Abstract
Background
Diagnosis of atrial fibrillation (AF) can be difficult, requiring cumbersome investigations. We aimed to determine the association of established whole‐blood gene expression scores with prevalent AF and to evaluate their performance for the identification of AF in a SIRS (Steroids in Cardiac Surgery) trial cohort.
Methods and Results
Whole‐blood, transcriptome‐wide gene expression profiling was performed using the Illumina HumanHT‐12 Expression BeadChip in 416 participants (65% men) before surgery, including 91 with a diagnosis of AF. An AF gene score (GS) calculated from 7 genes reported to be upregulated in AF and a validated GS for biological age based on 1254 genes related to aging were both independently associated with AF diagnosis before surgery in multivariate logistic regression analyses adjusting for known risk factors (P=0.0006 and P=0.003). Addition of AF and biological age GSs to clinical risk factors led to significant improvement in area under the receiver operating characteristic curve (from 0.77 to 0.80; P=0.03), continuous net reclassification improvement index (P<0.0001), and integrated discrimination improvement index (P=0.0002). When stratifying AF by subtype, AF GS was mainly associated with paroxysmal AF (P=0.003), whereas the biological age GS was mainly associated with permanent AF (P=0.017).
Conclusions
We validated the existence of a blood gene expression signature for prevalent AF and showed that biological age derived from gene expression is significantly associated with prevalent AF. These findings suggest a potential utility of blood gene expression for the identification of patients with AF, particularly paroxysmal AF. This result could have implications for the prevention and management of cryptogenic stroke.
Clinical Trial Registration
URL: http://www.clinicaltrials.gov. Unique identifier: NCT00427388.
Keywords: atrial fibrillation, discrimination, gene expression
Subject Categories: Arrhythmias, Genetics, Gene Expression & Regulation
Clinical Perspective
What Is New?
We validated a blood gene expression signature of the presence of atrial fibrillation.
Biological age derived from blood gene expression is also significantly associated with the presence of atrial fibrillation.
Blood gene expression improves discrimination for the presence of atrial fibrillation over traditional risk factors.
What Are the Clinical Implications?
Our results highlight the potential of blood gene expression for the identification of the biological signature of disease.
Blood gene expression could be part of a noninvasive strategy to identify patients with atrial fibrillation, for example, as part of the investigation of cryptogenic stroke.
Introduction
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia and its prevalence is increasing worldwide.1 The association of AF with long‐term morbidity and mortality is now well established. Individuals with AF are at increased risk of stroke (5‐fold), heart failure (3‐fold), and all‐cause mortality (1.5‐ to 2‐fold).2, 3 Anticoagulation therapy has been demonstrated to reduce the risk of stroke in patients with AF.4, 5 Nevertheless, detection of AF can be difficult, especially in patients with intermittent forms such as paroxysmal AF. Randomized trials evaluating long‐term rhythm monitoring in patients with cryptogenic stroke have shown >5‐fold increase in AF detection, which was identified in up to 15% of cases6, 7; however, these types of investigation are costly and cumbersome, which limits their application on a large scale.
A whole‐blood gene expression signature of prevalent AF has recently been identified in the Framingham Heart Study, consisting of 7 upregulated genes including PBX1, a gene involved in cardiovascular development.8 Transcriptome‐wide gene expression also allows the assessment of biological age. A recent study based on a meta‐analysis of 14 893 persons proposed a calculation of “transcriptomic age” from 1497 genes differentially expressed with aging. Transcriptomic age was associated with blood pressure, cholesterol levels, fasting glucose, and body mass index independent of chronological age.9 Considering the strong relationship between AF and chronological age, we hypothesized that biological age determined from gene expression could be a potential marker of AF.
This study aimed to verify the association of the previously reported AF gene signature and a biological age gene score (BA‐GS) with prevalent AF in a SIRS (Steroids in Cardiac Surgery) trial cohort and to evaluate their performance to identify AF and its subtypes.
Methods
Participants
SIRS was an international randomized control trial including 7507 high‐risk adult patients undergoing cardiac surgery involving the use of cardiopulmonary bypass. Participants were randomized to receive either methylprednisolone (250 mg at anesthetic induction and 250 mg at initiation of cardiopulmonary bypass) or placebo. Information on patient selection and eligibility criteria was published previously.10, 11 A subset of 525 participants from 12 centers located in 4 countries (Canada, United States, Australia, and Belgium) agreed to participate in the genomics substudy and provided a fasting blood sample before the surgery for genetic analyses. The study complies with the Declaration of Helsinki and was approved by the Hamilton Integrated Research Ethics Board. All participants gave written informed consent. We selected only participants of white race because they represented the vast majority (497/525; 94.7%) and excluded patients with end‐stage renal failure requiring dialysis (n=7) because of a potential impact on gene expression.12
Data Collection
Study personnel collected baseline characteristics including the following AF risk factors: height; weight; smoking status, categorized as never, recent (within 12 months), and former (>12 months); systolic and diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease (previous angina or myocardial infarction); dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; and chronic obstructive pulmonary disease.10 ECGs were done preoperatively, at 24 hours postoperatively, and at hospital discharge or on postoperative day 4, whichever was earlier. Diagnosis of AF was recorded based on history and preoperative ECG and divided into 3 subtypes: paroxysmal AF, defined as AF episodes that terminate spontaneously; persistent AF, defined as AF episodes that do not terminate spontaneously but convert with either electrical or pharmacological cardioversion; and permanent AF, defined as AF that does not terminate either spontaneously or with electrical or chemical conversion or for which cardioversion has not been attempted. Onset of AF following surgery was recorded at discharge and at 30 days (assessed during a follow‐up visit).
Gene Expression
Peripheral fasting whole blood was collected in PAXgene blood tubes the day before or the morning of the surgery and frozen at −70°C until analysis. RNA was extracted using the QiaSymphony (Qiagen) automated large‐sample nucleic acid purification system. RNA concentration was assessed using RiboGreen, and quality was evaluated using a BioAnalyzer lab‐on‐chip (Agilent) with the 18S/28S ratio and RNA integrity score. RNA was then hybridized to the Human HT‐12 BeadChip (Illumina), which interrogates 47 323 transcripts. Samples from patients with AF were randomly distributed on the chips.
Samples with an outlier pattern of expression or a low proportion of expressed probes (defined as <1.5 times the interquartile range below the first quartile) were removed (n=77). Samples with sex discrepancy according to genotyping data were removed (n=6). Probes with low level of expression were removed (detection P value <0.05 for >50% of samples; n=33 196). Raw data were background corrected using a normal‐exponential convolution model based on negative control probes, quantile normalized, and log‐2 transformed.13 After quality control was applied, 416 samples and 14 127 probes remained.
Statistical Analyses
The Student t test, Mann–Whitney test, and χ2 test were used as appropriate. A 2‐sided P<0.05 was considered significant. Preprocessing and quality control of gene expression data were performed using the limma package available through Bioconductor.14 Probes for genes previously reported to be upregulated in prevalent AF were isolated (PBX1, GID4, PNP, TPGS2, SLC7A1, SPTB, ANKH).8 For genes represented by >1 probe, the average expression was used. A meta‐analysis using Stouffer's Z score method15 was performed to combine the results of Lin et al8 with those in the SIRS trial cohort. P values obtained for the 7 genes in each cohort were transformed in Z scores, which were then combined using weights determined as the square root of the sample size of the cohort. An AF gene score (AF‐GS) was calculated as the sum of the expression value for each gene weighted according to the effect sizes (β coefficients) reported by Lin et al.8
A BA‐GS was determined using the method reported by Peters et al.9 The score was calculated from 1497 genes significantly associated with chronological age in a meta‐analysis of 14 983 persons of European ancestry. Genes represented in the expression data (n=1254) were selected using their ENTREZ gene identifier. When multiple probes had the same gene identifier, the average was used. Biological age (expressed in years) was calculated by rescaling BA‐GS based on the chronological age observed in the population (normal transformation from means and standard deviations). A third score was created by combining normalized AF‐GS and BA‐GS. Post hoc power analyses were performed to evaluate, for both scores, (1) the power to detect the observed effect size and (2) the effect sizes that could be detected with 80% and 90% power based on the sample size and the observed standard deviations at a type I error rate fixed at 5% (2‐sided).
Univariate and multivariate logistic regressions were performed with prevalent AF as the dependent variable. Models were adjusted for age and sex (model 1) and then for AF risk factors (model 2): body mass index, height, weight, smoking status (in the past 12 months and ever), systolic blood pressure, diastolic blood pressure, hypertension, diabetes mellitus, coronary artery disease (defined as previous angina or myocardial infarction), dyslipidemia, congestive heart failure, valvular heart disease, chronic renal failure, and chronic obstructive pulmonary disease. To exclude a potential confounding effect of blood cell composition, model 2 was also adjusted for the proportion of white blood cells (neutrophils, lymphocytes, and monocytes), as determined using cell deconvolution.16 Odds ratios (ORs) for AF per 1‐SD increase in the gene scores were calculated. As a sensitivity analysis to eliminate the effect of other potential confounders, residuals of the gene scores after adjustment for recruitment center and expression chip were used in the regression models. Sensitivity analyses were also performed after excluding patients with known congestive heart failure.
Analyses were then stratified by AF type: paroxysmal AF (n=31) and permanent AF (n=50). The number of participants with persistent AF (n=10) was too low to allow reliable analysis. Each subtype was compared with the group without AF. Additional analyses were performed with onset of AF following surgery as the outcome.
We evaluated the predictive performance of the models using the area under the receiver operating characteristic curve, which were compared using DeLong's test. Continuous net reclassification improvement and integrated discrimination improvement indexes were calculated to determine the increase in performance when AF‐GS and BA‐GS were added. Calibration was evaluated using Hosmer–Lemeshow goodness‐of‐fit test.
All analyses were performed using R software (version 3.2.2).
Results
After quality control, transcriptome‐wide gene expression data for 416 participants were available for analysis, including 91 with a diagnosis of AF (Figure 1). The median age was 75.3 years, and 65.4% were men. Main indications for surgery were cardiac valve replacement (70.2%) and coronary artery bypass grafting (56.3%). Participants with AF had lower systolic blood pressure and higher prevalence of congestive heart failure, valvular heart disease, chronic obstructive pulmonary disease, and chronic renal failure (Table 1).
Figure 1.
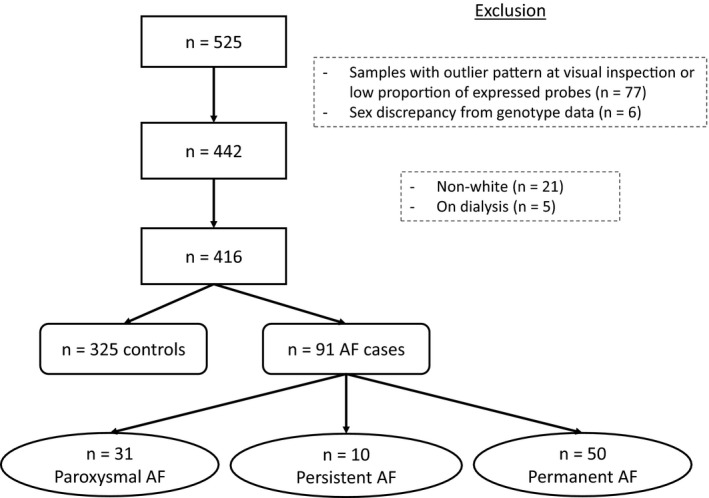
Flowchart of study participants. AF indicates atrial fibrillation.
Table 1.
Clinical Characteristics of Participants
| Prevalent AF | No AF | P Valuea | |
|---|---|---|---|
| n | 91 | 325 | ··· |
| Age, y | 75.8 (71.9–79.8) | 75.3 (68.4–79.9) | 0.052 |
| Male sex | 53 (58.2) | 219 (67.4) | 0.135 |
| Indication for surgery | |||
| Any valve | 77 (84.6) | 215 (66.2) | 0.001 |
| Any CABG | 47 (51.6) | 187 (57.5) | 0.378 |
| Isolated valve | 25 (27.5) | 81 (24.9) | 0.721 |
| Isolated CABG | 7 (7.7) | 66 (20.3) | 0.008 |
| Valve and CABG | 35 (38.5) | 98 (30.2) | 0.169 |
| BMI, kg/m2 | 27.5 (24.4–31.5) | 28.2 (25.4–32.0) | 0.420 |
| Height, cm | 170.0 (163.5–177.0) | 170.0 (163.0–177.0) | 0.808 |
| Weight, kg | 81.0 (71.0–90.4) | 83.0 (72.0–94.3) | 0.583 |
| Smoking (recent) | 9 (9.9) | 42 (13.1) | 0.518 |
| Smoking (ever) | 59 (64.8) | 194 (60.6) | 0.544 |
| SBP, mm Hg | 128.0 (117.0–141.0) | 134.0 (120.0–150.0) | 0.002 |
| DBP, mm Hg | 70.0 (64.5–80.0) | 72.0 (65.0–79.0) | 0.855 |
| Hypertension | 72 (79.1) | 259 (79.7) | 1.000 |
| Diabetes mellitus | 27 (29.7) | 98 (30.2) | 1.000 |
| Coronary artery disease | 40 (44.0) | 178 (54.8) | 0.088 |
| Congestive heart failure | 41 (45.1) | 59 (18.2) | <0.0001 |
| Valvular disease | 83 (91.2) | 260 (80.0) | 0.020 |
| COPD | 24 (26.4) | 49 (15.1) | 0.019 |
| Dyslipidemia | 58 (63.7) | 219 (67.4) | 0.599 |
| Chronic renal failure | 14 (15.4) | 25 (7.7) | 0.043 |
| Lymphocytes (relative) | 0.32 (0.28–0.37) | 0.34 (0.29–0.39) | 0.022 |
| Neutrophils (relative) | 0.51 (0.47–0.55) | 0.51 (0.47–0.54) | 0.400 |
| Monocytes (relative) | 0.14 (0.13–0.16) | 0.14 (0.12–0.16) | 0.012 |
Data are given as median and interquartile range for continuous variables and n and % for categorical variables. Coronary artery disease is defined as a previous history of myocardial infarction or angina. AF indicates atrial fibrillation; BMI, body mass index; CABG, coronary artery bypass grafting; COPD, chronic obstructive pulmonary disease; DBP, diastolic blood pressure; SBP, systolic blood pressure.
Student t test or χ2 test, as appropriate.
Gene Scores
All 7 genes previously reported to be overexpressed in AF had higher mean expression in AF cases than controls, including 6 with statistically significant higher expression (P<0.05; Table 2 and Figure 2). Expression was significantly increased in prevalent AF for the 7 genes when results from Lin et al8 and the SIRS trial cohort were combined in a meta‐analysis using Stouffer's Z score method (Figure 3). The mean weighted gene score (AF‐GS) was 3.33 (95% confidence interval [CI], 3.28–3.37) in participants with AF and 3.21 (95% CI, 3.19–3.24) in controls (P<0.0001; Table 2 and Figure 2).
Table 2.
Normalized Expression for the 7 Previously Reported Genes and AF‐GS
| Prevalent AF | No AF | P Valuea | |
|---|---|---|---|
| PBX1 | 5.57±0.57 | 5.43±0.51 | 0.034 |
| GID4 | 5.62±0.47 | 5.47±0.45 | 0.007 |
| PNP | 7.66±0.72 | 7.34±0.71 | 0.0003 |
| TPGS2 | 6.44±1.06 | 6.11±0.95 | 0.008 |
| SLC7A1 | 7.04±0.62 | 6.81±0.60 | 0.002 |
| SPTB | 5.43±0.72 | 5.17±0.62 | 0.002 |
| ANKH | 5.40±0.61 | 5.35±0.66 | 0.503 |
| AF‐GS | 3.33±0.23 | 3.21±0.22 | <0.0001 |
Data are given as mean±SD. Gene expression is given after background correction, quantile normalization, and log‐2 transformation. AF‐GS is a weighted gene score based on the 7 previously reported genes. AF indicates atrial fibrillation; AF‐GS, atrial fibrillation gene score.
Student t test.
Figure 2.
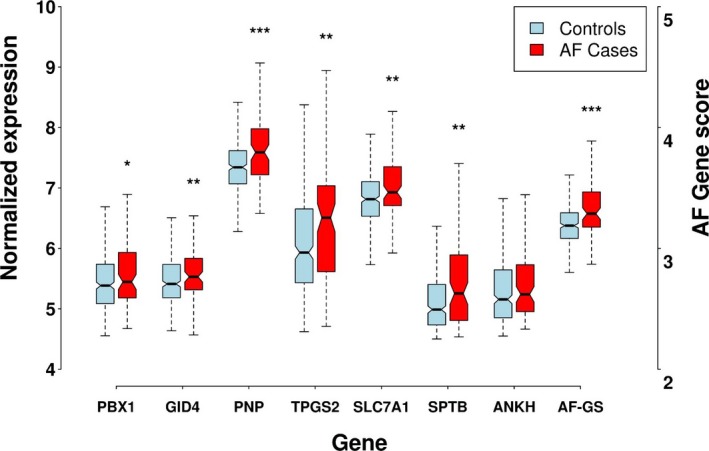
Normalized expression of the 7 genes included in the AF gene score in cases and controls. *P<0.05; **P<0.01; ***P<0.001. AF indicates atrial fibrillation; AF‐GS, atrial fibrillation gene score.
Figure 3.
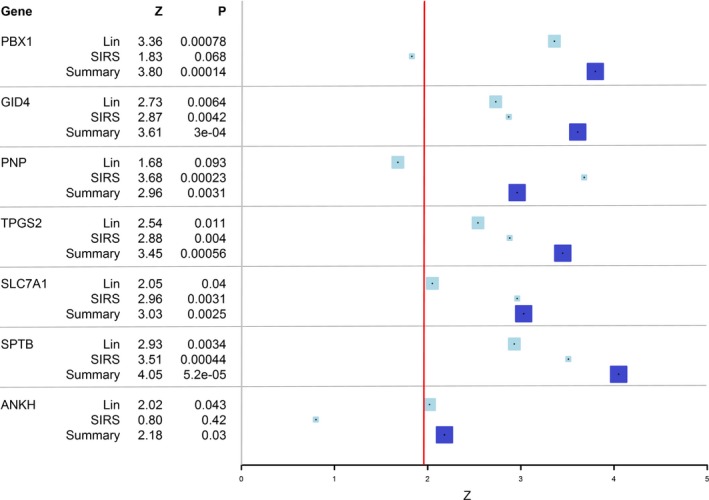
Meta‐analysis of the association between AF and the expression of 7 genes. P value is 2‐sided. Results are from the study by Lin et al.8 and from the SIRS trial cohort. Results were obtained in each cohort using a multivariable model including age, sex, smoking, height, weight, systolic blood pressure, diastolic blood pressure, prevalent diabetes mellitus, prevalent myocardial infarction, prevalent heart failure, and antihypertensive treatment. Meta‐analysis was performed using Stouffer's Z score method. Each box represents a Z score; light blue boxes represent individual studies, and dark blue boxes represent the summary statistic; the size of the box is proportional to the weight given in the meta‐analysis. The gray line represents a null effect (Z=0); the red line represents a significant effect (Z=1.96 corresponding to a 2‐sided P value of 0.05). SIRS indicates Steroids in Cardiac Surgery.
Mean biological age (obtained from the expression of 1254 genes) was significantly higher in participants with AF (76.3 [95% CI, 74.6–78.1] versus 72.5 [95% CI, 71.5–73.5]; P=0.0003). Notably, chronological age was not statistically different between the 2 groups (Table 3 and Figure 4).
Table 3.
Chronological and Biological Age
| Prevalent AF | No AF | P Valuea | |
|---|---|---|---|
| Chronological age, y | 74.8±7.9 | 72.9±9.8 | 0.052 |
| Biological age, y | 76.3±8.6 | 72.5±9.5 | 0.0003 |
Data are given as mean±SD. AF indicates atrial fibrillation.
Student t test.
Figure 4.
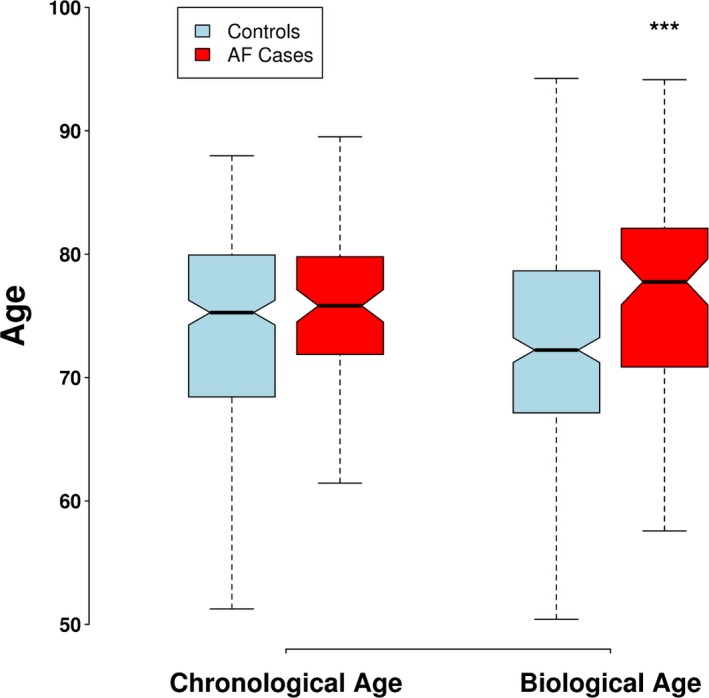
Distribution of chronologic and biological age in cases and controls. ***P<0.001. AF indicates atrial fibrillation.
Post hoc power analyses showed power of 98% for AF‐GS and 93% for biological age to detect the observed difference between the mean score in the 2 groups. There was 80% power to detect a difference of 0.33 SD (0.07 for AF‐GS and 3.1 years for biological age) and 90% power to detect a difference of 0.38 SD (0.09 for AF‐GS and 3.6 years for biological age).
Multivariable Models
In models adjusted for age, sex, and AF risk factors, both gene scores were significantly associated with prevalent AF (for 1‐SD increase in score, AF‐GS: OR: 1.63 [95% CI, 1.23–2.16], P=0.0006; BA‐GS: OR: 2.11 [95% CI, 1.28–3.48], P=0.003; Table S1). A third score combining AF‐GS and BA‐GS was associated with an OR of 2.20 (95% CI, 1.53–3.17; P<0.0001) for prevalent AF (Figure 5). The associations remained significant when using residuals of the gene scores after adjustment for recruitment center and expression chip. Associations remained significant when participants with congestive heart failure (n=100) were removed (P=0.015 for AF‐GS, P=0.040 for BA‐GS, and P=0.0014 for both scores; Figure S1).
Figure 5.
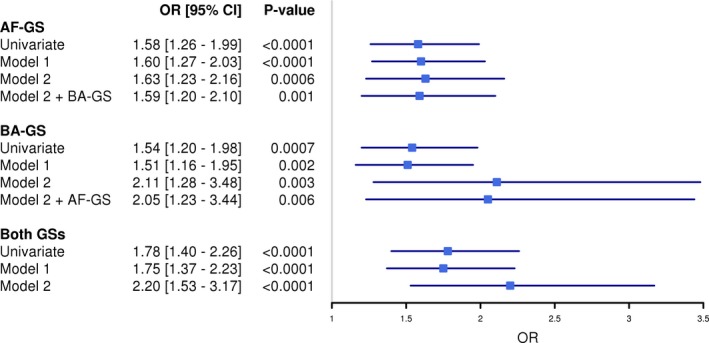
Multivariate models for the prediction of prevalent atrial fibrillation. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; CI, confidence interval; GS, gene score; OR, odds ratio.
Discrimination
Area under the receiver operating characteristic curve for identification of AF was 0.77 (95% CI, 0.72–0.82) using only clinical risk factors, 0.79 (95% CI, 0.74–0.84; P=0.06) when clinical risk factors were combined with AF‐GS, 0.78 (95% CI, 0.73–0.84; P=0.22) when clinical risk factors were combined with BA‐GS, and 0.80 (95% CI, 0.75–0.85; P=0.03) when clinical risk factors were combined with both gene scores (Table 4 and Figure 6).
Table 4.
Predictive Performance, Reclassification, and Discrimination
| AUC (95% CI) | P Value | cNRI (95% CI) | P Value | IDI (95% CI) | P Value | Calibration χ2 (P Value) | |
|---|---|---|---|---|---|---|---|
| Compared with RF only | |||||||
| RF only | 0.769 (0.716–0.823) | ··· | ··· | ··· | ··· | ··· | 5.80 (0.67) |
| RF+AF‐GS | 0.791 (0.741–0.841) | 0.055 | 0.468 (0.240–0.696) | <0.0001 | 0.029 (0.009–0.049) | 0.005 | 2.69 (0.95) |
| RF+BA‐GS | 0.784 (0.732–0.835) | 0.222 | 0.309 (0.079–0.538) | 0.009 | 0.022 (0.006–0.038) | 0.007 | 4.65 (0.79) |
| RF+both GSs | 0.803 (0.754–0.853) | 0.031 | 0.524 (0.298–0.751) | <0.0001 | 0.050 (0.024–0.076) | 0.0002 | 3.60 (0.89) |
| Compared with RF+BA‐GS | |||||||
| RF+both GSs | 0.803 (0.754–0.853) | 0.048 | 0.474 (0.247–0.702) | <0.0001 | 0.028 (0.009–0.047) | 0.003 | 3.60 (0.89) |
| Compared with RF+AF‐GS | |||||||
| RF+both GSs | 0.803 (0.754–0.853) | 0.219 | 0.252 (0.021–0.483) | 0.032 | 0.021 (0.007–0.036) | 0.004 | 3.60 (0.89) |
RFs for atrial fibrillation from model 2: age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF‐GS indicates atrial fibrillation gene score; AUC indicates area under the receiver operating characteristic curve; BA‐GS, biological age gene score; cNRI, continuous net reclassification improvement index; GS, gene score; IDI, integrated discrimination improvement index; RF, risk factor.
Figure 6.
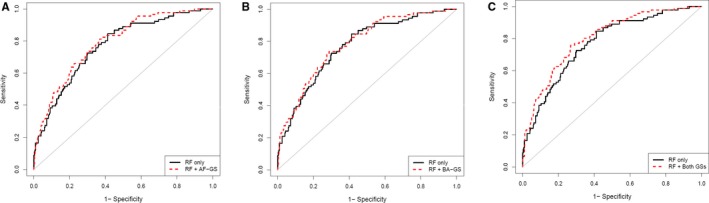
Receiver operating characteristic curves for the identification of prevalent atrial fibrillation. A, Model including AF‐GS compared with a model including clinical RFs alone. B, Model including BA‐GS compared with a model including clinical RFs alone. C, Model including both gene scores compared with a model including clinical RFs alone. RFs were age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; RF, risk factors
Continuous net reclassification improvement index (0.52±0.23, P<0.0001) and integrated discrimination improvement index (0.050±0.019, P=0.0002) indicated moderate improvement in discrimination when the 2 gene scores were added to clinical risk factors (Table 4). In total, 54 of the 91 participants with AF were reclassified as having a higher probability of the outcome, and 214 of 320 participants without AF and with data available for all covariables were reclassified as having lower probability of the outcome, for an overall excess of 125 participants reclassified in the right direction (Table S2). Calibration was adequate in the final model including both gene scores, as shown by the nonsignificant Hosmer–Lemeshow goodness‐of‐fit test (χ2=3.60, P=0.89; Table 4).
When stratified by AF subtype, AF‐GS was significantly associated with paroxysmal AF (P=0.003), whereas BA‐GS was significantly associated with permanent AF (P=0.017) in fully adjusted models (Figures S2 and S3). Area under the receiver operating characteristic curves for identification of paroxysmal and permanent AF were, respectively, 0.69 (95% CI, 0.59–0.79) and 0.85 (95% CI, 0.80–0.91) using clinical risk factors only and 0.75 (95% CI, 0.66–0.84; P=0.17) and 0.86 (95% CI, 0.81–0.92; P=0.42) when clinical risk factors were combined with both gene scores (Tables S3 and S4, Figure S4). Continuous net reclassification improvement and integrated discrimination improvement indexes showed moderate improvement in the models including the 2 gene scores (Tables S3 and S4). Finally, neither gene score was associated with the development of AF following cardiac surgery either at discharge or at 30 days (P>0.05).
Discussion
We report independent associations of a previously described AF gene expression signature and a gene expression score of biological age with prevalent AF in a cohort of well‐characterized patients undergoing cardiac surgery. The 2 gene scores significantly improved prevalent AF discrimination compared with traditional risk factors. AF‐GS was mainly associated with paroxysmal AF, whereas BA‐GS was mainly associated with permanent AF.
We observed a consistent direction of effect for all 7 genes reported to be upregulated in AF in the Lin et al study8 and confirmed the directionality of and association with 6 of 7. Expression of all 7 genes was significantly associated with AF when results from the 2 studies were combined in a meta‐analysis.
Of note, this previous study was performed in a different setting (community‐based observational cohort) in a younger population (mean of 66 years) with a lower prevalence of AF and other comorbidities such as hypertension, diabetes mellitus, coronary artery disease, and heart failure. The study also used a different platform for the determination of gene expression. We show that a weighted score using the reported effect sizes adds predictiveness to traditional risk factors for the identification of AF. The mechanisms by which these 7 genes are linked to AF remain unknown. PBX1 is involved in cardiovascular development,17 and polymorphisms in SLC7A1 were associated with hypertension and endothelial dysfunction,18 whereas SPTB, ANKH and TPGS2 are expressed in the atrial appendage.19 A deletion in the region containing GID4 (17p11.2) leads to Smith–Magenis syndrome, which is associated with congenital heart defects.20 PNP codes for purine nucleoside phosphorylase, a key enzyme in the conditioning response to myocardial ischemia.21 Interestingly, the strongest association was seen for paroxysmal AF, which suggests that these genes could be upregulated in early AF stages.
We also report an association between a whole‐blood transcriptomic measure of biological aging and AF independent of chronological age, which, to our knowledge, has never been reported. The 1254 genes included in the score are mainly involved in DNA and RNA metabolism, immune function, mitochondrial function, and regulation of actin cytoskeleton, lysosome, and fatty acid metabolism.9 The score was previously associated with blood pressure, cholesterol levels, fasting glucose, and body mass index, independent of age.9 Considering the strong association between chronological age and AF, it is reasonable to believe that mechanisms related to aging, such as atrial fibrosis and autonomic nervous system dysfunction, could be involved.22, 23 Such phenomena could be better estimated by a measure of biological age such as the one reported. Interestingly, shorter leukocyte telomere length was previously shown to be associated with prevalent AF and with the risk of cardioembolic stroke in patients with AF.24, 25
Both scores added complementary information for the identification of prevalent AF and moderately but significantly improved discrimination over traditional risk factors. The reported prediction improvement is comparable to what has been observed for blood biomarkers such as NT‐proBNP (N‐terminal prohormone of brain natriuretic peptide), midregional pro atrial natriuretic peptide, and high‐sensitivity troponins.26, 27 Combining other biomarkers with the reported scores could further increase the performance, as the mechanisms involved are likely to be different.
This study suggests a potential value of using blood gene expression for AF screening. It could be used as a noninvasive first‐tier test to better select patients for rhythm monitoring for an extended period of time. It could be particularly useful for the investigation of cryptogenic stroke, in which identification of AF, especially paroxysmal AF, remains challenging. Early detection of AF followed by early intervention including anticoagulation and rate and rhythm control has been proven to reduce the risk of adverse outcomes and to improve quality of life.5, 28, 29
Our study has some limitations. First, we limited our analysis to the 7 genes previously associated with AF—larger studies could identify a more accurate signature, including a higher number of genes and more precise estimates of effect sizes—however, the fact that we replicated a previously reported score eliminates the risk of overfitting and multiple hypothesis testing. Second, we included only white participants, which limits the generalizability to other populations. Third, the SIRS trial was not primarily designed to study AF and was restricted to patients requiring cardiac surgery; therefore, results should be replicated in other populations, including poststroke patients. Further studies should also evaluate performance to identify subclinical paroxysmal AF detected by Holter monitoring. Fourth, we cannot evaluate causality because data on long‐term incident cases were not available; the reported genes could be differentially expressed as a consequence of AF. Fifth, circulating biomarkers such as NT‐proBNP were not available and could potentially improve performance for the identification of AF.
Our results also highlight the potential of gene expression for the identification of the biological signature of disease. We showed that whole‐blood transcriptome‐wide gene expression adds discriminative value over traditional risk factors for the identification of prevalent AF. The measurement of biological aging using gene expression is particularly interesting because it could help predict the broad range of diseases influenced by aging, including AF. Although improvements in the diagnostic performance of gene scores are likely with yet larger and more comprehensive studies, our results suggest that gene expression is a promising avenue for improving screening of persons at high risk and management of cryptogenic stroke.
Sources of Funding
Thériault was supported by grants from the Canadian Institutes of Health Research and Université Laval (Québec, Qc, Canada). Paré is supported by a Canada Research Chair in Genetic and Molecular Epidemiology and the CISCO Professorship in Integrated Health Biosystems. SIRS trial and Genomics sub‐study were funded by the Canadian Institutes of Health Research and the Canadian Network and Centre for Trials Internationally.
Disclosures
None.
Supporting information
Table S1. Parameters of the Multivariate Model Including Both Gene Scores for the Prediction of Prevalent Atrial Fibrillation
Table S2. Detailed Reclassification of Participants When Adding the Gene Scores to the Clinical Model
Table S3. Predictive Performance, Reclassification, and Discrimination for Paroxysmal Atrial Fibrillation
Table S4. Predictive Performance, Reclassification, and Discrimination for Permanent Atrial Fibrillation
Figure S1. Multivariate models for the prediction of AF excluding patients with heart failure. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S2. Multivariate models for the prediction of paroxysmal AF. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S3. Multivariate models for the prediction of permanent AF. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S4. Receiver operating characteristic curves for the identification of prevalent AF by subtypes. A, Model including AF‐GS compared with a model including clinical RFs alone for the identification of paroxysmal AF. B, Model including both GSs compared with a model including clinical RFs alone for the identification of paroxysmal AF. C, Model including BA‐GS compared with a model including clinical RFs alone for the identification of permanent AF. D, Model including both GSs compared with a model including clinical risk factors alone for the identification of permanent AF. RFs were age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; RF, risk factor.
(J Am Heart Assoc. 2017;6:e006057 DOI: 10.1161/JAHA.117.006057.)28666990
References
- 1. Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, Gillum RF, Kim YH, McAnulty JH Jr, Zheng ZJ, Forouzanfar MH, Naghavi M, Mensah GA, Ezzati M, Murray CJ. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation. 2014;129:837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krahn AD, Manfreda J, Tate RB, Mathewson FA, Cuddy TE. The natural history of atrial fibrillation: incidence, risk factors, and prognosis in the Manitoba Follow‐Up Study. Am J Med. 1995;98:476–484. [DOI] [PubMed] [Google Scholar]
- 3. Zoni‐Berisso M, Lercari F, Carazza T, Domenicucci S. Epidemiology of atrial fibrillation: European perspective. Clin Epidemiol. 2014;6:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hart RG, Pearce LA, Aguilar MI. Meta‐analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med. 2007;146:857–867. [DOI] [PubMed] [Google Scholar]
- 5. Lip GY, Lane DA. Stroke prevention in atrial fibrillation: a systematic review. JAMA. 2015;313:1950–1962. [DOI] [PubMed] [Google Scholar]
- 6. Gladstone DJ, Spring M, Dorian P, Panzov V, Thorpe KE, Hall J, Vaid H, O'Donnell M, Laupacis A, Cote R, Sharma M, Blakely JA, Shuaib A, Hachinski V, Coutts SB, Sahlas DJ, Teal P, Yip S, Spence JD, Buck B, Verreault S, Casaubon LK, Penn A, Selchen D, Jin A, Howse D, Mehdiratta M, Boyle K, Aviv R, Kapral MK, Mamdani M. Atrial fibrillation in patients with cryptogenic stroke. N Engl J Med. 2014;370:2467–2477. [DOI] [PubMed] [Google Scholar]
- 7. Sanna T, Diener HC, Passman RS, Di Lazzaro V, Bernstein RA, Morillo CA, Rymer MM, Thijs V, Rogers T, Beckers F, Lindborg K, Brachmann J. Cryptogenic stroke and underlying atrial fibrillation. N Engl J Med. 2014;370:2478–2486. [DOI] [PubMed] [Google Scholar]
- 8. Lin H, Yin X, Lunetta KL, Dupuis J, McManus DD, Lubitz SA, Magnani JW, Joehanes R, Munson PJ, Larson MG, Levy D, Ellinor PT, Benjamin EJ. Whole blood gene expression and atrial fibrillation: the Framingham Heart Study. PLoS One. 2014;9:e96794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, Wilson YA, Kobes S, Tukiainen T, Ramos YF, Goring HH, Fornage M, Liu Y, Gharib SA, Stranger BE, De Jager PL, Aviv A, Levy D, Murabito JM, Munson PJ, Huan T, Hofman A, Uitterlinden AG, Rivadeneira F, van Rooij J, Stolk L, Broer L, Verbiest MM, Jhamai M, Arp P, Metspalu A, Tserel L, Milani L, Samani NJ, Peterson P, Kasela S, Codd V, Peters A, Ward‐Caviness CK, Herder C, Waldenberger M, Roden M, Singmann P, Zeilinger S, Illig T, Homuth G, Grabe HJ, Volzke H, Steil L, Kocher T, Murray A, Melzer D, Yaghootkar H, Bandinelli S, Moses EK, Kent JW, Curran JE, Johnson MP, Williams‐Blangero S, Westra HJ, McRae AF, Smith JA, Kardia SL, Hovatta I, Perola M, Ripatti S, Salomaa V, Henders AK, Martin NG, Smith AK, Mehta D, Binder EB, Nylocks KM, Kennedy EM, Klengel T, Ding J, Suchy‐Dicey AM, Enquobahrie DA, Brody J, Rotter JI, Chen YD, Houwing‐Duistermaat J, Kloppenburg M, Slagboom PE, Helmer Q, den Hollander W, Bean S, Raj T, Bakhshi N, Wang QP, Oyston LJ, Psaty BM, Tracy RP, Montgomery GW, Turner ST, Blangero J, Meulenbelt I, Ressler KJ, Yang J, Franke L, Kettunen J, Visscher PM, Neely GG, Korstanje R, Hanson RL, Prokisch H, Ferrucci L, Esko T, Teumer A, van Meurs JB, Johnson AD. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitlock R, Teoh K, Vincent J, Devereaux PJ, Lamy A, Paparella D, Zuo Y, Sessler DI, Shah P, Villar JC, Karthikeyan G, Urrutia G, Alvezum A, Zhang X, Abbasi SH, Zheng H, Quantz M, Yared JP, Yu H, Noiseux N, Yusuf S. Rationale and design of the steroids in cardiac surgery trial. Am Heart J. 2014;167:660–665. [DOI] [PubMed] [Google Scholar]
- 11. Whitlock RP, Devereaux PJ, Teoh KH, Lamy A, Vincent J, Pogue J, Paparella D, Sessler DI, Karthikeyan G, Villar JC, Zuo Y, Avezum A, Quantz M, Tagarakis GI, Shah PJ, Abbasi SH, Zheng H, Pettit S, Chrolavicius S, Yusuf S. Methylprednisolone in patients undergoing cardiopulmonary bypass (SIRS): a randomised, double‐blind, placebo‐controlled trial. Lancet. 2015;386:1243–1253. [DOI] [PubMed] [Google Scholar]
- 12. Scherer A, Gunther OP, Balshaw RF, Hollander Z, Wilson‐McManus J, Ng R, McMaster WR, McManus BM, Keown PA. Alteration of human blood cell transcriptome in uremia. BMC Med Genomics. 2013;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ritchie ME, Dunning MJ, Smith ML, Shi W, Lynch AG. Beadarray expression analysis using bioconductor. PLoS Comput Biol. 2011;7:e1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high‐throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lipták T. On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Közl. 1958;3:171–197. [Google Scholar]
- 16. Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang CP, Stankunas K, Shang C, Kao SC, Twu KY, Cleary ML. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development. 2008;135:3577–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Z, Venardos K, Jones E, Morris BJ, Chin‐Dusting J, Kaye DM. Identification of a novel polymorphism in the 3′UTR of the L‐arginine transporter gene SLC7A1: contribution to hypertension and endothelial dysfunction. Circulation. 2007;115:1269–1274. [DOI] [PubMed] [Google Scholar]
- 19. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue‐based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 20. Myers SM, Challman TD. Congenital heart defects associated with Smith‐Magenis syndrome: two cases of total anomalous pulmonary venous return. Am J Med Genet A. 2004;131:99–100. [DOI] [PubMed] [Google Scholar]
- 21. Aberg AM, Ronquist G, Haney M, Waldenstrom A. Effects of some modulators on purine nucleoside phosphorylase activity in myocardial tissue. Scand J Clin Lab Invest. 2010;70:8–14. [DOI] [PubMed] [Google Scholar]
- 22. Chen PS, Chen LS, Fishbein MC, Lin SF, Nattel S. Role of the autonomic nervous system in atrial fibrillation: pathophysiology and therapy. Circ Res. 2014;114:1500–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dzeshka MS, Lip GY, Snezhitskiy V, Shantsila E. Cardiac fibrosis in patients with atrial fibrillation: mechanisms and clinical implications. J Am Coll Cardiol. 2015;66:943–959. [DOI] [PubMed] [Google Scholar]
- 24. Carlquist JF, Knight S, Cawthon RM, Le VT, Jared Bunch T, Horne BD, Rollo JS, Huntinghouse JA, Brent Muhlestein J, Anderson JL. Shortened telomere length is associated with paroxysmal atrial fibrillation among cardiovascular patients enrolled in the Intermountain Heart Collaborative Study. Heart Rhythm. 2016;13:21–27. [DOI] [PubMed] [Google Scholar]
- 25. Allende M, Molina E, Gonzalez‐Porras JR, Toledo E, Lecumberri R, Hermida J. Short leukocyte telomere length is associated with cardioembolic stroke risk in patients with atrial fibrillation. Stroke. 2016;47:863–865. [DOI] [PubMed] [Google Scholar]
- 26. Patton KK, Ellinor PT, Heckbert SR, Christenson RH, DeFilippi C, Gottdiener JS, Kronmal RA. N‐terminal pro‐B‐type natriuretic peptide is a major predictor of the development of atrial fibrillation: the Cardiovascular Health Study. Circulation. 2009;120:1768–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schnabel RB, Wild PS, Wilde S, Ojeda FM, Schulz A, Zeller T, Sinning CR, Kunde J, Lackner KJ, Munzel T, Blankenberg S. Multiple biomarkers and atrial fibrillation in the general population. PLoS One. 2014;9:e112486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ha AC, Breithardt G, Camm AJ, Crijns HJ, Fitzmaurice GM, Kowey PR, Le Heuzey JY, Naditch‐Brule L, Prystowsky EN, Schwartz PJ, Torp‐Pedersen C, Weintraub WS, Dorian P. Health‐related quality of life in patients with atrial fibrillation treated with rhythm control versus rate control: insights from a prospective international registry (registry on cardiac rhythm disorders assessing the control of atrial fibrillation: RECORD‐AF). Circ Cardiovasc Qual Outcomes. 2014;7:896–904. [DOI] [PubMed] [Google Scholar]
- 29. Prystowsky EN, Padanilam BJ, Fogel RI. Treatment of atrial fibrillation. JAMA. 2015;314:278–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Parameters of the Multivariate Model Including Both Gene Scores for the Prediction of Prevalent Atrial Fibrillation
Table S2. Detailed Reclassification of Participants When Adding the Gene Scores to the Clinical Model
Table S3. Predictive Performance, Reclassification, and Discrimination for Paroxysmal Atrial Fibrillation
Table S4. Predictive Performance, Reclassification, and Discrimination for Permanent Atrial Fibrillation
Figure S1. Multivariate models for the prediction of AF excluding patients with heart failure. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S2. Multivariate models for the prediction of paroxysmal AF. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S3. Multivariate models for the prediction of permanent AF. ORs (95% CIs) for AF for a 1‐SD increase in the score. Model 1 was adjusted for age and sex. Model 2 was adjusted for age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; OR, odds ratio.
Figure S4. Receiver operating characteristic curves for the identification of prevalent AF by subtypes. A, Model including AF‐GS compared with a model including clinical RFs alone for the identification of paroxysmal AF. B, Model including both GSs compared with a model including clinical RFs alone for the identification of paroxysmal AF. C, Model including BA‐GS compared with a model including clinical RFs alone for the identification of permanent AF. D, Model including both GSs compared with a model including clinical risk factors alone for the identification of permanent AF. RFs were age; sex; body mass index; height; weight; smoking status (in the previous 12 months and ever); systolic blood pressure; diastolic blood pressure; hypertension; diabetes mellitus; coronary artery disease; dyslipidemia; congestive heart failure; valvular heart disease; chronic renal failure; chronic obstructive pulmonary disease; and relative lymphocytes, neutrophils, and monocytes. AF indicates atrial fibrillation; AF‐GS indicates atrial fibrillation gene score; BA‐GS, biological age gene score; GS, gene score; RF, risk factor.


