Abstract
Cornelia de Lange syndrome (CdLS) is a complex multisystem developmental disorder caused by mutations in cohesin subunits and regulators. While its precise molecular mechanisms are not well defined, they point toward a global deregulation of the transcriptional gene expression program. Cohesin is associated with the boundaries of chromosome domains and with enhancer and promoter regions connecting the three-dimensional genome organization with transcriptional regulation. Here, we show that connected gene communities, structures emerging from the interactions of noncoding regulatory elements and genes in the three-dimensional chromosomal space, provide a molecular explanation for the pathoetiology of CdLS associated with mutations in the cohesin-loading factor NIPBL and the cohesin subunit SMC1A. NIPBL and cohesin are important constituents of connected gene communities that are centrally positioned at noncoding regulatory elements. Accordingly, genes deregulated in CdLS are positioned within reach of NIPBL- and cohesin-occupied regions through promoter–promoter interactions. Our findings suggest a dynamic model where NIPBL loads cohesin to connect genes in communities, offering an explanation for the gene expression deregulation in the CdLS.
Keywords: chromosome architecture, transcription regulation, epigenomics, noncoding regulatory regions, transcriptional networks
CORNELIA de Lange syndrome (CdLS; Mendelian Inheritance in Man (MIM) #122470, 300590, 610759, 614701, and 300882) is a developmental disorder characterized by a typical facial dysmorphism in association with growth and mental retardation, upper limb anomalies, hirsutism, and other systemic involvement (Liu and Krantz 2008; Mannini et al. 2013; Boyle et al. 2015). CdLS is caused by mutations in genes coding for regulators or subunits of the cohesin complex (NIPBL, SMC1A, SMC3, RAD21, and HDAC8) (Krantz et al. 2004; Tonkin et al. 2004; Musio et al. 2006; Deardorff et al. 2007, 2012a,b). SMC1A, SMC3, and RAD21 are core subunits of cohesin, while NIPBL loads the complex and HDAC8 deacetylates SMC3 to favor protein recycling (Michaelis et al. 1997; Ciosk et al. 2000; Deardorff et al. 2012b). NIPBL is the most frequently mutated gene in CdLS with up to 65% of patients showing heterozygous mutations, while mutations in the other four causal genes account for 11% of patients (Mannini et al. 2013; Watrin et al. 2016). Although the genetic causes of CdLS are well defined, the molecular mechanisms remain to be fully understood.
Cohesin is an evolutionarily conserved protein complex essential for maintaining sister chromatid cohesion and transcriptional regulation (Nasmyth and Haering 2009; Dorsett and Merkenschlager 2013; Remeseiro et al. 2013; Singh and Gerton 2015; Hnisz et al. 2016). While showing some genomic instability (Revenkova et al. 2009), CdLS patient-derived cell lines are not prone to cohesion defects (Castronovo et al. 2009), arguing for changes in transcriptional regulation to explain the pathoetiology (Dorsett and Merkenschlager 2013; Remeseiro et al. 2013; Singh and Gerton 2015). Interestingly, other congenital malformation disorders sharing phenotypic similarities with CdLS, such as the Wiedemann–Steiner syndrome (MIM #605130) and the CHOPS syndrome (C for cognitive impairment and coarse facies, H for heart defects, O for obesity, P for pulmonary involvement, and S for short stature and skeletal dysplasia; MIM #616368), are also associated with mutations in transcriptional regulators (Jones et al. 2012; Izumi et al. 2015; Yuan et al. 2015). Moreover, similarities were found in the transcriptomic profiles of CHOPS syndrome and CdLS patients (Izumi et al. 2015). Taken together, these observations suggest that transcriptional regulation is a major contributor to the phenotypes observed in CdLS and the related developmental syndromes.
The emergence of genome-wide chromosome conformation capture methods have revealed an intricate interplay between chromosome architecture and the control of the gene expression program (Gómez-Díaz and Corces 2014; Dekker and Mirny 2016; Hnisz et al. 2016). In eukaryotes, transcription by RNA polymerase II (Pol II) is a multistep process. In particular, initiation of transcription is stimulated by transcription factors bound at distal regulatory sequences such as enhancers (Maston et al. 2006; Ong and Corces 2011). To achieve this function, distal regulatory genomic elements are brought in close proximity to their target genes through chromatin interactions (Sexton and Cavalli 2015; Spurrell et al. 2016). NIPBL, cohesin, and the coactivator complex mediator have been shown to play a pivotal role in the stabilization of these interactions (Kagey et al. 2010). In addition to long-range enhancer-promoter contacts, cohesin is also associated with topologically associating domains (TADs), which represent the building blocks of the genome’s organization (Lupiáñez et al. 2016). These self-associating domains are composed of complex networks of chromatin interactions that are restricted by domain boundaries enriched for CTCF and cohesin binding (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Nora et al. 2012). Interestingly, depletion of cohesin creates widespread gene expression changes, maintaining architectural compartments but modifying the TAD insulation function and sub-TAD interactions, such as those between enhancer and promoter regions (Seitan et al. 2013; Sofueva et al. 2013; Zuin et al. 2014a). In fact, TADs, in addition to CTCF-occupied regions, tend to be conserved through cell types and evolution (Vietri Rudan et al. 2015). These results highlight a pivotal role of cohesin in connecting the chromosome architecture with the control of transcriptional regulation.
Transcription is spatially compartmentalized in the mammalian nucleus (Sutherland and Bickmore 2009). Indeed, Pol II is observed in distinct foci dispersed throughout the nucleus where multiple genes converge to be cotranscribed (Osborne et al. 2004; Mitchell and Fraser 2008; Schoenfelder et al. 2010; Ghamari et al. 2013). Accordingly, chromatin interactions surrounding Pol II are involved in extensive promoter-centered contacts that create multigene complexes (Li et al. 2012). This three-dimensional (3D) organization centered on Pol II is connected but distinct from the TAD architecture (Tang et al. 2015). Interestingly, these connected genes are compartmentalized by biological functions (Sandhu et al. 2012). This system is reminiscent of genes found in prokaryotic operon systems, where a single promoter controls the transcription of multiple adjacent genes in response to an environmental cue (Jacob et al. 1960). Whether or not the specific physical association of genes or their transcriptional coregulation are biologically significant in human cells remains to be fully understood.
Current models suggest a major role of the 3D chromosomal architecture in the transcriptional output of connected genes. Accordingly, genes with promoters physically clustered within a single TAD are cotranscribed during early development (Nora et al. 2012). In addition, genes sharing a TAD corespond to hormone-induced changes in gene activity (Le Dily et al. 2014). Interestingly, loop-mediated contacts are required for transcriptional coregulation in a multigene complex. Indeed, disruption of loop-mediated contact between NF-κB-regulated genes alters the transcriptional status of interacting genes (Fanucchi et al. 2013). These results suggest that coordinated regulation of gene expression in mammalian cells is driven by the physical interactions between genes.
To gain molecular insights into the transcriptional deregulation observed in CdLS, we focused on the link between the chromosome architecture and gene expression changes associated with NIPBL and SMC1A mutations. Here, we show that connected gene communities, which are a combination of physically-associated genes (minimum two) and noncoding regulatory elements, provide a molecular explanation for gene expression changes observed in CdLS. Indeed, deregulated genes in CdLS were found in close proximity to NIPBL- and SMC1A-occupied regions within connected gene communities. We suggest that the organization of genes in connected communities underlies the pathoetiology of CdLS.
Materials and Methods
Gene expression data sets
Differentially expressed genes for NIPBL- and SMC1A-mutated lymphoblastoid cell lines (LCLs) from Liu et al. (2009) and Mannini et al. (2015) were used. To uniformize the nomenclature, genes were reannotated using the hgu133plus2.db Biocondutor package or reinferred from their RefSeq, ENSEMBL, or GenBank identifiers. The uniformized gene symbols used throughout the manuscript are provided (Supplemental Material, Table S1 and Table S2).
Cell culture
The GM12878 normal lymphoblastoid cells were obtained from the National Institute of General Medical Sciences Human Genetic Cell Repository at the Coriell Institute for Medical Research (Catalog ID: GM12878) and cultured in RPMI-1640 medium (MT10040CV; Fisher Scientific, Pittsburgh, PA) supplemented with 15% fetal bovine serum (qualified 12483020; Invitrogen, Carlsbad, CA), 2 mM L-glutamine (25030-081; GIBCO [Grand Island Biological], Grand Island, NY), 1× MEM nonessential amino acids (25-0250; Cellgro) and 1× Penicillin/Streptomycin (15170-063; GIBCO).
Chromatin immunoprecipitation sequencing (ChIP-Seq)
ChIP-Seq experiments were performed in duplicates as described previously (Bilodeau et al. 2009; Kagey et al. 2010; Fournier et al. 2016). Briefly, 50 million cells were cross-linked for 10 min with 1% formaldehyde and quenched with 125 mM glycine for 5 min. Cells were then washed with PBS, pelleted, flash frozen, and stored at −80°. Sonicated DNA fragments were immunoprecipitated with antibodies directed against NIPBL (A301-779A; Bethyl Laboratories), SMC1A (A300-055A; Bethyl Laboratories) and MED1 (A300-793A; Bethyl Laboratories). Library preparation and high-throughput sequencing were performed at the McGill University and Génome Québec Innovation Centre (MUGQIC), Montréal, Canada. Analysis of raw sequencing reads was performed using the MUGQIC ChIP-Seq pipeline (version 2.2.0). The data discussed in this publication have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO Series accession number GSE93080 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93080). Public ChIP-Seq data sets for CTCF, Pol II, and NIPBL in patient-derived LCLs (Table S3) were processed using the same ChIP-Seq pipeline.
To generate genomic visualizations, the BAM files for a given factor were pooled and their reads extended to 225 bp. Coverage was calculated using genomecov from bedtools v2.17.0 (http://bedtools.readthedocs.io). Tracks images were generated using the University of California, Santa Cruz (USCS) Genome Browser (Kent et al. 2002).
Overlap with genomic features
To calculate the overlap between the NIPBL- and SMC1A-occupied regions and chromatin states, the ChromHMM 18-state model was used (Kundaje et al. 2015). To simplify representations, chromatin states were grouped per functional type as follows: (a) transcription start site (TSS)-associated: 01_TssA, 02_TssFlnk, 03_TssFlnkU, 04_TssFlnkD, and 14_TssBiv; (b) transcribed: 05_Tx and 06_TxWk; (c) enhancer: 07_EnhG1, 08_EnhG2, 09_EnhA1, 10_EnhA2, 11_EnhWk, and 15_EnhBiv; (d) repressed: 12_ZNF/Rpts, 13_Het, 16_ReprPC, 17_ReprPCWk; and (e) quiescent: 18_Quies. Pairwise overlaps between the regions of interest and the chromatin states were determined using the findOverlaps function from the GenomicRanges package (Lawrence et al. 2013).
The closest gene for all NIPBL- and SMC1A-occupied regions were obtained using the annotatePeak function from the ChIPSeeker (Yu et al. 2015) and TxDb.Hsapiens.UCSC.hg19.knownGene packages (Carlson M, R package version 3.2.2). All regions within 1 kb of the TSS were considered as TSS-proximal regions. The types of regions identified using ChIPseeker were simplified as follows: (a) TSS-proximal: promoter; (b) gene-body: 5′-UTR, exon, intron, and 3′-UTR; and (c) intergenic: downstream and distal intergenic. If a target region overlapped more than one type of genomic region, it was assigned a type using the following priority order: TSS-proximal, gene-body, and intergenic.
Annotation of interaction points
Publicly available Pol II Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET) interactions (Tang et al. 2015) (GEO accession: GSM1872887; file GSM1872887_GM12878_RNAPII_PET_clusters.txt.gz.) were used as input regions to define the connected gene communities in GM12878 cells. Interactions involving the mitochondrial chromosomes were removed. Then, overlapping interaction regions were combined into single interaction points and annotated using the ChIPseeker package. Each gene with an interaction point within 1 kb from a TSS was attributed to this region. In the event of multiple regions fulfilling this criterion, the one involved in the most interactions was selected. Gene expression levels for each gene in GM12878 cells were obtained from the ENCODE Project (Bernstein et al. 2012) (Table S3) and mean Fragments Per Kilobase of transcript per Million mapped reads (FPKM) were calculated for each gene. A gene was considered actively transcribed if its mean FPKM was ≥1.
All interacting regions were attributed a single chromatin state as described above. If an interaction point overlapped more than one chromatin state, the following priorities for state attribution were used: TSS-associated, transcribed, enhancer, repressed, or quiescent. For transcription factor and cofactor occupancy, narrowPeaks files from the ENCODE Project (Bernstein et al. 2012) were obtained for the GM12878 cells (Table S3). For each factor, replicates were combined and overlapping regions were kept. The number of occupied regions for each transcription factor or cofactor found within each interacting region was then added to its annotations.
The same analysis was repeated using the interaction points of the promoter Capture Hi-C data set from Mifsud et al. (2015). However, the number of interactions in this data set was an order of magnitude larger than those in the Pol II ChIA-PET experiment (1,777,526 vs. 113,591). Therefore, only the 150,000 most significant promoter Capture Hi-C interactions were kept for the analysis to maintain comparable complexities and topologies of the two networks.
Identification of connected gene communities
To identify connected gene communities, the annotated interaction points were used as the vertices of a graph, which was modeled using the R igraph package (Csárdi and Nepusz 2006). Components bearing no TSS-proximal nodes were filtered out, and the remaining nodes were split into communities using the cluster_fast_greedy function (Clauset et al. 2004). Components without at least two TSS-proximal nodes were then filtered out and interchromosomal edges forming bridges between subcomponents of >10 nodes were removed. The remaining components formed the connected gene communities.
To determine the centrality of each vertex in the communities, two metrics were calculated: their degree and their closeness (Freeman 1978). These metrics were scaled separately for each network component and a centrality score averaging both metrics was attributed to each vertex. Within a community, vertices with a centrality score above the 95th percentile were labeled as central nodes. Graphical representations of connected gene communities were generated using Cytoscape (v3.4.0, http://www.cytoscape.org/).
Enrichment within connected gene communities
The proportions of base pairs for all chromatin states throughout the genome and within connected gene communities were calculated. The same proportions were also calculated using NIPBL- and SMC1A-occupied regions. The ratio (log2) between the proportions of chromatin states in a list of regions vs. the genome represented the chromatin state enrichment for the given set of regions.
To determine the enrichment of a factor in connected gene communities, the available genome, which represents the union of all occupied regions for all factors profiled in GM12878 cells, was used. While the entire genome is often used as a reference, the available genome is more stringent as it narrows the information to accessible regions. The enrichment of each factor was calculated by dividing the ratio of occupancy within connected gene communities and the available genome. The statistical significance of each enrichment was assessed using a hypergeometric test.
Coherency within a connected gene community
The number of upregulated and downregulated genes in CdLS was calculated for each community. The level of coherence, between 0.5 and 1, was calculated as coherence = max(# upregulated, # downregulated) / (# upregulated + # downregulated). A threshold of 0.75 was used to label coherent communities. To determine if the number of coherent communities was larger than expected by chance, we assumed that if the fold-change and community membership of misregulated genes were unrelated, upregulated and downregulated genes would be randomly distributed among the communities. Thus, the set of fold-changes associated with each data set was resampled across all deregulated genes 10,000 times and the proportion of coherent gene communities calculated for each resampling. Resampling the fold-changes preserved the directionality of gene expression variations, which has a large impact on the chosen coherence metric. P-values of overrepresentation were inferred from the resulting empirical cumulative distribution.
Proximity of NIPBL- and SMC1A-occupied nodes to CdLS-misregulated genes
To determine if the neighborhood of misregulated genes contained more NIPBL- and SMC1A-occupied regions than expected by chance, a number of nodes equal to the total number of NIPBL- and SMC1A-occupied nodes were randomly sampled from the connected gene communities. The distances between all misregulated genes and their closest selected nodes were then calculated. The process was repeated 10,000 times and distance distributions were inferred from the simulated values.
Data availability
The data discussed in this publication have been deposited in NCBI’s GEO (Edgar et al. 2002) and are accessible through GEO Series accession number GSE93080 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93080). The software implementing the methods described in this paper is available upon request.
Results
Variable occupancy of NIPBL and cohesin at CdLS-deregulated genes
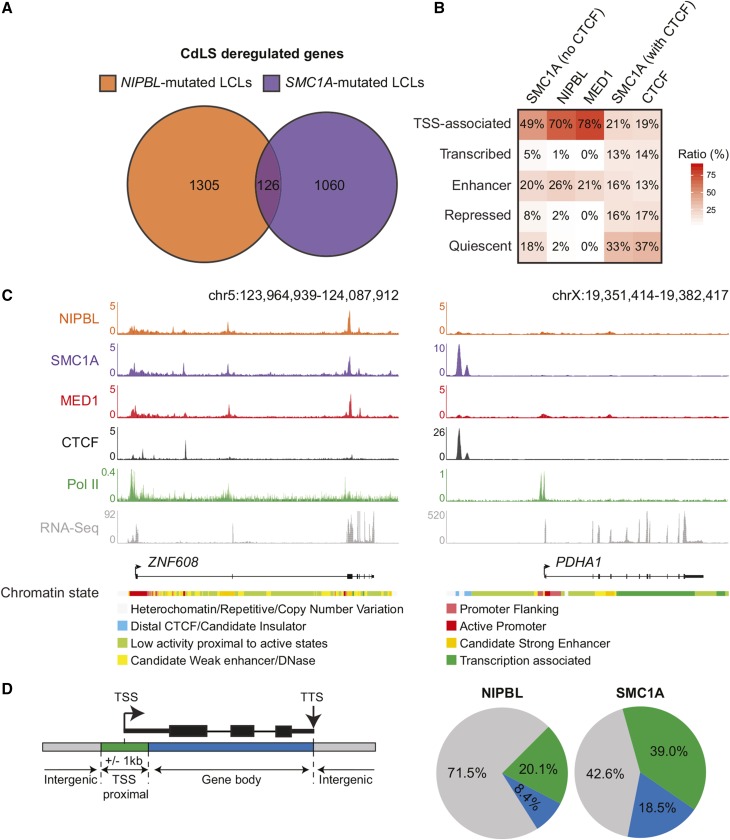
To gain insight into the molecular mechanism underlying CdLS, we compared the gene expression profiles of patient-derived lymphoblastoid cell lines (LCLs) with mutations in NIPBL and SMC1A. While a total of 1431 and 1186 genes were found to be significantly misregulated in NIPBL-mutated and SMC1A-mutated proband-derived lymphoblastoid cell lines, respectively (Liu et al. 2009; Mannini et al. 2015) (Table S1 and Table S2), only 126 differentially expressed genes were shared between the two gene signatures (Figure 1A). To identify the direct targets of NIPBL and SMC1A, we used ChIP coupled with massively parallel DNA sequencing (ChIP-Seq) in GM12878 normal lymphoblastoid cells. In mammalian cells, cohesin is typically associated with CTCF at TAD boundaries and with Mediator (MED1) and NIPBL at connecting enhancer–promoter regions (Kagey et al. 2010; Fournier et al. 2016; Merkenschlager and Nora 2016). Accordingly, SMC1A-occupied regions (without CTCF), NIPBL, and MED1 were found mostly at noncoding regulatory elements such as TSSs and enhancer regions (Figure 1B). In contrast, cohesin regions cooccupied by CTCF were found distributed throughout the genome with a predominance in quiescent regions. The genomic distribution of cohesin subunits SMC3 and RAD21 confirmed these results (Figure S1 in File S1). In addition, another NIPBL antibody (Zuin et al. 2014b) also identified TSSs and enhancer regions in LCLs (Figure S1 in File S1). Close examination of density profiles confirmed the occupancy of NIPBL and cohesin at predicted enhancer and promoter regions of the ZNF608 locus, a gene deregulated in CdLS patient-derived LCLs (Figure 1C). Therefore, NIPBL and cohesin occupy noncoding regulatory regions in lymphoblastoid cells.
Figure 1.
NIPBL and cohesin occupy a fraction of CdLS-deregulated genes. (A) Genes affected by mutations in NIPBL and SMC1A are different. Venn diagram representation of differentially expressed genes in CdLS patient-derived LCLs with mutations in NIPBL and SMC1A. A total of 1431 and 1186 genes were identified in NIPBL- and SMC1A-mutated cell lines, respectively, while only 126 genes were shared (see Table S1 and Table S2). (B) Heatmap showing the percentage of overlap between regions occupied by SMC1A (no CTCF), NIPBL, MED1, SMC1A (with CTCF), and CTCF and the functional genome. A simplified version of the ChromHMM 18-state model in GM12878 cells (see Materials and Methods) was used to represent the functional genome. TSS-associated and enhancer regions are occupied by SMC1A (no CTCF) and NIPBL. The color scale indicates the ratio of overlap. (C) ChIP-Seq occupancy profiles of NIPBL, SMC1A, MED1, CTCF, and Pol II at the ZNF608 and PDHA1 loci, two CdLS-deregulated genes in GM12878 cells. RNA-Seq profiles show that both ZNF608 and PDHA1 genes are transcribed. The chromatin states are displayed below the gene tracks. Noncoding regulatory regions of the ZNF608 locus are occupied by SMC1A and NIPBL while they are not for PDHA1. The scales of ChIP-Seq and RNA-Seq profiles are displayed in reads per million. (D) NIPBL and SMC1A occupancy of deregulated genes in CdLS. Percentage of deregulated genes in NIPBL-mutated or SMC1A-mutated cells occupied by NIPBL or SMC1A respectively. Regions associated to genes were defined as: TSS proximal (a ±1 kb region surrounding the TSS), gene body (from +1 kb to the TTS), and intergenic (not TSS proximal nor gene body). Overall, 71.5% of genes deregulated in NIPBL-mutated cells are not occupied by NIPBL, while 42.6% of genes deregulated in SMC1A-mutated cells are not occupied by SMC1A. CdLS, Cornelia de Lange syndrome; ChIP-Seq, chromatin immunoprecipitation sequencing; chr, chromosome; LCLs, lymphoblastoid cell lines; Pol II, RNA polymerase II; RNA-Seq, RNA sequencing; TSS, transcription start site; TTS, transcription termination site.
To assess whether deregulated genes are occupied by NIPBL and SMC1A, we investigated their distribution surrounding CdLS-deregulated genes. Strikingly, merely 20.1 and 39.0% of TSS proximal regions of genes deregulated in NIPBL-mutated and SMC1A-mutated cells were occupied by NIPBL and SMC1A, respectively, in normal conditions (Figure 1D and Table S4). For example, the PDHA1 gene is deregulated in CdLS, but was unoccupied by NIPBL and SMC1A in opposition to ZNF608 (Figure 1C). These results suggest that a large fraction of the transcriptional control exerted by NIPBL and cohesin on genes deregulated in CdLS extends beyond local promoter effect.
NIPBL and cohesin are constituents of noncoding regulatory regions within connected gene communities
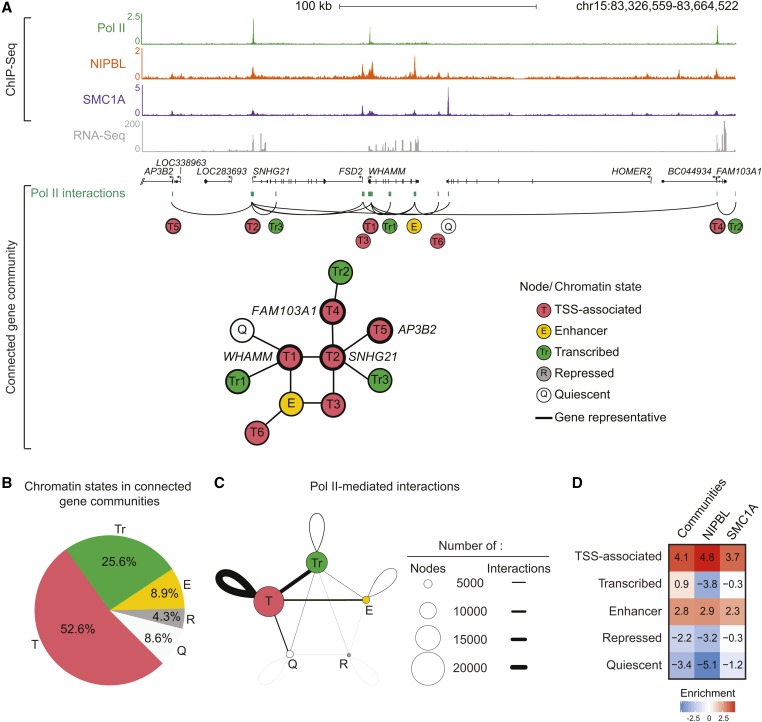
To understand the mechanism by which NIPBL and cohesin indirectly control gene expression in CdLS, we investigated their relationship with the chromosome architecture. Indeed, genes and noncoding regulatory regions are associated in the 3D space to create connected gene communities. To define these communities, we integrated and annotated Pol II ChIA-PET in GM12878 lymphoblastoid cells (Tang et al. 2015). We defined connected gene communities as networks of interacting regions (or nodes) containing at least two genes. A total of 1290 communities were found averaging 34.5 nodes and 5.9 genes (Table S5). To validate the analysis, we looked at the histone cluster 1 (HIST1) gene family, which was shown to be organized into interaction clusters (Li et al. 2012; Sandhu et al. 2012). Accordingly, 21 of the 58 HIST1 genes were found structured into a single connected gene community in GM12878 cells (Figure S2A in File S1). A connected gene community of 11 nodes is depicted in Figure 2A. Of these nodes, five were occupied by either NIPBL or SMC1A. Moreover, a total of six nodes were associated with a TSS chromatin state (T1–T6), while four of them (T1, T2, T4, and T5) overlapped an annotated TSS and were therefore labeled as gene representative (WHAM, SNHG21, FAM103A1, and AP3B2). In addition, three nodes were annotated as transcribed regions (Tr1–Tr3), one as an enhancer region (E), and one as a quiescent region (Q). As expected, gene representatives connected through Pol II interactions at the TSS were transcribed (with the exception of AP3B2). In accordance with previous reports (Li et al. 2012), genes found within connected gene communities were mostly transcribed (86%) and expressed at a higher level (4.63-fold) than nonconnected genes (Figure S2C in File S1 and Table S5). Furthermore, among interactions assigned to TADs [(Rao et al. 2014), 83% of all interactions], most were intra-TAD (88%) and rarely (12%) crossed TAD boundaries. These observations suggest that connected gene communities are found within larger chromosome domains like TADs.
Figure 2.
Active noncoding regulatory regions are occupied by NIPBL and cohesin within connected gene communities. (A) Representation of a connected gene community containing the WHAM, SNHG21, FAM103A1, and AP3B2 loci. First, ChIP-Seq occupancy profiles of Pol II, NIPBL, SMC1A and RNA-Seq are shown in GM12878 cells. The scales of ChIP-Seq and RNA-Seq profiles are displayed in reads per million. Connected gene communities were created by integrating the Pol II ChIA-PET interaction data (green boxes) (see Materials and Methods and Tang et al. (2015)). The represented community contains 11 nodes individually annotated using the simplified chromatin state model [pink: TSS-associated (T), yellow: enhancer (E), green: transcribed (Tr), dark gray: repressed (R), and light gray: quiescent (Q)]. Interacting regions overlapping an annotated TSS were defined as gene representatives. (B) Distribution of chromatin states within connected gene communities. The pie chart shows the average percentage of each simplified chromatin state (pink: T, yellow: E, green: Tr, dark gray: R, and light gray: Q) within a connected gene community. (C) Pol II-mediated interactions between chromatin states within connected gene communities. Each circle represents a simplified chromatin state (pink: T, yellow: E, green: Tr, dark gray: R, and light gray: Q). The size of the circle corresponds to the frequency of the chromatin state within connected gene communities. The thickness of the lines represents the frequency of interactions between the different chromatin states. TSS-associated nodes are the most prevalent and involved in the highest frequency of interactions. (D) Heatmap showing the enrichment of simplified chromatin states within connected gene communities compared to all NIPBL- and SMC1A-occupied regions. The enrichment fold was calculated relative to the genome. NIPBL and SMC1A are enriched at TSS-associated and enhancer regions, similar to connected gene communities. The color scale indicates the enrichment vs. the genome. CdLS, Cornelia de Lange syndrome; ChIA-PET, Chromatin Interaction Analysis by Paired-End Tag Sequencing; ChIP-Seq, chromatin immunoprecipitation sequencing; chr, chromosome; Pol II, RNA polymerase II; RNA-Seq, RNA sequencing; TSS, transcription start site.
In addition to genes, connected communities are composed of noncoding regulatory elements. On average, TSS-associated regions accounted for 52.6% of nodes within connected gene communities, while enhancer regions and repressed elements represented 8.9 and 4.3% of nodes, respectively (Figure 2B). Within communities, TSS–TSS interactions were the most frequent, followed by interactions between TSS and transcribed regions and TSS–enhancer interactions (Figure 2C). These observations confirm that connected gene communities are formed from the interactions of multiple types of regulatory regions.
The cohesin complex, and by association its loader NIPBL, have been associated with chromosome domains and enhancer–promoter interactions (Bonev and Cavalli 2016; Merkenschlager and Nora 2016). Consequently, we reasoned that NIPBL and cohesin could be enriched in connected gene communities. Accordingly, TSS-associated and enhancer regions were found enriched in NIPBL- and SMC1A-occupied regions, similarly to the chromatin states found enriched in connected gene communities (Figure 2D). Moreover, regions occupied by NIPBL and SMC1A were more frequently observed (4.4-fold, P < 0.001 and 2.1-fold, P < 0.001, respectively) within connected gene communities compared to the available genome (see Materials and Methods). These results suggest that NIPBL and SMC1A are components of regulatory regions within connected gene communities.
NIPBL and cohesin are central to connected gene communities
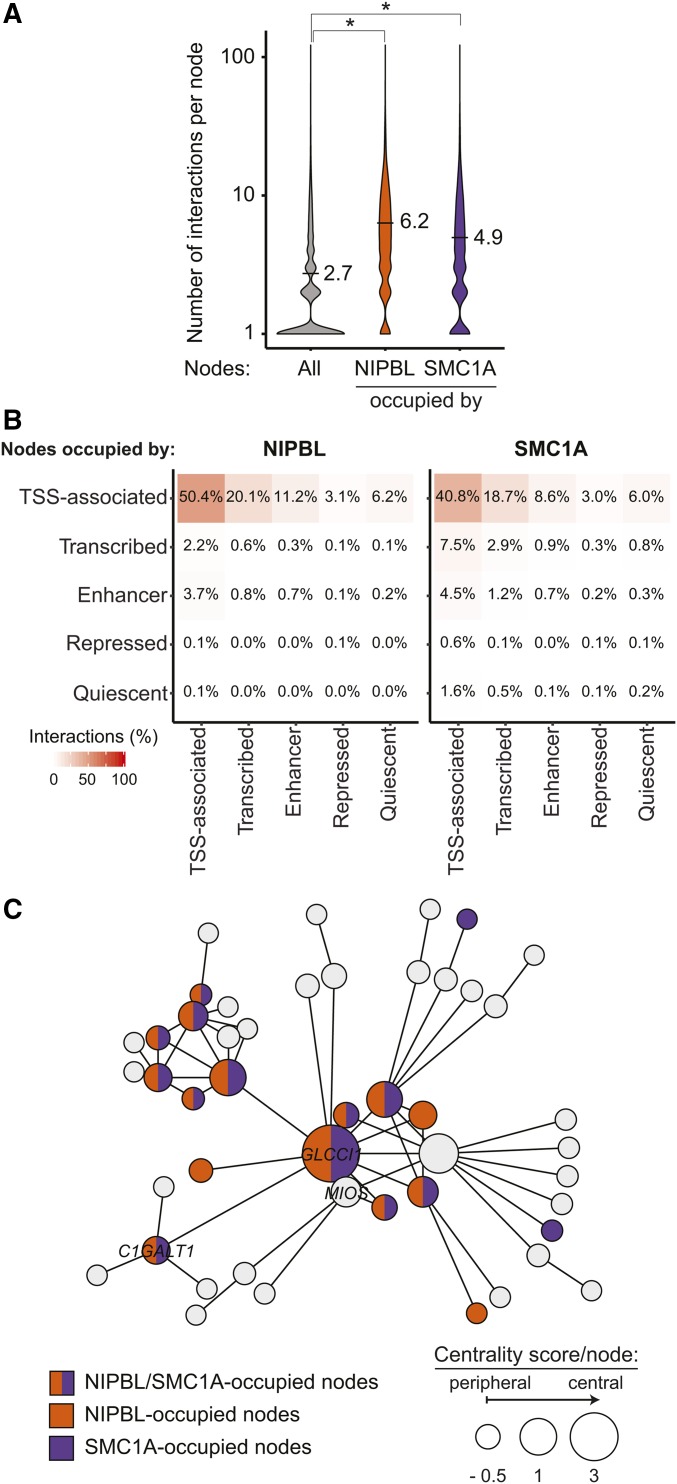
The role of cohesin in the maintenance of the chromosome architecture and its presence in connected gene communities suggest an important role in gene regulation. We postulated that if NIPBL and cohesin represented major constituents of connected gene communities, they would be found at highly interacting nodes. Indeed, within connected gene communities, nodes made an average of 2.7 contacts, while those occupied by NIPBL and SMC1A were implicated in an average of 6.2 (P ≤ 2.2e−16, Wilcoxon rank sum test) and 4.9 (P ≤ 2.2e−16, Wilcoxon rank sum test) interactions, respectively (Figure 3A). As expected, among the different regulatory elements occupied by NIPBL and SMC1A, TSS regions were forming the most contacts with other TSS elements in addition to transcribed and enhancer regions (Figure 3B). These results establish that regions occupied by NIPBL and cohesin are well connected within a gene community.
Figure 3.
Nodes occupied by NIPBL and cohesin create more interactions. (A) Violin plots representing the connectivity of NIPBL- and SMC1A-occupied nodes. The number of interactions for all nodes is displayed in gray while those for NIPBL- and SMC1A-occupied nodes are displayed in orange and purple, respectively. On average, nodes occupied by NIPBL and SMC1A are involved in 6.2 (P ≤ 2.2e−16, Wilcoxon rank sum test) and 4.9 (P ≤ 2.2e−16, Wilcoxon rank sum test) interactions compared to 2.7 for all nodes within connected gene communities. (B) Nodes occupied by NIPBL and SMC1A are mostly involved in inter-TSS interactions. Contact heatmap representing the proportion of nodes occupied by NIPBL and SMC1A involved in chromosome interactions between each simplified chromatin states. The color scale indicates the percentage of interactions. (C) NIPBL and SMC1A occupy central nodes. Representation of a connected gene community where the size of each node is proportional to the centrality score. The bigger the circle, the more central the node is. Gene names are indicated in nodes overlapping a TSS-proximal (±1 kb) region. Nodes are colored in function of their occupancy: NIPBL (orange), SMC1A (purple), both (orange and purple), or none (gray). TSS, transcription start site.
NIPBL loads cohesin at the promoter of active genes from which cohesin is translocated using Pol II (Lengronne et al. 2004; Busslinger et al. 2017). Therefore, we reasoned that a central position of NIPBL would provide an opportunity for cohesin to reach the entire gene community. Node centrality metrics identify vertices of importance within a biological network (Ma and Zeng 2003; Zotenko et al. 2008). Using a combination of node degree and closeness (Freeman 1978), we attributed a centrality score to each node (Figure S3 in File S1). Central nodes (nodes with a centrality score in the top five percentile of their component) were found enriched in NIPBL (3.6-fold, P < 0.001) and SMC1A (2.4-fold, P < 0.001). For example, the GLCCI1 gene is the most central node within a connected community of 48 nodes and was occupied by NIPBL and SMC1A (Figure 3C). In addition, 8 out of the 10 most central nodes in the community were also occupied by NIPBL and SMC1A. These results suggest that NIPBL and cohesin occupy a central position within connected gene communities.
To eliminate the possibility of a bias created by the use of Pol II-centric data in the definition of the connected gene communities, we aimed to confirm our conclusions using orthogonal data. We used promoter Capture Hi-C data, which consists of 3D interactions converging on promoter regions (Mifsud et al. 2015). Analyses of the data confirmed our main observations (Figure S4 in File S1). Indeed, promoter Capture Hi-C-defined connected gene communities were enriched in noncoding regulatory elements occupied by NIPBL and SMC1A (Figure S4A in File S1). Furthermore, nodes occupied by NIPBL and SMC1A featured more contacts than average (Figure S4B in File S1). Therefore, the important position of NIPBL and cohesin within connected gene communities is confirmed independently from the type of 3D chromosome information used to infer the communities.
Gene communities connect deregulated genes in CdLS
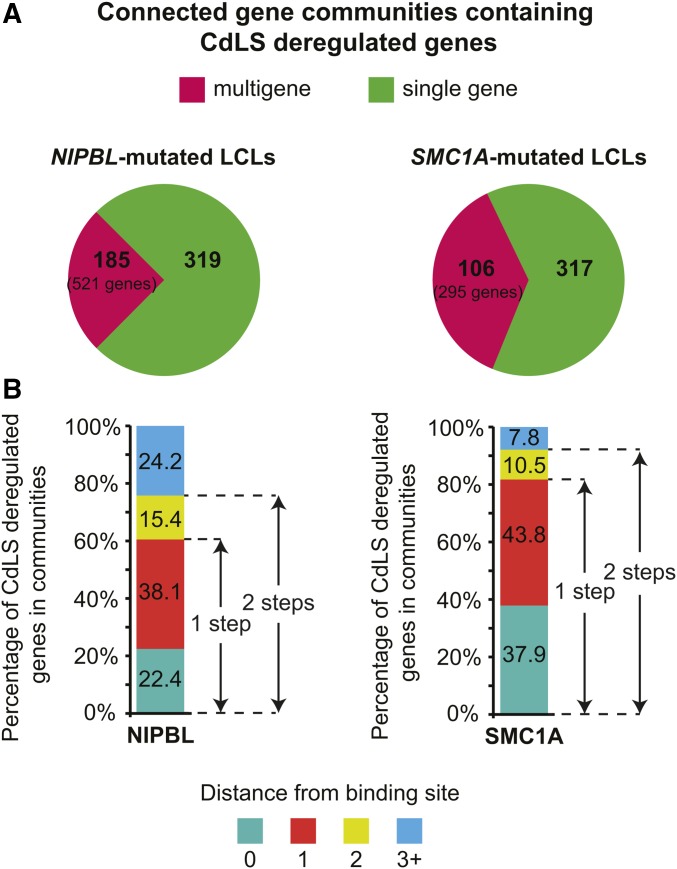
Whether or not the central position of NIPBL and SMC1A within connected gene communities is responsible for gene expression changes observed in CdLS is unknown. Genes deregulated in CdLS were 46% more prevalent in connected gene communities than expected by chance in GM12878 cells (P ≤ 6.2e−81, hypergeometric test). For NIPBL-mutated LCLs, 840 misexpressed genes were distributed in 504 connected gene communities (185 were multigenic, including two or more misexpressed genes), while 612 genes were found in 423 communities (106 were multigenic) in SMC1A-mutated cells (Figure 4A). These numbers were consistent with those found in promoter Capture Hi-C-defined connected gene communities (Figure S4C in File S1). Among these multigenic communities, 55 were shared between NIPBL- and SMC1A-mutated cells (29.7 and 51.8%, respectively) in Pol II ChIA-PET-defined connected gene communities while 61 were shared in promoter Capture Hi-C-defined communities (37.0 and 46.9%, respectively). These results suggest that connected communities organize genes deregulated in CdLS.
Figure 4.
Deregulated genes in CdLS are within reach of NIPBL- and cohesin-occupied regions. (A) Distribution of CdLS-deregulated genes within connected gene communities. Genes deregulated in NIPBL-mutated LCLs are found in 504 connected gene communities (185 multigene). Genes deregulated in SMC1A-mutated cells are found in 423 connected gene communities (106 multigene). (B) Genes deregulated in CdLS are connected to NIPBL- and SMC1A-occupied nodes. Graphical representation of the proportion of CdLS-deregulated genes as a function of the distance from a NIPBL- or SMC1A-occupied node. A distance of 0 corresponds to the gene locus deregulated in CdLS being directly occupied by NIPBL or SMC1A ± 1 kb from the TSS. A distance of 1, 2, or 3 corresponds to the number of steps from the occupied node. CdLS, Cornelia de Lange syndrome; LCLs, lymphoblastoid cell lines; TSS, transcription start site.
If connected gene communities control the gene expression changes associated with CdLS, deregulated genes should be within reach of nodes occupied by NIPBL and cohesin. In a network, the distance represents a measure of the number of steps required to reach a specific node. We computed the distance between deregulated genes and nodes occupied by NIPBL and SMC1A within connected gene communities. A random distribution analysis of NIPBL- and SMC1A-occupied regions showed that one step was sufficient to connect a significantly greater number of deregulated genes to a NIPBL- or SMC1A-occupied node than would be expected by chance [60.5%, P < 0.002, simulated 95% C.I. of (50.4, 56.0) and 81.7%, P < 0.002, simulated 95% C.I. of (72.9, 78.1), respectively] (Figure 4B). Once again, these observations were supported by the promoter Capture Hi-C-defined communities reaching 55.9% of NIPBL- [P < 0.05, simulated 95% C.I. of (42.4, 48.0)] and 87.3% of SMC1A- [P < 0.05, simulated 95% C.I. of (82.0, 86.7)] occupied nodes one step from a deregulated gene (Figure S4D in File S1). NIPBL- and SMC1A-occupied regions connected to unoccupied deregulated genes were typically associated with a promoter/TSS region (Table S6) supporting the role of some promoters as functional enhancers (Dao et al. 2017; Diao et al. 2017). Therefore, these results are consistent with the possibility that connected deregulated genes in CdLS are within reach of NIPBL- and cohesin-occupied noncoding regulatory regions. Altogether, our findings point toward a role of NIPBL and cohesin in the maintenance of the transcriptional integrity of connected gene communities.
NIPBL mutations lead to coordinated gene expression changes within communities
Patients with mutations in NIPBL and SMC1A share phenotypic characteristics, but differ in the severity of their symptoms (Mannini et al. 2013). In addition, NIPBL loads cohesin at the promoter of active genes from which cohesin is translocated (Lengronne et al. 2004; Busslinger et al. 2017). These observations led us to directly compare the distribution and function of NIPBL and cohesin within connected gene communities. First, NIPBL occupied the promoter of genes expressed at a higher level than SMC1A [4.12 and 3.26 log2(FPKM) respectively, P < 2.2e−16]. In addition, genes occupied by NIPBL were more central than those occupied by SMC1A (centrality measures of 1.74 and 1.36, respectively, P < 7.4e−15). These results suggest that the preponderant function of NIPBL is to load cohesin at the promoter of highly active central genes.
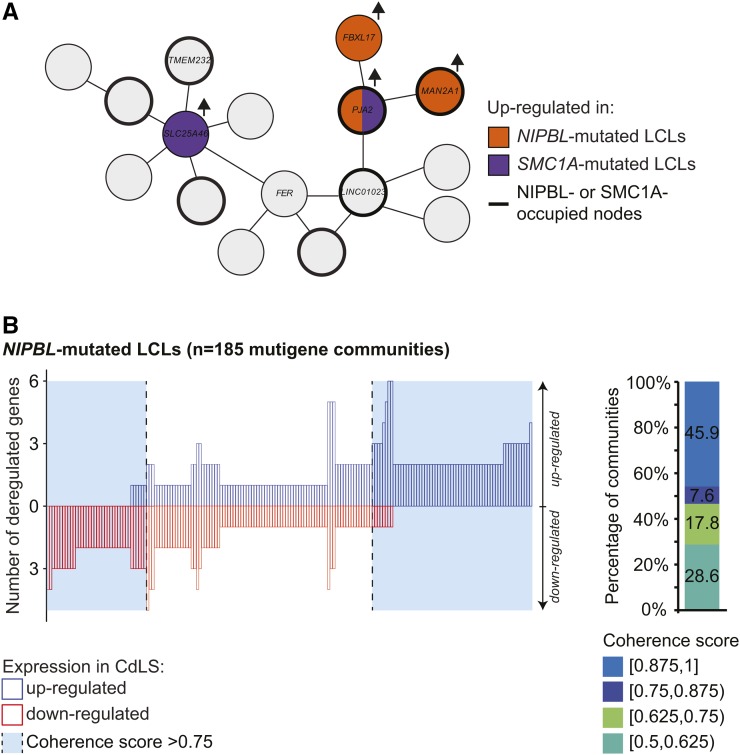
Whether connected gene communities offer a physical structure to coordinate gene expression changes in human diseases is an interesting question. While exploring multigenic connected gene communities, we observed coherent and noncoherent gene expression changes. For example, Figure 5A represents a connected community containing seven genes (TMEM232, SLC25A46, FER, LINC01023, PJA2, MAN2A1, and FBXL17) in which four were upregulated in CdLS (three in NIPBL-mutated and two in SMC1A-mutated cells; one gene was shared). Using a minimal requirement of three-quarter of CdLS-deregulated genes modulated in the same direction to define coherency, more than half of the genes in NIPBL-mutated [53.5%, P = 0.0013, 95% C.I. of (37.8, 49.2); P-value and C.I. obtained by resampling fold-changes within the networks] communities showed coherent changes in gene expression (Figure 5B). These results were corroborated by Capture Hi-C-defined connected gene communities [56.3%, P < 0.002, 95% C.I. of (37.8, 49.2)] (Figure S4E in File S1), but not for genes deregulated in SMC1A-mutated cells. These results suggest that connected gene communities can function as a transcriptional unit, coordinating gene expression changes following mutations of a major constituent like NIPBL.
Figure 5.
Coordinated deregulation of gene expression is associated with NIPBL mutations. (A) Example of a connected gene community containing seven genes (TMEM232, SLC25A46, FER, LINC01023, PJA2, FBXL17, and MAN2A1). Gene names are indicated in nodes overlapping an annotated TSS. Genes upregulated in NIPBL-mutated (orange), SMC1A-mutated (purple), or both (orange and purple) LCLs are highlighted. Nodes occupied by NIPBL or SMC1A are identified by a thicker line. (B) Coordinated gene expression changes in connected gene communities are associated with mutations in NIPBL. Left panel: quantification of the number of up- and downregulated genes in CdLS within individual multigene connected gene communities. Communities with at least 75% of coherency are highlighted in light blue. Right panel: distribution of coherence score of connected gene communities. A coherence score of 1 represents that all CdLS-modulated genes are deregulated in the same direction. CdLS, Cornelia de Lange syndrome; LCLs, lymphoblastoid cell lines; TSS, transcription start site.
Discussion
Global transcriptional disturbances have been associated with many human diseases. Here, we integrated the chromosome architecture surrounding transcriptional regulation to shed new light on the pathoetiology of CdLS, a complex multisystem developmental disorder associated with a perturbation in transcriptional mechanisms. While a large fraction of CdLS-deregulated genes are unoccupied by NIPBL and SMC1A (Figure 1), the majority were within one step of NIPBL- and SMC1A-occupied nodes within connected gene communities (Figure 4 and Table S6). Accordingly, nodes occupied by NIPBL and SMC1A were central to connected gene communities (Figure 2 and Figure 3), with genes deregulated in NIPBL-mutated cells being more expressed and centrally located than those deregulated in SMC1A-mutated cells. These results argue that the chromosome architecture provides essential information to explain gene expression changes associated with transcriptional disturbances in CdLS.
Integration of published studies with our observations allows the proposition of a working model illustrating the dynamic environment of connected communities in which active genes are found. NIPBL would predominantly load cohesin at the promoter of highly active, central, and connected genes. Then, using active transcription, Pol II would distribute cohesin on chromosomes, extruding DNA in the process to form loops and TADs (Busslinger et al. 2017). This model is corroborated by other network analyses suggesting a primary role for active Pol II and cohesin in the formation of chromatin–chromatin interactions (Kruse et al. 2013; Pancaldi et al. 2016; Azofeifa and Dowell 2017). Interestingly, NIPBL seems to favor promoter–promoter interactions (Pancaldi et al. 2016). Accordingly, loading of cohesin at the promoter of highly active genes would provide a mode of transportation for cohesin within connected gene communities to reach more distal regions. In agreement, genes deregulated in NIPBL-mutated cells are more central than those deregulated in SMC1A-mutated cells. Therefore, our results support a model where NIPBL loading of cohesin at central active genes is the epicenter of connected communities’ control.
NIPBL mutations decrease cohesin occupancy (Liu et al. 2009) while SMC1A mutations increase cohesin affinity with chromatin (Revenkova et al. 2009). In that context, mutations in NIPBL would decrease cohesin loading, globally affecting the chromosome architecture of connected communities leading to coordinated gene expression changes. On the other hand, mutations in SMC1A rendering cohesin more stable could lead to Pol II-dependent transportation problems or accumulation at distal sites. Similar to a WAPL loss-of-function (Busslinger et al. 2017; Haarhuis et al. 2017), SMC1A-mutated cohesin complexes could accumulate at distal sites, including CTCF and connected genes, where most of the differentially expressed genes in SMC1A-mutated cells were found. Therefore, NIPBL and SMC1A mutations likely modify the connections within gene communities at different levels leading to specific transcriptional changes.
Coordination and coregulation of genes is an emerging concept for normal and disease development. Noncoding regulatory regions, including promoters and enhancers, are implicated in multiple functional interactions with numerous genes to control their transcriptional responses (Maston et al. 2006; Ong and Corces 2011; Sexton and Cavalli 2015; Spurrell et al. 2016). Those central regulatory regions are occupied by NIPBL and cohesin predicting that mutations could lead to coordinated transcriptional effects. This model is supported by gene expression analyses in CdLS animal models where genes, including some linear clusters, show low to moderate expression changes (Kawauchi et al. 2009; Muto et al. 2014). For example, during limb development, NIPBL is required for the regulation of long-range chromosomal interactions and collinear expression of hox genes (Muto et al. 2014). This collinearity is associated with a switch between topological domains (Andrey et al. 2013). Our model postulates that NIPBL and cohesin are organizing gene communities inside those domains. In our B-lymphocytes model, deregulated genes in cells with NIPBL and SMC1A mutations are associated with hematological and immune functions (Liu et al. 2009) consistent with humoral immunity defects observed in CdLS patients (Jyonouchi et al. 2013). Interestingly, physically interacting genes have been suggested to be involved in related cellular functions (Li et al. 2012; Sandhu et al. 2012). Therefore, we propose the consistent model that, through the connected gene communities, the chromosome architecture provides the backbone to organize genes necessary for normal and consequently pathological functions.
How different deregulated genes associated with specific mutations lead to similar phenotypes in CdLS is unknown. The prevalent model is that the collective effects of gene expression changes associated with each mutation create the birth defects associated with CdLS (Muto et al. 2011). We are proposing an alternative model where controlling the integrity of the chromosome architecture of connected gene communities could play an important role during differentiation. Indeed, active noncoding regulatory elements and chromatin interactions surrounding Pol II are, in part, cell type-specific (Li et al. 2012; Kieffer-Kwon et al. 2013; Tang et al. 2015). Accordingly, the chromosome conformation is extensively reorganized to accommodate the creation of new cell states (Dixon et al. 2015). Interestingly, while the gene signatures associated with NIPBL and SMC1A mutations were different (Figure 1A), many connected communities were shared. Mutations in NIPBL and cohesin subunits could destabilize (or stabilize) the architecture of connected gene communities leading to cellular misreading of environmental cues, creating developmental timing problems. In agreement with this model, embryonic stem cells rapidly differentiate when NIPBL and SMC1A levels are decreased (Kagey et al. 2010). Taken together, these observations led us to propose that NIPBL and SMC1A maintain the structural integrity of connected gene communities, which represent an important feature of normal differentiation mechanisms.
In summary, integration of the chromosome architecture is essential to understand the mechanisms behind transcription-based diseases like CdLS. While our study focused on B-lymphocytes, our transcriptional model is applicable to all cell types encompassing all CdLS-related phenotypic observations. The clinical manifestations will be dependent on the biological role of the cell type in relation to the importance of maintaining the integrity of the transcriptional program for the cellular function. Future studies will reveal how the connected gene communities are formed and restructured depending on the genetic profiles of CdLS patients.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202291/-/DC1.
Acknowledgments
We thank Anne-Marie Pulichino and Samer Hussein for critical review of the manuscript, and the members of our laboratories for insightful discussions. This work was supported by funds from the Canada Research Chair in Transcriptional Genomics (grant #950-228321 to S.B.); from the Natural Sciences and Engineering Research Council of Canada (grant #436266-2013 to S.B.), and the Canadian Institutes for Health Research (grant #MOP-126058 to S.B.). The authors declare that they have no competing interests.
Footnotes
Communicating editor: C. D. Kaplan
Literature Cited
- Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D., et al. , 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Azofeifa J. G., Dowell R. D., 2017. A generative model for the behavior of RNA polymerase. Bioinformatics 33: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Birney E., Dunham I., Green E. D., Gunter C., et al. , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B., Young R. A., 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23: 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Cavalli G., 2016. Organization and function of the 3D genome. Nat. Rev. Genet. 17: 661–678. [DOI] [PubMed] [Google Scholar]
- Boyle M. I., Jespersgaard C., Brondum-Nielsen K., Bisgaard A.-M., Tümer Z., 2015. Cornelia de Lange syndrome. Clin. Genet. 88: 1–12. [DOI] [PubMed] [Google Scholar]
- Busslinger G. A., Stocsits R. R., van der Lelij P., Axelsson E., Tedeschi A., et al. , 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castronovo P., Gervasini C., Cereda A., Masciadri M., Milani D., et al. , 2009. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res. 17: 763–771. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., et al. , 2000. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5: 243–254. [DOI] [PubMed] [Google Scholar]
- Clauset A., Newman M. E. J., Moore C., 2004. Finding community structure in very large networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70: 66111. [DOI] [PubMed] [Google Scholar]
- Csárdi G., and T. Nepusz, 2006 The igraph software package for complex network research. InterJournal Complex Syst. Available at: https://pdfs.semanticscholar.org/1d27/44b83519657f5f2610698a8ddd177ced4f5c.pdf.
- Dao L. T. M., Galindo-Albarrán A. O., Castro-Mondragon J. A., Andrieu-Soler C., Medina-Rivera A., et al. , 2017. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 49: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Deardorff M. A., Kaur M., Yaeger D., Rampuria A., Korolev S., et al. , 2007. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 80: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff M. A., Wilde J. J., Albrecht M., Dickinson E., Tennstedt S., et al. , 2012a RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 90: 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff M. A., Bando M., Nakato R., Watrin E., Itoh T., et al. , 2012b HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Mirny L., 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y., Fang R., Li B., Meng Z., Yu J., et al. , 2017. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., et al. , 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., et al. , 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D., Merkenschlager M., 2013. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol. 25: 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S., Shibayama Y., Burd S., Weinberg M. S., Mhlanga M. M., 2013. Chromosomal contact permits transcription between coregulated genes. Cell 155: 606–620. [DOI] [PubMed] [Google Scholar]
- Fournier M., Bourriquen G., Lamaze F. C., Côté M. C., Fournier É., et al. , 2016. FOXA and master transcription factors recruit mediator and cohesin to the core transcriptional regulatory circuitry of cancer cells. Sci. Rep. 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L. C., 1978. Centrality in social networks. Soc. Networks 1: 215–239. [Google Scholar]
- Ghamari A., van de Corput M. P. C., Thongjuea S., van Cappellen W. A., van Ijcken W., et al. , 2013. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 27: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Díaz E., Corces V. G., 2014. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 32: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis J. H. I., Van Der Weide R. H., Blomen V. A., Brummelkamp T. R., De Wit E., et al. , 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Day D. S., Young R. A., 2016. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167: 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K., Nakato R., Zhang Z., Edmondson A. C., Noon S., et al. , 2015. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 47: 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Perrin D., Sanchez C., Monod J., 1960. Lʼopéron : groupe de gènes à expression coordonnée par un opérateur. C. R. Acad. Sci. 250: 1727–1729. [PubMed] [Google Scholar]
- Jones W. D., Dafou D., McEntagart M., Woollard W. J., Elmslie F. V., et al. , 2012. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyonouchi S., Orange J., Sullivan K. E., Krantz I., Deardorff M., 2013. Immunologic features of Cornelia de Lange syndrome. Pediatrics 132: e484–e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., et al. , 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S., Calof A. L., Santos R., Lopez-Burks M. E., Young C. M., et al. , 2009. Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange syndrome. PLoS Genet. 5: e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K.-R., Tang Z., Mathe E., Qian J., Sung M.-H., et al. , 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz I. D., McCallum J., DeScipio C., Kaur M., Gillis L. A., et al. , 2004. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36: 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse K., Sewitz S., Madan Babu M., 2013. A complex network framework for unbiased statistical analyses of DNA-DNA contact maps. Nucleic Acids Res. 41: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., et al. , 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., et al. , 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dily F., Baù D., Pohl A., Vicent G. P., Serra F., et al. , 2014. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 28: 2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., et al. , 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., et al. , 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Krantz I. D., 2008. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 9: 303–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang Z., Bando M., Itoh T., Deardorff M. A., et al. , 2009. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez D. G., Spielmann M., Mundlos S., 2016. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32: 225–237. [DOI] [PubMed] [Google Scholar]
- Ma H. W., Zeng A. P., 2003. The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19: 1423–1430. [DOI] [PubMed] [Google Scholar]
- Mannini L., Cucco F., Quarantotti V., Krantz I. D., Musio A., 2013. Mutation spectrum and genotype–phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 34: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L., Lamaze F. C., Cucco F., Amato C., Quarantotti V., et al. , 2015. Mutant cohesin affects RNA polymerase II regulation in Cornelia de Lange syndrome. Sci. Rep. 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston G. A., Evans S. K., Green M. R., 2006. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7: 29–59. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M., Nora E. P., 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17: 17–43. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K., 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. [DOI] [PubMed] [Google Scholar]
- Mifsud B., Tavares-Cadete F., Young A. N., Sugar R., Schoenfelder S., et al. , 2015. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47: 598–606. [DOI] [PubMed] [Google Scholar]
- Mitchell J. A., Fraser P., 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22: 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A., Selicorni A., Focarelli M. L., Gervasini C., Milani D., et al. , 2006. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 38: 528–530. [DOI] [PubMed] [Google Scholar]
- Muto A., Calof A. L., Lander A. D., Schilling T. F., 2011. Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange syndrome. PLoS Biol. 9: e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Ikeda S., Lopez-Burks M. E., Kikuchi Y., Calof A. L., et al. , 2014. Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet. 10: e1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H., 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43: 525–558. [DOI] [PubMed] [Google Scholar]
- Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., et al. , 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.-T., Corces V. G., 2011. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C. S., Chakalova L., Brown K. E., Carter D., Horton A., et al. , 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36: 1065–1071. [DOI] [PubMed] [Google Scholar]
- Pancaldi V., Carrillo-de-Santa-Pau E., Javierre B. M., Juan D., Fraser P., et al. , 2016. Integrating epigenomic data and 3D genomic structure with a new measure of chromatin assortativity. Genome Biol. 17: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., et al. , 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Losada A., 2013. Cohesin in development and disease. Development 140: 3715–3718. [DOI] [PubMed] [Google Scholar]
- Revenkova E., Focarelli M. L., Susani L., Paulis M., Bassi M. T., et al. , 2009. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 18: 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu K. S., Li G., Poh H. M., Quek Y. L. K., Sia Y. Y., et al. , 2012. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep. 2: 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Sexton T., Chakalova L., Cope N. F., Horton A., et al. , 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan V., Faure A., Zhan Y., McCord R., Lajoie B., et al. , 2013. Cohesin-based chromatin interactions enable regulated gene expression within pre-existing architectural compartments. Genome Res. 23: 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Cavalli G., 2015. The role of chromosome domains in shaping the functional genome. Cell 160: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Singh V. P., Gerton J. L., 2015. Cohesin and human disease: lessons from mouse models. Curr. Opin. Cell Biol. 37: 9–17. [DOI] [PubMed] [Google Scholar]
- Sofueva S., Yaffe E., Chan W.-C., Georgopoulou D., Vietri Rudan M., et al. , 2013. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32: 3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurrell C. H., Dickel D. E., Visel A., 2016. The ties that bind: mapping the dynamic enhancer-promoter interactome. Cell 167: 1163–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H., Bickmore W. A., 2009. Transcription factories: gene expression in unions? Nat. Rev. Genet. 10: 457–466. [DOI] [PubMed] [Google Scholar]
- Tang Z., Luo O. J., Li X., Zheng M., Zhu J. J., et al. , 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin E. T., Wang T.-J., Lisgo S., Bamshad M. J., Strachan T., 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36: 636–641. [DOI] [PubMed] [Google Scholar]
- Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D. T., et al. , 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E., Kaiser F. J., Wendt K. S., 2016. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Curr. Opin. Genet. Dev. 37: 59–66. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y., 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. [DOI] [PubMed] [Google Scholar]
- Yuan B., Pehlivan D., Karaca E., Patel N., Charng W., et al. , 2015. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Invest. 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotenko E., Mestre J., O’Leary D. P., Przytycka T. M., 2008. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J., Dixon J. R., van der Reijden M. I. J. A., Ye Z., Kolovos P., et al. , 2014a Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 111: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J., Franke V., van Ijcken W. F. J., van der Sloot A., Krantz I. D., et al. , 2014b A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 10: e1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s GEO (Edgar et al. 2002) and are accessible through GEO Series accession number GSE93080 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93080). The software implementing the methods described in this paper is available upon request.