required end-processing activity due to the extraction process. The possibility that an end-processing activity has been partially excluded from the final PTN A extract is currently under investigation, however, no such activity has yet been identified.
Figure 7.
Differential repair of bleomycin-induced DSB ends in comparison with StuI-induced blunt ends. Standard repair reactions were performed with either 100 ng substrate DNA linearized by 0.5 µg bleomycin/ml or StuI digestion. The repair reactions contained either 15 µg PTN A, 2 U E.coli endonuclease IV (Endo IV), 5 U T4 DNA ligase (T4 Ligase) or combinations as indicated. Reactions were incubated at 17°C for 18 h. (A) The reactions were as follows: lane 1, bleomycin (blm)-damaged DNA negative control; lane 2, blm-damaged DNA + PTN A; lane 3, blm-damaged DNA + endo IV; lane 4, blm-damaged DNA + T4 ligase; lane 5, blm-damaged DNA + endo IV + T4 ligase; lane 6, StuI cut (blunt-end) DNA negative control; lane 7, StuI cut DNA + PTN A; lane 8, StuI cut DNA + endo IV; lane 9, StuI cut DNA + T4 ligase; lane 10, StuI cut DNA + endo IV + T4 ligase. DNA substrate and product forms are indicated as linear (L), circular (C), dimer (D), trimer (T) and high molecular weight multimers (HM) to the right of the gel. (B) The data in the gel were plotted as a percentage of linear substrate DNA converted to rejoined products. Reactions performed with bleomycin-linearized DNA substrates are indicated by gray bars. Reactions conducted with the StuI-linearized blunt-end DSB substrates are indicated by black bars.
DSB structure-dependent effects on end joining
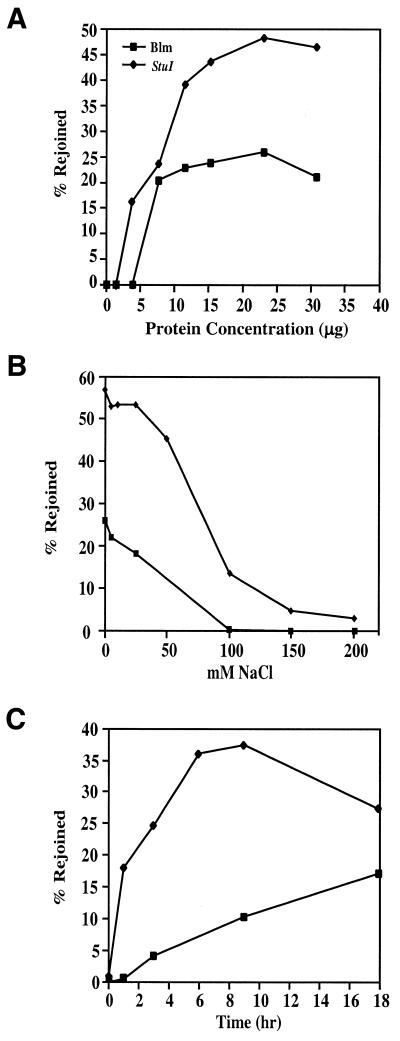
As with StuI-linearized substrate DNA, reaction parameters, including PTN A protein concentration, were examined with the bleomycin-linearized substrate and differences were observed between the two substrates. In contrast to the results obtained with StuI-linearized DNA, repair of bleomycin-linearized DNA reached near maximum activity with as little as 7.5 µg PTN A (Fig. 8A). As with StuI-linearized substrate DNA, maximum activity was reached at 23 µg PTN A, but the amount of rejoined products obtained at this point only exceeded the amount observed at 7.5 µg by 6%. Also, as observed with the StuI cut DNA, low PTN A concentrations resulted in substrate degradation with the bleomycin-linearized substrate, displaying this effect at up to 3.9 µg PTN A, in contrast to StuI-linearized DNA, where this effect was only observed below 3.9 µg PTN A.
Figure 8.
Dependence of repair reaction parameters on DSB end group structure. (A) Protein concentration dependence. Reactions were conducted at 17°C for 18 h with 0, 1.5, 3.9, 7.7, 11.6, 15.4, 23 or 30 µg PTN A. Repair of 0.5 µg/ml bleomycin-linearized DNA is indicated by closed squares. Data obtained for rejoining of StuI-linearized DNA (Fig. 5) are plotted for comparison (closed diamonds). (B) Ionic strength effects. Reactions were conducted under standard conditions with 100 ng bleomycin- (0.5 µg/ml) or StuI-linearized DNA at 17°C for 18 h. Each reaction contained 15 µg PTN A and increasing concentrations of NaCl as indicated in the figure. (C) Time course reactions for PTN A-mediated repair of bleomycin-induced DSBs and StuI-induced blunt-end DSBs. Reactions were conducted under standard conditions of DNA content and temperature as indicated above.
The effect of ionic strength on the end joining reaction was assessed by increasing the NaCl concentration within the reaction (Fig. 8B). StuI-linearized DNA rejoining was relatively tolerant to increased ionic strength between 0 and 25 mM NaCl, but began to be inhibited at concentrations ≥50 mM NaCl. Inhibition was ∼75% at 100 mM NaCl and reached nearly 95% at 200 mM NaCl. In contrast to the relatively high tolerance to increased ionic strength observed with the StuI-linearized substrate, reactions conducted with bleomycin-linearized DNA were sensitive to inhibition by NaCl even at low concentrations (Fig. 8B). Repair continuously decreased with increasing salt concentration and inhibition was nearly complete at 100 mM NaCl.
Time course analysis of the repair reactions for StuI-linearized and bleomycin-linearized DNAs indicate that the initial rate of repair for the two substrates is different. Bleomycin-induced DSBs are repaired with a 6-fold lower initial rate (measured at 3 h) than blunt-ended DSBs produced by StuI (Fig. 8C). In addition, repair of bleomycin-induced DSBs appears to involve a lag phase between 0 and 1 h that is not evident in the end joining reactions for blunt-ended DNA. These results may explain why PTN A consistently produces more product from the StuI-linearized substrate DNA than from bleomycin-linearized DNA. The initial lag in repair of the bleomycin-linearized DNA is of particular interest and may reflect end group processing, suggesting that this might be a rate limiting step in repair of this substrate.
No difference in pH optima was observed with the bleomycin-linearized DNA substrate and the blunt-ended StuI-linearized substrate. Maximum repair activity was obtained at pH 8.2 with both substrates (data not shown).
DISCUSSION
In this report we present an in vitro DSB end joining assay designed to permit biochemical analysis for the repair of bona fide oxidatively induced DNA DSBs. This assay has a number of advantages over current in vitro DSB repair assays. First, the assay is easy to set up and perform and it allows relatively high throughput with the fast turnover necessary for biochemical separation procedures. The sensitivity of VG staining followed by FluorImager detection permits direct observation of repair products in agarose gels. This method of detecting repair products allows us to overcome several technical challenges associated with other end joining assays. Most DSB end joining assays currently in use rely on either ethidium bromide detection of substrates and products, repair of 32P-labeled substrate DNA followed by autoradiography, or Southern blotting to detect repair products (13–16,18,31,39,40). Due to the limited sensitivity of ethidium bromide staining, assays using this method of detection require large quantities (≥1 µg) of substrate DNA in each reaction in order to permit detection of repair products. This approach limits the DSB induction method used to create linear plasmid substrate DNA to methods that can efficiently produce purified linear DNA in quantities of hundreds of micrograms. Therefore, the primary method used to produce substrate DNA for assays employing ethidium bromide detection is restriction enzyme cleavage. However, as discussed above, restriction enzyme-induced DSBs only replicate the physical discontinuity of naturally occurring DSBs and not their chemical complexity or physical structure.
Post-electrophoretic VG staining also permits us to avoid problems associated with handling and use of 32P-labeled substrates, including time limitations upon production and use of the substrate imposed by isotope half-life, as well as continued secondary damage produced by 32P decay. Finally, assays that require Southern blotting to detect repair products are slow and complex to complete. Consequently, they do not permit the fast turn-around times needed during protein purification and activity assessment.
Several assays currently in use to analyze restriction enzyme-induced DSB repair include dNTPs in the reaction buffer. Due to the 3′-OH and 5′-PO4 structure of restriction enzyme-induced DSB ends, the presence of dNTPs may permit polymerization reactions to occur. While this may or may not actually happen in vivo during the repair of oxidatively induced DSBs, it does add another level of complexity to analysis of the end joining reaction (20). By excluding dNTPs from our reactions, we limit the repair to end joining only.
Finally, although we present comparative results for DSB repair of restriction enzyme- and bleomycin-damaged linear plasmid substrates, substrates produced by any DSB-inducing agent can easily be substituted and compared. Therefore, this assay may be useful to assess the relative cytotoxicity of DNA double-strand-breaking agents based upon the relative repairability of the DSBs produced by these agents
Comparative analysis of oxidatively induced DSB and blunt-ended restriction enzyme-induced DSB repair demonstrates damage-dependent differences in the repair reaction. These results show the chemical and physical structures of the DSB end group to be necessary and important considerations when investigating NHEJ repair. Therefore, to accurately model NHEJ repair in vitro, repair reactions should use substrates containing end groups that simulate naturally occurring DSBs as closely as possible. This approach may allow the complete complement of enzymes and overall reaction mechanism involved in the repair of naturally occurring DSBs to be determined. Developing information of this type may also help to explain the differential cytotoxicity of various DNA-strand-breaking agents.
Acknowledgments
ACKNOWLEDGEMENT
The authors would like to thank Dr Irina Panyutin for her assistance during the development of procedures related to the methods and analysis of data presented in the current work.
References
- 1.Povirk L.F. (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res., 355, 71–89. [DOI] [PubMed] [Google Scholar]
- 2.Dahm-Daphi J., Sass,C. and Alberti,W. (2000) Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int. J. Radiat. Biol., 76, 67–75. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry M.A. and Weinfeld,M. (1997) Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem., 272, 15650–15655. [DOI] [PubMed] [Google Scholar]
- 4.Ames B.N. (1999) Cancer prevention: novel nutrient and pharmaceutical developments. Ann. N. Y. Acad. Sci., 889, 87–106. [DOI] [PubMed] [Google Scholar]
- 5.Pfeiffer P. (1998) The mutagenic potential of DNA double-strand break repair. Toxicol. Lett., 96/97, 119–129. [DOI] [PubMed] [Google Scholar]
- 6.Hagmann M., Adlkofer,K., Pfeiffer,P., Bruggmann,R., Georgiev,O., Rungger,D. and Schaffner,W. (1996) Dramatic changes in the ratio of homologous recombination to nonhomologous DNA-end joining in oocytes and early embryos of Xenopus laevis. Biol. Chem. Hoppe Seyler, 377, 239–250. [DOI] [PubMed] [Google Scholar]
- 7.Thacker J. (1999) Repair of ionizing radiation damage in mammalian cells. Alternative pathways and their fidelity. C. R. Acad. Sci. III, 322, 103–108. [DOI] [PubMed] [Google Scholar]
- 8.Jeggo P.A. (1998) Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res., 150, S80–S91. [PubMed] [Google Scholar]
- 9.Jeggo P.A., Tesmer,J. and Chen,D.J. (1991) Genetic-analysis of ionizing-radiation sensitive mutants of cultured mammalian-cell lines. Mutat. Res., 254, 125–133. [DOI] [PubMed] [Google Scholar]
- 10.Lees-Miller S.P., Godbout,R., Chan,D.W., Weinfeld,M., Day,R.S., Barron,G.M. and Allalunis-Turner,J. (1995) Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science, 267, 1183–1185. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y.S., Jin,S.F., Gao,Y.J., Weaver,D.T. and Alt,F.W. (1997) Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA, 94, 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riballo E., Critchlow,S.E., Teo,S.H., Doherty,A.J., Priestley,A., Broughton,B., Kysela,B., Beamish,H., Plowman,N., Arlett,C.F., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol., 9, 699–702. [DOI] [PubMed] [Google Scholar]
- 13.North P., Ganesh,A. and Thacker,J. (1990) The rejoining of double-strand breaks in DNA by human cell extracts. Nucleic Acids Res., 18, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason R.M., Thacker,J. and Fairman,M.P. (1996) The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic Acids Res., 24, 4946–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann E., Schmiemann,V., Goedecke,W., Reichenberger,S. and Pfeiffer,P. (2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res., 28, 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D.S., Herman,T. and Demple,B. (1991) Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res., 19, 5907–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu X.Y., Bennett,R.A. and Povirk,L.F. (1996) End-joining of free radical-mediated DNA double-strand breaks in vitro is blocked by the kinase inhibitor wortmannin at a step preceding removal of damaged 3′ termini. J. Biol. Chem., 271, 19660–19663. [DOI] [PubMed] [Google Scholar]
- 19.Suh D., Wilson,D.M. and Povirk,L.F. (1997) 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res., 25, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King J.S., Fairley,C.F. and Morgan,W.F. (1996) DNA end joining by the Klenow fragment of DNA polymerase I. J. Biol. Chem., 271, 20450–20457. [DOI] [PubMed] [Google Scholar]
- 21.McGall G.H., Rabow,L.E., Ashley,G.W., Wu,S.H., Kozarich,J.W. and Stubbe,J. (1992) New insight into the mechanism of base propenal formation during bleomycin-mediated DNA-degradation. J. Am. Chem. Soc., 114, 4958–4967. [Google Scholar]
- 22.Dizdaroglu M., von Sonntag,C. and Schulte-Frohlinde,D. (1975) Letter: Strand breaks and sugar release by gamma-irradiation of DNA in aqueous solution. J. Am. Chem. Soc., 97, 2277–2278. [DOI] [PubMed] [Google Scholar]
- 23.Beesk F., Dizdaroglu,M., Schulte-Frohlinde,D. and von Sonntag,C. (1979) Radiation-induced DNA strand breaks in deoxygenated aqueous solutions. The formation of altered sugars as end groups. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med., 36, 565–576. [DOI] [PubMed] [Google Scholar]
- 24.Henner W.D., Rodriguez,L.O., Hecht,S.M. and Haseltine,W.A. (1983) Gamma-ray induced deoxyribonucleic acid strand breaks. J. Biol. Chem., 258, 711–713. [PubMed] [Google Scholar]
- 25.Henner W.D., Grunberg,S.M. and Haseltine,W.A. (1983) Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem., 258, 15198–15205. [PubMed] [Google Scholar]
- 26.Burger R.M., Projan,S.J., Horwitz,S.B. and Peisach,J. (1986) The DNA cleavage mechanism of iron-bleomycin—kinetic resolution of strand scission from base propenal release. J. Biol. Chem., 261, 5955–5959. [PubMed] [Google Scholar]
- 27.Jones G.D.D., Boswell,T.V., Lee,J., Milligan,J.R., Ward,J.F. and Weinfeld,M. (1994) A comparison of DNA damages produced under conditions of direct and indirect action of radiation. Int. J. Radiat. Biol., 66, 441–445. [DOI] [PubMed] [Google Scholar]
- 28.Bertoncini C.R. and Meneghini,R. (1995) DNA strand breaks produced by oxidative stress in mammalian cells exhibit 3′-phosphoglycolate termini. Nucleic Acids Res., 23, 2995–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Povirk L.F., Han,Y.H. and Steighner,R.J. (1989) Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5′ extensions. Biochemistry, 28, 5808–5814. [DOI] [PubMed] [Google Scholar]
- 30.Ganesh A., North,P. and Thacker,J. (1993) Repair and misrepair of site-specific DNA double-strand breaks by human cell extracts. Mutat. Res., 299, 251–259. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Trujillo,K., Sung,P. and Tomkinson,A.E. (2000) Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem., 275, 26196–26205. [DOI] [PubMed] [Google Scholar]
- 32.Winters T.A., Russell,P.S., Kohli,M., Dar,M.E., Neumann,R.D. and Jorgensen,T.J. (1999) Determination of human DNA polymerase utilization for the repair of a model ionizing radiation-induced DNA strand break lesion in a defined vector substrate. Nucleic Acids Res., 27, 2423–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winters T.A., Weinfeld,M. and Jorgensen,T.J. (1992) Human HeLa-cell enzymes that remove phosphoglycolate 3′-end groups from DNA. Nucleic Acids Res., 20, 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dar M.E., Winters,T.A. and Jorgensen,T.J. (1997) Identification of defective illegitimate recombinational repair of oxidatively-induced DNA double-strand breaks in ataxia-telangiectasia cells. Mutat. Res., 384, 169–179. [DOI] [PubMed] [Google Scholar]
- 35.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 36.O’Dell S.D., Chen,X.H. and Day,I.N.M. (2000) Higher resolution microplate array diagonal gel electrophoresis: application to a multiallelic minisatellite. Hum. Mutat., 15, 565–576. [DOI] [PubMed] [Google Scholar]
- 37.Madabhushi R.S., Vainer,M., Dolnik,V., Enad,S., Barker,D.L., Harris,D.W. and Mansfield,E.S. (1997) Versatile low-viscosity sieving matrices for nondenaturing DNA separations using capillary array electrophoresis. Electrophoresis, 18, 104–111. [DOI] [PubMed] [Google Scholar]
- 38.Weinfeld M., Lee,J., Gu,R.Q., Karimi-Busheri,F., Chen,D. and Allalunis- Turner,J. (1997) Use of a postlabelling assay to examine the removal of radiation-induced DNA lesions by purified enzymes and human cell extracts. Mutat. Res., 378, 127–137. [DOI] [PubMed] [Google Scholar]
- 39.Pfeiffer P. and Vielmetter,W. (1988) Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res., 16, 907–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labhart P. (1999) Nonhomologous DNA end joining in cell-free systems. Eur. J. Biochem., 265, 849–861. [DOI] [PubMed] [Google Scholar]