Atilano et al. present a Drosophila post-infection survival screen that takes advantage of a library of miRNA mutant flies. Using genome wide microarray..
Keywords: innate immunity, immune response regulation, miRNAs, Drosophila melanogaster, host–pathogen interactions, Genetics of Immunity
Abstract
Small noncoding RNAs called microRNAs (miRNAs) have emerged as post-transcriptional regulators of gene expression related to host defenses. Here, we have used Drosophila melanogaster to explore the contribution of individual or clusters of miRNAs in countering systemic Candida albicans infection. From a total of 72 tested, we identify 6 miRNA allelic mutant backgrounds that modulate the survival response to infection and the ability to control pathogen number. These mutants also exhibit dysregulation of the Toll pathway target transcripts Drosomycin (Drs) and Immune-Induced Molecule 1 (IM1). These are characteristics of defects in Toll signaling, and consistent with this, we demonstrate dependency for one of the miRNA mutants on the NF-κΒ homolog Dif. We also quantify changes in the miRNA expression profile over time in response to three pathogen types, and identify 13 mature miRNA forms affected by pathogens that stimulate Toll signaling. To complement this, we provide a genome-wide map of potential NF-κB sites in proximity to miRNA genes. Finally, we demonstrate that systemic C. albicans infection contributes to a reduction in the total amount of branch-chained amino acids, which is miRNA-regulated. Overall, our data reveal a new layer of miRNA complexity regulating the fly response to systemic fungal infection.
DROSOPHILA melanogaster has led to some significant discoveries regarding the innate immune pathways involved in countering a wide range of microbial infections [reviewed in Kounatidis and Ligoxygakis (2012) and Bouchon et al. (2014)]. Microbial detection by pattern recognition receptors (PRRs) or sensors of “danger signals” leads to the activation of signaling cascades that result in expression of immune effectors and regulators.
The Toll and Immune Deficiency (IMD) signaling pathways regulate the activation of the nuclear factor κB (NF-κB) transcription factor homologs Dif/Dorsal and Relish, respectively [reviewed in Kounatidis and Ligoxygakis (2012) and Bouchon et al. (2014)]. Following Toll activation, a receptor–adaptor complex, which includes the death domain-containing proteins dMyD88, Pelle, and Tube, transmits the signal to the IκB homolog Cactus. The latter is then targeted for degradation, which leaves NF-κB proteins Dif/Dorsal free to translocate to the nucleus and regulate hundreds of genes (De Gregorio et al. 2001). Among others, Toll signaling triggers the transcription of several effector molecules including antimicrobial peptide (AMP) genes, such as Drosomycin (Drs). Dif enters the nucleus 20 min after infection (Ligoxygakis et al. 2002). Approximately 40 min following Dif nuclear entry, expression of the Toll pathway PRR (and target of Dif), PGRP-SA (peptidoglycan recognition protein-SA) is 10 times that of the basal level (De Gregorio et al. 2001, 2002). However, intracellular homeostasis of the pathway remains relatively unexplored. This is especially pertinent in the light of recent data that indicate that upon Toll induction, Pelle phosphorylates Cka of the STRIPAK (Striatin-interacting phosphatase and kinase) complex thus releasing Hippo signaling, which in turn prevents cactus transcription leading to the sustained activation of the Toll pathway (Liu et al. 2016).
MicroRNAs (miRNAs) are small noncoding RNAs, which act as post-transcriptional regulators of gene expression. MiRNAs are initially transcribed in the nucleus as a primary miRNA transcript by polymerase II, which are then cleaved into precursor miRNAs (pre-miRNAs) by the protein complex made up by nuclear RNase III protein Drosha and the double-stranded RNA-binding protein Pasha. The pre-miRNAs are then exported to the cytoplasm by the protein Exportin 5, where they are further processed by RNase III enzyme Dicer-1 with the help of double-stranded RNA-binding protein Loquacious to produce ∼22 nucleotide-long miRNA–miRNA* duplexes. Finally, one of the miRNA strands is loaded into the RNA-induced silencing complex (Risc), containing Argonaut-1 (Ago-1), for targeting of gene expression (Ha and Kim 2014).
Approximately 60% of all protein-coding genes have been estimated to be under miRNA control (Grun et al. 2005). A given miRNA can regulate several target transcripts, while an mRNA transcript might be targeted by multiple miRNAs. These RNAs play crucial roles in key biological processes, such as cell proliferation, cell fate, differentiation, apoptosis, and survival (Cayirlioglu et al. 2008; Bejarano et al. 2010; Weng and Cohen 2012; Morante et al. 2013; Chen et al. 2014). miRNAs have also emerged as important regulators of the host innate immune response and host–pathogen-interactions. Garbuzov and Tatar revealed that in Drosophila, 20-HE may regulate expression of the antimicrobial peptide Dpt via let-7 miRNA by interacting with the 3′-UTR of the target gene and repressing its translation (Garbuzov and Tatar 2010). These authors proposed that let-7, induced at the same time as Dpt by 20-HE, may set the limit on expression levels of Dpt by negatively regulating it to set a threshold for the AMP following immune induction, thus avoiding overstimulation of the innate immune system. Two other studies in Drosophila have indicated that miRNA-8 plays a role in maintaining the innate immune response (Choi and Hyun 2012; Lee and Hyun 2014). In these studies, miRNA-8 has been shown to regulate the levels of basal expression of AMPs such as Drs and Dpt. They revealed that, in the absence of infection, miRNA-8 targets the Toll receptor and Dorsal. By targeting these proteins, miRNA-8 ensures that, when the immune system is not stimulated, the levels of AMPs such as Drs are kept low. The miRNA cluster 959–964 has also been associated with immune response regulation in Drosophila (Vodala et al. 2012). In this case, the miRNA 959–964 cluster mutants revealed altered levels of mRNAs involved in immune responses, including Drs.
In addition, an in silico study by Fullaondo and Lee (2012) predicted that several other miRNAs may be involved in regulating the innate immune system, although, as yet, their predictions have not been tested due to the lack of available mutants. In fact, little is known about the in vivo role of miRNAs in innate immunity across species. Recently, the construction of a large collection of D. melanogaster miRNA allelic mutants was reported (Chen et al. 2014). Here, we utilize this collection to show that individual or clusters of miRNAs alter the ability of flies to cope with systemic infection, impacting on survival, the control of pathogen number, and the production of Toll-dependent transcripts.
Materials and Methods
Fly stocks
Flies obtained from the Bloomington Drosophila Stock Centre: c564 > Gal4 driver (#6982); UAS-myr_mRFP (#7118); Dif1 loss-of-function allele (#36559); UAS-miR-277Sponge (#61408); the control backgrounds w1118 (#5905), y1w1 (#1495), and the isogenic wild-type strain (#25174; originally from the Drosophila Genetics Reference Panel); and the collection of miRNA allelic mutants (Chen et al. 2014). Fly lines carrying UAS (upstream activating sequence)-RNA interference (RNAi) constructs were obtained from the Vienna Drosophila RNAi Centre: PGAP5RNAi (#47953); Exp5RNAi (#31706 and #31707); Dcr-1RNAi (#24667); LoqsRNAi (#22453); and Myd88RNAi (#25402). Yolk > Gal4 was kindly provided by Bruno Lemaitre. We also used the previously described strains UAS-Ago-1 and UAS-Dcr-1 (Dietzl et al. 2007), and miR-124Δ6 (Sun et al. 2012). All flies were raised on maize malt molasses food in a light-dark (12-hr cycle) incubator at 25° and 60% humidity. Flies were shifted to 30° and 60% humidity following infection for microarray, branch-chained amino acid (BCAA) quantification experiments, and those involving miRNA allelic mutants. Flies were shifted to 30° and 60% humidity 2 days prior to and then during infection for UAS-RNAi/UAS-miR-277Sponge/UAS-protein overexpression experiments.
Pathogens and preparation
Candida albicans strain SC5314 was streaked intermittently from a −80° stock onto Sabouraud Agar (SGA), grown at 30° for 36 hr, and stored at 4° for up to a week. The inoculant was prepared from a single colony grown in 10 ml Sabouraud Broth (SGB) for 18–21 hr at 30° and shaken at 200 rpm; this was washed and resuspended in PBS to an OD600 of 0.95–1.05 (readings taken with a NanoDrop 1000 spectrophotometer; Thermo Fisher Scientific) and further diluted fourfold in PBS for injection into flies. Staphylococcus aureus strain NCTC-8325 was streaked intermittently from a −80° stock onto Tryptic Soy Agar, grown at 30° for 36 hr, and stored at 4° for up to a week. The inoculant was prepared from a single colony grown in 10 ml Tryptic Soy Broth for 16–18 hr at 30° and shaken at 200 rpm; this was washed and resuspended in PBS to an OD600 of 0.36 and further diluted 1000-fold in PBS for injection into flies. Escherichia coli strain 1106 was cultured in a manner similar to S. aureus, but Luria-Bertani agar/broth was used and the OD600 0.36 suspension was not further diluted prior to injection.
Screening the collection of miRNA allelic mutants
Two to three-day-old female flies were anesthetized with CO2, and 13.8 nl of C. albicans inoculant (∼800 yeast cells) injected into the dorsolateral region of the thorax using a Nanoject II Injector (Drummond Scientific) coupled to a fine glass needle. Flies were kept at 30° following infection; a compromise between conditions for C. albicans growth and fly resistance to temperature. Survival was monitored every 24 hr over a 3-day period; lethality during the first 6 hr was attributed to effects associated with the injection procedure and was subtracted from the survival data. The miRNA allelic mutants were infected as several groups with each group including a background control, either w or yw. The survival profile of a particular mutant background was compared to that of the group control via the Log-rank test, and P-values adjusted for multiple comparisons using the Holm–Šidák method. For miRNA mutants whose survival profile statistically separated from their group control, a second round of C. albicans infection was performed; for further comparison, additional backgrounds were included that did not separate from their group control. In addition, most second-round miRNA mutant backgrounds were injected with 13.8 nl PBS and selected with 13.8 nl E. coli. All confirmed second-round miRNA mutants were tested for a third time, confirming the differences in estimated survival probabilities found previously.
To estimate changes in pathogen number during the course of infection six, infected female flies were homogenized in SGB at specific time points (0, 24, and 48 hr); the homogenates were serially diluted, plated on SGA, and incubated for 36 hr at 30° to enable suitable colony growth for counting.
To determine changes in the levels of Drs and Immune-induced Molecule 1 (IM1) transcripts during the course of infection, total RNA was extracted from six female flies at specific time points (0, 24, and 48 hr) using the Total RNA Purification Plus Kit (Cat. No. 48400; Norgen Biotek); this uses a column that efficiently removes genomic DNA. Purity and concentration of the RNA samples were checked using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific), and 500 ng of RNA reverse transcribed in a total volume of 20 µl using the SensiFAST cDNA Synthesis Kit (Cat. No. BIO-65054; Bioline, London, UK). Template cDNA—2 µl diluted 10-fold—was amplified using the SensiFast SYBR No-ROX Kit (Cat. No. BIO-98005; Bioline) and primer pairs against Drs, IM1, or TATA-Binding Protein (dTBP) at a concentration of 400 nM per primer; cycling was performed in a Rotor-Gene Q real-time PCR machine (QIAGEN, Valencia, CA). The amount of transcript was expressed as 2(−ΔCT), where ΔCT is the average threshold cycle (CT) value for the gene of interest (Drs/IM1) subtracted from the average CT value for the internal control (dTBP) (Schmittgen and Livak 2008). Primer pairs (5′–3′): Drs (+) GTACTTGTTCGCCCTCTTCG and Drs (−) TTAGCATCCTTCGCACCAG; IM1 (+) GAAATTCTTCTCAGTCGTC and IM1 (−) CATCAATGGCGATTGCAG; and dTBP (+) GGCAAAGAGTGAGGACGACT and dTBP (−) GAGCCGACCATGTTTTGAAT.
Identifying potential NF-κB-binding sites in close proximity to miRNA genes
The current version of the D. melanogaster genome (R6.12) was used to perform the binding site analysis. The transcription start sites (TSSs) for miRNA genes are yet to be comprehensively described, but predicted or experimentally determined TSSs generally lie within 1.5 kb of the miR-RM transcript (Qian et al. 2011; Ryazansky et al. 2011); these are the pre-miRNAs embedded in the primary transcripts. Therefore, the initial base (+ 1) of the miR-RM transcript was used as a reference for the promoter region of the corresponding miRNA gene. A Position Weight Matrix (PWM) approach was taken to identify potential NF-κB-binding sites within 2-kb regions flanking the (+ 1) of the miR-RM transcript; evidence from a study using a human cell line identified functional NF-κB-binding sites upstream and downstream of certain miRNA genes, and these sites were located up to 3 kb from the TSS (Zhou et al. 2010). The Toll and IMD PWMs were constructed using 15 sequences identified as overrepresented motifs upstream of Toll and IMD responsive genes, respectively (Busse et al. 2007). The Rel PWM was constructed from 31 sequences derived from a SELEX assay using a recombinant fragment of the D. melanogaster Rel protein, and the DIF/Rel PWM constructed from the base frequency table derived from a SELEX assay using a combination of recombinant DIF and Rel proteins (Senger et al. 2004).
miRNA profiling
To identify changes in the miRNA profile over time, a microarray experiment was performed. Wild-type flies from the same population were injected with 13.8 nl of either C. albicans, S. aureus, E. coli, or PBS, and transferred to 30°. At 0, 1, 7.5, 18, and 60 hr following injection, total RNA was purified from 10 females using the Total RNA Purification Plus Kit (Cat. No. 48400; Norgen Biotek). Next, 5 μg of total RNA (quantified using a NanoDrop 1000 spectrophotometer) from each sample was suspended in 200 μl of precipitation solution (3 M NaOAc, pH 5.2 and 100% ethanol) and immediately frozen in liquid nitrogen; these were stored at −80° until shipment for the miRNA microarray experiment (LC Sciences, Houston, TX). Samples were labeled with a fluorescent dye and hybridized to a µParaflo Microfluidic Chip that contained complementary sequences for 425 unique mature miRNA sequences, derived from 241 D. melanogaster miRNA genes (miRBase version 19.0). The background signal was subtracted and data normalized prior to analysis; ANOVA was applied to identify statistical differences. All data processing was performed by LC Sciences and is presented as clustered heat maps. Experiments were repeated independently on three occasions (n = 3), except for the E. coli infection, which was performed twice (n = 2).
Quantification of branched-chain amino acids (BCAAs)
Ten flies injected with PBS or C. albicans (weighing 10 mg) were flash-frozen in liquid nitrogen at time 0–24 hr following injection. Frozen flies were homogenized in 200 µl assay buffer. Homogenates were centrifuged for 10 min at 12,000 rpm, and 10 µl of supernatant used to measure the BCAA levels (BCAA Colorimetric Assay Kit; Cat. No. K564-100; Biovision); color intensity was quantified using a microplate reader (CLARIOstar; BMG Labtech).
Statistical analysis
Data were analyzed using GraphPad Prism (version 7.01) or the R-platform. To visualize survival data, estimated survival curves were constructed; in figures with estimated survival probabilities, C.I.s were excluded for clarity of image. The Log-rank test was used to identify statistical differences between survival curves and P-values were adjusted for multiple comparisons via the Holm–Šidák method. Data involving a time series (excluding the microarray data) were analyzed using repeated-measures two-way ANOVA, and P-values adjusted for multiple comparisons using Bonferroni correction. Single time point data were compared via one-way or two-way ANOVA, and P-values adjusted using the Holm–Šidák method.
Data availability
The miRNA mutant strain collection from the Cohen Lab and the mir sponge lines are available from the Bloomington Drosophila Stock Center. All P-values for Figure 6 can be found in Supplemental Material, Table S2. The microarray data are presented in Figure 4 for miRNA that presented statistically significant regulation and in Figure S3 for all others; Table S5 includes a list of P-values for the microarray analysis. Table S4 has a list of all miRNAs that have at least one consensus NF-κB-binding site, while Figure S4 summarizes the numbers of miRNAs with such site(s).
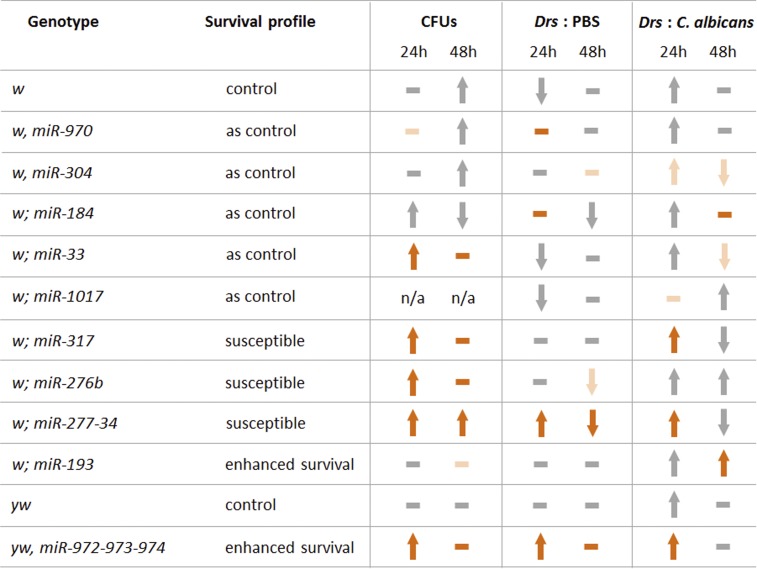
Figure 6.
Summary of phenotypes for selected miRNA mutant backgrounds. The table presents a summary for each miRNA mutant regarding changes to pathogen number (CFUs) and level of Drs transcript over time (see Figure 2, B and C). Direction of arrows in the CFUs column and both Drs columns (PBS and C. albicans) indicate how the measured variable changes over time for the particular miRNA background; a dash indicates there was not a significant change. The arrow/dash color represents whether the change is significantly greater (orange) than that of the given w or yw control (gray) or significantly below (light orange) that of the given control; if the variable does not separate significantly from the given control the arrow/dash is given as gray. For details regarding P-values see Table S2. CFU, colony forming unit; miRNA, microRNA; n/a, not applicable.
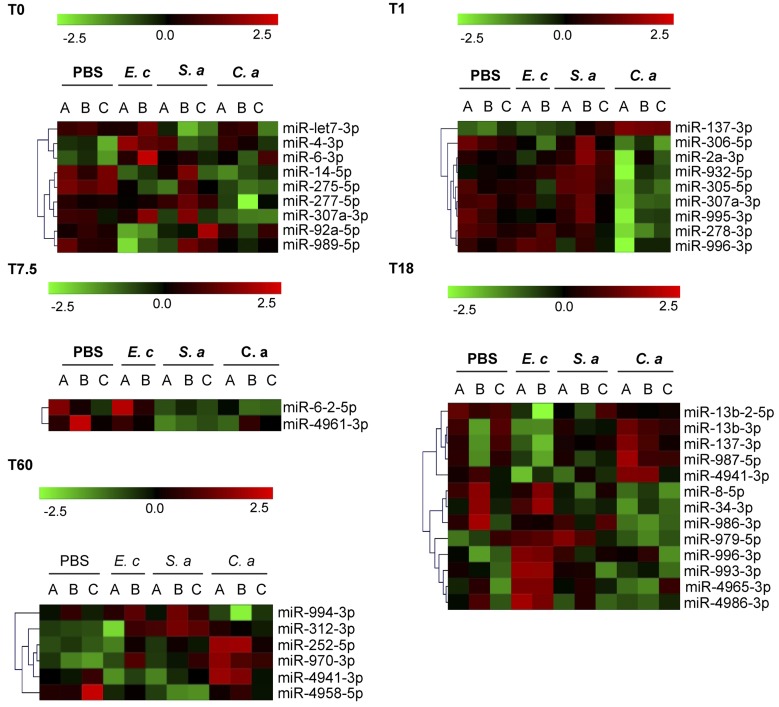
Figure 4.
Differential production of mature miRNA forms. Wild-type flies were injected with PBS or one of the given pathogens: E. coli (E. c), S. aureus (S. a), or C. albicans (C. a). Their miRNA complement was quantified at time points T0, T1, T7.5, T18, and T60 (numbers refer to hours following injection). Data are compared for all factors at each individual time point to identify mature miRNA forms whose production is differentially regulated; ANOVA-selected miRNAs (P < 0.1) are presented as clustered heat maps, where negative Z-values (green) indicate downregulation relative to the data set and positive Z-values (red) upregulation. Except with E. coli (n = 2), the infection experiments were repeated independently on three occasions (n = 3), indicated above the heat maps as A, B, and C. miRNA, microRNA.
Results
Disrupting miRNA biogenesis causes susceptibility to systemic C. albicans infection and potentially influences Toll signaling
To explore the contribution of miRNAs in controlling the response to infection at a whole-organism level, we initially examined the requirement for miRNA biogenesis in protecting the host against infection. To perturb this process, we used RNAi against Exp5, which transports pre-miRNAs from the nucleus to the cytoplasm, and against Loqs and Dcr-1, which together process the various pre-miRNAs to produce particular miRNA duplexes that are subsequently loaded onto Ago-1. We also examined the consequences of increasing the levels of Ago-1, since this protein is involved in production of the miRNA strand that guides translational repression/destabilization of target mRNAs (Ha and Kim 2014). Manipulating these components has been shown to alter the miRNA profile in human cell lines (Lee et al. 2004; Okamura et al. 2004; Hata and Kashima 2016; Jakob et al. 2016; Kim et al. 2016).
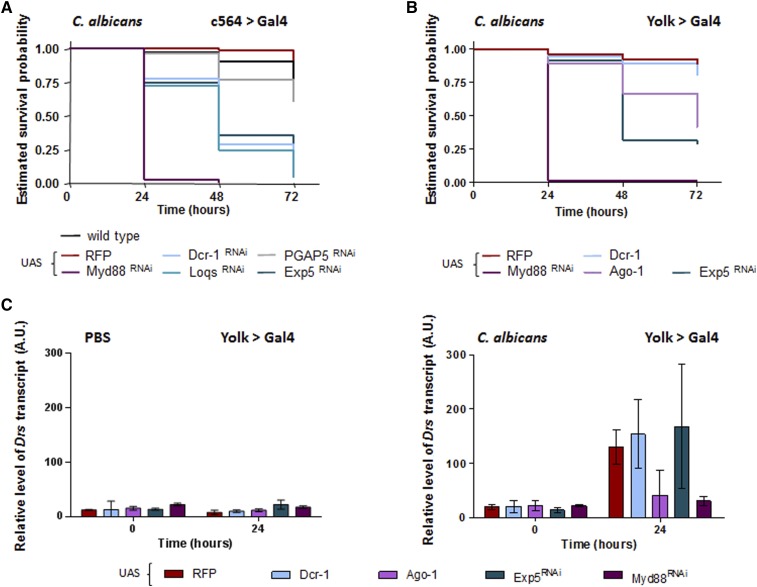
The c564 > Gal4 driver was used to express UAS-RNAi constructs in the major immune-responsive tissues (fat body, hemocytes, and gut) throughout development and into adulthood (Hrdlicka et al. 2002; Ryu et al. 2006). Female c564 > RNAi flies were injected with either C. albicans or PBS—the latter serving as a control for the injection procedure—and survival monitored. None of the c564 > RNAi flies exhibited physical abnormalities, nor did they die significantly following injection with PBS (data not shown). Those impaired for miRNA biogenesis (c564 > Exp5RNAi/c564 > LoqsRNAi/c564 > Dcr-1RNAi) succumbed to systemic C. albicans infection more rapidly than a wild-type strain (see Materials and Methods), and more rapidly than flies expressing either red fluorescent protein (RFP) (c564 > RFP) or a control RNAi construct (c564 > PGAP5RNAi) (Figure 1A). PGAP5 was chosen as a control since it has no defined role in immunity—likewise regarding the vertebrate ortholog metallophosphoesterase-1—and did not exhibit resistance/tolerance or susceptibility to C. albicans infection during a large-scale UAS-RNAi screen (data not shown); consistent with this, the survival of c564 > PGAP5RNAi flies was inseparable from the wild-type strain or from those expressing RFP (Figure 1A). However, flies impaired for miRNA biogenesis were less susceptible to infection than those deficient in Toll signaling (c564 > Myd88RNAi) (Figure 1A). These observations suggest that having the correct complement of miRNAs within immune tissues is necessary for flies to cope with systemic yeast infection. It may therefore be that dysfunctional miRNA biogenesis affects the output of Toll signaling, the major respondent to fungal infections. To examine this, we used Yolk > Gal4 to overexpress Ago-1 or Dcr-1 protein, or the Exp5RNAi construct, specifically in the fat body of female flies (Yolk > Ago-1/Yolk > Dcr-1/Yolk > Exp5RNAi); the driver is expressed ∼4 days after emergence of the adult and therefore bypasses potential effects that may occur during development (Georgel et al. 2001). Flies were again injected with either PBS or C. albicans and a subset of each population assessed for survival; the level of Drs transcript was measured in the surviving populations to determine an output of Toll signaling. There were no significant differences in the survival dynamics for flies injected with PBS (data not shown). Consistent with that observed with c564 > Exp5RNAi, Yolk > Exp5RNAi flies succumbed to systemic C. albicans infection more rapidly than those of the control population (Yolk > RFP), but not as rapidly as those deficient for Toll signaling (Yolk > Myd88RNAi) (Figure 1B). The survival profile for Yolk > Ago-1 flies was similarly positioned, and inseparable to that of Yolk > Exp5RNAi flies (Figure 1B). Although susceptibility to infection was equivalent, there were differences in their level of Drs transcript: for Yolk > Ago-1 flies this did not detectably change over the course of infection—mirroring that observed with a Toll pathway mutant—but it increased within Yolk > Exp5RNAi flies comparably to that observed with the Yolk > RFP control (Figure 1C). An additional feature unique to Yolk > Exp5RNAi flies was the significant increase relative to the control of the Drs transcript induced through PBS injection alone (Figure 1C).
Figure 1.
Perturbing miRNA biogenesis increases the susceptibility of flies to systemic C. albicans infection and affects production of the Drs transcript. (A) RNAi knockdown of Exp5, Dcr-1, or Loqs in the major immune tissues results in flies becoming more susceptible to systemic C. albicans infection relative to a wild-type strain, or RFP and PGAP5 controls, but they are less susceptible than immune-defective flies; P < 0.001 for the wild-type strain or either control compared to any miRNA biogenesis deficient background, and when the latter are compared to the Myd88 mutant. The survival profile of the wild-type strain is inseparable to either control (P > 0.1 in both cases). (B) Likewise, when Exp5 function is reduced or Ago-1 overexpressed specifically in the fat body, flies become more sensitive to systemic C. albicans infection relative to an RFP control (P < 0.001 in both cases), but are less sensitive than Myd88-deficient flies (P < 0.001 in both cases). The survival profiles for Yolk > Dcr-1 and control RFP flies are inseparable (P > 0.2). For clarity of image, 95% C.I.s have been omitted. (C) At the point of injection (time 0), there are generally no differences in the level of Drs transcript (PBS injected, P > 0.6 for any comparison, except for those with Yolk > Myd88RNAi flies where the transcript is slightly elevated; C. albicans injected, P > 0.05 for any comparison), implying that disrupting miRNA biogenesis per se does not alone affect its production. The amount of transcript is inseparable for Yolk > Ago-1 and Yolk > Myd88RNAi flies 24 hr following infection (P > 0.9), and does not detectably change for either from the point of injection (P > 0.9 in both cases). However, the level of Drs transcript increases during the course of infection within Yolk > Dcr-1 and Yolk > Exp5RNAi flies (P < 0.001 in both cases) to that of the RFP control (P > 0.2 for both comparisons to control). Only Yolk > Exp5RNAi flies exhibit a change in Drs transcript over time from the point of PBS injection (P < 0.01). Survival profiles were compared using the Log-rank test (n = 35–104, combined data from three independent infections) and P-values adjusted for multiple comparisons via the Holm–Šidák method. Quantification of Drs transcript was performed independently three times (n = 3) and is presented as mean values with 95% C.I.s; differences in relative transcript level were identified using repeated-measure two-way ANOVA and P-values adjusted for multiple comparisons through Bonferroni correction. miRNA, microRNA; RNAi, RNA interference; RFP, red fluorescent protein; UAS, upstream activating sequence; 95% CI, 95% Confidence interval.
Therefore, it seems likely that correct processing of miRNAs within immune tissues is necessary to protect against systemic C. albicans infection and, in some capacity, to control the output of Toll signaling. To identify specific miRNAs that contribute to either function, we used a collection of targeted knockout miRNA mutants recently constructed in D. melanogaster (Chen et al. 2014).
Screening for miRNAs that contribute to controlling the fly’s response to systemic C. albicans infection
To date, the miRNA mutants generated by the Cohen Lab have been assayed for changes in a variety of phenotypes but not their response to infection (Chen et al. 2014). Therefore, we injected the majority of available lines with C. albicans and compared their survival profiles to that of a background control: in total, we evaluated 72 miRNA mutant backgrounds covering 95 miRNAs. For miRNA mutants whose survival profile statistically separated from their control (P < 0.05), two repeat rounds of injection were performed; included in this were mutants whose survival profiles were close to but did not separate from their control (0.1 ≥ P > 0.05), and for further comparison, additional lines whose survival profiles were inseparable from their control (P > 0.1) (Table S1). We identified four mutant backgrounds that were more susceptible (miR-317, miR-276b, miR-277-34, and miR-310-311-312-313) to systemic C. albicans infection, and two mutant backgrounds that exhibited enhanced survival (miR-193 and miR-972-973-974) (Figure 2A and Table S1). In addition, none of the flies carrying the miRNA-137 mutation succumbed to infection (see Table S1); however, due to the low number of obtainable flies at the time of injection, the enhanced survival was not considered significant. Finally, none of the flies for the six mutant backgrounds exhibiting altered survival profiles had obvious defects to their external morphology, as all appeared healthy, nor did they exhibit differences in survival dynamics following PBS injection (Table S1).
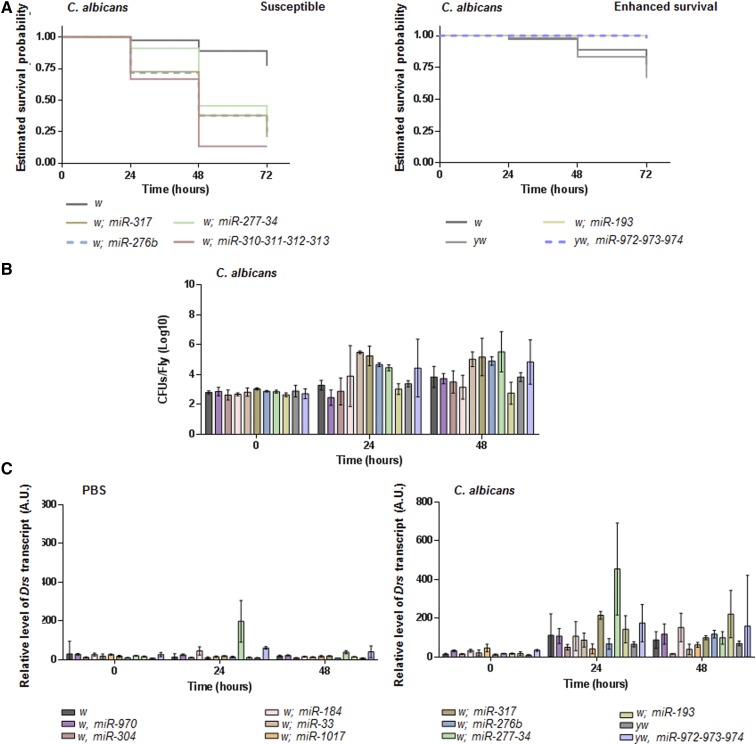
Figure 2.
Assaying selected miRNA allelic mutants for characteristics indicative of defects to immune signaling. (A) Survival profiles for miRNA mutant backgrounds shown to exhibit susceptibility or enhanced survival in response to systemic C. albicans infection; the estimated survival curves represent combined data for the repeat infections (n = 29–62, see Table S1). The w and yw curves were constructed using survival data taken from the groups within which a miRNA mutant background was originally compared: for w this is combined data from five groups (n = 153), and for yw combined data from two groups (n = 54). Survival profiles for the miRNA mutants separate significantly from their given w or yw control (P < 0.001 for any mutant compared to control). For clarity of image, 95% C.I.s have been omitted. (B) Changes in pathogen number during the course of systemic C. albicans infection. The initial inoculation (time 0) is equivalent for all miRNA mutants and the w and yw controls (P > 0.05 for any comparison). (C) Changes in the level of Drs transcript following PBS injection or infection. For each miRNA mutant, conclusions regarding the dynamics of the two variables over time and how this compares to the given control are summarized in Figure 6. CFUs and Drs quantification were performed independently on three occasions (n = 3) and are presented as mean values with 95% C.I.s. CFU, colony forming unit; miRNA, microRNA; 95% CI, 95% Confidence interval.
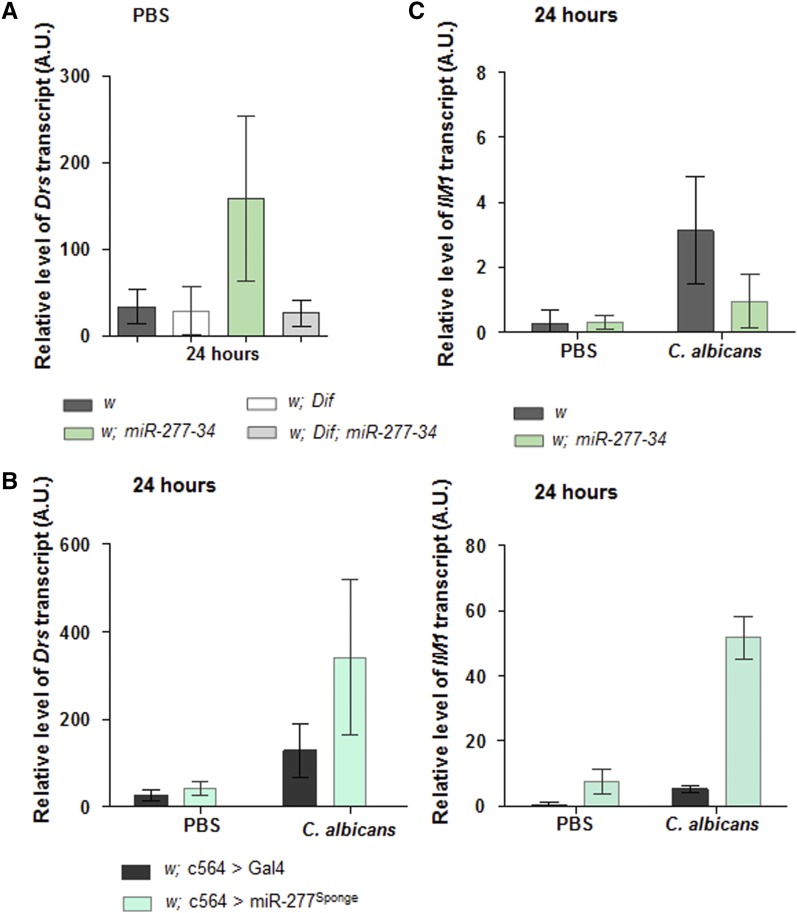
We next assessed the ability of the six miRNA mutants to control pathogen number by quantifying their colony forming units (CFUs) over time following inoculation with C. albicans, and for their ability to modulate an output of Toll signaling. These are two assays that reveal defects in classic innate immune function, and regarding CFUs, distinguish between mechanisms of resistance and tolerance toward infection (Schneider and Ayres 2008). For comparison, we also evaluated some of the miRNA mutants whose survival profiles were inseparable from their controls. During the first 24 hr, pathogen number increased within the susceptible miRNA mutants to a level greater than that observed with the w background control (Figure 2B and Figure 6). For the miR-317 and miR-276b susceptible mutants, pathogen number did not detectably change during a further 24 hr of infection, but continued to increase within the miR-277-34 mutant; at this 48-hr time point there was more C. albicans within the susceptible miRNA mutants than the w control (Figure 2B and Figure 6). For miRNA backgrounds whose survival did not differ from the w control there was, in general, no separation of pathogen number (Figure 2B and Figure 6). The most notable exception to this being for flies carrying the miR-33 mutation, whose CFU profile was akin to that of the susceptible mutants (Figure 2B and Figure 6). Within the miR-972-973-974 mutant, which exhibited enhanced survival, the amount of C. albicans significantly increased over time relative to the yw background control, for which there was no detectable change in CFUs (Figure 2B and Figure 6). Finally, miR-193 mutants that exhibited enhanced survival did not show any significant change from the initial inoculate over the 48-hr period (Figure 2B and Figure 6).
There were significant differences in the levels of Drs transcripts between some of the miRNA mutants at the point of injection (time 0 PBS and time 0 C. albicans, Figure 2C). However, for mutants carrying a w allele, these were inconsistent between those injected with PBS and those injected with C. albicans—the assumption being that differences should be consistent at time 0 if there is an effect due to mutation—and thus likely reflect natural variation rather than meaningful differences resulting from inherent biological changes. An exception to this may be the miR-304 mutant, where the level of transcript was reproducibly low across the course of infection (Figure 2C and Figure 6); this suggests that production of Drs mRNA may actually be compromised. All miRNA mutants (except miR-1017) and both controls experienced an elevation in their levels of Drs transcripts 24 hr following systemic C. albicans infection. For the susceptible miR-317 and miR-277-34 mutants and enhanced survival miR-972-973-974 mutant, this was above that of the respective w and yw control, being particularly notable for miR-277-34; these increases resolved to the level of control during the subsequent 24-hr infection period (Figure 2C and Figure 6). At this 48-hr time point, miRNA mutants with survival profiles similar to their control had either maintained their elevated levels of Drs transcripts (miR-970 and miR-184) or experienced a decrease to levels below that of the w background (miR-304 and miR-33) (Figure 2C and Figure 6). The miR-276b and miR-193 mutants continued to increase the level of transcript over the 48-hr infection period; for the susceptible miR-276b mutant this was at a level comparable to the w control, whereas the enhanced survival miR-193 mutant was accumulating significantly more (Figure 2C and Figure 6). For flies injected with PBS, only the susceptible miR-277-34 and enhanced survival miR-972-973-974 mutants exhibited a significant increase in the levels of Drs transcripts 24 hr following injection, and this was above that of the respective control (Figure 2C and Figure 6). The miR-972-973-974 mutant also had elevated transcript levels at the point of injection, suggesting that basal production of Drs is, to some degree, heightened in this mutant.
It is clear that individual or small clusters of miRNAs contribute to control of the response to systemic C. albicans infection through modulating the survival outcome, in general, limiting the increase in pathogen number, and influencing the production of the Drs transcript. These are traits associated with changes to Toll signaling. Therefore, we explored this potential connection.
miR-277 and miR-34 potentially have opposing effects on the output of Toll signaling
A defining feature of the susceptible miR-277-34 mutant is the strong Drs response observed 24 hr following PBS injection. We eliminated the increase in temperature—applied since PBS-injected flies serve as the control for infection, which proceeds at 30°—as being a causal factor, and demonstrated that sterile pricking is sufficient to invoke the response (Figure S1). This implies participation of Toll/IMD signaling, since pricking alone induces rapid production of numerous AMP transcripts at a low level (Lemaitre et al. 1997); presumably, in the miR-277-34 mutant, this pricking effect is not so efficiently downregulated. To substantiate a requirement for Toll signaling, we repeated the PBS injection of the miR-277-34 mutant while simultaneously reducing the function of Dif, the major Toll pathway transcription factor in the context of immunity. This suppressed the strong Drs response (Figure 3A), indicating that Toll signaling is required to produce the pool of transcript modulated by miR-277 and/or miR-34. To evaluate whether miR-277 alone was sufficient to regulate the level of Drs transcript in response to PBS injection and/or infection, we used an antisense RNA to knock its function down within the immune tissues (c564 > miR-277Sponge). For an independent readout of Toll signaling, we also quantified the relative level of IM1, a gene whose infection response is exclusively dependent on the Toll ligand Spz (De Gregorio et al. 2001, 2002). Although 24 hr following PBS injection the level of Drs transcript was slightly elevated within c564 > miR-277Sponge flies compared to those of the control, the difference was not significant; however, there was significantly more transcript following infection (Figure 3B). The level of IM1 transcript was higher than that observed for the control population 24 hr following either PBS or C. albicans injection (Figure 3B); an effect equivalent to that observed with Drs transcript in the miR-277-34 mutant (Figure 2C). This suggests that miR-277 functions to reduce signaling through the Toll pathway, and that its loss in the miR-277-34 mutant may be the causal factor underlying the response to infection. However, if a repression of Toll signaling has been removed, it raises a question regarding the susceptibility of the mutant. Therefore, we measured the level of IM1 transcript 24 hr following PBS injection or infection; in contrast to Drs, the level of transcript was comparable to the control following injection of PBS, and significantly lower in response to infection (Figure 3C). This suggests that miR-34 functions to promote signaling through the Toll pathway and acts downstream to the inhibitory effect of miR-277. The elevated level of Drs transcript within the miR-277-34 mutant, if signaling through Toll is reduced, may be a consequence of either miR-277 and/or miR34 directly targeting the transcript. Indeed, members of the miR-310-311-312-313 cluster and miR-964 target the 3′-UTR of Drs to reduce transcript abundance (Vodala et al. 2012; Li et al. 2017b). Therefore, we used freely available algorithms—PITA/TargetScan/miRanda-mirSVR (Enright et al. 2003; Kertesz et al. 2007; Ruby et al. 2007; Betel et al. 2010; Agarwal et al. 2015)—to scan the 3′-UTR of Drs and identified a potential binding site for miR-277 (Table S3), suggesting direct targeting of the transcript as a potential regulatory mechanism. There were no potential sites for miR-277 in the 3′-UTR or open reading frame (ORF) of IM1. The latter may be significant given that conserved miRNA-binding sites in the ORF of D. melanogaster mRNAs lead to downregulation of transcript levels (Schnall-Levin et al. 2010). There were also no potential binding sites for miR-34 in either the Drs or IM1 transcripts (Table S3). Therefore, we speculate that loss of miR-277 in the miR-277-34 mutant results in derepression of accumulated Drs transcript, and that this compensates for reduced signaling through the Toll pathway due to loss of miR-34.
Figure 3.
miR-277 and miR-34 potentially influence signaling through the Toll pathway. (A) When Dif function is also compromised, the elevation of Drs transcript observed within the miR-277-34 mutant 24 hr following PBS injection is abolished; P < 0.001 for any comparison to w; miR-277-34, and P > 0.9 for all other comparisons. (B) Knocking down the function of miR-277 in immune tissues (c564 > miR-277Sponge) leads to an increase in the level of Drs transcript 24 hr following infection relative to the control (c564 > Gal4, this is the driver crossed to the w background, P < 0.001), and for IM1 transcript, this is true also for PBS injection (P < 0.001 for PBS injection and infection). (C) In contrast to that observed with Drs, the level of IM1 transcript within the miR-277-34 mutant is lower than that of the control 24 hr following infection (P < 0.001). The data represents the mean of three independent biological repeats (n = 3/95% C.I.s) and was compared using one-way ANOVA (A) or two-way ANOVA (B and C); P-values were adjusted via the Holm–Šidák method. 95% CI, 95% Confidence interval.
From the miRNA mutants assessed for traits indicative of defects in innate immunity (Figure 6), other than miR-277, only miR-33 and miR-972 have potential binding sites in the 3′-UTR of the Drs transcript; none of the miRNAs are predicted to target its ORF (Table S3). This is consistent with studies demonstrating that miRNAs function in positive or negative feedback loops that modulate signaling through NF-κB pathways (Olarerin-George et al. 2013; Staedel and Darfeuille 2013; Teng et al. 2013; Maudet et al. 2014; Kumar et al. 2015; Xiong et al. 2016; Li et al. 2017b). Accordingly, immune-induced expression (or repression) of particular miRNAs has been shown to depend on NF-κB-binding sites (Taganov et al. 2006; O’Hara et al. 2010; Zhou et al. 2010; Wang and Cambi 2012; Thompson et al. 2013). In an attempt to identify miRNAs regulated by NF-κB activity, we assessed the potential for NF-κB occupancy in proximity to miRNA genes, and evaluated changes in the miRNA profile following systemic infection.
Distribution of potential NF-κB sites in proximity to miRNA genes and miRNA microarray analysis following systemic infection
A PWM approach was used to identify potential NF-κB-binding sites within 2-kb regions located either side of the first base (+) of the miR-RM transcripts; four PWMs were constructed (Table S4). Of the 256 miRNA genes in the annotated D. melanogaster genome, 36% had at least three strong NF-κB-binding sites (Figure S2), suggesting that certain miRNAs may indeed be regulated by innate immune signaling at the level of transcription.
To determine whether stimulating the Toll or IMD pathways altered the miRNA profile over time following systemic immune challenge, we performed a miRNA microarray experiment. Wild-type flies were infected with C. albicans or S. aureus (both Toll signaling), or E. coli (IMD signaling); the baseline was PBS injection. Samples were collected at time points (0, 1, 7.5, 18, and 60 hr following injection) a few hours prior to when significant changes occurred in Toll/IMD signaling, the assumption being that modulation of the miRNA profile must precede regulation of mRNAs. There were changes (P < 0.1, an acceptable threshold for exploratory data) in the profile over time for 38 and 33 mature miRNAs following C. albicans or S. aureus infection, respectively, and 15 for PBS injection (Figure S3 and Table S5). Of note, 13 of those modulated by S. aureus infection were also affected by systemic C. albicans (Table S5), supporting the notion their expression may indeed be regulated by Toll signaling. However, when PBS and pathogen were compared at the particular time points (Figure 4), from the 13 mature forms, only miR-275-5p and miR-307a-3p (time 0), and miR-305-5p/miR-306-5p/miR-307a-3p (1 hr) significantly separated (Table S5), and this was mostly due to there being lower levels in response to C. albicans infection. Among the miRNA profiles that changed over time were some required to control the response to systemic C. albicans infection (Figure 6, and here we include miR-137): C. albicans (miR-34-5p/miR-137-3p/miR-313-5p); S. aureus (miR-34-5p/miR-277-5p/miR-310-3p and miR-310-5p/miR-311-3p); and PBS (miR-277-5p). However, again, these differences did not come through strongly at the particular time points; in fact, only mature form miR-137-3p was shown to separate, being higher at two time points following C. albicans infection. In addition, miR-312-3p was at a higher level (60 hr) and miR-34-3p at a lower level (18 hr) in response to C. albicans and S. aureus infection. The signal intensity values for miR-34 have been plotted as an example of what the data generally looks like (Figure S4). This highlights three observations: (1) miRNAs whose profiles change significantly across time for a particular factor tend to do so at the later time points and (2) do not separate so much from the PBS baseline (miR-34-5p); and (3) where a difference is found between PBS and pathogen at a particular time point it tends to result from small error between biological repeats (miR-34-3p).
Systemic C. albicans infection promotes degradation of BCAAs
To assess potential functions of the miRNAs shown to contribute in controlling the response to systemic infection, we used DIANA TOOLS mirPath v.3 to predict the pathways and genes they are likely to regulate (Vlachos et al. 2015) (Table S6). Three of the most striking predictions were for miR-972 in modulating Hippo signaling and endocytosis, and for miR-277 in regulating catabolism of BCAAs (valine, leucine, and isoleucine); these are important nutrient signals that affect metabolism, for example, through activating the mammalian target of rapamycin complex 1 or serving as substrates for the tricarboxylic acid (TCA) cycle (Yoon 2016). Given that miR-277 does indeed regulate BCAA catabolism in D. melanogaster (Esslinger et al. 2013), we first examined whether infection can affect the process, and second whether this is modified in the miR-277-34 mutant.
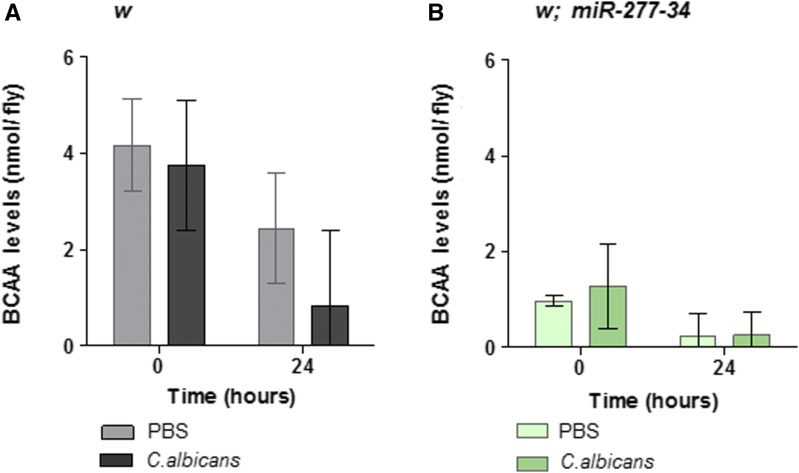
We injected w control flies and those of the miR-277-34 mutant with PBS or C. albicans and quantified their BCAA content after 24 hr. This decreased significantly in control flies during the 24-hr infection period whether they were injected with PBS or infected (Figure 5); we attribute the former to changes in metabolism that are likely to occur as a result of keeping flies at 30° during the infection period or alternatively, as a result of the injection per se. Nevertheless, within control flies the BCAA content was significantly lower following infection relative to PBS injection after 24 hr (Figure 5). In contrast, within the miR-277-34 mutant, BCAA levels were indistinguishable at 24 hr (Figure 5), although reduced compared to the levels at the point of injection (time 0, Figure 5). However, we note that miR-277-34 mutant flies had a significantly lower BCAA content relative to the w control at the point of time of injection (time 0, Figure 5). This suggests that BCAA catabolism was elevated in the mutant in the absence of infection, consistent with a previous study that demonstrated that overexpression of miR-277 negatively impacted on BCAA catabolism (Esslinger et al. 2013). Therefore, we propose that BCAA catabolism is constitutively active in the miR-277-34 mutant, and that resolvable differences relating to infection are consequently lost. However, the data demonstrate that systemic infection stimulates the catabolism of BCAAs.
Figure 5.
Systemic C. albicans infection contributes to lowering the total level of BCAAs. The total level of BCAAs within the w background and miR-277-34 mutant were measured at 0–24 hr following injection of PBS or C. albicans. (A) There is a decrease over time when w flies are injected with either PBS or C. albicans (P < 0.01 in both cases), and this resolves to a significant difference in levels at 24 hr, being more extensive following infection (P < 0.01). (B) This is similar to that observed with the w background (over time P < 0.05 in both cases), except there is no difference in BCAA levels come 24 hr (P > 0.2). Data presented as the mean of three independent biological experiments (n = 3/95% C.I.s); repeated-measure two-way ANOVA and Bonferroni correction were applied to determine statistical differences. BCAA, branch-chained amino acid; 95% CI, 95% Confidence interval.
Discussion
In animal tissues, miRNAs regulate many aspects of innate and adaptive immunity, functioning within a variety of network motifs that provide robustness to biological processes through buffering or setting thresholds for protein production or cell signaling (Ebert and Sharp 2012; Chen et al. 2013; Mehta and Baltimore 2016). Here, we have used D. melanogaster as a model system to explore the relationship between miRNAs and the response to systemic infection at the whole-organism level.
The miRNA biogenesis pathway
We first observed that RNAi knockdown of Exp5 or overexpression of Ago-1, manipulations that alter the miRNA profile of human cell lines (Maudet et al. 2014; Hata and Kashima 2016), led to immune deficiency phenotypes. Although survival profiles were indistinguishable when either component was manipulated specifically in fat body tissue, differences were seen regarding their effect on the level of Drs transcript; with Ago-1 overexpression, Drs was not induced in response to infection—behaving equivalently to a Toll pathway mutant—whereas Drs transcript level was comparable to the control with RNAi knockdown of Exp5. This suggests that the mechanism through which they exert their effect is different. One possibility is that Ago-1 impacts directly on a component of the Toll pathway, as has been described for Dcr-2, a key factor in the biogenesis of small interfering RNAs (Wang et al. 2015). These authors showed that Dcr-2 binds directly to the mRNA of Toll and promotes production of the receptor; consequently, Dcr-2 mutant flies are more sensitive to infection and exhibit reduced stimulation of Toll. The fact that knockdown of Exp5 causes enhanced susceptibility to systemic C. albicans infection, without appearing to affect the output of Toll signaling, suggests that other factors are dysregulated that otherwise contribute to promoting survival; this may be a consequence of broad changes to the miRNA profile leading to widespread dysfunction of host physiology.
Immune phenotypes of miRNA mutants
Our assessment regarding the contribution of individual or small clusters of miRNAs in regulating events during the infection process revealed that miR-277 and miR-34 are likely to have opposing effects on Toll signaling (Figure 6). Compromising the function of miR-277 within the immune tissues led to significant overproduction of the Toll-dependent transcripts Drs and IM1 in response to systemic C. albicans infection, and regarding the latter, this was also true for PBS injection. Given that infection-induced production of the IM1 transcript is exclusively dependent on the Toll receptor ligand Spz (De Gregorio et al. 2001, 2002), and that the transcript does not contain binding sites for miR-277 in its 3′-UTR or ORF, it seems most likely that Toll signaling is overactive in the absence of miR-277; this implies that the miRNA functions to reduce signaling through the pathway. Thus, the strong IM1 response following PBS injection when miR-277 function is compromised (or strong Drs response following sterile injury of the miR-277-34 mutant) may result from an inability to downregulate a known stimulation in the production of AMP transcripts that occurs early, and at a low level, in response to pricking alone (Lemaitre et al. 1997). This suggests that miR-277 may set a threshold to prevent robust stimulation of Toll signaling in response to low-level stimuli. In this regard, it could participate in a mechanism similar to that described for miR-146, which is proposed to regulate signaling through Toll-like receptor (TLR)4 at subinflammatory levels (Schulte et al. 2013). In contrast, production of IM1 transcript within the miR-277-34 mutant was comparable to that of the control following PBS injection, and actually compromised in response to infection, suggesting that miR-34 functions downstream of miR-277 to promote signaling through the Toll pathway. This would explain why the miR-277-34 mutant is susceptible to infection and does not efficiently control pathogen number compared to a control strain. An explanation as to why the level of the Drs transcript is overproduced in this mutant following PBS injection or infection may be direct targeting of the transcript, since it has a putative binding site for miR-277. Therefore, in the absence of miR-34, signaling through the Toll pathway would be compromised but the Drs transcript would accumulate due to concomitant loss of miR-277-dependent degradation. An alternative explanation is that other dysregulated pathways converge on the Toll pathway to induce Drs expression. For example, in Drosophila S2* cell culture, stimulating the Ecdysone receptor in the presence of diaminopimelic acid-type peptidoglycan induces production of Drs mRNA, and this requires the transcription factor Br-C (Rus et al. 2013). Relevant here is the recent observation that overexpression of miR-34 leads to reduced abundance of the Br-C protein (Xiong et al. 2016). Therefore, it may be that the level of Br-C is elevated in the miR-277-34 mutant, and that this is somehow provoked to induce the Drs transcript in response to pricking and/or infection.
One reason why miR-277 and miR-34 would exert an opposing influence on Toll signaling may be that they have coevolved to regulate the response to infection, since miR-277 originates from miR-34 (Marco et al. 2013). This is also true for miR-317, the mutant of which responds in a manner equivalent to that of the miR-277-34 mutant regarding infection. The elevated level of the Drs transcript observed within the miR-317 mutant in response to systemic C. albicans is consistent with data showing that overexpression of miR-317 leads to reduced production of the transcript following Micrococcus luteus inoculation, and with miR-317 being strongly predicted to target Pellino, a negative regulator of the Toll pathway (Ji et al. 2014; Li et al. 2017b). That miRNAs do indeed regulate signaling through the Toll pathway has been demonstrated for miR-958 (also identified in our microarray analysis), which when overexpressed in flies leads to a reduction in the levels of Toll and Dif protein and delays the immune response (Li et al. 2017a). Only within the miR-193 mutant did the Drs transcript continue to increase across the course of infection to a level higher than that of the control, and this was associated with enhanced survival and lack of pathogen growth, traits indicative of increased resistance. Given its impact on suppressing the level of the Drs transcript, it may be that miR-193 acts as a broad limiter after an inflammatory response in a manner analogous to miR-155 and signaling through TLR4 (Schulte et al. 2013). The miR-972-973-974 mutant has a complex phenotype, in that (following both PBS injection and infection) it exhibits enhanced survival, pathogen number, and a level of Drs above that of the control across the course of infection, and even at the point of injection. It may be that members of this cluster regulate Toll homeostasis and coordinate its response to infection. Involvement in Toll homeostasis is akin to the role of mir-8, which has been shown to suppress Toll signaling in adult fat body tissue under steady-state conditions, potentially by blocking the production of Toll (Choi and Hyun 2012; Lee and Hyun 2014). Regarding response to infection, miR-972 is predicted to target 18 genes from the Hippo signaling pathway, which is involved in regulating the level of cactus mRNA and promoting Drs production and survival in response to S. aureus infection (Liu et al. 2016).
On a final note, all the susceptible mutants exhibited an increase in pathogen number despite an elevated level of Drs transcript (except miR-276b regarding Drs, which was equivalent to the control). For the miR-277-34 mutant, this may be explained by a confounding interaction between miR-277 and miR-34, but for miR-317 and miR-276b only a single miRNA is deleted. One aspect to consider when interpreting miRNA data is that changes to even a single miRNA can affect the levels of thousands of proteins, often in a complementary manner (Selbach et al. 2008). This phenomenon explains how a single miRNA can affect a plethora of biological functions. For example, mir-8 has been shown to modulate the activity of the EGFR, Notch, and Toll pathways, act as a link between ecdysone and body growth, and affect motor neuron innervations of muscle (Carthew et al. 2017). Therefore, the disassociation between pathogen number and Toll signaling may be a consequence of a global dysfunction, where pathways that work in combination with Toll to protect the host from systemic infection are disrupted. Alternatively, other pathways that work independently of Toll to maintain general homeostasis/health may be affected.
We show, within the susceptible miR-277-34 mutant, that the total amount of BCAAs is reduced compared to the control, consistent with previous findings implicating miR-277 as a key factor in controlling the catabolism of BCAAs (Esslinger et al. 2013). After 24 hr following PBS injection or infection, the amount of BCAAs was further reduced to a nominal level, thereby precluding conclusions regarding the latter. A possible explanation for this is that the shift in temperature—from 25 to 30°—accelerates metabolism, thereby driving the catabolism of BCAAs, given that its breakdown products are substrates for the TCA cycle (2016). In control flies, systemic C. albicans infection contributes to acceleration of the breakdown of BCAAs. Therefore, it is possible that signaling through Toll regulates BCAA metabolism by exerting an inhibitory effect on miR-277, thereby stimulating metabolism to provide energy to fight the infection; we note that miR-277-5p continually declines following S. aureus inoculation, which would theoretically release the repression on BCAA catabolism during infection. Alternatively, it may be that releasing a brake on metabolism, through the removal of miR-277, impacts positively on the outcome of Toll signaling; it has been seen in both mammals and Drosophila that alterations to metabolism can modulate immune signaling, with the two processes being antagonistic. A simple test for the latter would be to reduce energy production through manipulating the TCA cycle in a miR-277 mutant, which, if the model is correct, should decrease the output of Toll signaling. Consistent with the connection between BCAAs and immune function, evidence shows that when they are lacking in the human diet, many aspects of immunity are impaired and susceptibility to pathogens is enhanced (Zhang et al. 2017).
Genomic approaches to decipher miRNA involvement in systemic infection
In an attempt to identify miRNAs responsive to systemic infection, we performed a time course experiment on flies infected with different pathogens or injected with PBS, quantifying the level of miRNAs from whole-body extracts via microarray analysis. Approximately 90% of fold changes for infected over PBS were >0.5 and <2, including for the statistically separated (P < 0.1) miRNAs. Although it is not unusual to see significant changes of this magnitude, for example, in cultured cells following infection with different pathogens (Jin et al. 2014; Siddle et al. 2015), changes greater in magnitude are not uncommon. For example, the NF-ĸB-dependent expression of miR-155 (Ceppi et al. 2009), which participates in an incoherent feedforward loop to regulate TLR signaling in hematopoietic cells, increased >20-fold in response to a variety of immune stimuli (Ryu et al. 2006; O’Connell et al. 2007; Siddle et al. 2015). More pertinent, a small-RNA-sequencing study identified 12 mature miRNA forms that were differentially expressed [greater than twofold (log2)] following systemic M. luteus infection in D. melanogaster; overall, 32 miRNAs were shown to be downregulated and 61 upregulated, with the vast majority of changes occurring 24 hr following infection (Li et al. 2017b). Notably, miR-34-3p/miR-277-3p/miR-311-3p/miR-973-3p were included in the upregulated miRNAs, and miR-34-5p/miR-276b-5p/miR-277-5p/313-5p/miR-317-3p and miR-317-5p were included in those that were downregulated. Regarding our miRNA microarray data, in general, we did not find consistency between miRNAs whose profiles changed over time for a particular pathogen and those differentially expressed relative to PBS. Therefore, we speculate that a critical aspect in the response to infection for some miRNAs is to maintain a relatively constant slope of production over time. This seems reasonable considering the time series heat maps, where the majority of statistically separated miRNA profiles switch from upregulated to downregulated (based on Z-score, thus changes given relative to the associated data set), or vice versa, at the later time points. Another consideration is that PBS injection may have an inconsistent effect on miRNA profiles; as such, their production may be similar to that for infected flies, but they will not be defined over time. It would be useful to know how miRNA profiles change in untreated flies. An interesting aside is that the switching at later time points (or general direction of the slope over time) may be a priming mechanism for a rapid response to further infection, once production of immune-stimulated transcripts has returned to basal level. For example, miR-958-5p and miR-34-5p tend to slope downward and upward, respectively, over the course of infection. Regarding the action of miR-958, this would in theory lead to increased production of Toll and Dif protein, whereas for miR-34 this would increase signaling through the Toll pathway.
The majority of mature miRNA forms (297 of 425) were not detected above background across the time course for PBS injected or infected flies, including both arms for members of the miR-972-973-974 and miR-959-964 clusters. This is notable given that deleting the former, or four members of the latter (Vodala et al. 2012), alters the response to systemic infection. More specifically, members of the miR-959-964 cluster were shown to cycle – albeit at a relatively low abundance – via circadian transcriptional control, where expression was most prominent in the adult head fat body. Significantly, when this cluster was partially deleted, the time during the light:dark cycle at which flies were most susceptible to gram-negative infection was shifted (Vodala et al. 2012). That we did not observe enhanced susceptibility of this mutant to C. albicans may be partly explained therefore, by the timing of our infections; however, this phenomenon is likely to be a rare event, since most miRNAs do not exhibit circadian oscillations (Vodala et al. 2012). These observations bring to attention the dynamism and specificity of miRNA expression, and that having mature miRNAs at low abundance does not preclude important functional output. Considering this, using quantifying techniques with greater sensitivity (for example Small-RNA-Sequencing) and analyzing particular tissues or cells from infected flies, is likely to be more revealing. More so, if immune deficient flies are analyzed in parallel. Reporter constructs with rapid-turnover fluorescent proteins would also be a useful tool in the study of miRNAs. Therefore, in relation to immunity, we have provided a genome-wide map of potential NF-κB-binding sites that reside in close proximity to miRNA genes. However, we should be cautious when relating strength of those potential sites and role in immunity as there was no such correlation in our findings.
Conclusions
We have identified six miRNA allelic mutant backgrounds that affect survival in response to systemic infection, the ability to control pathogen number, and production of Toll pathway-dependent transcripts. Furthermore, we reveal infection contributes to promoting BCAA catabolism, and through the susceptible miR-277-34 mutant, identify a potential connection between BCAA catabolism and Toll signaling. Future studies carefully detailing the expression, tissue-specific requirement, and functional interactions between these miRNAs and their targets, will undoubtedly reveal new insight into the mechanisms required to cope with systemic infection.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.196584/-/DC1.
Acknowledgments
We thank Stephen Cohen, Eric Lai, Bruno Lemaitre, and the Bloomington Drosophila Stock Centre for fly stocks. This work was supported by a European Molecular Biology organization Postdoctoral Fellowship [Long-Term-Fellowship (ALTF) 782-2012 to M.L.A.], a Consolidator Grant from the European Research Council (grant no 310912 “Droso-parasite” to P.L.), and grant BB/K003569/1 from the Biotechnology and Biological Sciences Research Council (to P.L.).
Author contributions: P.L., M.L.A., and M.G. designed experiments; M.L.A., M.G., and A.M. conducted experiments; M.L.A., M.G., A.M., and P.L. analyzed data; R.C. contributed the data in Figure S2 and Table S4; and M.G. wrote the manuscript with input from P.L.
Footnotes
Communicating editor: B. P. Lazzaro
Literature Cited
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P., 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F., Smibert P., Lai E. C., 2010. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 338: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C., Leslie C., 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon N., Silverman N., Cherry S., 2014. Immunity in Drosophila melanogaster: from microbial recognition to whole organism physiology. Nat. Rev. Immunol. 14: 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse M. S., Arnold C. P., Towb P., Katrivesis J., Wasserman S. A., 2007. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. 26: 3826–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Agbu P., Giri R., 2017. MicroRNA function in Drosophila melanogaster. Semin. Cell Dev. Biol. 65: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Kadow I. G., Zhan X., Okamura K., Suh G. S., et al. , 2008. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., et al. , 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 106: 2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Z., Schaffert S., Fragoso R., Loh C., 2013. Regulation of immune responses and tolerance: the microRNA perspective. Immunol. Rev. 253: 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. W., Song S., Weng R., Verma P., Kugler J. M., et al. , 2014. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell 31: 784–800. [DOI] [PubMed] [Google Scholar]
- Choi I. K., Hyun S., 2012. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 37: 50–54. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P. T., Rubin G. M., Lemaitre B., 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 98: 12590–12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P. T., Tzou P., Rubin G. M., Lemaitre B., 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21: 2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Sharp P. A., 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., John B., Gaul U., Tuschl T., Sander C., et al. , 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger S. M., Schwalb B., Helfer S., Michalik K. M., Witte H., et al. , 2013. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 10: 1042–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullaondo A., Lee S. Y., 2012. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 36: 267–273. [DOI] [PubMed] [Google Scholar]
- Garbuzov A., Tatar M., 2010. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly (Austin) 4: 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P., Naitza S., Kappler C., Ferrandon D., Zachary D., et al. , 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1: 503–514. [DOI] [PubMed] [Google Scholar]
- Grun D., Wang Y. L., Langenberger D., Gunsalus K. C., Rajewsky N., 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Kim V. N., 2014. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Hata A., Kashima R., 2016. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 51: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlicka L., Gibson M., Kiger A., Micchelli C., Schober M., et al. , 2002. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis 34: 51–57. [DOI] [PubMed] [Google Scholar]
- Jakob L., Treiber T., Treiber N., Gust A., Kramm K., et al. , 2016. Structural and functional insights into the fly microRNA biogenesis factor Loquacious. RNA 3: 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S., Sun M., Zheng X., Li L., Sun L., et al. , 2014. Cell-surface localization of Pellino antagonizes Toll-mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat. Commun. 5: 3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Ibeagha-Awemu E. M., Liang G., Beaudoin F., Zhao X., et al. , 2014. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genomics 15: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E., 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Kim B., Kim V. N., 2016. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 113: E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidis I., Ligoxygakis P., 2012. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2: 1200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Sahu S. K., Kumar R., Subuddhi A., Maji R. K., et al. , 2015. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 17: 345–356. [DOI] [PubMed] [Google Scholar]
- Lee G. J., Hyun S., 2014. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev. Comp. Immunol. 45: 245–251. [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Nakahara K., Pham J. W., Kim K., He Z., et al. , 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Reichhart J. M., Hoffmann J. A., 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94: 14614–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li Y., Shen L., Jin P., Chen L., et al. , 2017a miR-958 inhibits Toll signaling and Drosomycin expression via direct targeting of Toll and Dif in Drosophila melanogaster. Am. J. Physiol. Cell Physiol. 312: C103–C110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Li R., Xu J., Jin P., et al. , 2017b Genome-wide miRNA screening reveals miR-310 family members negatively regulate the immune response in Drosophila melanogaster via co-targeting Drosomycin. Dev. Comp. Immunol. 68: 34–45. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P., Bulet P., Reichhart J.-M., 2002. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defence of Drosophila. EMBO Rep. 3: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zheng Y., Yin F., Yu J., Silverman N., et al. , 2016. Toll receptor-mediated Hippo signaling controls innate immunity in Drosophila. Cell 164: 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A., Ninova M., Ronshaugen M., Griffiths-Jones S., 2013. Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res. 41: 7745–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudet C., Mano M., Eulalio A., 2014. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 588: 4140–4147. [DOI] [PubMed] [Google Scholar]
- McKean K. A., Lazzaro B. P., 2011. The costs of immunity and the evolution of immunological defense mechanisms, pp. 299–310 in Mechanisms of Life History Evolution: The Genetics and Physiology of Life History Traits and Trade-Offs, edited by Flatt T., Heyland A. Oxford University Press, New York. [Google Scholar]
- Mehta A., Baltimore D., 2016. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 16: 279–294. [DOI] [PubMed] [Google Scholar]
- Morante J., Vallejo D. M., Desplan C., Dominguez M., 2013. Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev. Cell 27: 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D., 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104: 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara S. P., Splinter P. L., Gajdos G. B., Trussoni C. E., Fernandez-Zapico M. E., et al. , 2010. NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J. Biol. Chem. 285: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M. C., 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarerin-George A. O., Anton L., Hwang Y. C., Elovitz M. A., Hogenesch J. B., 2013. A functional genomics screen for microRNA regulators of NF-kappaB signaling. BMC Biol. 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Zhang Z., Liang J., Ge Q., Duan X., et al. , 2011. The full-length transcripts and promoter analysis of intergenic microRNAs in Drosophila melanogaster. Genomics 97: 294–303. [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., et al. , 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus F., Flatt T., Tong M., Aggarwal K., Okuda K., et al. , 2013. Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J. 32: 1626–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazansky S. S., Gvozdev V. A., Berezikov E., 2011. Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 12: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J. H., Ha E. M., Oh C. T., Seol J. H., Brey P. T., et al. , 2006. An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 25: 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schnall-Levin M., Zhao Y., Perrimon N., Berger B., 2010. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc. Natl. Acad. Sci. USA 107: 15751–15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D. S., Ayres J. S., 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte L. N., Westermann A. J., Vogel J., 2013. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 41: 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., et al. , 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Senger K., Armstrong G. W., Rowell W. J., Kwan J. M., Markstein M., et al. , 2004. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13: 19–32. [DOI] [PubMed] [Google Scholar]
- Siddle K. J., Tailleux L., Deschamps M., Loh Y. H., Deluen C., et al. , 2015. Bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS Genet. 11: e1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedel C., Darfeuille F., 2013. MicroRNAs and bacterial infection. Cell Microbiol. 15: 1496–1507. [DOI] [PubMed] [Google Scholar]
- Sun K., Westholm J. O., Tsurudome K., Hagen J. W., Lu Y., et al. , 2012. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 8: e1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K. D., Boldin M. P., Chang K. J., Baltimore D., 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G. G., Wang W. H., Dai Y., Wang S. J., Chu Y. X., et al. , 2013. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS One 8: e56709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. C., Vardinogiannis I., Gilmore T. D., 2013. Identification of an NF-kappaB p50/p65-responsive site in the human MIR155HG promoter. BMC Mol. Biol. 14: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I. S., Zagganas K., Paraskevopoulou M. D., Georgakilas G., Karagkouni D., et al. , 2015. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 43: W460– W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodala S., Pescatore S., Rodriguez J., Buescher M., Chen Y. W., et al. , 2012. The oscillating miRNA 959–964 cluster impacts Drosophila feeding time and other circadian outputs. Cell Metab. 16: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Cambi F., 2012. MicroRNA expression in mouse oligodendrocytes and regulation of proteolipid protein gene expression. J. Neurosci. Res. 90: 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu D., Liu Y., Xia X., Gong W., et al. , 2015. Drosophila Dicer-2 has an RNA interference-independent function that modulates Toll immune signaling. Sci. Adv. 1: e1500228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R., Cohen S. M., 2012. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development 139: 1427–1434. [DOI] [PubMed] [Google Scholar]
- Xiong X. P., Kurthkoti K., Chang K. Y., Li J. L., Ren X., et al. , 2016. miR-34 modulates innate immunity and ecdysone signaling in Drosophila. PLoS Pathog. 12: e1006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. S., 2016. The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients 8: E405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zeng X., Ren M., Mao X., Qiao S., 2017. Novel metabolic and physiological functions of branched chain amino acids: a review. J. Anim. Sci. Biotechnol. 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Hu G., Gong A. Y., Chen X. M., 2010. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 38: 3222–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The miRNA mutant strain collection from the Cohen Lab and the mir sponge lines are available from the Bloomington Drosophila Stock Center. All P-values for Figure 6 can be found in Supplemental Material, Table S2. The microarray data are presented in Figure 4 for miRNA that presented statistically significant regulation and in Figure S3 for all others; Table S5 includes a list of P-values for the microarray analysis. Table S4 has a list of all miRNAs that have at least one consensus NF-κB-binding site, while Figure S4 summarizes the numbers of miRNAs with such site(s).
Figure 6.
Summary of phenotypes for selected miRNA mutant backgrounds. The table presents a summary for each miRNA mutant regarding changes to pathogen number (CFUs) and level of Drs transcript over time (see Figure 2, B and C). Direction of arrows in the CFUs column and both Drs columns (PBS and C. albicans) indicate how the measured variable changes over time for the particular miRNA background; a dash indicates there was not a significant change. The arrow/dash color represents whether the change is significantly greater (orange) than that of the given w or yw control (gray) or significantly below (light orange) that of the given control; if the variable does not separate significantly from the given control the arrow/dash is given as gray. For details regarding P-values see Table S2. CFU, colony forming unit; miRNA, microRNA; n/a, not applicable.
Figure 4.
Differential production of mature miRNA forms. Wild-type flies were injected with PBS or one of the given pathogens: E. coli (E. c), S. aureus (S. a), or C. albicans (C. a). Their miRNA complement was quantified at time points T0, T1, T7.5, T18, and T60 (numbers refer to hours following injection). Data are compared for all factors at each individual time point to identify mature miRNA forms whose production is differentially regulated; ANOVA-selected miRNAs (P < 0.1) are presented as clustered heat maps, where negative Z-values (green) indicate downregulation relative to the data set and positive Z-values (red) upregulation. Except with E. coli (n = 2), the infection experiments were repeated independently on three occasions (n = 3), indicated above the heat maps as A, B, and C. miRNA, microRNA.