ABSTRACT
Latitudinal clines in circadian rhythms have consistently been described in various plant species, with the most recent examples appearing in soybean cultivars and in monkey flower natural populations. These latitudinal clines provide evidence that natural variation in circadian rhythms is adaptive, but it is still unclear what adaptive benefits this variation confers, particularly because circadian rhythms are not usually measured in day/night conditions that reflect those experienced by organisms in nature. Here, we report that daily rhythms of GIGANTEA expression respond to day length in a way that depends on the latitude of origin of Arabidopsis accessions. We additionally extend previous findings by confirming that natural variation in GI expression affects growth related traits, and alters the expression of different target genes. The results support the idea that natural variation in daily rhythms of expression have broad effects on plant development and are of potential adaptive value.
KEYWORDS: Circadian clock, day night cycles, GIGANTEA, latitudinal cline, PSEUDO RESPONSE REGULATOR 9, rhythms of gene expression, stem growth
Most organisms have adapted to the daily rotations of the earth by evolving temporal regulation mechanisms that generate internal circadian rhythms. Circadian rhythms provide an advantage in cycling environmental conditions as they allow for biologic processes to be promoted at the most favorable times of the day.1,2 Circadian rhythms have been detected in all kingdoms of life, and the extensive list of processes known to be under their control is still expanding. In plants, temporally regulated processes include traits of agricultural value such as flowering time, growth and stress resistance.3 Manipulating circadian rhythms might therefore be a way of improving crop performance in different environments.4 Natural variation in circadian rhythms has been described in natural genotypes of several plant species,5,6,7 and this variation was shown to follow a latitudinal gradient in Arabidopsis thaliana.5 A new study now describes similar latitudinal gradients in soybean (Glycine max) and the monkey flower (Mimulus guttatus).7 These latitudinal clines provide evidence that circadian rhythms are adaptive, but several questions still need to be addressed before we understand how natural variation in circadian rhythms provides benefits in specific environments. Rhythms need to be studied in day/night conditions that resemble the conditions that plants experience in nature, not only in artificial continuous conditions usually used to determine circadian parameters. Moreover, a broader view of the molecular outputs and phenotypes that vary as a consequence of these changes in rhythms is required to know what benefits changes in rhythms confer.
Within day/night cycles, rhythms are precisely controlled by the circadian clock and by external inputs from the environment, e.g. light and temperature. These external inputs can directly influence rhythms by, for instance, regulating the timing and amplitude of gene expression during the day. External inputs can also influence rhythms indirectly through complex interactions with the circadian system. An example of these interactions is the control by light and temperature of the pace of the circadian clock (circadian period length). Another example is the resetting of the circadian system by environmental transitions at dawn and dusk. Resetting ensures that the period of an oscillation is synchronized to the 24-hour external cycle. A consequence of resetting is that period length and other properties of the oscillator can only be determined in artificial constant conditions in which plants are relieved from the influence of environmental transitions that reset the circadian system. Latitudinal clines in period length have been reported,5,7 but interpreting these clines requires assessing the impact of changes in period length on circadian rhythms measured during day/night cycles where the influence of external inputs plays an important role. Studies that have extensively surveyed natural variation in the timing (phase) of biologic rhythms during day/night cycles are scarce,8 and tend to agree with data showing that phase and period do not necessarily correlate in natural genotypes.5,8 Why phase and period can vary independently might at least partly be explained by the direct influence of external inputs on particular rhythms,8 a phenomenon that in circadian terminology is called masking.9,10,11 These observations do not imply that changes in period length never influence phase, but they nevertheless emphasize the need to measure natural variation of circadian rhythms during day/night cycles that more closely reflect what plants experience in their natural environment.
Natural variation in circadian rhythms during the day was extensively surveyed in a collection of 77 Arabidopsis accessions transformed with a circadian reporter that allowed monitoring of rhythms at high temporal resolution and in different photoperiods.8 The marker consisted of the GIGANTEA promoter fused to the firefly Luciferase cDNA (GI::LUC). GIGANTEA is a pleiotropic gene whose rhythm of expression is characterized by a peak in the evening that is delayed by long day photoperiods (Fig. 1A).12,13,14 GI::LUC expression precisely follows the temporal expression pattern of the endogenous gene, and is an established circadian marker that displays robust rhythms in a variety of conditions.15,16,17 Natural variation in the peak time of GI expression (hereafter called GI peak time or GI phase) was mostly detected when the accessions were grown in long day (LD) photoperiods of 12 to 16 hours, but was reduced when the accessions were grown in short days (SD).8 In the current study, we used the same rationale than in previous works describing natural variation of circadian rhythms,5,7 and asked whether the timing of GI expression measured in LDs depended on the geographical origin of Arabidopsis accessions. For this analysis we took into account that a major feature of the regulation of GI was its sensitivity to photoperiod, defined as the delay of GI expression by long day lengths (Fig. 1A). We reasoned that the sensitivity to photoperiod better reflects what plants experience in their natural habitats where photoperiod is not constant but changes dynamically. The rate of day length change varies depending on the latitude, with day length increasing faster during spring in northern latitudes compared with latitudes closer to the equator. Natural variation in the sensitivity to photoperiod of agronomical traits has been described in plants, and was proposed to contribute to yield improvement in different environments.18,19 Here, we tested if the sensitivity of GI to photoperiod depended on the latitude of origin of Arabidopsis accessions.
Figure 1.
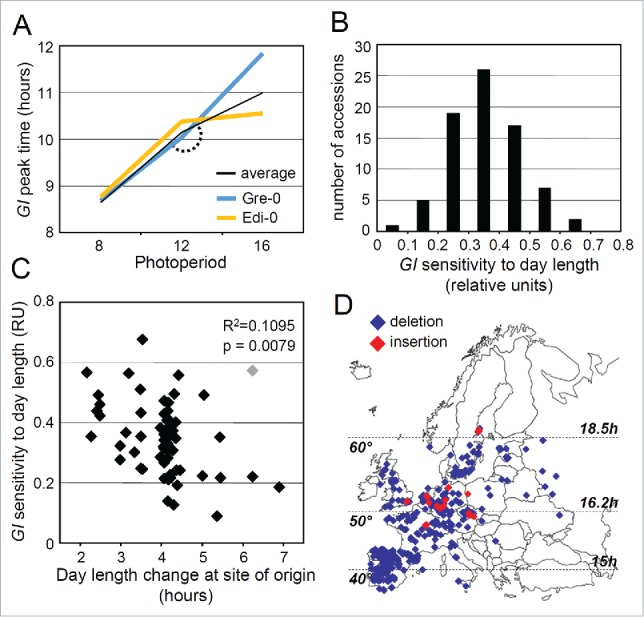
Arabidopsis accessions from northern latitudes show reduced sensitivity of GI expression to changing photoperiods. The measurement of GI peak time in the accessions and the description of how the transgenic lines were obtained are described in.8 (A) GI peak time in the 2 accessions Gre-0 and Edi-0 grown in SDs of 8 h, LDs of 12 h and LDs of 16 h. These 2 accessions were chosen to illustrate the variation in the response of GI to photoperiod changes from LDs of 12 h to LDs of 16 h. (B) For each accession, the sensitivity of GI to photoperiod (from LDs of 12 to 16 h) was estimated by calculating the absolute peak time difference between both day lengths, but also by calculating the relative peak time variation and the angle of the response as described in Table 1. The angle of the response is illustrated by the arc in A. The distribution of the relative GI peak time variation is shown in B. (C) Relative GI peak time variation negatively correlates with the day length changes that the accessions experience in spring at their site of origin (between the 21st of March and the 21st of June). Results of the correlations presented on the plot include one outlier (in gray on the plot). When the outlier was removed the results of the Pearson test were R2 = 0.1936 and p = 0.0003. The day length data were determined based on the geographical coordinates of each accession obtained from.44 Only the 63 accessions whose geographical origin had been tested in44 were included in the analysis. (D) Geographical distribution of the PHYB indel. The accessions used in this analysis included the 77 accessions described in,8 accessions from the Hapmap collection,45 and iberian accessions described in.26 In total 576 accessions were tested for the presence of the insertion, out of which 228 were from latitudes below 47° N. Only European accessions were considered, with the coordinates of the most eastern accessions being 38°28′ E. In house genotyping was necessary to verify the presence of the insertion because the available resequencing data of Arabidopsis accessions (http://www.1001genomes.org/) in most cases failed to detect the deletion. The presence of the insertion was tested by PCR using primers PHYBfw (5′–3′: TTCACCCTAAATCCTTCCTTGTCTC) and PHYBrev (5′–3′: CGTCGTCGTTTTGAGTGATTGTG). Horizontal dashed lines indicate latitudes 40°, 50° and 60°. The corresponding day lengths at summer solstice are indicated on the right of the panel.
Natural variation of GI phase in the accession had originally been detected in LD photoperiods but not in SDs,8 so we estimated the sensitivity of GI to lengthening spring photoperiods by calculating the delay of GI peak time from 12 to 16 hour photoperiods. 12 hours is the day length that the 77 accessions would experience in their native European environment at the spring equinox (21st of March), and 16 hours lies within the range of photoperiods that the accessions would experience at the summer solstice (21st of June). The sensitivity of GI expression to changing day lengths was estimated in multiple ways for each accession. First, we calculated the absolute peak time difference between LDs of 16 h and LDs of 12 h. The lowest and highest values of absolute peak time differences were detected in the Edi-0 accession (0.18 h) and in Gre-0 (1.80 h), respectively (Fig. 1A). Second, we calculated a relative estimate of photoperiod sensitivity based on a method described elsewhere,18,19 and which consisted in normalizing the peak time difference between 12 h and 16 h day lengths to the total peak time difference between 8 h and 16 h day lengths. The distribution of this relative estimate is shown in Fig. 1B. Finally, we calculated the angle of the response, as represented by the arc in Fig. 1A. We found that GI sensitivity to day length significantly correlated with latitude and with the day length variation that each accession experiences at its site of origin in spring (calculated between the 21st of March and the 21st of June) (Fig. 1C, Table 1). The correlations were significant with the 3 estimates of GI sensitivity to day length but were stronger with the relative estimate and the angle of the response, suggesting that these estimates were better predictors of how GI expression responds to lengthening photoperiods (Table 1). In summary, latitudinal clines in circadian period length have been observed in various plant species, but we now show that the temporal regulation of a rhythm measured during day/night cycles can also vary with latitude in an extensive collection of Arabidopsis accessions.
Table 1.
GI sensitivity to day length significantly correlates with the latitude or with the day length variation at the site of origin of the accessions. The correlation coefficient (R) and the p value (p) were determined with the Pearson test. The absolute GI peak time difference is the absolute difference between GI peak times measured in LDs of 16 h and LDs of 12 h. The relative GI peak time variation was obtained by normalizing the peak time differences between LDs of 12 and LDs of 16 h to the total variation from SDs of 8 h to LDs of 16 h (illustrated in Fig. 1A). The angle of the response was determined as represented in Fig. 1A. The latitudinal data were obtained from.44 Only the 63 accessions whose geographical origin had been verified were included in the analysis.44 The day length data were determined based on the geographical coordinates of each accession, and the day length variation at the site of origin was calculated between the spring equinox and the summer solstice (21st of March and 21st of June, respectively).
| Angle | Relative GI peak time variation from LDs of 12 to 16 h | Absolute GI peak time variation from LDs of 12 to 16 h | ||
|---|---|---|---|---|
| Latitude of origin | R p |
−0.361 0.004 |
−0.336 0.007 |
−0.263 0.037 |
| Day length variation at the site of origin | R p |
−0.347 0.005 |
−0.331 0.008 |
−0.25 0.049 |
The interpretation of the latitudinal cline is that the delay of GI expression in response to lengthening spring photoperiods is enhanced in southern accessions despite the rate of day length change being slower in the south. On the contrary, northern accessions tend to limit the delay of GI despite day length changing faster in the north. Arabidopsis accessions from northern latitudes might have evolved mechanisms to compensate for the faster rate of day length change that they experience at their site of origin. Such patterns of phenotypic variation known as counter-gradient variation have been widely documented in the literature.20,21 We next tested if the geographical distribution of a natural allele that regulates the timing of GI expression was consistent with the tendency of northern accessions to limit GI sensitivity to day length. The Col-0 allele of PHYTOCHROME B (PHYB) bears an insertion that advances the timing of GI expression in LDs of 16 h but not in LDs of 12 h,8 and in effect reduces the sensitivity of GI to day length. Genotyping of a large panel of accessions revealed that the insertion is exclusively present in central and northern genotypes, and is absent in southern accessions coming from below 47° N (Fig. 1D). The phenotypic effect of the Col-0 PHYB allele is weak,8,22,23 and the distribution of this allele alone cannot fully explain the geographical distribution of GI expression patterns (Fig. 1C). More experiments and data are needed to determine whether the PHYB locus is under selection, but these results nonetheless show that the Col-0 PHYB insertion is tolerated in northern accessions, and that its effect is consistent with the latitudinal cline. Most Arabidopsis accessions descend from a lineage that originates from an unknown glacial refugium, and this lineage has invaded the habitats once occupied by older “relict” populations.24,25 Relict populations can still be found in the Iberian peninsula where they co-exist with “non-relict” genotypes. The uncommon PHYB insertion was not detected in any of the 162 Iberian genotypes that were tested here and that include 20 relict accessions.26 This polymorphism likely appeared after the spread of the non-relict genotypes in Europe.
The need for accessions to reduce GI sensitivity to day length in the north and increase it in the south might be explained by deleterious phenotypic consequences of GI peak time occurring outside of an optimal time window. The peak time of GI expression is evolutionarily conserved among plant species,27,28,29 suggesting that evolutionary forces have acted to maintain the peak time of GI within an appropriate range. It is possible that maximum expression levels of GI too early in the day or during the night could have deleterious effects on phenotypes controlled by GI. Fully understanding the ecological significance of temporal GI expression patterns will require unravelling the molecular targets and phenotypes that vary as consequence of the observed changes in GI expression. This was addressed by searching for GI-regulated phenotypes that co-vary with changes in GI expression induced by natural loci.
Alterations of GI function via artificial mutations affect a wide range of phenotypes,12 but connecting changes in GI expression with downstream phenotypic variation in natural accessions is challenging due to the genetic complexity of this material. To circumvent this issue, a NIL population was generated in which combinations of QTLs that precisely regulate GI expression were introgressed in a homogeneous genetic background.8 Significant variation in the phase of GI expression was associated with precise alterations of GI expression levels in the evening, that in turn strongly correlated with hypocotyl growth.8 We looked for other traits associated with changes in GI expression in the NILs, and found that evening GI expression levels significantly correlated with the growth of the stem (Fig. 2A). Height was measured for each NIL on the day that the first flower opened, which corresponds to stem elongation before resources are allocated to flowers and fruits. This measurement of height was expected to be dependent on the flowering interval (number of days between bolting and flowering), a character that could potentially be regulated independently of growth. To distinguish between effects of GI on growth and on flowering time, we determined the growth rate of the stem by expressing height at flowering relative to the flowering interval. GI expression correlated more strongly with growth rate than with height and did not correlate with the flowering interval (Fig. 2A). Changes in GI expression therefore seem to influence growth per se and not the time available for growth before the opening of the first flower. Growth rate measurements in the gi-2 mutant further confirmed that GI is a repressor of stem growth (Fig. 2B), which is consistent with GI being a repressor of hypocotyl elongation.30 In conclusion, the stem growth experiments generally support that natural variation in GI expression is correlated with growth related traits.
Figure 2.
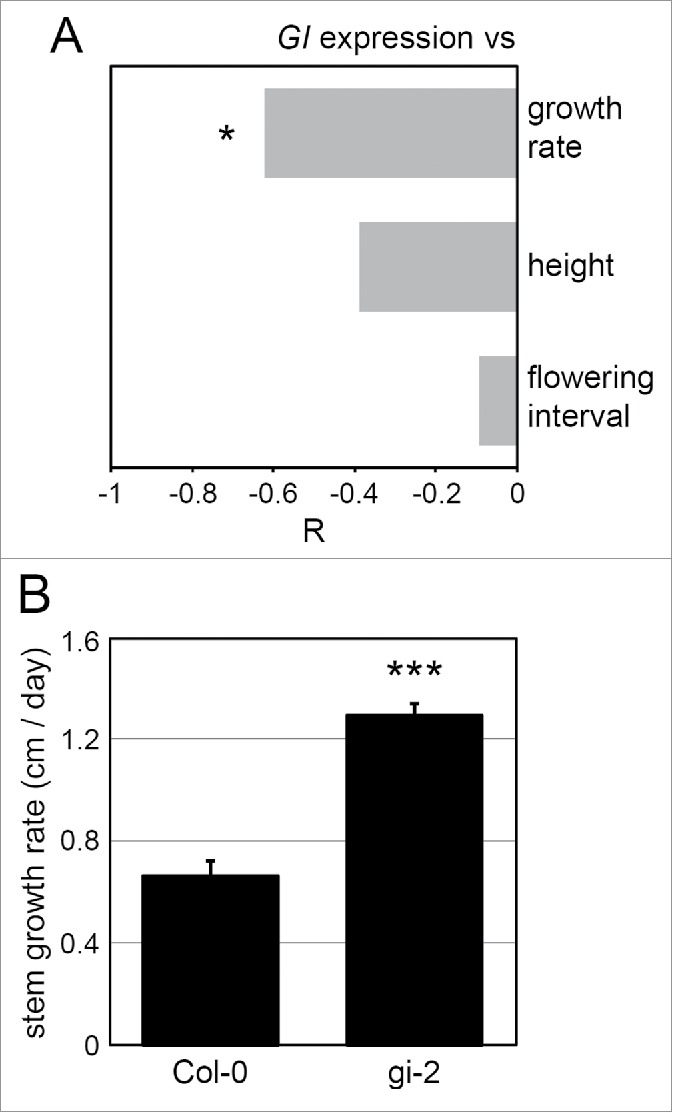
Variation of GI expression correlates with stem growth rate. Generation of the 12 NILs and measurements of GI expression with the luciferase system are described in.8 GI::LUC expression levels at the time of peak (or GI maximum expression) were obtained for each NIL from 5 independent experiments, with 12 individuals per genotype per experiment. The height of each NIL grown in LDs of 16 h was measured on the day that the first flower opened. The flowering interval, defined as the number of days between bolting and the opening of the first flower, was used to normalize the height data and to calculate stem growth rate for each line. 10 plants per NIL were used to determine stem growth. (A) GI maximum expression significantly correlates with stem growth rate but not with height or with the flowering interval. The Pearson test was used to determine the significance of the correlations. The Pearson correlation coefficient (R) indicates the strength of the correlations, 1 and −1 indicating perfect positive and negative correlations respectively. *: p ≤ 0.05. (B) Stem growth rate was determined in wild type Col-0 and in the gi-2 mutant in the same way than in the NILs (n = 10). ***: p ≤ 0.001 with a 2 tailed Student t-test (α = 0.05). Error bars represent SE of the mean.
In parallel to screening for phenotypes that co-vary with GI expression in the NILs, another approach was to identify the genes whose expression correlates with changes in temporal GI expression patterns. This approach demonstrated that changes in GI expression modify hypocotyl growth through the repression of PHYTOCHROME INTERACTING 4 (PIF4) expression,8 a transcription factor that promotes growth.31 Causality between changes in GI expression, PIF4 expression and growth was supported by genetic and molecular analyses of artificial mutants.8 It was proposed that GI might regulate PIF4 through its interaction with components of the Evening Complex (EC),32 a protein complex that represses PIF4 during the night. We now report that GI expression in the NILs also correlates with the nighttime expression of PSEUDO RESPONSE REGULATOR 9 (PRR9) (Fig. 3A), another direct transcriptional target of the EC.33,34 This tendency was confirmed by quantifying the expression of PRR9 in null mutants. Genetic experiments had demonstrated a synergistic effect of the gi-2 and phyB-9 mutations on the activation of PIF4 expression,8 and quantification of PRR9 expression in the mutants support that GI and PHYB interact to regulate PRR9 in a similar manner (Fig. 3B). Overall, the results reveal that PRR9 and PIF4 are regulated by a common GI-dependent mechanism, and argue that PRR9 is another GI regulated gene whose expression responds to precise changes in GI expression. PRR9 is a circadian clock component implicated in the regulation of metabolism and cold tolerance,35,36 and it will be interesting to test whether changes in GI expression alter these processes as well. From a broader perspective, a deeper analysis of all the genes and phenotypes regulated by GI should provide insights on how variation in daily rhythms of expression of a single gene generally impacts on downstream pathways and phenotypes. As GI is a pleiotropic gene tightly regulated by the circadian clock and external inputs, it will be a useful model gene to address such questions.
Figure 3.
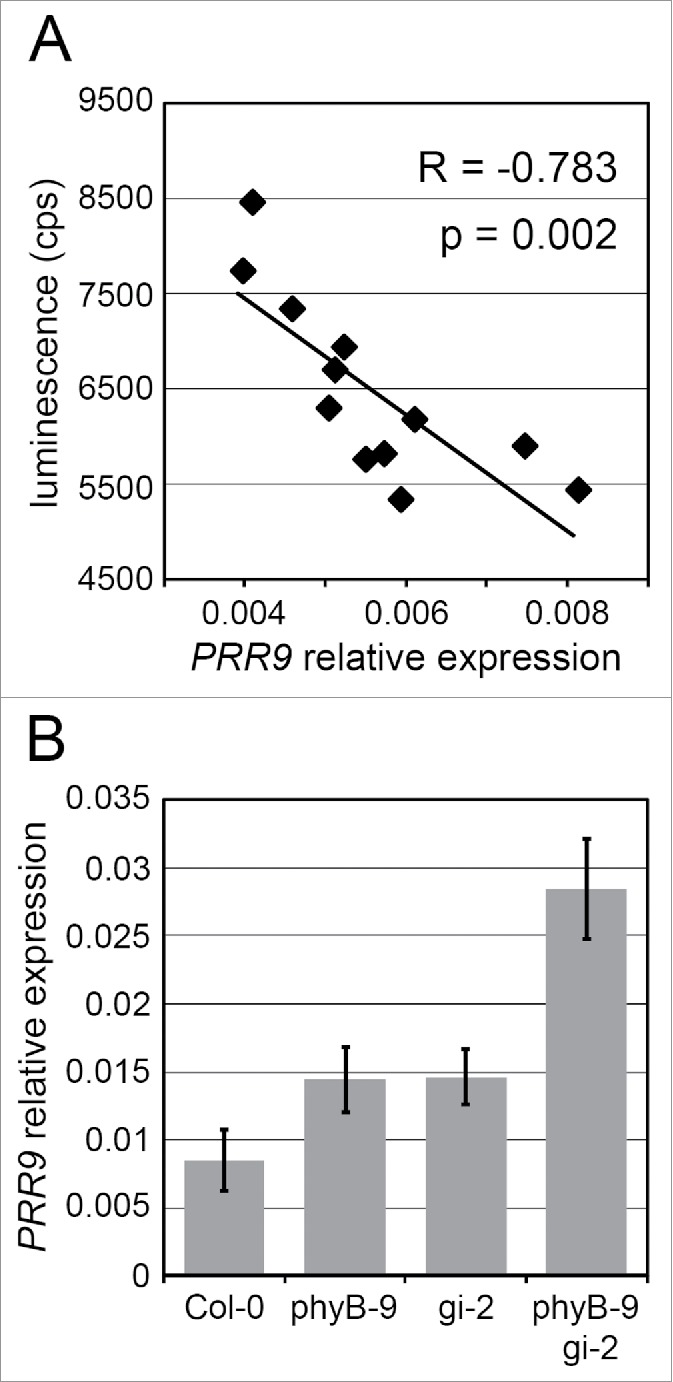
Variation of GI expression correlates with PRR9 expression. Generation of the 12 NILs and measurements of GI expression with the luciferase system are described in.8 GI::LUC expression levels at the time of peak (or GI maximum expression) were obtained for each NIL from 5 independent experiments, with 12 individuals per genotype per experiment. As described previously for PIF4,8 PRR9 mRNA levels were quantified in the different genotypes after 10 d of entrainment in LDs of 16 h, and samples were harvested 20 hours after dawn. (A) Correlation of GI maximum expression in the NILs with PRR9 mRNA. cps: counts per second. (B) PRR9 mRNA levels quantified by qRT-PCR in Col-0, phyB-9, gi-2 and phyB-9 gi-2. PCR conditions and cycles were as described in,8 and PRR9 expression was quantified by qRT-PCR using primers PRR9fw (ATCAAAAGCTTAGCCTCTCTG) and PRR9rev (CTGTGGACTGAAGAACTTGGTTAC). The isopentenyl pyrophosphate / dimethylallyl pyrophosphate isomerase (IPP2) was used as a housekeeping gene for normalization. Erros bars represent SD of 4 technical replicates.
Collectively, our data not only show that the temporal regulation of gene expression can vary depending on the latitude, but also illustrate how natural variation in temporal expression waveforms can impact on various phenotypes and on the transcription of different genes. An important aspect of our experiments is that rhythms were measured during ecologically relevant day/night cycles, in different photoperiods and in a large collection of natural accessions. Due to this combination of conditions and material, and because the timing of GI expression is sensitive to photoperiod, it was possible to estimate the sensitivity of a rhythm to changes in day length, a condition that more accurately reflects what organisms experience in their natural habitat. The latitudinal cline in the temporal regulation of GI expression is largely consistent with the growing body of evidence supporting that circadian rhythms vary with latitude. To demonstrate that temporal patterns of GI expression are adaptive, it will be necessary to find all the phenotypes that vary as a consequence of these changes in rhythms, and then ask if the phenotypic variation induced by GI provides a benefit in specific environments. Intriguingly, CONSTANS expression and flowering did not significantly correlate with GI expression in the NILs, emphasizing that not all downstream pathways are equally sensitive to changes in gene expression waveforms.8 Different sensitivities of GI outputs to alterations of GI function had already been reported in gi mutant alleles.37 Transcriptomics approaches will help identify the downstream pathways that most strongly respond to changes in daily expression patterns of GI and of other model circadian-regulated genes.6,38 Future studies will additionally need to address how much the post-translational regulation of GI and of other circadian-regulated proteins contribute to changes in the activity of downstream molecular pathways.39,40,41 Finally, ecological approaches consisting of testing the impact of natural variation of rhythms in populations grown in natural field conditions will greatly contribute to enhance our understanding of the adaptive value of circadian rhythms.42,43
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Elisa de Ansorena for skilful technical assistance.
Funding
This work was supported by core funding from the Max Planck Society.
References
- 1.Millar AJ. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 2016; 67:595-618; PMID:26653934; https://doi.org/ 10.1146/annurev-arplant-043014-115619 [DOI] [PubMed] [Google Scholar]
- 2.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 2015; 16:598-610; PMID:26370901; https://doi.org/ 10.1038/nrg3976 [DOI] [PubMed] [Google Scholar]
- 3.de Montaigu A, Tóth R, Coupland G. Plant development goes like clockwork. Trends Genet 2010; 26:296-306; PMID:20483501; https://doi.org/ 10.1016/j.tig.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 4.Gehan MA, Greenham K, Mockler TC, McClung CR. Transcriptional networks-crops, clocks, and abiotic stress. Curr Opin Plant Biol 2015; 24:39-46; PMID:25646668; https://doi.org/ 10.1016/j.pbi.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 5.Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 2003; 302:1049-53; PMID:14605371; https://doi.org/ 10.1126/science.1082971 [DOI] [PubMed] [Google Scholar]
- 6.Muller NA, Wijnen CL, Srinivasan A, Ryngajllo M, Ofner I, Lin T, Ranjan A, West D, Maloof JN, Sinha NR, et al.. Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet 2016; 48:89-93; PMID:26569124; https://doi.org/ 10.1038/ng.3447 [DOI] [PubMed] [Google Scholar]
- 7.Greenham K, Lou P, Puzey JR, Kumar G, Arnevik C, Farid H, Willis JH, McClung CR. Geographic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms 2016; 32:26-34; PMID:27920227; https://doi.org/ 10.1177/0748730416679307 [DOI] [PubMed] [Google Scholar]
- 8.de Montaigu A, Giakountis A, Rubin M, Tóth R, Cremer F, Sokolova V, Porri A, Reymond M, Weinig C, Coupland G. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc Natl Acad Sci 2015; 112:905-10; PMID:25548158; https://doi.org/ 10.1073/pnas.1422242112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muranaka T, Oyama T. Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci Adv 2016; 2:e1600500; PMID:27453946; https://doi.org/ 10.1126/sciadv.1600500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aschoff J. Exogenous and endogenous components in circadian rhythms. Cold Spring Harb Symp Quant Biol 1960; 25:11-28; PMID:13684695; https://doi.org/ 10.1101/SQB.1960.025.01.004 [DOI] [PubMed] [Google Scholar]
- 11.Roenneberg T, Dragovic Z, Merrow M. Demasking biological oscillators: Properties and principles of entrainment exemplified by the Neurospora circadian clock. Proc Natl Acad Sci 2005; 102:7742-7; PMID:15899977; https://doi.org/ 10.1073/pnas.0501884102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra P, Panigrahi KC. GIGANTEA – an emerging story. Front Plant Sci 2015; 6:8; PMID:25674098; https://doi.org/ 10.3389/fpls.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 1999; 18:4679-88; PMID:10469647; https://doi.org/ 10.1093/emboj/18.17.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999; 285:1579-82; PMID:10477524; https://doi.org/ 10.1126/science.285.5433.1579 [DOI] [PubMed] [Google Scholar]
- 15.Onai K, Okamoto K, Nishimoto H, Morioka C, Hirano M, Kami-Ike N, Ishiura M. Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J 2004; 40:1-11; PMID:15361136; https://doi.org/ 10.1111/j.1365-313X.2004.02191.x [DOI] [PubMed] [Google Scholar]
- 16.Ding Z, Millar AJ, Davis AM, Davis SJ. TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 2007; 19:1522-36; PMID:17496120; https://doi.org/ 10.1105/tpc.106.047241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palágyi A, Terecskei K, Adám E, Kevei E, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 2010; 153:1834-45; PMID:20530216; https://doi.org/ 10.1104/pp.110.153031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao H, Jin M, Zheng XM, Chen J, Yuan D, Xin Y, Wang M, Huang D, Zhang Z, Zhou K, et al.. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci U S A 2014; 111:16337-42; PMID:25378698; https://doi.org/ 10.1073/pnas.1418204111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-Ariza J, Galbiati F, Goretti D, Brambilla V, Shrestha R, Pappolla A, Courtois B, Fornara F. Loss of floral repressor function adapts rice to higher latitudes in Europe. J Exp Bot 2015; 66:2027-39; PMID:25732533; https://doi.org/ 10.1093/jxb/erv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conover DO, Schultz ET. Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol Evol 1995; 10:248-52; PMID:21237029; https://doi.org/ 10.1016/S0169-5347(00)89081-3 [DOI] [PubMed] [Google Scholar]
- 21.Pemberton JM. Evolution of quantitative traits in the wild: Mind the ecology. Philos Trans R Soc B Biol Sci 2010; 365:2431-8; PMID:20643732; https://doi.org/ 10.1098/rstb.2010.0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci 2008; 105:3157-62; PMID:18287016; https://doi.org/ 10.1073/pnas.0712174105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filiault DL, Maloof JN. A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet 2012; 8:e1002589; PMID:22438834; https://doi.org/ 10.1371/journal.pgen.1002589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W, et al.. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016; 166:481-91; PMID:27293186; https://doi.org/ 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C-R, Svardal H, Farlow A, Exposito-Alonso M, Ding W, Novikova P, Alonso-Blanco C, Weigel D, Nordborg M. On the post-glacial spread of human commensal Arabidopsis thaliana. Nat Commun 2017; 8:14458; PMID:28181519; https://doi.org/ 10.1038/ncomms14458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian Peninsula. Genetics 2008; 180:1009-21; PMID:18716334; https://doi.org/ 10.1534/genetics.108.089581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao XY, Liu MS, Li JR, Guan CM, Zhang XS. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol Biol 2005; 58:53-64; PMID:16028116; https://doi.org/ 10.1007/s11103-005-4162-2 [DOI] [PubMed] [Google Scholar]
- 28.Dunford RP, Griffiths S, Christodoulou V, Laurie DA. Characterisation of a barley (Hordeum vulgare L.) homologue of the Arabidopsis flowering time regulator GIGANTEA. Theor Appl Genet 2005; 110:925-31; PMID:15682288; https://doi.org/ 10.1007/s00122-004-1912-5 [DOI] [PubMed] [Google Scholar]
- 29.Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 2007; 144:648-61; PMID:17468223; https://doi.org/ 10.1104/pp.107.096818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci U S A 2000; 97:9789-94; PMID:10920210; https://doi.org/ 10.1073/pnas.170283997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007; 448:358-61; PMID:17589502; https://doi.org/ 10.1038/nature05946 [DOI] [PubMed] [Google Scholar]
- 32.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011; 475:398-402; PMID:21753751; https://doi.org/ 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al.. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012; 24:428-43; PMID:22327739; https://doi.org/ 10.1105/tpc.111.093807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 2012; 7:170-3; PMID:22307044; https://doi.org/ 10.4161/psb.18766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci U S A 2009; 106:7251-6; PMID:19359492; https://doi.org/ 10.1073/pnas.0900952106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 2009; 50:447-62; PMID:19131357; https://doi.org/ 10.1093/pcp/pcp004 [DOI] [PubMed] [Google Scholar]
- 37.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 2007; 143:473-86; PMID:17098855; https://doi.org/ 10.1104/pp.106.088757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota A, Shim JS, Imaizumi T. Natural variation in transcriptional rhythms modulates photoperiodic responses. Trends Plant Sci 2015; 20:259-261; PMID:25802094; https://doi.org/ 10.1016/j.tplants.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David KM, Armbruster U, Tama N, Putterill J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 2006; 580:1193-7; PMID:16457821; https://doi.org/ 10.1016/j.febslet.2006.01.016 [DOI] [PubMed] [Google Scholar]
- 40.Kim Y, Han S, Yeom M, Kim H, Lim J, Cha JY, Kim WY, Somers DE, Putterill J, Nam HG, et al.. Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev Cell 2013; 26:73-85; PMID:23830866; https://doi.org/ 10.1016/j.devcel.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 41.Kim J, Geng R, Gallenstein RA, Somers DE. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013; 140:4060-9; PMID:24004949; https://doi.org/ 10.1242/dev.096651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, Nagamura Y, Izawa T. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 2012; 151:1358-69; PMID:23217716; https://doi.org/ 10.1016/j.cell.2012.10.048 [DOI] [PubMed] [Google Scholar]
- 43.Salmela MJ, Greenham K, Lou P, McClung CR, Ewers BE, Weinig C. Variation in circadian rhythms is maintained among and within populations in Boechera stricta. Plant Cell Environ 2016; 39:1293-303; PMID:26514754; https://doi.org/ 10.1111/pce.12670 [DOI] [PubMed] [Google Scholar]
- 44.Anastasio AE, Platt A, Horton M, Grotewold E, Scholl R, Borevitz JO, Nordborg M, Bergelson J. Source verification of mis-identified Arabidopsis thaliana accessions. Plant J 2011; 67:554-66; PMID:21481029; https://doi.org/ 10.1111/j.1365-313X.2011.04606.x [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2010; 107:21199-204; PMID:21078970; https://doi.org/ 10.1073/pnas.1007431107 [DOI] [PMC free article] [PubMed] [Google Scholar]


