A single three-dimensional protein nanocrystal was used for structure determination by electron diffraction. Data were acquired using the rotation method with a Timepix hybrid pixel detector for low-dose data acquisition.
Keywords: electron crystallography, protein nanocrystals, hybrid pixel detector
Abstract
Three-dimensional nanometre-sized crystals of macromolecules currently resist structure elucidation by single-crystal X-ray crystallography. Here, a single nanocrystal with a diffracting volume of only 0.14 µm3, i.e. no more than 6 × 105 unit cells, provided sufficient information to determine the structure of a rare dimeric polymorph of hen egg-white lysozyme by electron crystallography. This is at least an order of magnitude smaller than was previously possible. The molecular-replacement solution, based on a monomeric polyalanine model, provided sufficient phasing power to show side-chain density, and automated model building was used to reconstruct the side chains. Diffraction data were acquired using the rotation method with parallel beam diffraction on a Titan Krios transmission electron microscope equipped with a novel in-house-designed 1024 × 1024 pixel Timepix hybrid pixel detector for low-dose diffraction data collection. Favourable detector characteristics include the ability to accurately discriminate single high-energy electrons from X-rays and count them, fast readout to finely sample reciprocal space and a high dynamic range. This work, together with other recent milestones, suggests that electron crystallography can provide an attractive alternative in determining biological structures.
1. Introduction
Electron crystallography can be used for structure determination of macromolecules from crystalline samples. Originally, the method concentrated on diffracting and imaging two-dimensional crystals (Raunser & Walz, 2009 ▸; Stahlberg et al., 2015 ▸), and resulted in important structures of membrane proteins (Unwin & Henderson, 1975 ▸; Gonen et al., 2005 ▸). Electron diffraction of three-dimensional crystals allowed the structure solution of organic and inorganic samples (Vainshtein, 1964 ▸; Dorset, 1995 ▸; Weirich et al., 1996 ▸; Mugnaioli et al., 2009 ▸; Kolb et al., 2010 ▸; Gorelik et al., 2012 ▸; Zou et al., 2011 ▸; Guo et al., 2015 ▸). Crystallographic data are most efficiently collected by continuously rotating the crystal (Dauter, 1999 ▸). The rotation method has been the standard approach for data collection in protein crystallography for the last four decades (Arndt & Wonacott, 1977 ▸). In electron crystallography, alignment of the crystal with the rotation axis is not always straightforward and the rotation stages are not always as accurate as desired, which prompted the enhancement of the method using either conical beam precession (Vincent & Midgley, 1994 ▸; Kolb et al., 2007 ▸, 2008 ▸; Gemmi et al., 2013 ▸) or beam tilt (Zhang et al., 2010 ▸; Wan et al., 2013 ▸; Yun et al., 2015 ▸). Recently, continuous three-dimensional data collection from protein nanocrystals was accomplished (Nederlof et al., 2013 ▸). The first protein structure of a micrometre-sized crystal was determined soon after, using discrete rotation steps (Shi et al., 2013 ▸). More recently, continuous rotation became the preferred method in protein electron crystallography (Nannenga, Shi, Leslie et al., 2014 ▸; Nannenga, Shi, Hattne et al., 2014 ▸; Yonekura et al., 2015 ▸). The attractiveness of electron crystallography for macromolecular samples is further encouraged by the observation that a large fraction of seemingly failed crystallization attempts contain nanocrystals (Stevenson et al., 2014 ▸, 2016 ▸). Nanocrystals may also contain fewer defects than micrometre-sized crystals and lead to better data quality (Cusack et al., 1998 ▸; de la Cruz et al., 2017 ▸).
The electrostatic scattering potential map, which is the basis for model building, is calculated by a Fourier transform of the phased structure-factor amplitudes and assumes kinematic scattering. Dynamic scattering affects (Cowley & Moodie, 1957 ▸; Dorset et al., 1992 ▸; Glaeser & Downing, 1993 ▸), but does not prevent, structure solution using electron diffraction data (Dorset, 1995 ▸; Glaeser & Downing, 1993 ▸; Palatinus et al., 2017 ▸). In the presence of multiple scattering, the diffraction data can no longer be interpreted using a purely kinematic approximation where I(hkl) ∝ |F(hkl)|2. Structure refinement against electron diffraction data using dynamical scattering theory (Jansen et al., 1998 ▸; Palatinus, Petříček et al., 2015 ▸; Palatinus, Corrêa et al., 2015 ▸; Palatinus et al., 2017 ▸) is not yet available for protein crystals. However, if the crystalline sample is sufficiently thin then this ensures that the measured data are predominantly kinematic and should not hamper structure solution too severely (Cowley & Moodie, 1957 ▸). The small crystal volume directly affects data acquisition; smaller crystals require longer exposure to obtain the same signal-to-noise ratio (SNR) as larger crystals, which results in more radiation damage. Radiation damage is a major limiting factor in the study of macromolecules (Henderson, 1995 ▸; Owen et al., 2006 ▸); thus, diffraction data need to be collected under low-dose cryoconditions and sensitive, low-noise electron detection is imperative.
Previously, we used a single quad Medipix detector (Georgieva et al., 2011 ▸; Nederlof et al., 2013 ▸) and a Timepix detector (van Genderen et al., 2016 ▸) of 512 × 512 pixels (55 × 55 µm pixel size). For very well ordered crystals this detector size is sufficient for resolving up to 50 orders of diffraction. However, for protein crystals with larger unit cells, preventing overlap between adjacent Bragg spots may impose a (virtual) detector distance that limits the resolution of the diffraction patterns1. Tiling of multiple Timepix quad detectors to give larger arrays can overcome these difficulties. Therefore, we developed a novel in-house-designed 1024 × 1024 pixel Timepix hybrid pixel detector (55 × 55 µm pixel size).
Detector features that are of particular interest for electron diffraction are the absence of readout noise, a high dynamic range and the ability to distinguish between the signal from diffracted electrons and that from the high X-ray background that is inherently present in any TEM (Georgieva et al., 2011 ▸; Nederlof et al., 2013 ▸; van Genderen et al., 2016 ▸). These features require a counting detector, a concept that has recently also been introduced in monolithic and CMOS detectors. Hybrid pixel detectors (such as the one employed here) only count high-energy electron hits in counting mode if the energy deposited in the silicon sensor layer for a single pixel is higher than a user-defined threshold during a clock cycle (Llopart et al., 2002 ▸, 2007 ▸). This allows a linear detection range of more than 106 electrons per pixel per second in counting mode. Monolithic and CMOS detectors count after the frame has been read out. So, for these detectors, the dynamic range per pixel in counting mode cannot exceed about one tenth of the number of frames that can be read out per second. This dynamic range is many orders of magnitude smaller than the dynamic range of hybrid pixel detectors.
Monolithic detectors are also more radiation-sensitive than hybrid pixel detectors because the electrons directly hit the integrating readout electronics of the detector. Since electron diffraction data can have spikes of high intensity at low resolution and in Bragg peaks, monolithic detectors are currently not used for measuring electron diffraction data. However, in hybrid pixel detectors the high-energy electrons are stopped by the silicon sensor layer that is bump-bonded to the counting and integration electronics (McMullan et al., 2007 ▸, 2009 ▸; Faruqi & McMullan, 2011 ▸). The integrating electronics of CMOS detectors can be shielded by a phosphor, at the expense of an increased point spread. Thus, hybrid pixel detectors sacrifice pixel size to achieve radiation hardness, a high dynamic range and megahertz counting modes. Pixel size is less important in diffraction data acquisition than in imaging, since the resolution of the data is not determined by the level of detail on the detector but by the number of diffraction orders that can be resolved (Nederlof et al., 2013 ▸).
Here, we present structure determination from a very thin single protein nanocrystal with a diffracting volume of only 0.14 µm3. Diffraction data were acquired using the rotation method on a novel Timepix hybrid pixel detector electron diffraction camera designed for electron crystallography. Standard data-processing procedures and software as commonly used in macromolecular X-ray crystallography were adopted for electron diffraction data with minor adaptations. We discuss instrumentation and data acquisition throughout structure solution, model building and refinement.
2. Methods
2.1. Data acquisition
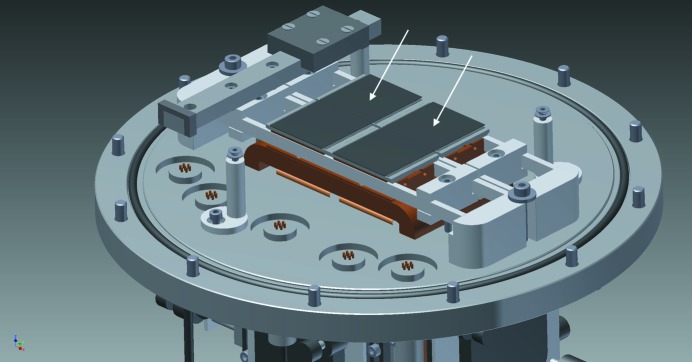
Electron diffraction data were acquired on an FEI Talos Arctica TEM (Center for Cellular Imaging and NanoAnalytics, Basel, Switzerland) and an FEI Titan Krios TEM (NeCEN, Leiden, The Netherlands). Both microscopes were equipped with a Timepix hybrid pixel detector (1024 × 1024 pixels, 55 × 55 µm pixel size). We developed a prototype of such a tiled detector camera of 2 × 2 Timepix quad detectors (Supplementary Fig. S1), which gave an effective array of 1024 × 1204 pixels (Fig. 1 ▸). The Timepix quad cannot be abutted without gaps of ∼35 pixels (horizontal) and ∼175 pixels (vertical). The former gap is imposed by the sensitive silicon layer being slightly larger than the pixel array, and the latter is imposed by the presence of the readout wire bonds on opposing sides of the detector chip.
Figure 1.
The flange design of the camera housing, including the Timepix hybrid pixel detector in the centre (Supplementary Fig. S1). The tiled detector assembly holds four Timepix quads (512 × 512 pixels each). The dark grey top layers pointed out by the arrows represent the sensitive silicon layers of a pair of Timepix quads and the light grey slabs below represent the chip board. The gaps between the chips are necessary to accommodate the wire bonds to the readout boards.
Because high electron fluxes may be focused in Bragg spots, the energy of the incident electron should be completely deposited in the sensor layer to prevent any damage to the readout ASIC that is underneath. For 200 and 300 keV electrons the potential scattering distances are approximately 225 and 450 µm, respectively (McMullan et al., 2007 ▸, 2009 ▸; Faruqi & McMullan, 2011 ▸). For the prototype, we used a 300 µm sensitive silicon layer. A thicker sensitive layer was considered which would allow the use of 300 keV electrons. However, because of the perpendicular impact of a 300 keV incident electron on the detector, on average the first pixel and the last pixel of its track receive the highest deposited dose. This means that at the energy threshold used for each pixel (∼60 keV), the electron is counted once (70%) or twice (30%) (McMullan et al., 2007 ▸, 2009 ▸; Faruqi & McMullan, 2011 ▸). This means that the Bragg spot is spread out over a larger area. To reduce this effect, we opted for 200 keV electrons.
Hen egg-white lysozyme nanocrystals were prepared as described previously (Nederlof et al., 2013 ▸). The microscope was operated at 200 kV and aligned for diffraction with a parallel beam that had a diameter of 2.0 and 1.7 µm in microprobe mode for the Talos Arctica and Titan Krios TEMs, respectively. EM grids were scanned for nanocrystals in imaging mode at 4k–10k magnification. Once a suitable crystal had been found, the crystal was centred on the rotation axis and the beam was centred on the crystal. Diffraction data were collected using the rotation method (Arndt & Wonacott, 1977 ▸), with continuous crystal rotation and shutterless data acquisition (Hasegawa et al., 2009 ▸). A constant rotation of the goniometer was set using the TADui (FEI) and TEMspy (FEI) interfaces of the Talos Arctica and the Titan Krios, respectively. Independently, a fixed frame-exposure time was set with the SoPhy software (Amsterdam Scientific Instruments) for controlling the detector readout. Hence, each frame received the same electron dose and captured a constant rotation increment, as in the rotation method for X-ray crystallography. Data sets were collected with different fixed frame-exposure times (Supplementary Table S1). The dead time of the detector during readout amounted to 4–10% of the exposure time. During data acquisition, the dose rate on the Talos Arctica was ∼0.017 e− Å−2 s−1. The electron flux on the Titan Krios was approximately 20 million electrons per second, amounting to a dose rate of ∼0.08 e− Å−2 s−1 on the crystal (Supplementary Table S1).
2.2. Data processing
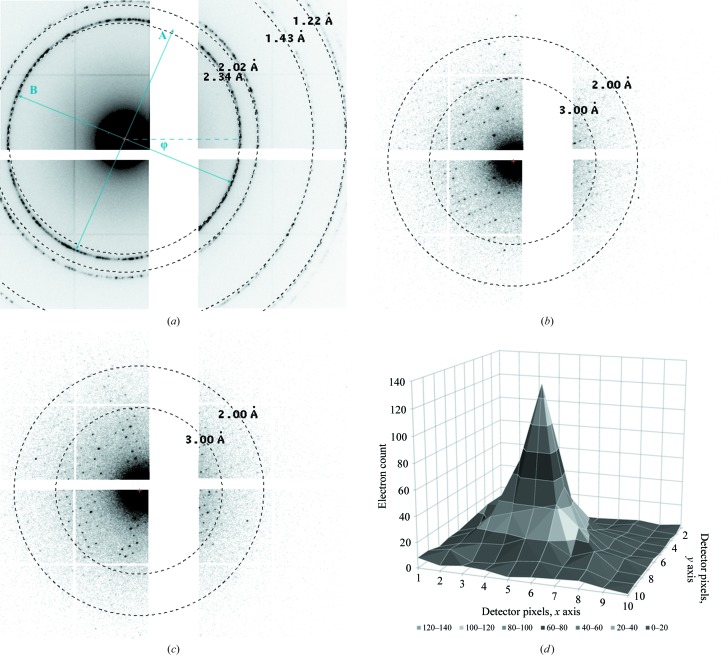
Output frames from the tiled detector were interpolated on an orthogonal grid and converted to PCK format (Abrahams, 1993 ▸) based on the positioning and orientation of the four individual Timepix quads (Figs. 1 ▸ and 2 ▸). We observed a small but significant elliptical distortion from powder diffraction patterns of an aluminium diffraction standard both before and after acquiring data. The distortion could not be modelled by a detector tilt. We determined the magnitude and orientation of the distortion (Fig. 2 ▸ a). Correction tables for XDS were generated by first creating a fake brass-plate pattern based on the distortion parameters using the program geocorr.f90 kindly provided by Dr Wolfgang Kabsch. The calculated geometric correction tables were used with the PILATUS template from XDS, with keywords X-GEO_CORR and Y-GEO_CORR (Kabsch, 2010 ▸).
Figure 2.
Electron diffraction data acquisition. (a) Measured powder pattern of an aluminium diffraction standard after correcting for the tiling offsets of the Timepix quad ASICs. An elliptical distortion can be observed with a deviation of 1.043 (= A/B) at an angle of φ = 21.3°. Diffraction from the single lysozyme crystal summed over 1.0° of rotation (b) from −17.0° to −16.0° and (c) from −6.0 to −5.0°. Crosses on individual quads are owing to corrections for larger border pixels as described by Nederlof et al. (2013 ▸) and van Genderen et al. (2016 ▸); these pixels were not taken into account for processing of the protein diffraction data. Note that owing to the radiation hardness of the detector, no backstop was required. Resolution rings were plotted with ADXV (http://www.scripps.edu/tainer/arvai/adxv.html). (d) A typical spot profile of a high-intensity peak at 16.33 Å resolution recorded on a single frame with an angular increment of 0.076° per frame at a dose rate of ∼0.01 e− Å−2 per frame, shown in a 10 × 10 pixel array with 0.055 × 0.055 mm pixel size.
The effective detector distance was calibrated using aluminium powder diffraction patterns after correcting for the elliptical distortion (Fig. 2 ▸ a). The orientation of the rotation axis was initially estimated by identifying reflections close to the rotation axis, which have a wider rocking curve. The angular frame width was assumed to be constant and was determined by dividing the total rotation range by the number of frames. Data were processed with XDS (Kabsch, 2010 ▸). Since the unit-cell parameters are unusual for lysozyme, and the quality indicators of electron diffraction data are very different to those for X-ray diffraction data, we confirmed the experimental parameters with RED (Wan et al., 2013 ▸), which enables the quick, routine inspection of electron diffraction patterns in three-dimensional reciprocal space. After applying corrections for the elliptical distortion, XDS found the unit-cell dimensions with sufficient accuracy for data processing. Without applying these corrections, the elliptical distortion was too large for XDS to home in on the correct unit cell. The rotation-axis parameters were refined during data integration. The angular frame width was refined by minimizing the deviation of the unit-cell angles from an orthorhombic cell (Supplementary Table S1). With this, XDS suggested Laue group mmm, consistent with space group P21212 (Supplementary Tables S2 and S3).
2.3. Structure solution
Data sets were scaled with XSCALE (Kabsch, 2010 ▸), converted to MTZ format with POINTLESS (Evans, 2006 ▸) and merged with AIMLESS (Evans & Murshudov, 2013 ▸). Structure-factor amplitudes were obtained with TRUNCATE (Winn et al., 2011 ▸). A polyalanine model of tetragonal lysozyme (PDB entry 2ybl; De la Mora et al., 2011 ▸) was created using CHAINSAW (Winn et al., 2011 ▸). The polyalanine monomer was used in a search in all orthorhombic primitive Sohncke groups in molecular replacement (MR) with Phaser (McCoy et al., 2007 ▸). The Matthews coefficient suggested that the crystal contained two monomers per asymmetric unit, and Phaser unequivocally identified the rotation and translation parameters of both monomers and confirmed the space group as P21212. Side chains were placed by automated model building with Buccaneer/REFMAC5 (Cowtan, 2006 ▸; Murshudov et al., 2011 ▸). For the merged data three side chains were missing after autobuilding, although in all three instances clear difference potential was observed in the map (Supplementary Fig. S3). Thus, after inspecting the model and map these three missing residues were fitted using Coot (Emsley et al., 2010 ▸). We did not further enhance the models by manual rebuilding in order to evaluate the extent to which refinement was able to correct errors in the model.
2.4. Refinement
The model was optimized using PDB_REDO (Joosten et al., 2014 ▸), in which electron scattering factors were set by placing ‘EXPDTA ELECTRON CRYSTALLOGRAPHY’ into the PDB header. The model was then refined with REFMAC5 (Murshudov et al., 2011 ▸) using NCS restraints. To ensure convergence, the input model was refined for 1000 cycles using REFMAC5 (Supplementary Fig. S4). Electron scattering factors were set in REFMAC5 with the keyword ‘SOURCE ELECTRON MB’. To calculate the map coefficients, REFMAC5 was set to not restore unobserved reflections with the keyword ‘MAPC FREE EXCLUDE’.
We validated refinement in REFMAC5 with R complete instead of R free. When considering data sets with less than about 10 000 unique reflections, as is the case for our data, calculating R complete is preferred (Brünger, 1997 ▸). The R complete validation method allows all reflections to be used in refinement, and thus our R work is equivalent to R1. R1 defines how well the model explains all observed reflections. Like R work, it is likely to be affected by model bias. R complete was calculated afterwards according to standard procedures with a 0.2% test-set size (Luebben & Gruene, 2015 ▸). Briefly, all nonmeasured observations were first removed from the reflection file with SFTOOLS (Winn et al., 2011 ▸). 500 separate, non-overlapping and unique test sets were then randomly created with FREERFLAG (Winn et al., 2011 ▸), each containing 0.2% of the observed structure-factor amplitudes. Thus, when combined, these test sets represent all data. 500 independent refinements were then performed until convergence, each time omitting a different test set. Each refinement started with the same (final) model from which R1 had been calculated. After each of the 500 validation refinement cycles had converged, the values of F c were calculated from the resulting model. Only F c(h) values corresponding to reflections that had been omitted from that particular cycle (and thus were not biased by that cycle) were extracted. All these extracted reflections from each of the 500 independent refinement cycles were then combined into a single reflection file representing the unbiased F c(h) values corresponding to all observed structure-factor amplitudes. Finally, R complete was calculated by comparing these excluded data with the observed structure-factor amplitudes. R complete is therefore not biased by the model, just like in standard R free calculations, yet it is a more robust measure of model bias, especially for incomplete and/or sparse data, because all reflections contribute to its value.
3. Results
3.1. Data integration
Data were acquired from a single cryocooled lysozyme nanocrystal with dimensions of 200 × 500 × 1400 nm (Fig. 3 ▸). The crystal was found in a thin layer of vitreous ice over a hole in the carbon support film of the EM grid. The crystal was continuously rotated for 38.2° with an angular increment of 0.076° per frame in a 2 µm diameter beam. The central beam was positioned such that during data collection only the tip of the crystal over the hole was illuminated, thus eliminating any background noise from the amorphous carbon in the support film. In our experience, it was favourable to collect data from crystals that were still attached at one end to the carbon support. Crystal bending upon exposure to the beam was observed in cases where the crystals were suspended in vitreous ice but not attached to the carbon, probably owing to charging effects. The total dose received by the crystal did not exceed ∼4.4 e− Å−2. Data from the single crystal were integrated to 2.1 Å resolution (Table 1 ▸, Fig. 2 ▸). The single-crystal data had a completeness of only ∼50%, but were sufficient for full structure solution (Table 1 ▸).
Figure 3.
Micrographs of a single three-dimensional lysozyme crystal (200 × 500 × 1400 nm) in a thin layer of vitreous ice across a hole in the Lacey carbon EM grid at (a) +20° tilt angle and (b) +50° tilt angle. Diffraction data were acquired with a 2.0 µm diameter parallel beam in microprobe mode, indicated by a circle in (a). During data collection only the tip of the crystal was kept in the central beam to limit noise from the carbon support. The width of the crystal at both tilt angles was used to derive its dimensions; the length was measured from the tip of the crystal to the edge of the carbon and was the maximum size of the crystal within the central beam at any point during rotation.
Table 1. Data integration and refinement statistics.
Values in parentheses are for the highest resolution shell; the data were truncated at I/σ(I) > 1.0 (Diederichs & Karplus, 2013 ▸).
| Single crystal (PDB entry 5o4w) | Merged data (PDB entry 5o4x)† | |
|---|---|---|
| Data integration | ||
| Space group | P21212 | |
| a, b, c (Å) | 104.56, 68.05, 32.05 | |
| α, β, γ (°) | 90.0, 90.0, 90.0 | |
| No. of crystals | 1 | 7 |
| Resolution (Å) | 41.46–2.11 (2.17–2.11) | 57.03–2.11 (2.17–2.11) |
| R merge (%) | 26.3 (56.6) | 39.8 (64.0) |
| 〈I/σ(I)〉 | 2.6 (1.0) | 2.7 (1.0) |
| Completeness (%) | 49.5 (49.8) | 61.7 (49.8) |
| Reflections | 12601 (1462) | 41191 (1462) |
| Unique reflections | 6749 (545) | 8560 (545) |
| Structure solution | ||
| Translation-function Z-score | 22.5 | 26.7 |
| LLG score | 395 | 535 |
| Refinement | ||
| Reflections | 6717 | 8503 |
| R1‡ (%) | 33.5 | 26.4 |
| R complete ‡ (%) | 35.0 | 27.9 |
| 〈B〉 (Å2) | 24.0 | 27.0 |
| R.m.s.Z, bonds | 0.92 | 0.85 |
| R.m.s.Z, angles | 1.27 | 0.97 |
| Ramachandran plot | ||
| Favoured (%) | 93.7 | 98.4 |
| Allowed (%) | 5.9 | 1.6 |
| Outliers (%) | 0.4 | 0.0 |
Data-integration statistics for the individual crystals used for merging are shown in Supplementary Table S1; data-merging statistics are presented in Supplementary Fig. S2 and Supplementary Table S4.
We present R1 and R complete instead of R work and R free. For less than 10 000 unique reflections R complete is preferred over R free, since it is calculated from all reflections (Brünger, 1997 ▸; Luebben & Gruene, 2015 ▸). Since all structure factors are used, this in turn leads to a more robust calculation than R free. Using this validation method, the actual refinement uses all reflections; hence, R work is equivalent to R1.
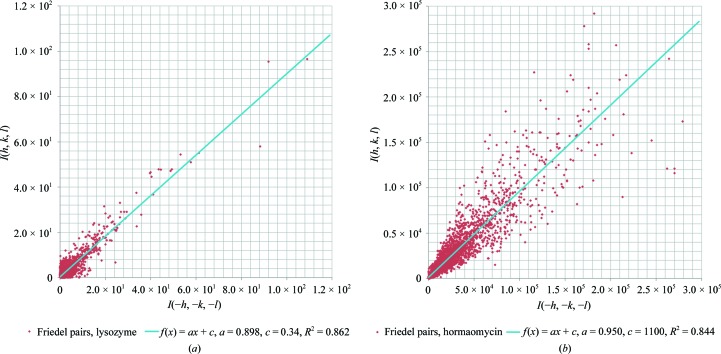
To investigate the inter-crystal consistency of the data with those of other nanocrystals, we collected additional diffraction data (Table 1 ▸). After merging with diffraction data from six other nanocrystals (Supplementary Table S1) that diffracted to 2.5–3.0 Å resolution rather than 2.1 Å, the overall completeness increased to ∼60% (Supplementary Fig. S2, Supplementary Table S4). The limiting factors were radiation damage and the preferred orientation of the crystals, combined with the limited rotation range of the goniometer holding the EM grid. At higher angles, the distance that the electrons have to travel through the surrounding amorphous ice and the protein crystal can become too large for accurate data acquisition. These limitations are inherent to current implementations of electron diffraction: others have collected to up to ∼44° (Nannenga, Shi, Leslie et al., 2014 ▸), ∼61° (Nannenga, Shi, Hattne et al., 2014 ▸) and ∼40° (Yonekura et al., 2015 ▸). These data were collected on crystals that were significantly larger than our nanocrystals, and in case of Yonekura and coworkers needed merging from 58 and 99 crystals (Table 2 ▸). Further, we compared the differences in the measured intensities of Friedel pairs after scaling but before merging of the single-crystal data set (Fig. 4 ▸). The variation in Friedel pair intensities for the single-crystal data is low, even when compared with X-ray data from a small macrocyclic depsipeptide crystal that could be solved by direct methods.
Table 2. Relative crystal volume used for structure determination in recent macromolecular electron diffraction studies.
| PDB code | Detector | d (Å) | Space group | Unit-cell dimensions (Å) | No. of crystals | Individual crystal size (µm) and total diffracted volume† | No. of unit cells‡ (×106) | Relative unique diffracted intensity§ (×106) | |
|---|---|---|---|---|---|---|---|---|---|
| Lysozyme | 5o4w | Hybrid pixel | 2.1 | P21212 | 105 × 68 × 32 | 1 | 0.2 × 0.5 × 1.4 (0.14 µm3) | 0.6 | 1.4 |
| Lysozyme (Nannenga, Shi, Leslie et al., 2014 ▸) | 3j6k | CMOS | 2.5 | P43212 | 76 × 76 × 37 | 1 | 0.5 × 2.0 × 2.0 (2 µm3) | 9.4 | 18 |
| Catalase (Nannenga, Shi, Hattne et al., 2014 ▸) | 3j7b | CMOS | 3.2 | P212121 | 68 × 172 × 182 | 1 | 0.15 × 4.0 × 6.0 (3.6 µm3) | 1.7 | 14 |
| Catalase (Yonekura et al., 2015 ▸) | 3j7u | CCD | 3.2 | P212121 | 69 × 173 × 206 | 58 | 0.1 × 2.0 × 2.0 (23 µm3) | 9.4 | 77 |
| Ca2+-ATPase (Yonekura et al., 2015 ▸) | 3j7t | CCD | 3.4 | C2 | 166 × 64 × 147 (β = 98°) | 99 | 0.1 × 2.0 × 2.0 (40 µm3) | 25 | 490 |
The illuminated crystal size used for data acquisition is estimated from the reported crystal dimensions and the aperture sizes used; for the structures with PDB codes 3j7u and 3j7t (Yonekura et al., 2015 ▸) we assumed that the plate-like crystals had a surface area of 2 × 2 µm. The total diffracted volume (indicated by the number in parentheses) takes the number of crystals required for the three-dimensional data set into account.
The required number of unit cells was calculated by dividing the total diffracted volume by the unit-cell volume.
We calculated the relative unique diffracted intensity by dividing the number of required unit cells (given in the previous column) by the number of asymmetric units in the unit cell and multiplying the result by the cube of the resolution of the data set.
Figure 4.
Differences in intensities of Friedel pairs after scaling plotted for (a) a single lysozyme crystal used for structure solution with R Friedel = 0.329 and (b) X-ray data for hormaomycin, a macrocyclic depsipeptide in space group P1 with R Friedel = 0.151 (Gruene et al., 2014 ▸).
3.2. Structure determination
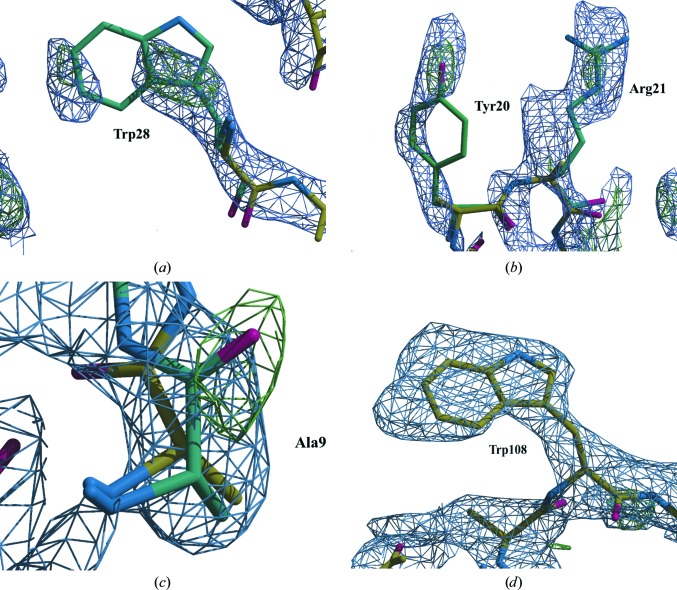
Molecular replacement with a monomeric polyalanine lysozyme model derived from a different, tetragonal space group successfully located a single monomer in the asymmetric unit. It also then placed the second monomer. A Z-score of 22.5 is sufficiently high above the threshold of 8.0, indicating a successful structure solution (McCoy et al., 2007 ▸). Automated model building with Buccaneer/REFMAC5 (Cowtan, 2006 ▸; Murshudov et al., 2011 ▸) was used to reconstruct the side chains (Figs. 5 ▸ a and 5 ▸ b). The densities that are shown were not refined. Hence, they look poor. However, they show that the molecular replacement was successful, as they demonstrate that the phases from a polyalanine MR solution allow the placement of side-chain density for atoms that were not included in the MR model. Subsequent refinement using only the observed reflections improved the quality of the map; for example, the refined density suggests that residue Ala9 is a cis-peptide, which differs from the tetragonal MR model (Fig. 5 ▸ c). However, the 1.9 Å resolution X-ray structure of the same orthorhombic polymorph confirms that the peptide is cis in this crystal form. This strongly validates the quality of our structure solution. Density that was refined according to standard, default protocols shows continuous, high-resolution density (Fig. 5 ▸ d).
Figure 5.
Automated model building using the single-crystal data. After molecular replacement with the polyalanine monomer (yellow C atoms), the difference map shows the position of bulky side-chain residues such as (a) Trp28 as placed during autobuilding by Buccaneer (turquoise C atoms) and (b) Tyr20 and Arg21. The map is stretched, which is typical for incomplete data; as always with poor map quality, careful interpretation of the region is required. The map improves after side-chain reconstruction with Buccaneer and refinement with REFMAC5. (c) The refined density suggests that Ala9 (yellow C atoms) is a cis-peptide; it is confirmed by the X-ray structure of the same polymorph (turquoise C atoms; PDB entry 4r0f) that the peptide is cis. Refinement using standard protocols can further improve the map and shows continuous density (d) for a Trp108 side-chain residue in chain A of the single-crystal model. All density is shown at a standard contour level of 1.2σ.
At 2.1 Å resolution, and in particular with incomplete data, maps are prone to model bias. To estimate how much information our data contain, we calculated r.m.s.d. values between an X-ray model of orthorhombic lysozyme in the same space group with a similar unit cell (PDB entry 4r0f; Sharma et al., 2016 ▸) and (i) our refined model with autobuilt side chains (r.m.s.d. = 0.7 Å) and (ii) our model with side-chain rotamers that are statistically preferred in proteins (r.m.s.d. = 1.1 Å) (Supplementary Table S5, Supplementary Fig. S5). This indicates that the placement of side-chain residues is based on real information contained in the single-crystal data. These results demonstrate the validity of the diffraction data, despite the relatively poor merging and model statistics compared with complete X-ray data (Table 1 ▸).
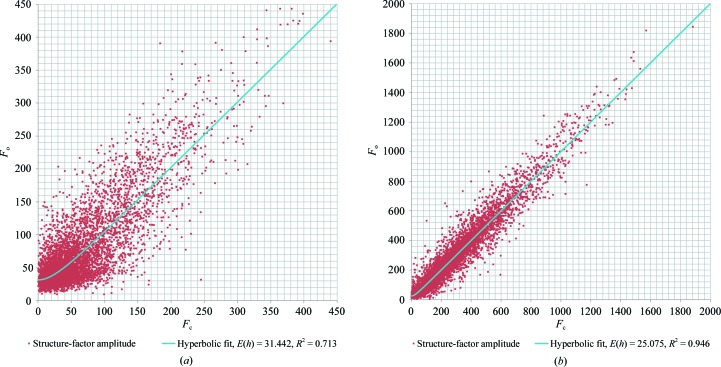
To assess the influence of dynamical scattering on our electron diffraction data, we plotted F o against the refined F c (Fig. 6 ▸ a). In the absence of dynamical scattering, a linear correlation between the measured and calculated structure-factor amplitudes is expected (Fig. 6 ▸ b). However, in our diffraction data the correlation between F o and F c is no longer linear for the lower intensity structure factors. Using least squares, we fitted a hyperbolic curve to the diffraction data describing the nonlinear flattening for the lower intensity part of the F o versus F c graph. We further fitted a hyperbolic curve to the merged diffraction data, showing similar fitting parameters as found for the single-crystal data set (Supplementary Fig. S6).
Figure 6.
F o versus F c graphs for (a) electron diffraction of a single lysozyme nanocrystal and (b) an X-ray data set for cubic (bovine) insulin at 1.6 Å resolution. The data were least-squares fitted with a hyperbolic function described by 〈|F o|〉 = [|F c|2 + 〈|E(h)|〉2]1/2. F o versus F c graphs for only the low-resolution part of the single-crystal data and for the merged crystal data are shown in Supplementary Fig. S6.
4. Discussion
Here, we show the structure determination from electron diffraction data of a single continuously rotated cryopreserved three-dimensional protein nanocrystal with a diffracted volume at least an order of magnitude smaller than was previously possible. For all steps of the structure elucidation, we used standard procedures and software that were originally developed for X-ray protein crystallography. The completeness of the data is low, but because there are two molecules in the asymmetric unit we could apply noncrystallographic symmetry restraints. This NCS was exploited during refinement, and the deleterious effects of data incompleteness could be mitigated. Completeness is also determined by crystallographic symmetry. For instance, if the lysozyme nanocrystal had had tetragonal symmetry, instead of orthorhombic symmetry, the completeness with the same rotation range would have been 84% or greater.
Dynamical scattering has been a longstanding argument against electron crystallography of three-dimensional protein crystals. It causes the intensity of each Bragg peak to be affected by the structure factors of the other Bragg peaks that are recorded in the same exposure. When recorded in a different crystal orientation, its measured intensity will therefore be different even after scaling and Lorentz corrections. This effect also causes differences between the measured intensities of symmetry-equivalent reflections (Glaeser & Downing, 1993 ▸). Dynamical scattering can compromise structure solution of crystals of macromolecules, since current phasing methods and refinement procedures do not account for its effects. Thin crystals minimize the effects of dynamic scattering, and on the basis of multi-slice simulations it has been suggested that the maximal thickness of a protein crystal that still allows structure solution is about 100 nm for 200 keV electrons (Subramanian et al., 2015 ▸), but these calculations ignore inelastic scattering, which is three times more prevalent than elastic diffraction for organic samples.
X-ray data in which the intensities of Friedel pairs correlated as poorly as in our electron diffraction data have been solved and refined using standard procedures (Fig. 4 ▸), indicating that the noise that our data suffered owing to dynamical scattering was tolerable. Furthermore, we show a F o versus F c graph of our electron diffraction data after model refinement (Fig. 6 ▸). It shows a linear correlation for the higher intensity, but at lower intensity the value of F o is overestimated. On average, dynamical diffraction is anticipated to affect weaker reflections more than strong reflections. Thus, on average, weak spots close to intense spots will become more intense, whereas intense spots close to weak spots will hardly be affected (Weirich et al., 2000 ▸). Assuming an expected complex-valued error E(h) that is uncorrelated to F(h), we can infer a hyperbolic relationship between the expected value of 〈|F o|〉 and |F c|,
Our data indeed show such a relationship (Fig. 6 ▸). Merging reduces the random errors of the data, and should also reduce some of the dynamical effect, provided the merged crystals have different orientations. However, the fitting parameters from the F o versus F c graph for the merged data are similar compared with the single-crystal data (Supplementary Fig. S6). The expected error increases at lower resolution (Supplementary Fig. S6), indicating an increased dynamic effect within this resolution range. These observations suggest that weak spots become relatively more affected by other sources of noise with increasing resolution. Nevertheless, although the data were very weak and were compromised by dynamical scattering, they were of sufficient quality for a realistic molecular-replacement solution.
Radiation damage and the small volume of the crystal presented here severely limit the SNR and make data acquisition more challenging. We could improve the SNR substantially with a more accurate and sensitive detector. Previously, we measured three-dimensional nanocrystals similar to the polymorph presented here using CCD detectors and image plates (Georgieva et al., 2007 ▸, 2011 ▸). For protein crystals that had a similar diffracting volume to that reported here, we could never measure more than a few diffraction patterns of high-resolution data with a CCD detector or image plate before radiation damage became too severe. A quantitative comparison between image plates and a Medipix hybrid pixel detector indicated a substantial improvement to be offered by the latter (Georgieva et al., 2011 ▸; Nederlof et al., 2013 ▸). Hybrid pixel detectors such as Medipix, Timepix and EIGER (Llopart et al., 2002 ▸, 2007 ▸; Johnson et al., 2012 ▸) are well suited for measuring high-energy electrons (McMullan et al., 2007 ▸), and can overcome difficulties in detecting weak peaks, for example for CCD and CMOS detectors (Hattne et al., 2016 ▸; Rodriguez & Gonen, 2016 ▸).
An inherent drawback of the detector design is the loss of information in the gaps between the individual tiles (each tile being a 512 × 512 quad Timepix). Because Timepix quads are connected by wire bonds to their readout electronics, these gaps are unavoidable. Without the gaps the data would have been more accurate, but not much more complete, as the geometry of the experiment allowed the data for the Friedel equivalents of most of the missing reflections to be collected (Figs. 1 ▸ and 2 ▸). The deleterious effect of the gap on data completeness can be further mitigated by aligning the rotation axis with the large gap. This would mainly lead to the loss of reflections with Lorentz factors that are so high that they would be discarded by the data-processing software anyway (Fig. 2 ▸).
The total illuminated volume of the single nanocrystal that we used for the data acquisition described here was only ∼0.14 µm3 (Fig. 3 ▸). The data provided sufficient information for structure solution, model building and refinement (Table 1 ▸, Fig. 5 ▸). The total diffracting volume of the crystal is no more than 6 × 105 unit cells (Table 2 ▸). A comparison with structures of macromolecules previously solved by electron diffraction recorded on CCD and CMOS detectors show that these used significantly larger crystals (Nannenga, Shi, Leslie et al., 2014 ▸; Nannenga, Shi, Hattne et al., 2014 ▸; Yonekura et al., 2015 ▸; Hattne et al., 2015 ▸). Since the quality of the diffraction data from protein crystals is determined in the limiting case by the crystallinity of the sample, these data need to be interpreted with great care and should only be used to infer trends. To correct for differences in unit-cell volumes, we determined the number of unit cells used for structure solution. Resolution and crystal symmetry will also affect the amount of unique data within a data set. After correcting for these effects, the hybrid pixel detector allowed structure solution using at the very least an order of magnitude less unique diffracted intensity than obtained previously with other detectors (Table 2 ▸).
Additional hardware modifications may further benefit electron diffraction studies of macromolecular compounds, for example a reliable and well integrated goniometer tilt (Yonekura et al., 2015 ▸) and using an in-column energy filter (Yonekura et al., 2015 ▸). The data presented here show that with a highly sensitive and accurate hybrid pixel detector, nanometre-sized crystals of macromolecules are now also possible targets for three-dimensional protein electron crystallography, which has the advantage of reducing the effects of dynamical diffraction. It is possible that data from micrometre-sized crystals may also be measured more accurately, although it needs to be investigated further whether data accuracy is limited by detector sensitivity or the amount of dynamical diffraction for such crystals. The introduction of hybrid pixel detectors has had a major positive impact on protein X-ray crystallography owing to their high speed, increased sensitivity and high dynamic range (Broennimann et al., 2006 ▸). Based on the results that we present here, we suggest that specialized hybrid pixel detectors may have a similar impact on electron diffraction studies of protein crystals.
5. Related literature
The following references are cited in the Supporting Information for this article: Diederichs & Karplus (1997 ▸), van Genderen (2015 ▸), Kleywegt (1996 ▸) and Visser et al. (2011 ▸).
Supplementary Material
PDB reference: lysozyme, 5o4w
PDB reference: 5o4x
Supplementary Tables and Figures.. DOI: 10.1107/S2059798317010348/tz5091sup1.pdf
Acknowledgments
We thank Wolfgang Kabsch for a program to correct for elliptical distortions and for discussions about data processing. We thank Kay Diederichs and Sven Hovmöller for discussions about data processing. We thank Henning Stahlberg and Kenneth Goldie for support and discussions. We thank Rishi Matadeen, Sacha de Carlo, Thorsten Blum and Igor Nederlof for assistance with sample preparation and data acquisition. We thank Robbie Joosten and Garib Murshudov for discussions on model refinement. We thank ASI (Amsterdam Scientific Instruments) for support with detectors, readout and software. We thank Jan Visser and Bas van der Heijden (NIKHEF, Amsterdam) for early support, design and testing.
Footnotes
Just as an example: assuming a unit cell with 100 Å axes in a well ordered protein crystal, and a preferred minimum distance of five pixels between adjacent Bragg peaks, the maximum attainable resolution for a single quad is approximately 3.5 Å if the direct beam is centred on the detector. For the same crystal, tiling four quads as presented here would increase the maximum resolution at which Bragg spots can be identified to beyond 1.0 Å, again assuming a central direct beam.
References
- Abrahams, J. P. (1993). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 28, 39–41.
- Arndt, A. W. & Wonacott, A. J. (1977). The Rotation Method in Crystallography. Amsterdam: North-Holland.
- Broennimann, C., Eikenberry, E. F., Henrich, B., Horisberger, R., Huelsen, G., Pohl, E., Schmitt, B., Schulze-Briese, C., Suzuki, M., Tomizaki, T., Toyokawa, H. & Wagner, A. (2006). J. Synchrotron Rad. 13, 120–130. [DOI] [PubMed]
- Brünger, A. T. (1997). Methods Enzymol. 277, 366–396. [DOI] [PubMed]
- Cowley, J. M. & Moodie, A. F. (1957). Acta Cryst. 10, 609–619.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Cruz, M. J. de la, Hattne, J., Shi, D., Seidler, P., Rodriguez, J., Reyes, F. E., Sawaya, M. R., Cascio, D., Weiss, S. C., Kim, S. K., Hinck, C. S., Hinck, A. P., Calero, G., Eisenberg, D. & Gonen, T. (2017). Nature Methods, 14, 399–402. [DOI] [PMC free article] [PubMed]
- Cusack, S., Belrhali, H., Bram, A., Burghammer, M., Perrakis, A. & Riekel, C. (1998). Nature Struct. Biol. 5, 634–637. [DOI] [PubMed]
- Dauter, Z. (1999). Acta Cryst. D55, 1703–1717. [DOI] [PubMed]
- De la Mora, E., Carmichael, I. & Garman, E. F. (2011). J. Synchrotron Rad. 18, 346–357. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (2013). Acta Cryst. D69, 1215–1222. [DOI] [PMC free article] [PubMed]
- Dorset, D. L. (1995). Structural Electron Crystallography. New York: Plenum Press.
- Dorset, D. L., Tivol, W. F. & Turner, J. N. (1992). Acta Cryst. A48, 562–568. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Faruqi, A. R. & McMullan, G. (2011). Q. Rev. Biophys. 44, 357–390. [DOI] [PubMed]
- Gemmi, M., Galanis, A. S., Karavassili, F., Das, P. P., Calamiotou, M., Gantis, A., Kollia, M., Margiolaki, I. & Nicolopoulos, S. (2013). Microsc. Anal. 27, 24–29.
- Genderen, E. van (2015). Doctoral thesis. Leiden University, The Netherlands. https://openaccess.leidenuniv.nl/handle/1887/36598.
- Genderen, E. van, Clabbers, M. T. B., Das, P. P., Stewart, A., Nederlof, I., Barentsen, K. C., Portillo, Q., Pannu, N. S., Nicolopoulos, S., Gruene, T. & Abrahams, J. P. (2016). Acta Cryst. A72, 236–242. [DOI] [PMC free article] [PubMed]
- Georgieva, D. G., Jansen, J., Sikharulidze, I., Jiang, L., Zandbergen, H. W. & Abrahams, J. P. (2011). J. Instrum. 6, C01033.
- Georgieva, D. G., Kuil, M. E., Oosterkamp, T. H., Zandbergen, H. W. & Abrahams, J. P. (2007). Acta Cryst. D63, 564–570. [DOI] [PubMed]
- Glaeser, R. M. & Downing, K. H. (1993). Ultramicroscopy, 52, 478–486. [DOI] [PubMed]
- Gonen, T., Cheng, Y., Sliz, P., Hiroaki, Y., Fujiyoshi, Y., Harrison, S. C. & Walz, T. (2005). Nature (London), 438, 633–638. [DOI] [PMC free article] [PubMed]
- Gorelik, T. E., van de Streek, J., Kilbinger, A. F. M., Brunklaus, G. & Kolb, U. (2012). Acta Cryst. B68, 171–181. [DOI] [PubMed]
- Gruene, T., Sheldrick, G. M., Zlatopolskiy, B. D., Kozhushkov, S. I. & de Meijere, A. (2014). Z. Naturforsch. B, 69, 945–949.
- Guo, P., Shin, J., Greenaway, A. G., Min, J. G., Su, J., Choi, H. J., Liu, L., Cox, P. A., Hong, S. B., Wright, P. A. & Zou, X. (2015). Nature (London), 524, 74–78. [DOI] [PubMed]
- Hasegawa, K., Hirata, K., Shimizu, T., Shimizu, N., Hikima, T., Baba, S., Kumasaka, T. & Yamamoto, M. (2009). J. Appl. Cryst. 42, 1165–1175. [DOI] [PMC free article] [PubMed]
- Hattne, J., Reyes, F. E., Nannenga, B. L., Shi, D., de la Cruz, M. J., Leslie, A. G. W. & Gonen, T. (2015). Acta Cryst. A71, 353–360. [DOI] [PMC free article] [PubMed]
- Hattne, J., Shi, D., de la Cruz, M. J., Reyes, F. E. & Gonen, T. (2016). J. Appl. Cryst. 49, 1029–1034. [DOI] [PMC free article] [PubMed]
- Henderson, R. (1995). Q. Rev. Biophys. 28, 171–193. [DOI] [PubMed]
- Jansen, J., Tang, D., Zandbergen, H. W. & Schenk, H. (1998). Acta Cryst. A54, 91–101.
- Johnson, I., Bergamaschi, A., Buitenhuis, J., Dinapoli, R., Greiffenberg, D., Henrich, B., Ikonen, T., Meier, G., Menzel, A., Mozzanica, A., Radicci, V., Satapathy, D. K., Schmitt, B. & Shi, X. (2012). J. Synchrotron Rad. 19, 1001–1005. [DOI] [PMC free article] [PubMed]
- Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. (2014). IUCrJ, 1, 213–220. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J. (1996). Acta Cryst. D52, 842–857. [DOI] [PubMed]
- Kolb, U., Gorelik, T. E., Kübel, C., Otten, M. T. & Hubert, D. (2007). Ultramicroscopy, 107, 507–513. [DOI] [PubMed]
- Kolb, U., Gorelik, T. E., Mugnaioli, E. & Stewart, A. (2010). Polym. Rev. 50, 385–409.
- Kolb, U., Gorelik, T. E. & Otten, M. T. (2008). Ultramicroscopy, 108, 763–772. [DOI] [PubMed]
- Llopart, X., Ballabriga, R., Campbell, M., Tlustos, L. & Wong, W. (2007). Nucl. Instrum. Methods Phys. Res. A, 581, 485–494.
- Llopart, X., Campbell, M., Dinapoli, R., San Segundo, D. & Pernigotti, E. (2002). IEEE Trans. Nucl. Sci. 49, 2279–2283.
- Luebben, J. & Gruene, T. (2015). Proc. Natl Acad. Sci. USA, 112, 8999–9003. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McMullan, G., Cattermole, D. M., Chen, S., Henderson, R., Llopart, X., Summerfield, C., Tlustos, L. & Faruqi, A. R. (2007). Ultramicroscopy, 107, 401–413. [DOI] [PubMed]
- McMullan, G., Chen, S., Henderson, R. & Faruqi, A. R. (2009). Ultramicroscopy, 109, 1126–1143. [DOI] [PMC free article] [PubMed]
- Mugnaioli, E., Gorelik, T. E. & Kolb, U. (2009). Ultramicroscopy, 109, 758–765. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nannenga, B. L., Shi, D., Hattne, J., Reyes, F. E. & Gonen, T. (2014). Elife, 3, e03600. [DOI] [PMC free article] [PubMed]
- Nannenga, B. L., Shi, D., Leslie, A. G. W. & Gonen, T. (2014). Nature Methods, 11, 927–930. [DOI] [PMC free article] [PubMed]
- Nederlof, I., van Genderen, E., Li, Y.-W. & Abrahams, J. P. (2013). Acta Cryst. D69, 1223–1230. [DOI] [PMC free article] [PubMed]
- Owen, R. L., Rudiño-Piñera, E. & Garman, E. F. (2006). Proc. Natl Acad. Sci. USA, 103, 4912–4917. [DOI] [PMC free article] [PubMed]
- Palatinus, L., Brázda, P., Boullay, P., Perez, O., Klementová, M., Petit, S., Eigner, V., Zaarour, M. & Mintova, S. (2017). Science, 355, 166–169. [DOI] [PubMed]
- Palatinus, L., Corrêa, C. A., Steciuk, G., Jacob, D., Roussel, P., Boullay, P., Klementová, M., Gemmi, M., Kopeček, J., Domeneghetti, M. C., Cámara, F. & Petříček, V. (2015). Acta Cryst. B71, 740–751. [DOI] [PubMed]
- Palatinus, L., Petříček, V. & Corrêa, C. A. (2015). Acta Cryst. A71, 235–244. [DOI] [PubMed]
- Raunser, S. & Walz, T. (2009). Annu. Rev. Biophys. 38, 89–105. [DOI] [PubMed]
- Rodriguez, J. A. & Gonen, T. (2016). Methods Enzymol. 579, 369–392. [DOI] [PMC free article] [PubMed]
- Sharma, P., Verma, N., Singh, P. K., Korpole, S. & Ashish (2016). Sci. Rep. 6, 22475. [DOI] [PMC free article] [PubMed]
- Shi, D., Nannenga, B. L., Iadanza, M. G. & Gonen, T. (2013). Elife, 2, e01345. [DOI] [PMC free article] [PubMed]
- Stahlberg, H., Biyani, N. & Engel, A. (2015). Arch. Biochem. Biophys. 581, 68–77. [DOI] [PubMed]
- Stevenson, H. P. et al. (2014). Proc. Natl Acad. Sci. USA, 111, 8470–8475.
- Stevenson, H. P. et al. (2016). Acta Cryst. D72, 603–615. [DOI] [PMC free article] [PubMed]
- Subramanian, G., Basu, S., Liu, H., Zuo, J. M. & Spence, J. C. H. (2015). Ultramicroscopy, 148, 87–93. [DOI] [PubMed]
- Unwin, P. N. T. & Henderson, R. (1975). J. Mol. Biol. 94, 425–440. [DOI] [PubMed]
- Vainshtein, B. K. (1964). Structure Analysis by Electron Diffraction. Oxford: Pergamon.
- Vincent, R. & Midgley, P. A. (1994). Ultramicroscopy, 53, 271–282.
- Visser, J., van der Heijden, B., Weijers, S. J. A., de Vries, R. & Visschers, J. L. (2011). Nucl. Instrum. Methods Phys. Res. A, 633, S22–S25.
- Wan, W., Sun, J., Su, J., Hovmöller, S. & Zou, X. (2013). J. Appl. Cryst. 46, 1863–1873. [DOI] [PMC free article] [PubMed]
- Weirich, T. E., Ramlau, R., Simon, A., Hovmöller, S. & Zou, X. (1996). Nature (London), 382, 144–146.
- Weirich, T. E., Zou, X. D., Ramlau, R., Simon, A., Cascarano, G. L., Giacovazzo, C. & Hovmöller, S. (2000). Acta Cryst. A56, 29–35. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yonekura, K., Kato, K., Ogasawara, M., Tomita, M. & Toyoshima, C. (2015). Proc. Natl Acad. Sci. USA, 112, 3368–3373. [DOI] [PMC free article] [PubMed]
- Yun, Y., Zou, X., Hovmöller, S. & Wan, W. (2015). IUCrJ, 2, 267–282. [DOI] [PMC free article] [PubMed]
- Zhang, D., Oleynikov, P., Hovmöller, S. & Zou, X. (2010). Z. Kristallogr. 225, 94–102.
- Zou, X., Hovmöller, S. & Oleynikov, P. (2011). Electron Crystallography. Oxford University Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: lysozyme, 5o4w
PDB reference: 5o4x
Supplementary Tables and Figures.. DOI: 10.1107/S2059798317010348/tz5091sup1.pdf