Abstract
Monolayers of single-stranded DNA on gold substrates were studied by scanning force microscopy. Complementary DNA probes labeled by gold nanoparticles were applied for contrast enhancement. Substrate regions modified with DNA could be visualized in a highly specific manner. The influence of the solution concentration on the surface density of adsorbed nanoparticles could be visualized. Because individual label particles can be easily detected, this labeling technique opens the way for characterization of DNA monolayers with a lateral resolution in the nanometer range.
INTRODUCTION
Devices for array-based genetic diagnostics show tremendous promise for medical, pharmaceutical, analytical and other applications. They rely on the specific interaction of immobilized (capture) DNA and free (target) DNA in solution. This interaction is strongly influenced by the conformation and distribution of the immobilized DNA and has been the subject of a variety of experimental studies.
Previous work on characterizing immobilized DNA has included the application of techniques like hydroxyl radical footprinting (1), fluorescence energy transfer (2), surface plasmon resonance (3), a combination of X-ray photoelectron spectroscopy (XPS) and 32P-radiolabeling experiments (4) and neutron reflectivity (5). These studies pointed to the importance of electrostatic interaction between substrate and immobilized DNA (1,2). Other factors influencing the DNA conformation are the DNA surface density (1,2) and the use of mixed monolayers (4). Most of these data are the result of integral measurements over large surface regions, lacking the lateral resolution which is needed for the characterization of microspots used in DNA chip technologies or the DNA-modified colloidal particles applied for colorimetric gene detection (6) and in nanotechnology (7). Scanning force microscopy (SFM) was applied in a microscopical approach which used partial mechanical removal of the DNA for thickness measurements of the monolayer (8). This study demonstrated the influence of an applied electrical field on the orientation of double-stranded DNA, but the binding (hybridization) behavior of immobilized single-stranded DNA, an important aspect for applications in chip technologies, was not covered. An investigation of the binding behavior (and especially the activity) of the capture DNA should be based on hybridization to target DNA with a complementary sequence. The use of target DNA with radioactive or fluorescence labels is a typical method for this approach (9). However, such experiments cannot provide the lateral resolution below the micrometer range which is needed to reveal, for example, the lateral distribution of active capture DNA or the defect density of the molecular layer.
Colloidal metal particles with diameters between 10 and 60 nm, known from electron microscopy, have already been applied in SFM for topological labeling of DNA (10) and as a calibration standard (11,12). SFM has also been used for the detection and manipulation of metal particles immobilized by specific or non-specific interactions (13). Nanoparticle DNA labels were used on silicon oxide or glass surfaces (14,15) and detection methods based on optical (16,17) and electrical (18) principles were proposed. Monomolecular layers on gold are an important model system for the study of the processes involved in the interaction of molecules at the solid–liquid interface (19), and a new experimental method aimed at these processes with a lateral resolution in the nanometer range will be beneficial in further elucidation of these processes.
In this paper we introduce the use of colloidal gold particles as topographic labels to characterize DNA hybridization on gold substrates with regard to homogeneity and density at a high lateral resolution. Applying SFM, we show how the nanoparticle can be used to characterize hybridization behavior on gold surfaces with a resolution in the nanometer range. We demonstrate that the specificity of the DNA–DNA interaction is high in the presence of the topographic labels and that this labeling method can be used to monitor the hybridization of labeled DNA on gold substrates.
MATERIALS AND METHODS
Immobilization of capture DNA oligonucleotides
Gold substrates were prepared by sputtering a 3 nm adhesion layer of chromium and ∼100 nm gold onto cleaned glass slides. The gold surfaces were plasma etched, heated for 5 min to 150°C and washed in absolute ethanol for 15 min.
Pure single-stranded (ss)DNA layers were prepared by incubation of 1 µM thiolated ssDNA in 0.1 M NaCl, 10 mM sodium phosphate buffer with the gold substrates for 48 h. After immobilization the surfaces were washed with water.
For the preparation of mixed layers of ssDNA and alkanethiol, the substrates were incubated with 100 µl of 1 µM thiolated ssDNA in 1 M potassium phosphate buffer pH 6.7, for 120 min with constant shaking at 600 r.p.m. Then the substrates were incubated in 100 µl of an aqueous solution of 1 mM 6-mercapto-1-hexanol (Fluka, Deisenhofen, Germany) for 60 min at room temperature in an argon atmosphere, prior to rinsing with water and drying.
Colloidal gold
Colloidal gold for electron microscopy (Plano/British Biocell, Wetzlar, Germany) with mean diameters of 15, 30 and 60 nm was used in the experiments. For comparison, colloidal gold for DNA labeling (Genogold; Plano/British Biocell, Wetzlar, Germany) with diameters in the range 17–20 nm was also investigated.
Preparation of gold-labeled target DNA
For preparation of gold–DNA complexes, 3′-alkylthiolated oligonucleotides (BioTeZ, Berlin, Germany) were cleaved from a CPG support and preincubated with gold nanoparticle solution (30 nm diameter) for 16 h at room temperature in a ratio of 0.33 nM gold and 200 nM DNA (20). Another incubation was carried out after adjusting the solution to 0.1 M NaCl, 10 mM sodium phosphate buffer, pH 7.0, for 40 h at room temperature. The DNA–nanoparticle complexes were repeatedly washed with buffer and redispersed in 0.3 M NaCl, 10 mM sodium phosphate buffer, pH 7.0.
Solutions with higher concentrations were prepared by centrifugation and redispersion in a corresponding lower volume of buffer.
Hybridization of target to capture DNA
Hybridization of immobilized capture DNA C with 0.66 nM (2 OD) DNA–gold complex TC or TN (cf. Fig. 1) was by incubation of the chips at 65°C for 10 min. Then the samples were cooled to room temperature over the course of several hours (overnight). After washing twice with washing buffer (see above) they were washed thoroughly with deionized water prior to air drying.
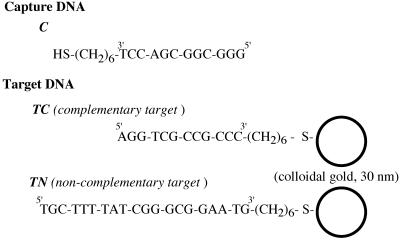
Figure 1.
DNA sequences used in the experiments. The thiolated capture DNA (C) was immobilized on gold substrates. Target DNA with a sequence complementary (TC) or non-complementary (TN) to the capture DNA was labeled with colloidal gold (30 nm diameter). Note that each gold colloid is covered by several DNA molecules, which is omitted in this scheme.
Microscopy
SFM (also known as atomic force microscopy, AFM) was conducted with a Dimension 3100 microscope (Digital Instruments, Santa Barbara, CA) in tapping mode in air. Usually a scan size of 4 µm was imaged and exported as a TIFF file.
RESULTS AND DISCUSSION
Gold colloids in SFM
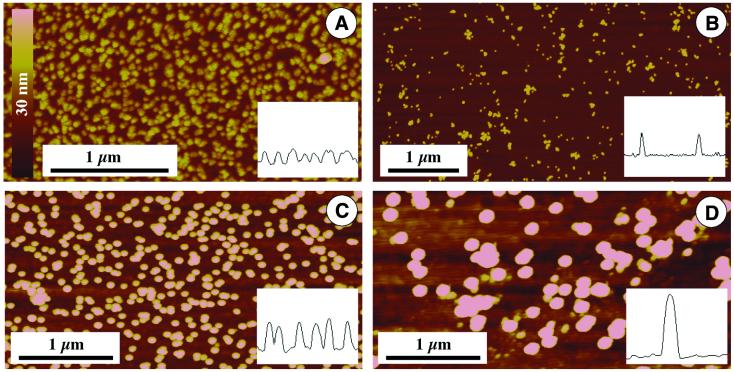
Different kinds of colloidal gold were characterized by SFM, investigating colloids with diameters ranging from 15 to 60 nm. All sizes can be clearly distinguished by SFM imaging (Fig. 2), because the roughness of the gold-sputtered substrate is below 5 nm peak-to-peak (cf. Fig. 3A), but the larger particles are more prominent. However, the stability of the colloidal solution decreased with increasing diameter. This stability, especially against an increase in salt concentration, is crucial for modification of the particles with biomolecules. Additionally, commercial stock solutions have a lower concentration with increasing particle size and observed non-specific binding (cf. Fig. 4) increases with diameter. Thus we decided to work with particles of 30 nm diameter (Fig. 2C), which appear to show high specificity of binding but are still large enough for convenient imaging by SFM and other standard microscopical methods like scanning electron microscopy.
Figure 2.
Various kinds of colloidal gold imaged by SFM. The height is brightness coded according to the bar in (A). Insets show cross-sections. (A) 15 nm nominal diameter; (B) Genogold 17–20 nm diameter; (C) 30 nm nominal diameter, as used for DNA labeling, shown in Figures 3–5; (D) 60 nm nominal diameter.
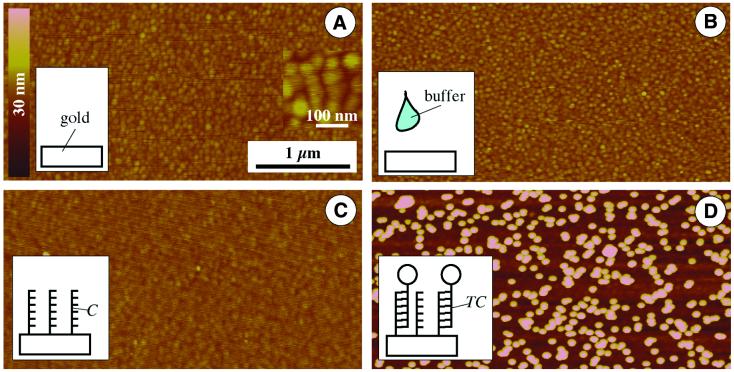
Figure 3.
SFM imaging of the chip surface showing different steps of substrate modification. Height and lateral scale according to the bars in (A). (A) Pure gold substrate. (Inset) Higher magnification revealing the grain size of the sputtered gold. (B) Control, gold substrate incubated with buffer solution without capture DNA. (C) Gold substrate after incubation with capture oligonucleotides, which results in a layer of capture probes. (D) A DNA-modified gold substrate (as shown in C) after incubation with 30 nm gold-labeled target probes.
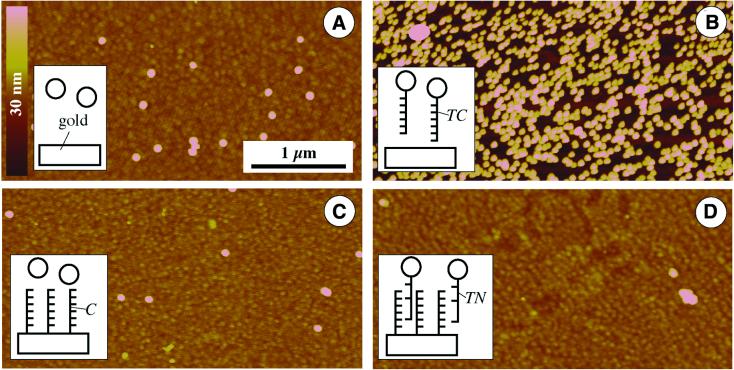
Figure 4.
Control experiments characterizing non-specific binding. Height and lateral scale according to the bars in (A). (A) Pure gold substrates incubated with pure gold colloid (without immobilized DNA). (B) Pure gold substrate after treatment with DNA-modified gold colloid. (C) DNA-modified surface after incubation with pure gold solution. (D) DNA-modified substrate treated with gold-labeled non-complementary DNA.
Gold substrates
Sputtered gold surfaces revealed a typical grainy surface topography in SFM (Fig. 3A). The observed grain size is usually in the 15–30 nm range; typical values for surface roughness are 5–6 nm peak-to-peak measured ∼4000 nm. Due to this relatively low roughness, colloidal gold particles with diameters down to 15 nm are still clearly resolvable by SFM, as mentioned before (cf. Fig. 2A). To maintain the surface smoothness through the surface modification steps, thorough washing is essential. This is demonstrated in a control experiment, which includes incubation of the surface with a buffer solution which was DNA-free. After washing and air drying, SFM imaging confirmed preservation of the smoothness (Fig. 3B) compared to the gold surface before incubation (Fig. 3A).
DNA modification of gold substrates
Gold substrates were incubated with the thiolated capture oligonucleotide C with the sequence given in Figure 1. SFM imaging of such DNA-modified surfaces resulted in a micrograph (Fig. 3C) comparable to images of unmodified gold substrates, taking the usual variation in image sharpness into account, which is due to typical variations in parameters such as tip sharpness (due to manufacturing, adsorption or breakage) or feedback control. Similar results were reported in the literature (8). Thus other means of surface characterization are needed to monitor this preparation step. Contact angle measurements can clearly detect changes in surface properties induced by DNA modification of gold substrates (21), but they do not provide lateral resolution in the submicron range.
Gold labeling: a new method for visualizing active hybridization sites
For application of DNA layers in chip technology, the lateral distribution and density of the active capture DNA with nanometer precision is important. A method that identifies and selectively visualizes DNA molecules on the surface is needed, so that only DNA that is able to hybridize with complementary target DNA molecules in solution will be detected. The use of fluorescence-labeled molecules is well established for characterization of immobilized DNA (22), but does not provide the required resolution. This approved combination of a specific reaction with a microscopic label has to be further developed to higher resolution. We think that the use of colloidal gold as label could provide this resolution for a range of applications. Hybridization of gold-labeled DNA should result in the gold label pinpointing the location of active capture probes. Using the high lateral resolution of SFM, a precision in the nanometer range seems feasible.
Hybridization of gold-labeled target DNA
Gold-labeled complementary target DNA TC was incubated with capture DNA-modified substrates. SFM imaging of these samples clearly shows the individual gold labels on the surface (Fig. 3D). The labels are usually separated from each other or situated in a chain-like pattern. This can be explained by electrostatic repulsion between the particles due to the DNA modification. This repulsion favors distancing between particles. In the case of contact between two particles, the lowest repulsion (and so the most favorable location for a third particle) is at both ends of the chain for a two-dimensional arrangement.
Control of non-specific binding
The results of control experiments are shown in Figure 4. Colloidal gold without DNA modification exhibits only a low binding to pure gold surfaces (Fig. 4A), resulting in <3 particles/µm2. The particles only occasionally appear clustered; usually they show an interparticle distance in the range of several diameters.
This image changes completely when the colloidal gold is modified with ssDNA. Then the incubated pure gold surface reveals a high coverage by the particles (∼140 particles/µm2), indicating a high extent of non-specific interactions between the gold substrate and the ssDNA on the surface of the colloidal gold. This observation is in accordance with the reported high degree of adsorption of ssDNA on a bare gold surface (3).
Assuming such a non-specific interaction, one would expect that a DNA-modified surface would exhibit a high degree of adsorption of pure gold colloids (without DNA modification). Surprisingly, this experiment resulted in very low adsorption (Fig. 4C). One explanation for this effect could be a higher negative charge on the particle surface, which would lead to repulsive electrostatic interactions with the negative charges of the surface-immobilized DNA molecules. Such a negative charge is typical for colloidal gold particles (23).
Specificity of complementary binding
Although this study of interactions between DNA and gold yielded interesting results, the most important question for possible applications in the field of DNA arrays or chips is the differentiation between complementary and non-complementary DNA. Thus chip-immobilized capture DNA was incubated with DNA-modified gold particles, whereby the sequences were non-complementary. The results of such an experiment are given in Figure 4D and clearly show low binding in this case compared to complementary sequences (cf. Fig. 3D). An average particle density of <1 particle/µm2 was determined, which was the lowest value of all control experiments. These experiments clearly show that the relatively large gold label (compared, for example, to fluorescence markers) does not significantly interfere with DNA hybridization, justifying the use of these labels for DNA chip applications.
Particle coverage
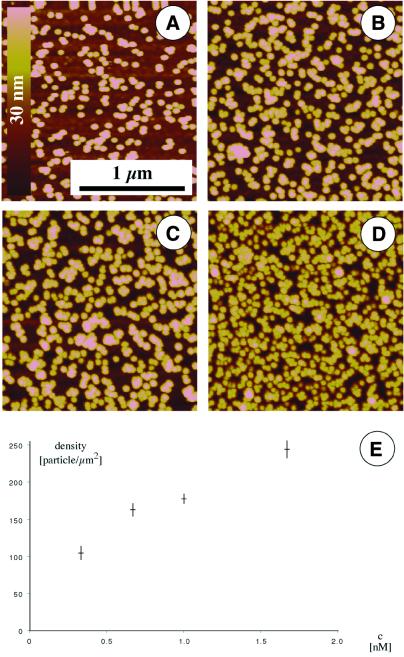
Independent of experimental conditions, different particle densities were achieved. Results from experiments with increasing concentrations of DNA–gold complexes in solution are shown in Figure 5A–D. The highest experimentally observed density was ∼250 particles/µm2. These experiments showed clearly that an increase in solution concentration resulted in a higher surface density (Fig. 5E). This is an important point for applications in DNA chip technology, because quantitative results can be obtained based on nanoparticle labeling. However, for these applications measurement speed (high throughput) is necessary, so that optical (16) or electrical (18) readout schemes are preferred to the more accurate but slower SFM.
Figure 5.
Quantification of hybridization of gold-labeled target DNA. Solutions containing different concentrations of labeled target DNA were incubated with surface-immobilized capture DNA, prior to washing, air drying and SFM imaging. Concentrations of 1 (A), 2 (B), 3 (C) and 5 OD (D) were used. (E) Surface density as a function of solution concentration of particles.
Conclusions
Nanoparticle labeling provides a new tool for the characterization of DNA layers on gold substrates. The sequence specificity allows defined labeling and provides the potential for high parallelization, as needed in such applications as DNA chips. The studied layers were homogeneous. Binding of the labeled complementary DNA is concentration dependent.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. Porwol and M. Sossna for microstructuring, K. Kandera for plasma etching, H. P. Saluz for advice regarding DNA hybridization, J. Reichert, U. Klenz and H. Stürmer for helpful discussions and S. Bechstedt, J. Krauße and R. Gerlach for assistance with sample preparation. This work was funded by the DFG (Fr-1348/3-1, 2).
References
- 1.Walker H.W. and Grant,S.B. (1995) Langmuir, 11, 3772–3777. [Google Scholar]
- 2.Charreyre M.-T., Tcherkasskaya,O., Winnik,M.A., Hiver,A., Delair,T., Cros,P., Pichot,C. and Mandrand,B. (1997) Fluorescence energy transfer study of the conformation of oligonucleotides covalently bound to polystyrene latex particles. Langmuir, 13, 3103–3110. [Google Scholar]
- 3.Peterlinz K.A., Georgiadis,R.M., Herne,T.M. and Tarlov,M.J. (1997) Observation of hybridization and dihybridization of thiol-thethered DNA using two-color surface plasmon resonance spectroscopy. J. Am. Chem. Soc., 119, 3401–3402. [Google Scholar]
- 4.Herne T.M. and Tarlov,M.J. (1997) Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc., 119, 8916–8920. [Google Scholar]
- 5.Levicky R., Herne,T.M., Tarlov,M.J. and Satija,S.K. (1998) Using self-assembly to control the structure of DNA monolayers on gold: a neutron reflectivity study. J. Am. Chem. Soc., 120, 9787–9792. [Google Scholar]
- 6.Elghanian R., Storhoff,J.J., Mucic,R.C., Letsinger,R.L. and Mirkin,C.A. (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science, 277, 1078–1080. [DOI] [PubMed] [Google Scholar]
- 7.Taton A.T., Mucic,R.C., Mirkin,C.A. and Letsinger,R.L. (2000) The DNA-mediated formation of supramolecular mono- and multilayered nanoparticle structures. J. Am. Chem. Soc., 122, 6305–6306. [Google Scholar]
- 8.Kelley S.O., Barton,J.K., Jackson,N.M., McPherson,L.D., Potter,A.B., Spain,E.M., Allen,M.J. and Hill,M.G. (1998) Orienting DNA helices on gold using applied electric fields. Langmuir, 14, 6781–6784. [Google Scholar]
- 9.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 10.Shaiu W.L., Larson,D.D., Vesenka,J. and Henderson,E. (1993) Atomic force microscopy of oriented linear DNA molecules labeled with 5 nm gold spheres. Nucleic Acids Res., 21, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S. and Arnsdorf,M.F. (1994) Calibration of the scanning (atomic) force microscope with gold particles. J. Microsc., 173, 199–210. [DOI] [PubMed] [Google Scholar]
- 12.Vesenka J., Manne,S., Giberson,R. and Marsh,T. (1993) Colloidal gold particles as an incompressible atomic force microscope imaging standard for assessing the compressibility of biomolecules. Biophys. J., 65, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer D.M., Reifenberger,R., Patil,A. and Andres,R.P. (1995) Fabrication of two-dimensional arrays of nanometer-size clusters with the atomic force microscope. Appl. Phys. Lett., 66, 1012–1014. [Google Scholar]
- 14.Möller R., Csáki,A., Köhler,J.M. and Fritzsche,W. (2000) DNA probes on chip surfaces studied by scanning force microscopy using specific binding of colloidal gold. Nucleic Acids Res., 28, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemeyer C.M., Ceyhan,B., Gao,S., Chi,L., Peschel,S. and Simon,U. (2001) Site-selective immobilization of gold nanoparticles funtionalized with DNA oligomers. Colloid Polymer Sci., 279, 68–72. [Google Scholar]
- 16.Reichert J., Csáki,A., Köhler,J.M. and Fritzsche,W. (2000) Chip-based optical detection of DNA-hybridization by means of nanobead labeling. Anal. Chem., 72, 6025–6029. [DOI] [PubMed] [Google Scholar]
- 17.Taton T.A., Mirkin,C.A. and Letsinger,R.L. (2000) Scanometric DNA array detection with nanoparticle probes. Science, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 18.Möller R., Csáki,A., Köhler,M.J. and Fritzsche,W. (2001) Electrical classification of the concentration of bioconjugated metal colloids after surface adsorption and silver enhancement. Langmuir, in press.
- 19.Nuzzo R.G. and Allara,D.L. (1983) Adsorption of bifunctional organic disulfides on gold surfaces. J. Am. Chem. Soc., 105, 4481–4483. [Google Scholar]
- 20.Storhoff J.J., Elghanian,R., Mucic,R.C., Mikron,C.A. and Letsinger,R.L. (1998) One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc., 120, 1959–1964. [Google Scholar]
- 21.Fritzsche W., Csáki,A., Möller,R., Bickel,R., Senz,S., Langer,J. and Köhler,J.M. (1999) Recent Research Developments in Vacuum Science and Technology. Transworld Research Networks, Vol. 1, pp. 81–91.
- 22.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baschong W. and Stierhof,Y.-D. (1998) Preparation, use and enlargement of ultrasmall gold particles in immunoelectron microscopy. Microsc. Res. Techniques, 42, 66–79. [DOI] [PubMed] [Google Scholar]