ABSTRACT
HD2D is one of the 4 plant specific HD2 family histone deacetylases (HDAC) identified in Arabidopsis and it is distantly related to the other HD2 members. At this time, little is known about the function of HD2D in plants. Here we provide evidence that HD2D is involved in the control of flowering time. Flowering was delayed in transgenic plants overexpressing HD2D and hd2d mutant plants flowered earlier compared with wild-type in both long day and short day conditions. Expression of several floral identity genes was altered in these plants. Taken together, our findings suggest that HD2D is a negative regulator of flowering that modulates the transition of vegetative to reproductive growth in a photoperiod independent manner.
KEYWORDS: Flowering, HD2D, histone deacetylases
Histone acetylation is one of the rapid and reversible post-translational modifications that regulate gene expression in plants.1,2 It is controlled by histone acetyltransferases (HAT) and histone deacetylases (HDACs). While HDACs remove the acetyl group from histones, resulting in a condensed chromatin configuration and repressing gene expression, acetylation of histone by HAT leads to a relaxed chromatin structure and activate gene expression.3,4 Among the 3 HDAC families the histone deacetylase 2 (HD2) family is unique to plants.5-7 In Arabidopsis, 4 HD2 type HDACs were identified, namely, HD2A, HD2B, HD2C and HD2D.8-10
Histone modification via HAT and HDACs plays important roles in regulating flowering. This transaction from vegetative growth to reproductive growth, is a critical development change during the life cycle of a plant, being regulated by several pathways that respond to external signals, such as photoperiod and temperature, and internal signals such as hormones and developmental cues.11,12,13 For example, HDA9 has been shown to associate with the AGAMOUS-LIKE (AGL19) locus, which is a known activator of flowering.14 hda9 mutants have higher amounts of H3K9K27 acetylation at the promoter region of AGL19 resulting in chromatin remodeling leading to increased AGL19 expression.14,15 However, HDA9 only affects flowering time under SD but not LD conditions,15 suggesting that the action of HDA9 is tightly controlled by the prevailing photoperiod. Another HDAC, HDA6 has been shown to interact with FLOWERING LOCUS D (FLD) and affects global H3K9K14 acetylation levels.16 Unlike hda9 mutants, the hda6 mutants showed delayed flowering under both long day (LD) and short day (SD) conditions,17 indicating HDA6 is involved in pathways other than photoperiod. The hda6/fld double mutant showed delayed flowering compared with either hda6 or fld single mutants, suggesting they operate separately from one another to regulate flowering.16 Recently, HDA5 has also been identified as a part of the HDA5-HDA6-FLD repression complex that regulates FLC expression by histone acetylation.18
Research has shown that transgenic plants overexpressing HD2A exhibited delayed flowering.10 However, little is known about the effects of other HD2s on flowering. Here, we present evidence demonstrating that HD2D is involved in regulating flowering time in Arabidopsis. Transgenic plants overexpressing HD2D (HD2DOE) were generated and an hd2d mutant was identified. Under SD conditions, hd2d-1 plants flowered 2 d earlier while flowering was delayed for 8 d compared with WT in HD2DOE plants (Fig 1A and B). Under LD condition, hd2d-1 plants flowered 4 d earlier while flowering of HD2DOE was delayed by 7 d comparing to WT (Fig 1C and D). These results demonstrate that HD2D affected flowering under both SD and LD conditions in a photoperiod independent manner.
Figure 1.
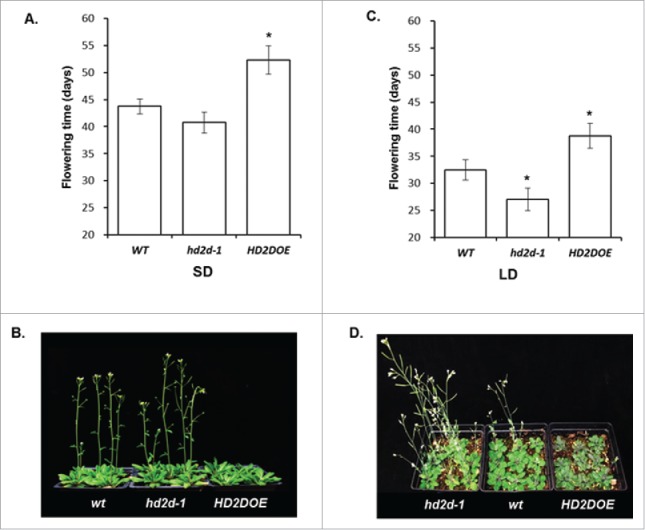
HD2D affects flowering time in Arabidopsis (A) Flowering time of WT, hd2d-1 and HD2DOE plants under short day (SD) conditions. (B) WT, hd2d-1 and HD2DOE plants 48 d post germination under SD conditions. (C) Flowering time of WT, hd2d-1 and HD2DOE plants under long day (LD) conditions. (D) WT, hd2d-1 and HD2DOE plants 37 d post germination under SD conditions. The experiment included 3 replicates (n>54). Asterisks labeled values are significantly different from the wild type (P <0.05).
The number of rosette leaves is an indication of the time length of the vegetative growth. Plants with longer vegetative growth time will exhibit greater number of rosette leaves at flowering, while plants that exhibit retarded growth will have the same number of rosette leaves at flowering but will still flower later.19 To investigate whether the delay in flowering was due to overall growth retardation or a longer vegetative growth phase, the number of rosette leaves was recorded for WT, hd2d-1¸ and HD2DOE plants at flowering. The HD2DOE lines had a greater number of rosette leaves comparing to WT and hd2d-1 (Fig 2A). Interestingly, even though WT and hd2d-1 plants did not show difference in the number of rosette leaves, WT plants had a significantly greater total leaf surface area comparing to hd2d-1 (Fig. 2B), indicating that hd2d-1 plants have smaller leaves comparing to WT plants. HD2DOE plants had greater total leaf surface area comparing to WT plants, which is not a surprise due to their greater numbers.
Figure 2.
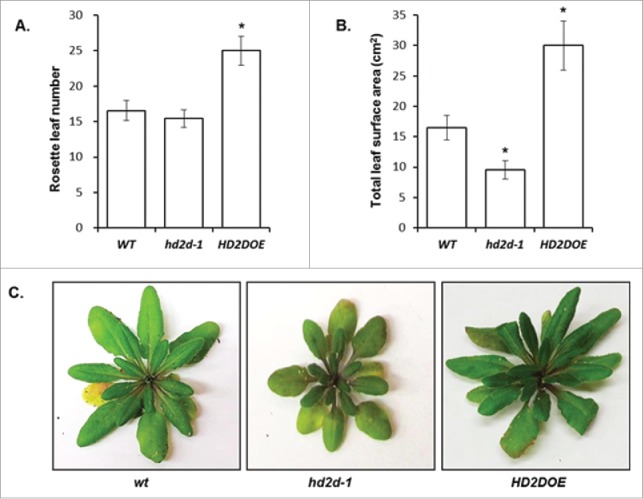
HD2D affects the number of rosette leaves and the leaf surface area. (A) Number of rosette leaves of WT, hd2d-1 and HD2DOE plants at flowering under SD conditions. (B) Total rosette leaf surface area of WT, hd2d-1 and HD2DOE plants at flowering under SD conditions. (C) WT, hd2d-1 and HD2DOE plants at flowering to show their rosette leaves.
The expression of several flowering related genes was investigated in WT, hd2d-1 and HD2DOE plants including: FLC (Flowering Locus C), SOC1(Suppressor of Overexpression of Constans 1), FUL (Fruitfull), and SEP3 (Sepallata 3). FLC is a MADS-box transcription factor that is a key negative regulator of flowering.20 Expression of FLC was increased in HD2DOE plants compared with WT, while it was lower in hd2d-1 plants (Fig. 3). SOC1, SEP3 and FUL are also transcription factors that are involved in promoting flowering and floral identity.21,22 Expression of SOC1, SEP3 and FUL was decreased in HD2DOE plants but increased in hd2d-1 plants comparing to WT (Fig. 3). These results indicated that the HD2D affects the transition from vegetative growth to reproductive growth by regulating the expression of flowering time-related genes.
Figure 3.
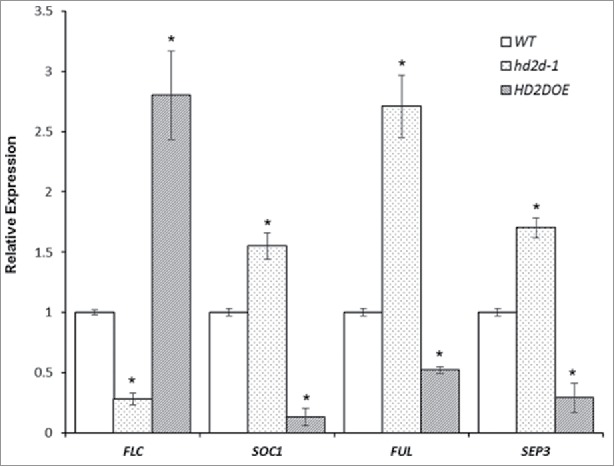
Expression of FLC, SOC1, FUL and SEP3 in WT, hd2d-1, and HD2DOE plants. Total RNA was isolated from leaf tissue of 2 week old seedlings and the expression of FLC, SOC1, FUL and SEP3 was determined by RT-qPCR. Asterisks labeled values are significantly different from the wild type (P<0.05). The experiment was repeated 3 times with similar results.
During development, flowering can be delayed either by overall growth retardation or the lengthening of the vegetative phase.19 Plants with longer vegetative phases have more rosette leaves while those exhibiting growth retardation do not.19 The number of rosette leaves in HD2DOE plants was greater than in the WT and hd2d-1 lines, suggesting that the HD2DOE plants exited the vegetative growth phase later than the WT and hd2d-1. These results indicate that HD2D delays bolting by prolonging the vegetative phase of plant development.
The flowering time in Arabidopsis is regulated via multiple pathways including photoperiod, autonomous, vernalization, and gibberellic acid (GA) pathways.23 SOC1 is a floral integrator and acts as a convergence point for all floral induction pathways, and as one would expect, its expression is affected by all flowering pathways.23 The expression of FUL and SEP3 is affected by the photoperiod, autonomous, and vernalization floral induction pathways, but not the GA pathway. FLC is regulated via the vernalization and autonomous pathways and acts upstream of SOC1, SEP3, and FUL.23 In this study, the late flowering HD2DOE plants showed decreased expression of SOC1, FUL, and SEP3, while these genes had increased expression in the hd2d-1 mutant plants. Conversely, HD2D overexpression lines showed increased expression of FLC, while the hd2d-1 mutant line had decreased expression of FLC. Also, HD2D affected flowering in a similar manner under both SD and LD conditions. These observations indicate that HD2D is not involved in the photoperiod floral induction pathway. The photoperiod floral induction pathway usually affect flowering under either SD or LD conditions, but not both.24 Taken together, these results suggest that HD2D may affect flowering through regulation of either the vernalization or autonomous pathways to promote FLC expression.
The data presented here indicates that HD2D affects flowering time in a manner distinctly different than how other HDACs act to regulate flowering time. Kim et al.15 found that HDA9 is a positive regulator of flowering solely under SD conditions and does not affect FLC expression, concluding that HDA9 is part of the photoperiod floral induction pathway. Zhou et al.10 observed that plants overexpression HD2A exhibited delayed flowering under LD conditions. However, flowering time under SD condition was not reported. HDA5 and HDA6 form a repression complex with the FLD that negatively regulate the expression of FLC by histone modification.16 This complex is not involved in the photoperiod pathway as plants overexpressing HDA6 flowering early under both SD and LD conditions, 16 similar to HD2D. Since HDA6 also interacts with HD2D,25 one possibility is that the interaction between HDA6 and HD2D disrupts the interaction or activities of the FLD-HDA6-HDA5 complex, preventing the complex from catalyzing histone modification at the FLC locus. This would explain the delay in flowering seen in HD2DOE plants and the early flowering in the hd2d-1 mutant plants. Alternatively, HD2D could reduce gene expression through histone deacetylation of genes within the autonomous or vernalization floral induction pathways, which would cause an increase FLC expression and delay flowering, separately from the FLD-HDA6-HDA5 complex.
Disclosure of potential confllicts of interest
No potentential conflicts of interest were disclosed.
Funding
We would like to acknowledge Canadian NSERC Discovery Program for the funding for this research.
References
- 1.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002; 12:142-8; PMID:11893486; https://doi.org/ 10.1016/S0959-437X(02)00279-4 [DOI] [PubMed] [Google Scholar]
- 2.Reyes JC, Hennig L, Gruissem W. Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiol 2002; 130:1090-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev 1998; 12:627-39; PMID:9499399; https://doi.org/ 10.1101/gad.12.5.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Tai R, Wu K. Transcriptional repression by histone deacetylases in plants. Mol Plant 2014; 7:764-72; PMID:24658416; https://doi.org/ 10.1093/mp/ssu033 [DOI] [PubMed] [Google Scholar]
- 5.Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 2006; 46:124-33; PMID:16553900; https://doi.org/ 10.1111/j.1365-313X.2006.02678.x [DOI] [PubMed] [Google Scholar]
- 6.Ma X, Lv S, Zhang C, Yang C. Histone deacetylases and their functions in plants. Plant Cell Rep 2013; 32:465-78; PMID:23408190; https://doi.org/ 10.1007/s00299-013-1393-6 [DOI] [PubMed] [Google Scholar]
- 7.Grandperret V, Nicolas-Francès V, Wendehenne D, Bourque S. Type-II histone deacetylases: elusive plant nuclear signal transducers. Plant Cell Environ 2014; 37:1259-69; PMID:24236403; https://doi.org/ 10.1111/pce.12236 [DOI] [PubMed] [Google Scholar]
- 8.Dangl M, Brosch G, Haas H, Loidl P, Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 2001; 213:280-5; PMID:11469594; https://doi.org/ 10.1007/s004250000506 [DOI] [PubMed] [Google Scholar]
- 9.Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 2002; 30:5036-55; PMID:12466527; https://doi.org/ 10.1093/nar/gkf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J 2004; 38:715-24; PMID:15144374; https://doi.org/ 10.1111/j.1365-313X.2004.02083.x [DOI] [PubMed] [Google Scholar]
- 11.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant cell 2004; 16 Suppl:S18-31; PMID:15037730; https://doi.org/ 10.1105/tpc.015958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 2008; 59:573-94; PMID:18444908; https://doi.org/ 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- 13.He Y. Chromatin regulation of flowering. Trends Plant Sci 2012; 17:556-62; PMID:22658650; https://doi.org/ 10.1016/j.tplants.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 14.Kang MJ, Jin HS, Noh YS, Noh B. Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol 2015; 206:281-94; PMID:25406502; https://doi.org/ 10.1111/nph.13161 [DOI] [PubMed] [Google Scholar]
- 15.Kim W, Latrasse D, Servet C, Zhou DX. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun 2013; 432:394-8; PMID:23237803; https://doi.org/ 10.1016/j.bbrc.2012.11.102 [DOI] [PubMed] [Google Scholar]
- 16.Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 2011; 156:173-84; PMID:21398257; https://doi.org/ 10.1104/pp.111.174417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 2008; 59:225-34; PMID:18212027; https://doi.org/ 10.1093/jxb/erm300 [DOI] [PubMed] [Google Scholar]
- 18.Luo M, Tai R, Yu CW, Yang S, Chen CY, Lin WD, Schmidt W, Wu K. Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J 2015; 82:925-36; PMID:25922987; https://doi.org/ 10.1111/tpj.12868 [DOI] [PubMed] [Google Scholar]
- 19.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 1991; 229:57-66; PMID:1896021; https://doi.org/ 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- 20.Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2009; 106:11661-6; PMID:19564609; https://doi.org/ 10.1073/pnas.0901367106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005; 309:1056-9; PMID:16099980; https://doi.org/ 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 2010; 61:2247-54; PMID:20413527; https://doi.org/ 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- 23.Corbesier L, Coupland G. The quest for florigen: a review of recent progress. J Exp Bot 2006; 57:3395-403; PMID:17030536; https://doi.org/ 10.1093/jxb/erl095 [DOI] [PubMed] [Google Scholar]
- 24.Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 2006; 20:898-912; PMID:16600915; https://doi.org/ 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 2012; 63:3297-306; PMID:22368268; https://doi.org/ 10.1093/jxb/ers059 [DOI] [PMC free article] [PubMed] [Google Scholar]


