SUMMARY
Targeted patch clamp recording is a powerful method for characterizing visually identified cells in intact neural circuits, but it requires skill to perform. We previously developed an algorithm that automates “blind” patching in vivo, but full automation of visually guided, targeted in vivo patching has not been demonstrated, with currently available approaches requiring human intervention to compensate for cell movement as a patch pipette approaches a targeted neuron. Here we present a closed-loop real-time imaging strategy that automatically compensates for cell movement by tracking cell position and adjusting pipette motion while approaching a target. We demonstrate our system’s ability to adaptively patch, under continuous two-photon imaging and real-time analysis, fluorophore-expressing neurons of multiple types in the living mouse cortex, without human intervention, with yields comparable to skilled human experimenters. Our “imagepatching” robot is easy to implement, and will help enable scalable characterization of identified cell types in intact neural circuits.
Keywords: Patch clamp, in vivo electrophysiology, fluorescent proteins, fluorescent object detection, automation, cell types, mouse, cortex, imaging, two-photon microscopy
INTRODUCTION
Targeted patch clamp recording of visually identified neurons (Dittgen et al., 2004; Kitamura et al., 2008; Margrie et al., 2003) is a powerful technique for electrophysiological characterization of cells of a given class in the living mammalian brain, and is in increasing demand for its ability to link a cell’s molecular and anatomical identity with its electrophysiological characteristics in the context of specific behaviors, states, and diseases (Chen et al., 2015; Li et al., 2015; Pala and Petersen, 2015; Runyan et al., 2010; van Welie et al., 2016). However, the manual labor and skill required to perform visually guided patching in vivo have limited widespread adoption of the technique. Previously, we discovered that non-image guided (i.e., ‘blind’) patching in vivo could be reduced to an algorithm, and we accordingly built a robot, which we called the “autopatcher”, that automatically performs blind patch-clamp recordings of single neurons in the intact brain by detecting cells based on changes in pipette tip impedance (Kodandaramaiah et al., 2012, 2016). Since then, several attempts have been made to automate visually guided patch clamp recordings of targeted neurons. Although these attempts have enabled automatic positioning of a patch pipette near a visually identified neuron, all currently available systems either need a human to perform the final patching process itself (Long et al., 2015) or require human adjustment of the patching process for about half of the trials (Wu et al., 2016). We realized that a system that can achieve the whole-cell patch clamp configuration from a targeted cell without human intervention needs to address a key technical challenge: as a patch pipette moves towards a target cell for patch clamping, the cell moves as well, causing the pipette to miss its mark without manual adjustments of pipette motion that compensate for cell movement.
We therefore designed a new kind of algorithm, which we call “imagepatching”, in which realtime imaging in a closed-loop fashion allows for continuous adaptation of the pipette trajectory in response to changes in cell position throughout the patching process. We constructed a simple robotic system and software suite implementing imagepatching that can operate on a conventional two-photon microscope with commercially available manipulators and amplifiers, and show that we can obtain in vivo patch clamp recordings from fluorescently labeled neurons, of multiple cell types, in the living mouse cortex without any human intervention, and with a quality and yield similar to or even exceeding that obtained by skilled human experimenters. Our imagepatching robot is easy to implement, and will help enable scalable electrophysiological characterization of identified cell types in intact neural circuits.
RESULTS
Closed-loop real-time imaging algorithm for compensation of target cell movement during image-guided patch clamping
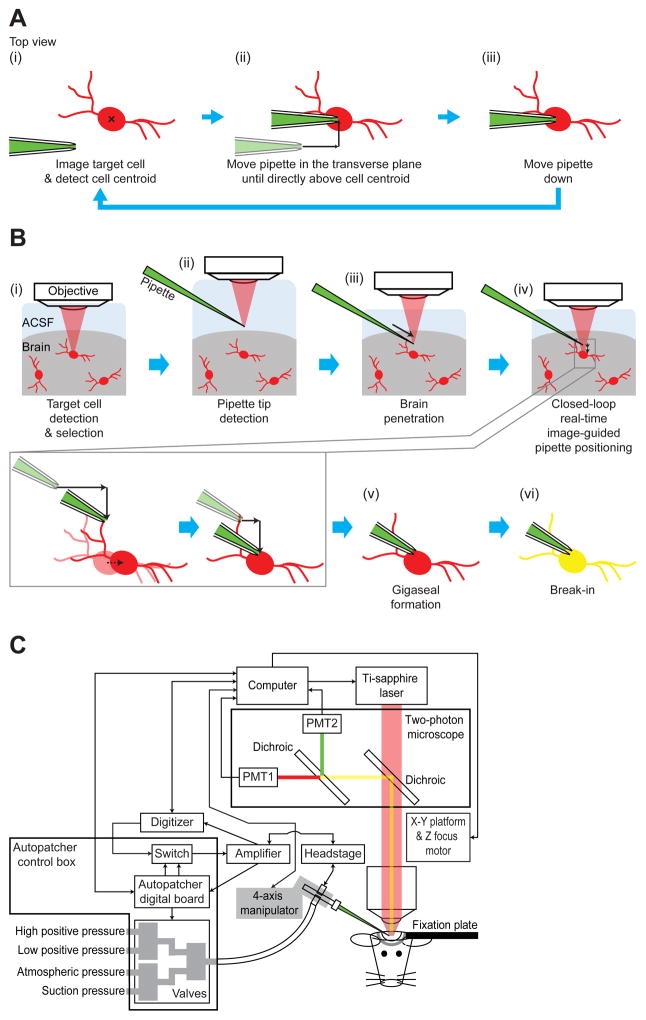
In the anesthetized mouse cortex, we found that moving a patch pipette by 300 – 400 μm from above the brain surface into layer 2/3 along the axial direction (i.e., parallel to the pipette axis, 30º below the horizontal) resulted in a target cell displacement of 6.8 ± 5.1 μm (mean ± standard deviation used throughout; n = 25 cells in 6 mice; Figure S1A) in the transverse plane. In addition, we observed that pipette navigations in the vicinity of a targeted cell (i.e., pipettes moving by ~5 – 10 μm when starting ~20 – 30 μm away from the cell) caused the targeted cell to move by 2.2 ± 1.4 μm (n = 27 cells in 17 mice; Figure S1B) in the transverse plane. These findings suggested that to correctly place the pipette tip on a targeted cell and patch it in a fully automated fashion, the displacement of the target cell resulting from pipette movement needs to be compensated for as the pipette is advanced towards the cell. Accordingly, we developed a closed-loop real-time image-guided algorithm that involves repeated target cell imaging followed by centroid detection (Figure 1A(i)) and pipette movement (Figure 1A(ii) and (iii)) stages, to continuously compensate for cell movement as the pipette approaches the target. We found that with the closed-loop algorithm supporting pipette navigation to a targeted cell, the entire image-guided patching process could be reduced to a six-stage “imagepatching” algorithm (Figure 1B; full flowchart in Figure S2). Imagepatching fuses closed-loop real-time image-guided pipette positioning with our earlier impedance-based cell detection strategy (Kodandaramaiah et al., 2012, 2016) to enable automated cell-attached or whole-cell patch clamp recording of visually identified cells in the intact mammalian brain.
Figure 1. Imagepatching: closed-loop real-time image-guided patch clamping in vivo.
(A) The closed-loop algorithm for continuous cell centroid localization and pipette position adjustment while approaching the targeted cell (for step-by-step flowchart, see Figure S2). Green, patch pipette filled with fluorescent dye; red, fluorescent cell targeted for patching; black x, target cell centroid; black arrows, pipette movement.
(B) The six stages of the image-guided automated patching algorithm (for step-by-step flowchart, see Figure S2). ACSF, artificial cerebrospinal fluid; red, fluorescent cells; green, patch pipette filled with fluorescent dye; light red, laser for two-photon imaging; black solid arrows, pipette movements; black dotted arrow, cell movement; yellow, target cell filled with the fluorescent dye from the pipette.
(C) Schematic of the imagepatcher hardware, composed of a conventional two-photon image-guided patch clamp rig and our previously developed autopatcher control box (Kodandaramaiah et al., 2012, 2016). Arrows indicate the direction of information flow. PMT, photomultiplier tube.
To implement imagepatching, we built a robotic system (“imagepatcher”) on a commercial two-photon microscope, which we controlled using ScanImage software (Pologruto et al., 2003) integrated with our MATLAB code that performs the real-time closed-loop image analysis. We chose ScanImage as the core software for the imagepatcher, since it works with two-photon systems from multiple vendors, and because its open code allowed us to incorporate real-time analysis of ScanImage-acquired images. The imagepatcher hardware shown in Figure 1C was assembled by augmenting a conventional two-photon image-guided patch clamp rig with an autopatcher control box (Kodandaramaiah et al., 2012, 2016) that was modified to provide a wide range of pressure values (see Methods for details of this, as well as other technical implementation points summarized in the following section). We validated the imagepatcher by using it to obtain targeted in vivo recordings from tdTomato-expressing cells in somatosensory and motor cortices of anesthetized Cre-dependent reporter mice, namely parvalbumin (PV)-positive interneurons in PV-Cre x Ai14 mice and calcium/calmodulin-dependent protein kinase II isoform alpha (CaMKIIα)-positive pyramidal neurons in CaMKIIα-Cre x Ai14 mice (Clarke, 1993; Hippenmeyer et al., 2005; Tsien et al., 1996). PV-positive and CaMKIIα-positive cells had different cortical densities (9.6 ± 6.3 tdTomato-expressing cells per volume of 200 × 200 × 100 μm3 in 9 PV-Cre x Ai14 mice vs 47.0 ± 31.8 tdTomato-expressing cells in this volume in 7 CaMKIIα-Cre x Ai14 mice) and morphologies (example two-photon images of tdTomato-expressing cells in layer 2/3 somatosensory cortex of each mouse line are shown in Figures 4A and 4B), and thus allowed us to explore the degree of generality that the imagepatcher offers to an end user.
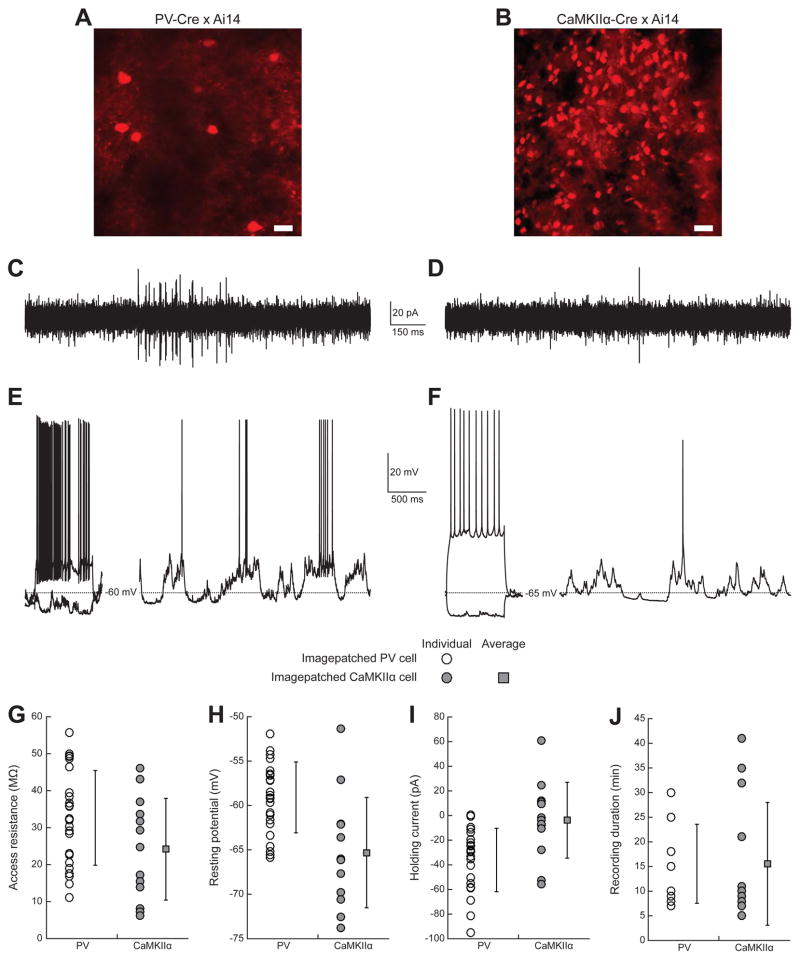
Figure 4. Imagepatching of multiple cell types in mouse cortex.
(A) Maximum intensity projection of a z-stack (20 images, 5 μm step size) of tdTomato (red)-expressing PV-positive cells in layer 2/3 somatosensory cortex of a PV-Cre x Ai14 mouse.
(B) Maximum intensity projection of a z-stack (20 images, 5 μm step size) of tdTomato (red)-expressing CaMKIIα-positive cells in layer 2/3 somatosensory cortex of a CaMKIIα-Cre x Ai14 mouse.
(C) Cell-attached current recording from an imagepatched PV-positive neuron.
(D) Cell-attached current recording from an imagepatched CaMKIIα-positive neuron.
(E) Whole-cell voltage recordings from an imagepatched PV-positive neuron under current injection (left, −100 and +200 pA), and at rest (right).
(F) Whole-cell voltage recordings from an imagepatched CaMKIIα-positive neuron under current injection (left, −100 and +200 pA), and at rest (right).
(G–J) Recording quality of imagepatched PV-positive neurons (white symbols; n = 24 cells from 14 PV-Cre x Ai14 mice for G–I; n = 9 cells from 5 PV-Cre x Ai14 mice for J) and imagepatched CaMKIIα-positive neurons (gray symbols; n = 13 cells from 7 CaMKIIα-Cre x Ai14 mice) in somatosensory and motor cortices of isoflurane-anesthetized mice. Square and error bars are mean ± standard deviation.
(G) Access resistance.
(H) Resting potential.
(I) Holding current.
(J) Recording duration.
Imagepatcher operation
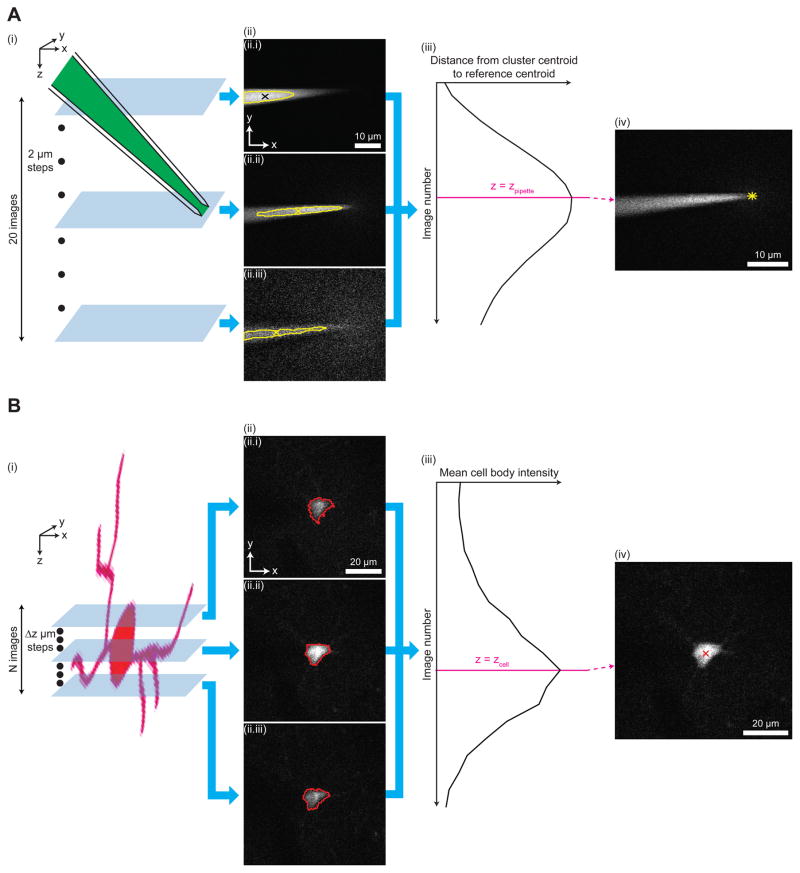
The imagepatcher starts by executing a target cell detection stage (Figure 1B(i)), in which two-photon images of the mouse brain are acquired and then analyzed to identify fluorescent cells. From these candidate cells, the end user can select a neuron of interest using the imagepatcher’s graphical user interface (see Methods S1 for details). The imagepatcher then moves on to the pipette tip detection stage (Figure 1B(ii)), where a dye (e.g., Alexa 488)-filled patch pipette is brought into the field-of-view above the brain, and the tip of the patch pipette is automatically located. The pipette tip is identified using a pipette tip detection algorithm (Figure 2A) derived from our finding that the cluster of bright pixels in the pipette image (Figure 2A(ii), area bounded by yellow outline), which represents the fluorescence from the dye inside the pipette, robustly changes its position as the focal plane of the microscope objective is moved downward from above the pipette tip. Accordingly, we developed a pipette tip detection algorithm to acquire a z-stack around the pipette tip (Figure 2A(i)) and to identify the image in the stack capturing the cluster of bright pixels that is furthest away from the far end of the pipette (represented by the centroid of the cluster in the topmost image in the stack; Figure 2A(ii.i), black x), assigning the z-coordinate of this farthest-cluster image as the z-coordinate of the pipette (Figure 2A(iii), zpipette). The portion of the pipette tip detection algorithm responsible for the identification of the pipette tip in the image at zpipette (Figure 2A(iv)) was developed based on the fact that an image focused on the pipette tip shows a triangular object corresponding to the pipette shank converging to a point (i.e., the pipette tip). We therefore designed the pipette tip detection algorithm to find the cluster of bright pixels that captures three vertices of the pipette from the image at zpipette (Figure S3A(ii.ii)), which is then analyzed to identify the pixel corresponding to the pipette tip (Figure S3A(iii)). When tested on 16 z-stacks (2 μm step size, 20 images, 17× zoom), each of which captured the tip of a separate Alexa 488-filled patch pipette (angled at 30º below the horizontal) at a distinct position within the stack, the pipette tip detection algorithm was capable of accurately extracting the pipette tip, with the tip location determined by the algorithm deviating from the visually assessed tip position by −1.0 ± 0.8 μm, −0.2 ± 0.4 μm, and 1.0 ± 2.4 μm in the x, y, and z directions respectively. The algorithm’s performance was similar for patch pipettes at different angles below the horizontal (see STAR Methods, Performance of the pipette tip detection algorithm at angles other than 30º below the horizontal for details).
Figure 2. Key algorithms for closed-loop real-time image analysis.
(A) Steps of the pipette tip detection algorithm. (i) A z-stack with 20 images and 2 μm step between consecutive images is acquired around a pipette filled with a dye (e.g., Alexa 488, green). (ii) Each image in the z-stack is analyzed to identify the cluster of bright pixels (area bounded by yellow outline, corresponding to the fluorescence from Alexa 488 inside the pipette) and the centroid of the cluster (x). The centroid in the topmost image of the z-stack ((ii.i), black x) is used as a reference location corresponding to the far end (i.e., end opposite to the pipette tip) of the pipette. Images 1 (ii.i), 10 (ii.ii), and 20 (ii.iii) of the z-stack, numbered from top to bottom, are shown as examples. (iii) The distance between the cluster centroid (x in (ii)) and the reference centroid (black x in (ii.i)) is calculated for each image in the z-stack. The image at which this distance is the largest is identified as the image focused on the pipette tip (magenta line). The z-coordinate of the focused image corresponds to that of the pipette tip (zpipette). (iv) The image focused on the pipette tip is analyzed to yield the location of the pipette tip in the transverse plane (yellow star). For image analysis steps used to locate the pipette tip in the transverse plane, see Figure S3A.
(B) Steps of the cell position detection algorithm. (i) A z-stack is acquired around a tdTomato-expressing cell (red), with N images and Δz step between consecutive images (N = 24, Δz = 3 μm for cell position detection in the brain penetration stage; N = 10, Δz = 2 μm for cell position detection in the closed-loop real-time image-guided pipette positioning stage). (ii) Each image in the z-stack is analyzed to detect the boundary of the cell body (red outline). Images 8 (ii.i), 12 (ii.ii), and 16 (ii.iii) of the z-stack, numbered from top to bottom, are shown as examples. (iii) The mean intensity of pixels representing the cell body (i.e., pixels surrounded by the detected boundary in (ii)) is calculated for each image in the z-stack. The image at which this mean intensity is the highest is identified as the image focused on the centroid of the cell body (magenta line). The z-coordinate of the focused image corresponds to that of the cell centroid (zcell). (iv) The image corresponding to the z-coordinate of the cell centroid is analyzed to yield the centroid position in the transverse plane (red x). For image analysis steps used to detect the boundary and the centroid of the cell body, see Figure S3B.
The pipette tip location determined during the pipette tip detection stage is used by the imagepatcher to compute the ideal trajectory to the target cell at the start of the brain penetration stage (Figure 1B(iii)), and also to calculate the pipette tip position in subsequent stages of imagepatching; we decided not to utilize the pipette tip detection algorithm (Figure 2A) to locate the pipette tip from the brain penetration stage onward, because a pipette that entered the brain without contamination ejected a plume of fluorescent dye that obscured the exact location of the pipette tip, which made it difficult to robustly resolve the pipette tip using an image-based algorithm. To enter the brain, the imagepatcher applies high positive pressure (600 mBar) to the pipette and moves it along the calculated trajectory at about 600 μm/s (i.e., at the maximum speed that our 4-axis micromanipulator can generate under software control; the same speed is used to move the pipette throughout the imagepatcher operation) until the pipette tip is within 75 μm from the initial target cell location. At this point, the pipette pressure is quickly reduced to 300 mBar to prevent excessive background fluorescence, but if little or no dye is ejected around the pipette tip, or a drastic resistance increase is observed, the pipette is deemed contaminated and brief pulses of positive pressure (>300 mBar) are applied to clean the pipette tip (as described in Komai et al., 2006). The pressure value of the pulse is increased until the pipette tip is cleared, but no more than 800 mBar is applied as a pipette ejecting the dye at this high pressure can cause excessive background fluorescence that interferes with cell detection in subsequent steps of imagepatcher operation. If the clogged state persists even after two pulses of high positive pressure, the contaminated pipette is automatically retracted. The imagepatcher applies a pipette pressure of 300 mBar while moving the clean pipette to the vicinity of the target, as we found this pressure value to be high enough to keep the pipette tip clean inside the brain, but not so high as to cause a lingering flood of dye that would lead to excessive background fluorescence (see STAR Methods, Derivation of pipette pressure for brain entry and cell approach for details).
Once the pipette tip is within 50 μm from the target cell’s initial location, the imagepatcher re-images and re-detects the target cell to account for cell movement resulting from pipette entry into the brain, using the cell position detection algorithm described in Figure 2B. The algorithm was derived based on the fact that in fluorescence microscopy, a fluorescent object looks the brightest when it is in focus (i.e., an image of a fluorescently labeled cell captures pixels corresponding to the fluorescence of the cell, and these pixels have higher intensities in an image focused on the cell compared to out-of-focus images). We thus built the cell position detection algorithm to detect the cell body in each image of a z-stack of the target cell (Figure 2B(ii)) and then to identify the image with the highest mean pixel intensity within the cell body (Figure 2B(iii), magenta line), which yields the z-coordinate of the target cell (Figure 2B(iii), zcell). We also designed the cell position detection algorithm to identify the centroid (i.e., center of mass) of the cell body in the image at zcell (Figure 2B(iv), red x), which is then assigned as the cell position in the transverse plane, because the cell body centroid is where investigators manually performing image-guided patching would aim with the tip of a patch pipette (Häusser and Margrie, 2014; Komai et al., 2006). When tested on 21 z-stacks (2 or 3 μm step size, 20 or 24 images, 17× zoom; from 5 mice), each capturing a PV-positive neuron at a different position within the cortex, the cell position detection algorithm correctly yielded x, y, and z coordinates of the cell centroid in all 21 stacks (visually assessed). Following cell position detection, the pipette is moved so that its tip is 25 μm above the updated target cell centroid, and the pipette tip is checked again for contamination.
With the clean pipette in place, the closed-loop real-time image-guided pipette positioning stage (Figure 1B(iv)) begins by lowering the pipette pressure (to 100 mBar) to prevent the target cell from being blown out of place and by performing another cell position detection (as in Figure 2B) to update the location of the target cell. The imagepatcher then repeatedly finds the target cell centroid (Figures 1A(i) and 2B(iv)) and repositions the pipette in the transverse plane according to the offset from the pipette tip to the cell centroid (Figure 1A(ii)) before each downward 3 μm z-step towards the target cell (Figure 1A(iii); example data from multiple steps of this closed-loop operation in Figures 3A and 3B, with corresponding imagepatching impedance trajectory in Figure 3C). Similar to manual image-guided patching in vivo (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2003), both visual (pipette tip within the boundary of the target cell soma; Figure 3A(iii)) and electrical (resistance increase that exceeds a certain threshold; Figure 3C(iii)) parameters are repeatedly checked while the pipette advances towards the target cell to determine when the pipette tip touches the target cell membrane. The imagepatcher maintains the pipette pressure at 100 mBar until the pipette tip makes contact with the cell membrane, because we found that this pressure helped prevent pipette tip clogging and allowed a detectable change in pipette resistance to be observed when the pipette tip touched the cell membrane (as in Figure 3C(iii)) while not blowing the target cell out of place and not resulting in excessive background fluorescence (see STAR Methods, Derivation of pipette pressure for brain entry and cell approach and Optimization of cell-pipette contact detection, gigaseal formation, and break-in for details). Once the pipette tip makes contact with the cell membrane, the imagepatcher dynamically changes the pipette pressure from 100 mBar to 30 mBar to prepare for gigaseal formation. We found that this lowering of pressure resulted in reduction of, and fluctuation of, the amplitude of current pulses that were observed in response to the application of voltage steps to the pipette tip, corresponding to the heartbeat modulation that has been reported previously (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2003). We also found that the amount of amplitude reduction, which we computed by comparing the pipette resistance before and after the pressure change, and the amount of amplitude fluctuation, which we quantified by calculating the standard deviation of the amplitude of current pulses, were useful predictors of gigaseal formation when they each exceeded a certain threshold (see STAR Methods, Optimization of cell-pipette contact detection, gigaseal formation, and break-in for details). The imagepatcher therefore checks if resistance change and the standard deviation of the current pulse amplitude following the lowering of pipette pressure to 30 mBar are high enough before advancing to the gigaseal formation stage (Figure 1B(v)).
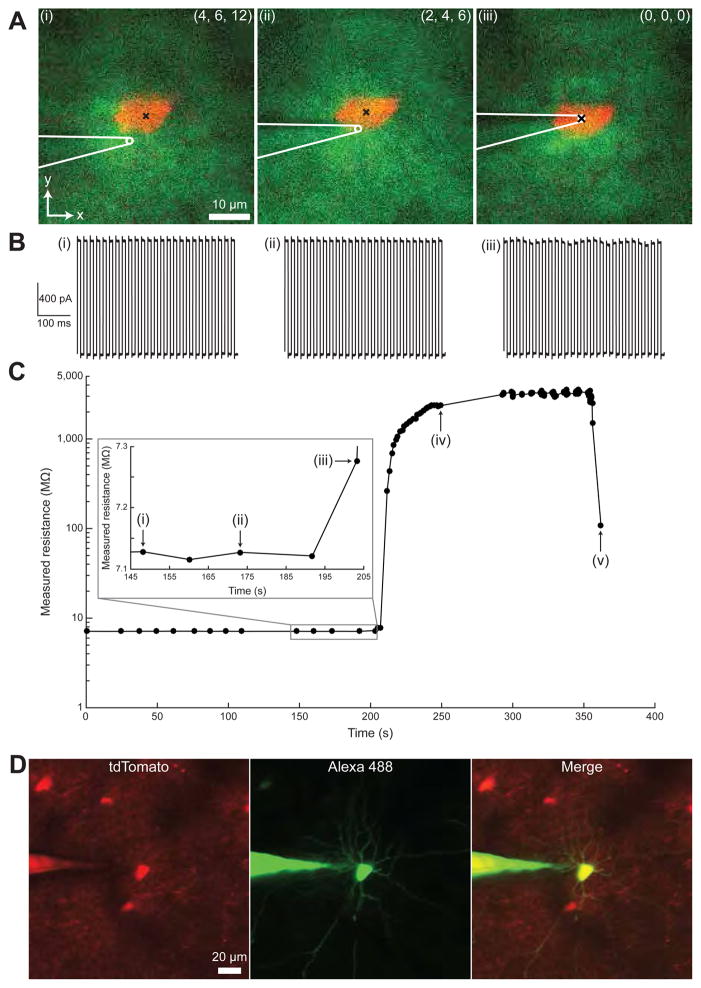
Figure 3. Imagepatcher operation.
(A) Two-photon images of a parvalbumin (PV)-positive neuron acquired at three different time points (indicated by Roman numerals for reference in (B) and (C)) during closed-loop real-time image-guided pipette positioning. White, sketch of pipette tip; green, Alexa 488; red, tdTomato; x, target cell centroid; numbers in the upper right, vector (x, y, z) from the pipette tip to the target cell centroid (in μm).
(B) Pipette current traces in response to 10 mV voltage pulses, with Roman numerals indicating the corresponding images in (A).
(C) Pipette resistance during imagepatching over time, with Roman numerals corresponding to time points referenced in (A) and (B): (i) pipette is 12 μm above the target cell centroid; (ii) pipette is 6 μm above the centroid; (iii) pipette is 0 μm above the centroid (i.e., in contact with the cell); (iv) gigaohm seal is established; (v) cell is broken-into.
(D) Maximum intensity projection (MIP) of a z-stack (48 images, 2 μm step size) of an imagepatched PV-positive neuron, showing tdTomato (left, red), Alexa 488 (middle, green) and overlay (right).
During gigaseal formation, the positive pressure is removed and 20 mBar suction is applied while hyperpolarizing the pipette. When a gigaohm seal is established (Figure 3C(iv)), the imagepatcher operation may be halted to obtain cell-attached extracellular recordings. For whole-cell mode, the imagepatcher advances to the break-in stage (Figure 1B(vi)), in which increasing pulses of suction (starting at 25 mBar and increased up to 350 mBar) are applied to achieve the whole-cell configuration (Figure 3C(v)). As done at the completion of manual image-guided patching in vivo (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2003), the imagepatcher checks if the dye from the pipette is filling the target cell, by first acquiring a z-stack around the target cell, then identifying pixels corresponding to the cell body (as in Figure 2B(ii)), and finally calculating the mean pixel intensity of the cell body in the microscope channel corresponding to the pipette dye, to verify successful break-in (example of a dye-filled cell at the end of successful imagepatching is shown in Figure 3D).
Imagepatcher performance
Using the imagepatcher, stable cell-attached extracellular and whole-cell intracellular recordings could be obtained from PV-positive neurons (example recordings in Figures 4C and 4E) and CaMKIIα-positive neurons (example recordings in Figures 4D and 4F) in layer 2/3 of somatosensory and motor cortices of anesthetized mice, enabling in vivo observations of supra- and subthreshold activities of these cells. Access resistance, resting potential, and holding current (Figures 4G–4I) of imagepatched cells (n = 24 PV-positive neurons from 14 PV-Cre x Ai14 mice and 13 CaMKIIα-positive neurons from 7 CaMKIIα-Cre x Ai14 mice) were comparable to those reported by previous studies involving two-photon image-guided patching of cortical neurons in vivo (Atallah et al., 2012; Gentet et al., 2010, 2012; Mateo et al., 2011; Pala and Petersen, 2015), and were not significantly different from the cells that we manually patched (n = 11 PV-positive neurons from 8 PV-Cre x Ai14 mice; Figures S4A-S4C; P = 0.49 for access resistance, P = 0.08 for resting potential, P = 0.19 for holding current when comparing imagepatched and manually patched PV-positive cells; two-sided Student’s t-test with 95% confidence level, assuming unknown and unequal variances). Other properties of imagepatched neurons, such as input resistance and spontaneous firing rate (Figures S4D and S4E; n = 9 PV-positive neurons from 5 PV-Cre x Ai14 mice and 13 CaMKIIα-positive neurons from 7 CaMKIIα-Cre x Ai14 mice), also showed distributions of values that overlapped with those obtained in previous in vivo studies of cortical neurons (Mateo et al., 2011; Pala and Petersen, 2015).
The imagepatcher obtained targeted patch clamp recordings in 10 ± 3 minutes from the brain penetration stage onwards (n = 24 PV-positive neurons from 14 PV-Cre x Ai14 mice and 13 CaMKIIα-positive neurons from 7 α-Cre x Ai14 mice; each of the two preceding stagesCaMKII of the algorithm takes around one to two minutes extra), with the recordings lasting for 7 – 30 minutes for PV-positive neurons (n = 9 cells, 5 PV-Cre x Ai14 mice; Figure 4J) and 5 – 41 minutes for CaMKIIα-positive neurons (n = 13 cells, 7 CaMKIIα-Cre x Ai14 mice; Figure 4J). When targeting PV-positive cells, the gigaohm seal was obtained 42 times out of 108 attempts, and 24 of the 42 gigaohm seals successfully led to the whole-cell configuration (from 17 PV-Cre x Ai14 mice, of which 16 yielded one or more gigaohm seals and 14 yielded one or more whole-cell configurations). For CaMKIIα-positive cells, from 65 trials, the gigaohm seal was achieved 19 times, out of which the whole-cell configuration was achieved 13 times (from 10 CaMKIIα-Cre x Ai14 mice, of which 10 yielded one or more gigaohm seals and 7 yielded one or more whole-cell configurations). These success rates (for PV-positive neurons, 38.9% for obtaining gigaohm seals, and 22.2% for the whole-cell configuration; for CaMKIIα-positive neurons, 29.2% for obtaining gigaohm seals, and 20.0% for the whole-cell configuration) are comparable to or higher than that obtained by manually performing two-photon image-guided patching of fluorescently labeled neurons in vivo (for us, 10.6% success rate for manual whole-cell patching of tdTomato-expressing PV-positive neurons; n = 11 out of 104 attempts, 19 PV-Cre x Ai14 mice; a 10 – 20% success rate for obtaining a whole-cell recording from an EGFP-labeled PV-positive neuron was reported in Margrie et al., 2003). During our imagepatching experiments, some pipettes were occluded after brain penetration (n = 22 out of 108 when targeting PV-positive neurons; 14 out of 65 when targeting CaMKIIα-positive neurons; detailed breakdown of unsuccessful patching attempts in Table S1), and were automatically retracted by the imagepatcher; focusing on trials that entered the closed-loop stage (Figure 1B(iv)), the rates of successfully achieving the gigaohm seal and the whole-cell configuration were 48.8% and 27.9% respectively (n = 42 gigaohm seals and 24 whole-cell configurations out of 86 trials) for PV-positive neurons, and 37.3% and 25.5% respectively (n = 19 gigaohm seals and 13 whole-cell configurations out of 51 trials) for CaMKIIα-positive neurons. These success rates did not vary substantially with target cell depth (Table S2) nor with the density of labeled cells around a target cell (Table S3), suggesting that the imagepatcher performance was consistent.
DISCUSSION
We developed an algorithm and a robotic system that fully automates targeted patch clamping of visually identified cells in vivo, by implementing closed-loop real-time imaging to dynamically adjust the pipette position to hone in on a cell of interest. Our strategy makes the imagepatcher the first system that enables fully hands-free navigation of a patch pipette to a targeted cell and subsequent automated patch clamping in the intact brain. Unlike other previous systems that heavily rely on human intervention for successful patch clamp recordings of visually identified cells (Long et al., 2015; Perin and Markram, 2013; Steinmeyer and Yanik, 2012; Wu et al., 2016), the imagepatcher eliminates the need for manual adjustments and corrections during the entire patching process, making the robot a powerful tool that can facilitate systematic electrophysiological characterizations of specific classes of cells. Certain factors that can prevent investigators performing manual patching from achieving the whole-cell patch clamp state, such as variations in pipette trajectory, pipette movement speed, and pipette pressure levels, are also reduced in our automated system. The reduction of variation in these factors provides consistency in patch clamping procedures that may be difficult to obtain manually and may prove particularly beneficial for studies targeting very sparse populations of cells.
With our software designed to work in parallel with ScanImage operation, and our hardware designed to augment a conventional two-photon microscope in a modular way, the imagepatcher may be adapted to work on any microscope that ScanImage (or another openly modifiable software package) supports. Although our current study focused on targeted patching guided by two-photon microscopy in the intact brain, the imagepatcher could, in principle, also be used to automate and enable experiments utilizing other imaging modalities (e.g., one-photon fluorescence microscopy) and/or other tissues or preparations – as long as a target provides fluorescence corresponding to its size and position. In case of experiments involving targeted patching of non-fluorescent, unlabeled cells (i.e., “shadowpatching” developed by Kitamura et al., 2008), the cell position detection algorithm (Figure 2B), as it currently stands, may lead to incorrect identification of the target cell’s z-coordinate, because we found from our shadow images that the mean pixel intensity of the cell body shadow does not vary with a defined pattern as a function of microscope focus (unlike cells labeled with a photostable fluorescent marker, which look the brightest when in focus). A new cell position detection algorithm that identifies the z-coordinate of a target cell based on its other properties (e.g., cell body size, cell body shape) might permit, in the future, automation of shadowpatching. The open nature of the imagepatcher code allows for integration of such an algorithm, in addition to tuning of software settings that might be required for different microscopes and imaging conditions.
Integrating our robot with patch pipette cleaning protocols for repeated patch pipette use (Kolb et al., 2016) may enable the elimination of some of the manual preparatory steps required to utilize imagepatching (e.g., filling a patch pipette with intracellular solution and inserting it into a pipette holder). By developing and using a bright pipette dye that has a fluorescence emission spectrum overlapping minimally (or ideally, not overlapping at all) with that of the target cells’ fluorescent marker, the high level of background fluorescence that results from multiple penetrations into the brain may have little or no effect on cell position detection and targeting by the imagepatcher, enabling many imagepatching trials and thus patch clamp recordings per animal. Further augmentation of the imagepatcher hardware (i.e., integration of multiple autopatcher control boxes, each linked to an individual pipette, with a single two-photon microscope) and refinement of the software (e.g., code development for simultaneous micromanipulator control in response to multiple pipette impedances and imaged positions of target neurons) may also enable multi-cell targeted patch clamp recordings in vivo (Jouhanneau et al., 2015; Pala and Petersen, 2015; van Welie et al., 2016), which will provide information on how cells communicate with each other in an intact brain network. Although we have not obtained patch clamp recordings in the awake brain using the imagepatcher, with an appropriate restraint habituation strategy (to reduce brain motion), a robust image analysis approach (which compensates for large motion artifacts), or a strategy for real-time switching of target cell identity (which enables targeting of an alternative cell, if present, when motion artifacts are large enough to displace the originally targeted cell out of the field-of-view), the imagepatcher may enable patch clamping of targeted neurons in awake animals.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
All code, schematics, and parts lists will also be posted to http://autopatcher.org at time of publication. Further requests and inquiries should be directed to, for fulfillment by, the corresponding author, Dr. Edward S Boyden (esb@media.mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experimental procedures were performed on 6 – 8 week old transgenic mice, male and female used equally, of PV-Cre (Jax strain #: 017320) x Ai14 (tdTomato; Jax strain #: 007914) genotype or CaMKIIα-Cre (Jax strain #: 005359) x Ai14 (tdTomato; Jax strain #: 007914) genotype, in accordance with protocols approved by the Massachusetts Institute of Technology (MIT) Committee on Animal Care (CAC). Six PV-Cre x Ai14 mice were used to quantify neuron movements in response to a pipette entering the brain (Figure S1A), and seventeen additional PV-Cre x Ai14 mice were used to measure how much neurons change their locations when a pipette is navigated towards them inside the brain (Figure S1B). Eight PV-Cre x Ai14 mice were used to find the set of optimal pipette pressure levels for brain entry and cell approach during patching (STAR Methods, Derivation of pipette pressure for brain entry and cell approach). Z-stacks of tdTomato-labeled parvalbumin (PV)-positive neurons, which were used to validate our cell detection algorithm (Figure 2B), were obtained from somatosensory and motor cortices of five PV-Cre x Ai14 mice. We used twenty PV-Cre x Ai14 mice to optimize the portion of the closed-loop image-guided pipette positioning stage responsible for resistance-based detection of the pipette tip-cell membrane contact, as well as the gigaseal formation and the break-in stages (STAR Methods, Optimization of cell-pipette contact detection, gigaseal formation, and break-in). Three PV-Cre x Ai14 mice were used to derive the dye-based pipette blockage test (STAR Methods, Derivation of dye-ejection based pipette blockage test). For comparing the cortical density of calcium/calmodulin-dependent protein kinase II alpha (CaMKIIα)-positive neurons to that of PV-positive neurons, z-stacks of tdTomato-expressing cells were obtained from somatosensory and motor cortices of seven CaMKIIα-Cre x Ai14 mice and nine PV-Cre x Ai14 mice. To validate the robotic system running the imagepatching algorithm (Figure 4), seventeen PV-Cre x Ai14 mice and ten CaMKIIα-Cre x Ai14 mice were used, while nineteen PV-Cre x Ai14 mice were used for the manual patching experiments (Figures S4A–S4C).
METHOD DETAILS
Surgical procedures
Throughout the surgery, mice were anesthetized with 1 – 2% (vol/vol) isoflurane in oxygen and maintained at 37ºC using a heating pad. After shaving the scalp, the mouse was placed in a custom-built stereotax, with its eyes covered with ophthalmic ointment. Betadine and 70% ethanol were then applied to the shave area for sterilization. A polycarbonate recording chamber was implanted on the skull using dental acrylic, and a 1 – 2 mm diameter craniotomy, contained inside a 3 mm diameter window of the recording chamber, was made 1.5 – 2 mm posterior to the bregma and 1.5 – 2 mm to the right of the midline. The dura was then carefully removed to expose the brain surface. Right before starting an imaging or a patch clamp experiment, 1.5% (vol/w) agar in HEPES buffered artificial cerebrospinal fluid (ACSF, containing 145 mM NaCl, 5.4 mM KCl, 10 mM HEPES, 1.8 mM CaCl2, 1 mM MgCl2 (Chen et al., 2015) or 150 mM NaCl, 2.5 mM KCl, 10 mM HEPES, 2 mM CaCl2 and 1 mM MgCl2 (van Welie et al., 2016); pH adjusted to 7.3 – 7.4 with NaOH) was applied on top of the brain to dampen pulsations caused by respiration and heartbeat, and then the craniotomy was covered with ACSF to keep the brain moist throughout the experiment. We took extra care to minimize bleeding throughout the surgery as blood on the cortical surface can greatly diminish optical clarity during two-photon imaging (Komai et al., 2006). In case of bleeding, the brain surface was irrigated with ACSF to stop the bleeding and remove as much blood as possible from the cortical surface. At the end of the experiment, mice were euthanized under anesthesia.
Electrophysiology
Cell-attached and whole-cell patch clamp recordings were performed on mice under 1 – 1.5% isoflurane anesthesia on a 37ºC heating pad (DC Temperature Control System, FHC). Patch pipettes with resistance values between 5 – 7.5 MΩ were prepared by pulling filamented borosilicate glass capillaries (Warner or WPI) using a micropipette puller (Flaming-Brown P97 model, Sutter Instruments or PC-10 vertical puller, Narishige). These pipettes were filled with an internal solution containing (in mM): 135 K-gluconate, 4 KCl, 10 HEPES, 10 Na2-phosphocreatine, 4 MgATP, 0.3 Na3GTP (pH adjusted to 7.3 – 7.4 with KOH; osmolarity 280 – 290 mOsm), and 50 μM Alexa 488 dye (ThermoFisher; for pipette visualization under the two-photon microscope) or 125 K-Methanesulfonate, 7 KCl, 10 HEPES, 2 MgATP, 2 Na2ATP, 0.5 Na2GTP, 0.05 EGTA (pH adjusted to 7.3 with KOH; osmolarity 280–290 mOsm), and 50 μM Alexa 488 dye. Fully manual patch clamp experiments (Figures S4A–S4C) were performed following previously reported protocols (Häusser and Margrie, 2014; Komai et al., 2006).
Hardware and software setup
We modified a standard two-photon image-guided in vivo patch clamp rig to construct the imagepatching system (Figure 1C). Hardware for the standard rig included a two-photon laser scanning microscope (Ultima moving IV, Prairie Technologies), a mode-locked Ti-sapphire laser (Mai Tai HP; Spectra-Physics), a water-immersion objective (CFI75 LWD 16× W NA 0.8 WD 3.0mm objective, Nikon), a programmable 4-axis micromanipulator comprised of a single-axis micromanipulator (SMX-SA, Sensapex) mounted at a 30º angle below the horizontal on a left-handed three-axis micromanipulator (SMX-L-RS-50-HL-US, Sensapex), a pipette holder (Warner) mounted on the single-axis micromanipulator and connected to the CV-7B headstage of a patch amplifier (Multiclamp 700B, Molecular Devices) via an intermediate cable (IM-SMB, Sensapex), and a digitizer (Digidata 1440A, Molecular Devices) relaying signals between the amplifier and a computer. The laser was set to emit 960 nm (~800 mW average output power), which could excite both tdTomato and Alexa 488. To minimize bleed-through, we replaced the user-exchangeable PMT filters in the Ultima (which are optimized for dual labeling using Alexa 594 and Alexa 488) with red (630/30 nm, Chroma) and green (510/10 nm, Semrock) filters. The 4-axis micromanipulator was connected to a rotary knob controller (SMXS-K-2-RS-US) that communicated with the computer through a USB port.
The patch amplifier and the digitizer of the standard rig were connected to our autopatcher control box as previously described (Kodandaramaiah et al., 2012, 2016) (Figure 1C). The autopatcher control box was constructed as previously described (Kodandaramaiah et al., 2012, 2016), with a slight modification; potentiometers mounted on the front panel of the original autopatcher control box, each of which is used to manually pre-set a pressure value to be used during the autopatcher operation (see Kodandaramaiah et al., 2016 for details), were replaced by analog outputs from a standard data acquisition (DAQ) device (PCIe-6343, National Instruments) that can be programmed to send a command voltage of an arbitrary value to electronic pressure regulators inside the autopatcher control box, thus enabling automated, real-time control of the pipette pressure to any desired level at any rate (e.g., in a ramp) during the imagepatcher operation (note that PCIe-6343 can be replaced by any programmable device that can generate analog output ranging from 0 to 5 VDC, with < 400 mA).
To operate the microscope with ScanImage 3.8, a MATLAB-based open-source software package, previously reported instructions (Wilson et al., 2013) were followed to make necessary connections between the microscope hardware and the computer. ScanImage was configured such that each acquired image has a single frame with 256 lines per frame and 256 pixels per line, and each line is scanned in 2.64 ms for a frame rate of 1.48 frames per second. To acquire high quality images of a neuron at the end of the imagepatching experiment (example image in Figure 3D), ScanImage was configured to produce 2048 lines per frame and 2048 pixels per line at a frame rate of 0.18 frames per second.
We used MATLAB R2013b (MathWorks) to write and run our program executing the imagepatching algorithm. Our program was divided into two main modules: (i) a graphical user interface (GUI) that allows the user to start the imagepatching algorithm and to view the results of image acquisition as well as analysis during the algorithm execution (see Methods S1, “find_cells_gui_SI.m”); (ii) image analysis code that is executed upon the completion of image acquisition to perform real-time image analysis (see Methods S1, “image_autopatcher_v1.m”; MATLAB scripts can be bound to one or more ScanImage events, such as the start of the ScanImage software and the completion of image acquisition, by adding them as user functions; see Methods S1, “Imagepatcher User Manual.docx” and the ScanImage website [https://openwiki.janelia.org/wiki/pages/viewpage.action?pageId=29524376] for details). A ScanImage function responsible for image frame generation, called makeFrameByStripes, was augmented with our code to direct raw image data acquired by the microscope to our image analysis code (see Methods S1, “makeFrameByStripes.m,” for the function with our code).
During the imagepatcher operation, when the pipette resistance had to be measured, the autopatcher digital board (USB-6211, National Instruments) was used to apply 50 Hz, 10 mV square waves to the pipette tip and to record the resulting current pulses at 20 kHz. 10 mV (i.e., amplitude of the applied voltage) was divided by the amplitude of each current pulse, and the average of the resulting values was assigned as the pipette resistance while the standard deviation of the resulting values was used as a metric quantifying the amount of heartbeat modulation. We performed whole-cell recordings of patched cells using Clampex 10.4 (Molecular Devices), acquiring data through a low-pass filter (Bessel filter, 10 kHz cutoff) at a rate of 40,000 samples per second. The acquired signals were analyzed using MATLAB 2013b (MathWorks) and Clampfit 10.5 (Molecular Devices).
Assessment of target cell movements in response to pipette navigations into and inside the brain
Cell movements following the pipette navigation into the brain (Figure S1A) were observed by first locating a tdTomato-labeled cell ~150 – 250 μm below the brain surface and recording the coordinates of the cell centroid (visually assessed). Using trigonometric functions in MATLAB, the pipette trajectory parallel to the pipette axis (i.e., 30º below the horizontal) was then calculated, setting the start and the end of the trajectory to the locations 25 μm above the brain surface and 50 μm directly above the cell centroid respectively. Subsequently, a patch pipette whose resistance value was between 5 – 7.5 MΩ was filled with the internal solution and installed into the pipette holder that was positioned on the left side of the craniotomy. While applying low positive pressure (~15 – 30 mBar), the pipette was moved into the ACSF covering the brain, positioning the pipette tip at the start position of the calculated trajectory. A high positive pressure (~200 – 300 mBar) was then applied to the pipette, and the single-axis micromanipulator (i.e., the micromanipulator whose axis is parallel to the pipette axis) was controlled using MATLAB code (Methods S1, “move_sensapex_manipulator_HJS.m”) interacting with a software development kit from Sensapex (the software development kit available on the Sensapex website [http://www.sensapex.com/support/downloads-updates/]) to automatically and accurately move the pipette along the calculated trajectory at ~600 μm/s. When the pipette movement was complete, a z-stack (20 or 24 images, 2 μm or 3 μm step size, 17× zoom) was acquired around the original cell centroid coordinates that were recorded before the pipette movement. The z-stack was analyzed post-hoc to determine the new coordinates of the cell centroid (visually assessed), and these coordinates were compared to those of the cell centroid before the pipette movement into the brain to quantify the amount of cell displacement.
To determine the amount of cell movement in response to the pipette navigation inside the brain (Figure S1B), a patch pipette (resistance value 5 – 7.5 MΩ) filled with the internal solution was first placed inside the brain, with its tip located ~20 – 30 μm above a tdTomato-labeled cell and ~5 – 13 μm away from the cell centroid in the transverse plane, simulating an offset from the pipette tip to the target cell that can result from the pipette entry into the brain. After recording the coordinates of the cell centroid (visually assessed), the micromanipulator was automatically moved using MATLAB code (Methods S1, “move_sensapex_manipulator_HJS.m”) in the x, y, and z directions to place the pipette tip ~10 – 20 μm directly above the cell centroid, which is where an investigator manually performing patching would aim to bring the pipette tip to approach the targeted cell in the vertical direction (Häusser and Margrie, 2014; Komai et al., 2006). Once the micromanipulator movement was complete, a z-stack (20 or 24 images, 2 μm or 3 μm step size, 17× zoom) was acquired around the cell centroid coordinates that were recorded prior to the pipette navigation. Post-hoc analysis of the z-stack was then performed to locate the new cell centroid (visually assessed), coordinates of which were compared to those of the cell centroid before the pipette navigation inside the brain to quantify the amount of cell movement.
Details of the pipette tip detection algorithm
The pipette tip detection algorithm first applies a 2D Gaussian filter (19 × 19 window; 9/2 variance) to each image in the z-stack of a patch pipette to remove the background noise. The filtered topmost image of the stack is then subjected to a range of threshold values (corresponding to 1 – 95% of the maximum pixel intensity of the filtered image) to determine the maximum threshold value at which the resulting cluster of bright pixels (Figure 2A(ii.i), area bounded by yellow outline) has a characteristic shape (i.e., has 3 endpoints when subjected to the bwmorph function with ‘endpoints’ operation in MATLAB); in case of multiple clusters for a single threshold value, the largest cluster (i.e., the cluster composed of the highest number of pixels) is analyzed for the endpoint detection. Subsequently, the cluster obtained from the filtered topmost image at this threshold value is analyzed to determine its area (i.e., number of pixels in the cluster; using MATLAB’s bwboundaries function) and centroid (Figure 2A(ii.i), black x; using MATLAB’s regionprops function with ‘centroid’ as an input argument). The detected centroid is also considered as the pixel corresponding to the far end of the pipette (i.e., the end opposite to the pipette tip) and used as the reference point in the subsequent stages of the algorithm. The rest of the filtered images in the stack are then subjected to a range of threshold values, identifying the threshold value for each image at which the resulting cluster has an area closest to that obtained from the topmost image (Figure 2A(ii.ii), (ii.iii), areas bounded by yellow outlines); in case of multiple clusters for a single threshold value, the largest cluster (i.e., the cluster composed of the highest number of pixels) is used for the area comparison. Each of the resulting cluster is subsequently subjected to MATLAB’s regionprops function (with ‘centroid’ as an input argument) to locate its centroid (Figure 2A(ii.ii), (ii.iii), yellow x’s). The distance between this cluster centroid and the reference point (i.e., cluster centroid in the topmost image of the stack; Figure 2A(ii.i), black x) is calculated for each image, and the calculated values are sorted according to the image number (Figure 2A(iii)), with the images in the stack numbered from top to bottom (i.e., the topmost image was image 1). After filtering the sorted distance values with a 5-point moving average filter, the algorithm identifies the image number at which the filtered distance value starts to flatten (Figure 2A(iii), magenta line) by first calculating the approximate derivative of the filtered values (i.e., difference between two consecutive filtered values), then filtering the approximate derivative using a 19-point moving average filter, and finally finding the first instance where the filtered derivative value exceeds the original derivative value. The algorithm assigns the z-coordinate of the corresponding image as the z-coordinate of the pipette tip (Figure 2A(iii), zpipette).
After assigning the z-coordinate of the pipette tip, the algorithm calculates the angle between the cluster centroid in image 1 (Figure 2A(ii.i), black x) and the cluster centroid in the image at zpipette, which is assigned as the pipette angle in the transverse plane. Subsequently, the image at zpipette, smoothed by a 2D Gaussian filter (19 × 19 window; 9/2 variance) (Figure S3A(i)), is segmented using a range of threshold values (corresponding to 5 – 75% of the maximum pixel intensity of the filtered image). The algorithm then analyzes the resulting clusters (Figure S3A(ii), white) for the endpoint detection (using the bwmorph function with ‘endpoints’ operation in MATLAB; Figure S3A(ii), yellow boxes); in case of multiple clusters for a single threshold value, the largest cluster (i.e., the cluster composed of the highest number of pixels) is analyzed for the endpoint detection. The lowest threshold value at which the resulting cluster has 3 vertices or endpoints (Figure S3A(ii.ii), yellow boxes) is considered optimal for isolating the pixels that accurately represent the entire body of the pipette tip, and the corresponding cluster of pixels (Figure S3A(ii.ii), white) is further analyzed for the pipette tip detection; other threshold values that are higher or lower than the optimal value result in clusters that have less or more than 3 endpoints, and are not further analyzed (example images of the clusters resulting from a threshold value higher and lower than the optimal value shown in Figure S3A(ii.i) and (ii.iii) respectively, with the yellow boxes representing the endpoints of these clusters). Out of all the pixels in the cluster resulting from the optimal threshold value, the one that is the furthest away from the centroid of the cluster is identified. Its distance to the centroid is then used as the length of a line (Figure S3A(iii), yellow dotted line) pointing in the direction of the pipette angle in the transverse plane (the angle that was determined earlier as described above) and emanating from the cluster centroid (Figure S3A(iii), yellow x). The pixel in the cluster closest to the endpoint of the line is assigned as the tip of the pipette in the transverse plane (Figure S3A(iii), yellow star; Figure 2A(iv), yellow star), and the location of the pixel in the image is assigned as the x and y coordinates of the pipette tip. See “find_one_pipette_HJS.m” and “pipette_tip_detection_HJS.m” in the Methods S1 for MATLAB codes running the algorithm.
Details of the cell position detection algorithm
The cell position detection algorithm begins its operation by subjecting each image in the z-stack of a tdTomato-expressing cell to a 2D Wiener filter (3 × 3 window), removing the background noise. Each filtered image (example filtered image shown in Figure S3B(i)) is then segmented using a range of threshold values (corresponding to 5 – 95% of the maximum pixel intensity of the filtered image), and the area of (i.e., the number of pixels in) the resulting clusters of pixels (Figure S3B(ii), white) is compared to a reference area (i.e., the area of the target cell chosen by the user during the target cell detection and selection stage); in case of multiple clusters for a single threshold value, the cluster whose centroid is the nearest to the image center and whose area is the closest to the reference area is used for the area comparison. The threshold value at which the resulting cluster of pixels has the area closest to the reference area is considered optimal for identifying the pixels accurately representing the target cell soma, and is isolated by the algorithm from other threshold values that lead to clusters of pixels that represent only a small portion of the cell body (example image of such a cluster shown in Figure S3B(ii.i)) or capture background pixels (example image of such a cluster shown in Figure S3B(ii.iii)). The cluster obtained by using the optimal threshold value (Figure S3B(ii.ii)) is then subjected to MATLAB’s bwboundaries function to determine the pixels that represent the boundary (Figure S3B(ii.ii), red outline; Figure 2B(ii), red outline) and the interior (Figure S3B(ii.ii), white; Figure 2B(ii), area inside the red outline) of the cross-section of the cell body captured by each image in the stack. The centroid of the cluster (Figure S3B(iii), red x) is also determined using MATLAB’s regionprops function (with ‘centroid’ as an input argument). The algorithm then calculates the mean intensity of pixels representing the interior of the cell body from each image and sorts the calculated values according to the image number, with the images in the stack numbered from top to bottom (i.e., the topmost image was image 1; example plot of the mean pixel intensity as a function of image number shown in Figure 2B(iii)). The image capturing the cross-section of the cell body with the highest mean pixel intensity (i.e., the brightest cross-section of the cell body; Figure 2B(iii), magenta line) is considered to be focused on the centroid of the cell body, and its z-coordinate is assigned as the z-coordinate of the cell centroid (Figure 2B(iii), zcell). Subsequently, the x and y coordinates of the cluster centroid in the image at zcell (Figure S3B(iii), red x; Figure 2B(iv), red x), which is determined along with those of the clusters in other images in the stack as described above, are assigned as the x and y coordinates of the cell centroid. See “soma_contour_detection.m” and “image_autopatcher_v1.m” in Methods S1 for MATLAB codes running the algorithm.
Micromanipulator-microscope platform calibration
Before performing imagepatching experiments, step sizes of motion and axis angles of the micromanipulator were automatically calibrated to those of the motorized platform of the two-photon microscope, which moved the microscope objective relative to the sample to be imaged, using the imagepatcher. In the first stage of calibration, which was performed once upon initial hardware setup, a patch pipette filled with 50 μM Alexa 488 dye (in deionized water) was installed into the pipette holder, and its tip was manually moved to the center of the field-of-view using the rotary knob controller of the 4-axis micromanipulator. The expected angle below the horizontal for each of the 4 micromanipulator axes was then specified by typing in a value in the corresponding text box on the imagepatcher GUI (for our micromanipulator, 30 for diagonal, 0 for x, 0 for y, and 90 for z-axis). Subsequently, the calibration of one of the micromanipulator axes was initiated by clicking one of four pushbuttons displayed on the GUI, each button corresponding to the calibration of each of the 4 axes of the micromanipulator. Pressing the pushbutton started the acquisition of a z-stack (20 images, 2 μm step size, 17× zoom) around the pipette. The location of the pipette tip was then automatically identified using the pipette tip detection algorithm (Figure 2A), and the imagepatcher program sent a command to the micromanipulator to move the axis being calibrated forward by a pre-set distance (30 μm for the diagonal axis; 25 μm for x, y, and z axes). Following the micromanipulator movement, the microscope objective was moved to the expected pipette tip position; the expected pipette tip position was calculated by first mapping the micromanipulator axis movement to the x, y, and z directional movements of the motorized platform of the microscope, using both the pipette angle in the transverse plane, which was obtained from the z-stack using the pipette tip detection algorithm (Figure 2A), and the expected angle below the horizontal of the moved axis (e.g., when a pipette facing dead-right moves along the diagonal axis by 30 μm, the pipette tip moves by 30·cos(30º)·cos(0º) ≈ 26 μm in the x-direction, 30·cos(30º)·sin(0º) = 0 μm in the y-direction, and 30·sin(30º) = 15 μm in the z-direction), and then adding the mapped values to the original pipette tip location. With the microscope objective at the expected pipette tip position, another z-stack (20 images, 2 μm step size, 17× zoom) was acquired, and the actual pipette tip location was determined using the pipette tip detection algorithm (Figure 2A). The angle and the distance between the new tip location and the original tip location were then calculated in the angle and distance units of the microscope’s motorized platform. Subsequently, the calculated values were assigned to the forward movement of the micromanipulator axis being calibrated. For the backward movement calibration, the program moved the axis of the micromanipulator backward by the pre-set distance (30 μm for the diagonal axis; 25 μm for x, y, and z axes) and then detected the resultant pipette tip location. As done for the forward movement calibration, the angle and the distance between the tip locations before and after the backward movement were calculated in the microscope’s motorized platform units, and the calculated values were assigned to the backward movement of the micromanipulator axis being calibrated. The forward and backward movement calibrations were then repeated for other pre-set distances (55, 150, 320, 350, 420, and 480 μm for the diagonal axis; 150, 325, 400, and 460 μm for x and y axes; 50, 150, 200, and 250 μm for the z axis). Once the calibration for the axis was complete, the calibration results were saved into a.m file, which we could load to our program for future imagepatching experiments. Without replacing the patch pipette, each of the rest of the micromanipulator axes was calibrated in the same way as described above.
The second stage of calibration was implemented to account for a variability in the locked position of the pipette, which stemmed from a lever-based locking mechanism of our micromanipulator. Unlike the first stage that was performed only once, the second stage of calibration was automatically executed for each imagepatching experiment at the start of the brain penetration stage (Figure 1B(iii)). Right before performing the second stage of calibration, the amount that each micromanipulator axis would have to move to reach the targeted position inside the brain (i.e., 50 μm directly above the target cell centroid) was calculated using the calibration results from the first stage. Subsequently, the new calibration results were obtained by moving each micromanipulator axis by the calculated amount and finding the new pipette tip location following the axis movement; the new pipette tip location was determined using the same procedure implemented for the first stage of calibration, except the expected pipette tip position after the micromanipulator movement was calculated using the calibration results from the first stage instead of the expected angle below the horizontal specified in the imagepatcher GUI. See “find_cells_gui_SI.m” in Methods S1 for MATLAB code executing the calibration.
Details of the imagepatcher operation
At the start of the imagepatching experiment, we opened ScanImage and imaged the brain inside the craniotomy to visually determine the location of the brain surface. The z-position of the objective corresponding to where we found the brain surface was denoted as the z-coordinate of the brain surface by the imagepatcher. We then specified the number of images (5 – 10), the step size between two consecutive images (5 – 10 μm), and the starting depth (100 – 250 μm) of a z-stack to be acquired inside the brain by using the corresponding text boxes on the imagepatcher GUI. The z-stack acquisition (and the target cell detection and selection stage; Figure 1B(i)) was started by pressing the corresponding pushbutton on the imagepatcher GUI, and a display window inside the GUI sequentially showed the most recently acquired image during the stack acquisition. At the end of the stack acquisition, the GUI also showed the list of acquired images, which could be used to select images to display in the display window and to run automated cell detection on. After choosing 1 – 3 images, each capturing at least one or two bright cells (visually assessed), by clicking the images in the list, we set the minimum brightness (specified as percentage of the maximum pixel intensity of each of the selected images) of cells to detect to any value between 10 and 25% while setting the desired minimum and maximum cell body radii to 3 and 15 μm respectively by using the corresponding text boxes on the GUI. Out of all the tdTomato-expressing PV-positive neurons captured by the images, only those that met our detection criteria were identified by the imagepatcher and shown with red outlines in the display window of the GUI; the imagepatcher detected the cells by smoothing each of the selected images using a 2D Wiener filter (3 × 3 window), then segmenting each of the filtered images using the minimum brightness as the threshold value, and finally determining the boundary of resulting objects that met our radii specifications (see “find_center_and_circle_soma_cell_radius_range.m” in Methods S1 for MATLAB code responsible for the cell detection). To conclude the target cell detection and selection stage, we chose a target cell to patch by clicking the interior of one of the outlined cells in the display window, which turned the outline of the selected cell to yellow and registered the information about the target cell (x, y coordinates of the target cell centroid, z-coordinate of the target cell depth, area of the target cell, minimum brightness threshold for the cell detection) in the imagepatcher.
Following the target cell selection, we initiated the pipette tip detection stage (Figure 1B(ii)) by clicking the corresponding pushbutton on the GUI. This stage began with the autopatcher control box outputting 15 mBar to the pipette while the imagepatcher moved the microscope objective vertically to position the objective away from the brain surface and to provide enough space for a patch pipette. We then filled a pipette with the internal solution, installed it into the pipette holder, and used the rotary knob controller of the 4-axis micromanipulator to manually move the pipette tip to the center of the objective field-of-view (FOV). While imaging the pipette via ScanImage, we used a slider presented on the imagepatcher GUI to adjust pipette pressure to a value that minimized a plume of dye at the pipette tip and subsequently made the pipette tip clearly visible (visually assessed; typically 6 to 8 mBar). A z-stack (20 images, 2 μm step size, 17× zoom) was then acquired around the pipette tip by pressing the corresponding pushbutton on the GUI. From this stack of images, the imagepatcher detected the pipette tip (using the pipette tip detection algorithm described in Figure 2A) and logged the maximum intensity of pixels representing the pipette in the image focused on the pipette tip.
Next, the brain penetration stage (Figure 1B(iii)) was started by clicking the corresponding pushbutton on the GUI. Before moving the pipette into the brain, step sizes of motion and axis angles of the micromanipulator were automatically calibrated to those of the motorized platform of the two-photon microscope (i.e., the second stage of micromanipulator calibration was performed; see “Micromanipulator-microscope platform calibration” section for details); in subsequent steps of the imagepatching process, the imagepatcher calculated the pipette tip location by adding calibrated micromanipulator axes displacements to the original pipette tip location before the micromanipulator movement. Following the calibration, the imagepatcher calculated a linear path along the diagonal axis of the micromanipulator (i.e., a trajectory parallel to the pipette axis) with the start and end points located 25 μm above the brain surface and 50 μm directly above the target cell centroid respectively. The micromanipulator was then automatically moved to bring the pipette tip to the start point of the calculated path, and 600 mBar was applied to the pipette by the autopatcher control box. At this point, the imagepatcher measured the pipette resistance for 5 seconds and displayed the result; if the displayed value was outside of an acceptable range (e.g., 5 – 7.5 MΩ), it was assumed that the pipette was clogged with some undesired particles (in case of resistance greater than 7.5 MΩ) or the pipette tip was broken (in case of resistance less than 5 MΩ), and we retracted the pipette to install a new one. For the pipette with a resistance value within the acceptable range, the imagepatcher logged the pipette resistance and then moved the pipette along the calculated path into the brain. Once the pipette tip entered the brain and was positioned 75 μm above the target cell centroid (i.e., 25 μm above the endpoint of the calculated trajectory), dye ejection at the pipette tip was examined by the imagepatcher to check the pipette tip quality; if an image (17× zoom) capturing the pipette tip at the image center had either the maximum pixel intensity at least 2 times higher than that logged at the end of the pipette tip detection stage (i.e., maximum intensity of pixels representing the pipette in the image focused on the pipette tip, acquired outside the brain), or the median of pixel intensities at least 40% of the maximum pixel intensity of the image and the maximum pixel intensity at least as high as the maximum intensity logged at the end of the pipette tip detection stage, the imagepatcher considered the pipette to be clean (see STAR Methods, Derivation of dye-ejection based pipette blockage test for derivation of these criteria). Once the clean pipette reached the end point of the calculated path (i.e., 50 μm above the target cell centroid), its pressure was automatically lowered to 300 mBar, and the imagepatcher checked the pipette tip for clogging by measuring the pipette resistance and comparing the measured value to the value obtained outside the brain (i.e., 25 μm above the brain surface), and by performing another evaluation of dye ejection. After this quality check, the imagepatcher acquired a z-stack (24 images, 3 μm step size, 17× zoom) around the original target cell centroid (i.e., target cell centroid logged at the end of the target cell detection and selection stage) and determined the target cell position (using the cell position detection algorithm described in Figure 2B) to update the target cell location. The imagepatcher then logged this new cell position and moved the x, y, and z axes of the micromanipulator such that the pipette tip would be 25 μm directly above the updated target cell centroid. After the micromanipulator movement, the dye ejection and the pipette resistance were again examined by the imagepatcher to check the pipette tip for clogging, concluding the brain penetration stage.
At the start of the closed-loop real-time image-guided pipette positioning stage (Figure 1B(iv)), the pipette pressure was automatically lowered to 100 mBar, and another z-stack (10 images, 2 μm step size, 17× zoom) was automatically acquired around the target cell centroid logged during the brain penetration stage. After finding the coordinates of the target cell centroid from the stack (using the cell position detection algorithm described in Figure 2B), the imagepatcher entered the closed-loop (Figure 1A), repeatedly updating the cell centroid location and positioning the pipette tip directly above the cell centroid following each 3 μm-pipette step in the z-direction. Every z-step of the pipette was followed by automatic acquisition of two images; to support repeated cell centroid detection while minimizing image acquisition time, one image, instead of a full z-stack, was captured at the z-coordinate of the cell centroid determined at the start of the stage, with an assumption that small z-steps used in the closed-loop would cause negligible movement of the cell in the z-direction; the second image was acquired at the calculated location of the pipette tip to check for dye ejection and subsequently verify the pipette tip quality as done in the preceding stage. The imagepatcher also measured the pipette resistance and logged the measured value after each pipette step in the z-direction to monitor changes in the pipette resistance. When the imagepatcher detected a small resistance increase while approaching the target cell, the pipette pressure was automatically lowered to 30 mBar and the current pulses at the pipette tip were checked for another increase in resistance as well as heartbeat modulation, both of which indicated tight contact between the pipette tip and the cell membrane.
Once this resistance increase and heartbeat modulation were detected, the imagepatcher initiated the gigaseal formation stage (Figure 1B(v)). In this stage, the pipette movement was stopped, and suction as well as hyperpolarizing voltage were automatically applied to form a gigaohm seal between the pipette and the cell membrane. Once a stable gigaseal was established (i.e., the pipette resistance stayed above a gigaohm and did not increase by more than 15% over a 15-second period), the imagepatcher GUI displayed a pushbutton for starting the break-in process. By clicking this pushbutton, we started the break-in stage (Figure 1B(vi)). At the start of the stage, the imagepatcher established a baseline of cell filling by the pipette dye (i.e., determined the amount of pipette dye inside the target cell) by acquiring a z-stack (10 images, 2 μm step size, 17× zoom) around the cell, identifying pixels corresponding to the cell body in the focused image of the stack using the cell position detection algorithm described in Figure 2B, and calculating the mean pixel intensity of the cell body in the microscope channel corresponding to the pipette dye (channel 2 for our microscope). Subsequently, suction pulses were applied in a ramp by the autopatcher control box while monitoring the seal resistance. Once the resistance dropped below a value characteristic of the whole-cell state, the imagepatcher imaged the target cell again (at the cell depth and centroid determined from the previous z-stack; 1 image, 17× zoom) to calculate the mean pixel intensity within the cell boundary in channel 2. When the new mean value was at least 15% higher than the original value obtained before the suction pulses, the imagepatcher considered the cell to be filled sufficiently with the pipette dye and concluded the break-in stage. Once the imagepatcher operation was complete, we recorded signals from cells that had achieved a successful whole-cell state, which we defined as that requiring no more than 500 pA current injection to hold the cell at −65 mV (i.e., exhibiting holding current less than or equal to 500 pA) in the voltage-clamp mode, as we did previously (Kodandaramaiah et al., 2012). See Imagepatcher User Manual in Methods S1 for detailed description on how to interact with the imagepatcher GUI for an imagepatching experiment.
Performance of the pipette tip detection algorithm at angles other than 30º below the horizontal
15 pipettes all angled at 25º below the horizontal were each imaged using a z-stack (2 μm step size, 20 images, 17× zoom), with each z-stack capturing the pipette tip at a distinct position within the stack. By applying the pipette tip detection algorithm (Figure 2A) to each of the 15 z-stacks, we found that the algorithm yielded pipette tip locations that were close to those determined visually, with the tip positions from the algorithm and from visual assessment differing by (mean ± s.d.) −1.5 ± 1.4 μm, 0.1 ± 1.0 μm, and 1.6 ± 3.0 μm in the x, y, and z directions respectively. The pipette tip detection algorithm also enabled accurate tip detection from another 15 z-stacks (2 μm step size, 20 images, 17× zoom) that each captured the tip of a separate pipette angled at 35º below the horizontal at a distinct position within the stack; the tip locations determined by the pipette tip detection algorithm from the 15 z-stacks were (mean ± s.d.) −0.7 ± 0.5 μm, −0.5 ± 0.8 μm, and 0.4 ± 1.4 μm off of the visually assessed tip positions in the x, y, and z directions respectively.
Derivation of pipette pressure for brain entry and cell approach
To determine the optimal pipette pressure for entering the brain during the brain penetration stage (Figure 1B(iii)), a few penetrations were performed with 100 mBar, 200 mBar, 400 mbar, 600 mBar, and 800 mBar pressure applied at the back of the pipette (these pressure values were chosen based on previously reported protocols for fully manual two-photon image-guided or blind patch clamp recordings in vivo (Häusser and Margrie, 2014; Komai et al., 2006; Lee et al., 2009; Margrie et al., 2002), in which pipette pressure ranging from 100 mBar to 800 mBar were typically used). For our experiments, we used Alexa 488-filled patch pipettes angled at ~30º below the horizontal, moving ~300 – 400 μm in the diagonal direction (i.e., parallel to the pipette axis), from outside the brain near the brain surface to inside the brain (i.e., ~150 – 200 μm deep in the cortex; layer 2–3 for adult mouse brain (Altamura et al., 2007; Mountcastle, 2003)), at ~600 μm/s (i.e., the maximum speed that our 4-axis micromanipulator could generate under software control). As expected, higher pressure (i.e., 600 mBar and 800 mBar) produced less pipette blockage compared to lower pressure values. However, 800 mBar led to much background signal that caused bleed-through of the Alexa 488 signal into the imaging channel used to visualize tdTomato, making it difficult to resolve tdTomato-expressing cells after brain penetration. As a result, we decided to focus on 600 mBar and found the pressure to cause a reasonably low pipette blockage rate (12.5%; 2 out of 16 trials; 3 PV-Cre x Ai14 mice) when used as the pipette pressure for brain penetration. We therefore chose to implement 600 mBar as the pipette pressure for brain entry during the brain penetration stage of the imagepatching algorithm (Figure 1B(iii)).
The optimal set of pipette pressure levels for approaching the cell once inside the brain was determined based on our manual patching experiments as well as the values provided in previously reported protocols for manual patching (Häusser and Margrie, 2014; Komai et al., 2006). During our manual slice and in vivo patching experiments, we had a few trials in which a gigaohm seal was quickly obtained with an Alexa 488-filled pipette that continuously ejected a plume of dye at its tip until right before it made contact with the target cell. From these trials, we inferred that one of the most critical prerequisites of gigaseal formation might be maintaining a “clean” pipette tip throughout its movement inside the tissue, all the way up to the point where it makes contact with the target cell (this assumption agrees with what has been briefly described in Margrie et al., 2002). During our initial experiments, we observed that the pipette would often cease to eject the dye at its tip (i.e., the pipette was contaminated, as described in Komai et al., 2006) while moving inside the tissue at 20 – 40 mBar, which is the range of pipette pressure values used in previously reported protocols for two-photon image-guided patching in vivo (Häusser and Margrie, 2014; Komai et al., 2006). We tried a few different pressure values that were higher than 40 mBar, and found 100 – 300 mBar to be high enough to keep the pipette tip blockage rate reasonably low (21.7%; 5 out of 23 trials; 5 PV-Cre x Ai14 mice) while navigating the pipette inside the tissue, but not so high as to cause bleed-through of the pipette dye fluorescence into the tdTomato imaging channel. However, we also noticed that when the pipette pressure was ~300 mBar, the cells were often “blown away” by the pipette even when the pipette tip was somewhat distant from the target cell (e.g., ~10 – 20 μm from the target cell membrane, which corresponded to ~20 – 30 μm distance between the pipette tip and the center of a ~20 μm diameter cell). To prevent the pipette blowing away the target cell, we tried a few combinations of distances and pressure values for approaching the target cell. As a result, we found a pressure of ~100 mBar to keep the cell in place right until the pipette tip made contact with the cell membrane (contact between the pipette tip and the cell membrane was visually assessed by observing the presence of heartbeat modulation as in Komai et al., 2006; n = 17 tdTomato-expressing PV-positive cells from 3 PV-Cre x Ai14 mice). We therefore decided to implement three different pressure values in the brain penetration (Figure 1B(iii)) and closed-loop real-time image-guided pipette positioning (Figure 1B(iv)) stages of the imagepatching algorithm while approaching the target cell: (i) 600 mBar for moving the pipette into the brain through upper cortical layers to 75 μm away from the center of the target cell; (ii) 300 mBar for moving the pipette to 25 μm away from the center of the target cell; (ii) 100 mBar for making the final approach to the cell.
Optimization of cell-pipette contact detection, gigaseal formation, and break-in
Akin to manual two-photon image-guided patching (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2003) and our previously developed blind autopatching method (Kodandaramaiah et al., 2012, 2016), imagepatching uses an increase in pipette resistance as a signal for when the pipette tip contacts the cell membrane. To determine the amount of resistance increase corresponding to a cell membrane-pipette tip contact amenable to a gigaohm seal, we tried forming a gigaseal (by applying slight suction to the pipette and hyperpolarizing the pipette) after observing resistance increases over consecutive z steps (3 μm step size). After a few trials on tdTomato-expressing PV-positive neurons in somatosensory and motor cortices of anesthetized PV-Cre x Ai14 mice, we found that a 20 – 50% increase in pipette resistance, which is the amount visually assessed during manual patching when the pipette tip-cell membrane contact is amenable to gigaseal formation (Häusser and Margrie, 2014; Margrie et al., 2002, 2003), was hard to observe when the increase in resistance was calculated over a single z step (i.e., over 3 μm; step size of 3 μm was used for single z steps throughout cell-pipette contact detection optimization described below) of a pipette at 100 mBar. This characteristic resistance increase was observable by taking one or more additional steps towards the target cell after seeing some resistance increase over 3 μm, but these extra steps sometimes damaged the cell membrane, resulting in cell lysis. Even when the cell seemed intact, releasing the positive pressure and applying light suction following the 20 – 50% resistance increase over 2 or more consecutive z steps (i.e., over 6 μm or more) did not improve the rate of forming a gigaseal compared to the same pressure modulation following a single z step with less resistance increase, maybe because the pipette tip was pushed too much into the target cell, damaging the cell membrane. As expected, there were instances in which analyzing the pipette resistance over a single z step seemed “noisy”, showing some resistance increases even when the pipette tip was far away from the target cell membrane. However, we could minimize the number of such false positives by analyzing the resistance increase only when the distance between the pipette tip and the cell centroid was small enough that a contact was likely (e.g., the pipette tip-cell centroid distance was less than the radius of the target cell). As a result, we decided to closely examine the resistance increase over a single z step of a patch pipette as the tip of the pipette at 100 mBar was brought in contact with the cell membrane, aiming to determine the amount of resistance increase that subsequently maximized the rate of forming a gigaseal. Since most of the single z steps led to a pipette resistance increase of less than 1% when the pipette tip was far away from the cell membrane, we studied the relationship between the amount of resistance increase of at least 1% over a single z step and the likelihood of successful gigaseal formation. By analyzing 46 gigaseal formation attempts (in 14 mice), in which we released positive pressure and applied suction using the autopatcher control box immediately after observing a resistance increase of 1% or more over a single z step, we found that resistance increases by 4% or more over a single z step led to successful gigaseal formation 22.2% of the time (n = 4 gigaseals out of 18 attempts), while single z steps with a resistance increase between 1% and 4% yielded a higher rate of successful gigaseal formation of 32.1% (n = 9 gigaseals out of 28 attempts). Out of the 28 attempts with a resistance increase between 1% and 4% over a single z step, we also found that single z steps with a resistance increase between 1 and 2% were more likely to result in successful gigaseal formation (50%; n = 4 gigaseals out of 8 attempts) compared to those with higher amounts of resistance increase (e.g., 27.3%, or n = 3 gigaseals out of 11 attempts, for single z steps with a 2 – 3% resistance increase; 22.2%, or n = 2 gigaseals out of 7 attempts, for single z steps with a 3 – 4% resistance increase). As a result, we decided to use a resistance increase by 1% over a single z step (i.e., an increase by ~60 – 70 kΩ over 3 μm for 6 – 7 MΩ pipettes) as the threshold in the imagepatching algorithm for the detection of contact between the pipette tip and the cell membrane.
Once the pipette tip made contact with the cell membrane, the micromanipulator was halted to keep the pipette stationary, and the pipette pressure was lowered from 100 mBar to 30 mBar to prepare the target cell for gigaseal formation. From a few manual patching experiments, we found that this reduction in pipette pressure led to an increase in pipette resistance by ~1.5% or more when the pipette tip was in close contact with the cell membrane, perhaps because the lower positive pressure allowed the cell membrane, which had been displaced away from the pipette tip due to the higher positive pressure, to spring back towards the pipette tip. In addition to the resistance increase, we could also observe heartbeat modulation of the pipette current pulses as described before (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2002). The metric used to quantify heartbeat modulation was derived based on our finding during manual patching that, when the pipette tip was in close contact with the cell membrane, current pulses resulting from 50 Hz square waves of injected voltage had a characteristic variation in their resistance values when obtained over 1 second. Specifically, we observed that when the pipette tip at ~30 mBar was in tight contact with the cell membrane, the resistance values of 50 current pulses (corresponding to a 1-second long measurement; resistance was calculated by dividing the amplitude of the injected voltage by the amplitude of an observed current pulse) showed a standard deviation of at least 0.1 MΩ, and a subsequent suction led to a gigaseal (n = 6 neurons, 6 mice). We therefore used the standard deviation of the current pulse resistances calculated over 1 second at 30 mBar pipette pressure as a second criterion for detecting contact between the pipette tip and the cell membrane. Even when the resistance increase was less than ~1.5% or the amount of heartbeat modulation was less than 0.1 MΩ, a tight contact amenable to gigaseal formation could be achieved by first increasing the pipette pressure back to 100 mBar (to clear the pipette tip in case it became partially occluded while at 30 mBar) and then advancing the pipette tip by one or more additional z steps until a resistance increase by ~1.5% or more was observed. We mimicked this resistance-based check for the pipette tip-cell membrane contact at 30 mBar in the imagepatching algorithm by first performing two consecutive resistance measurements in response to 50 Hz square waves, each for 1 second, then comparing each of the average resistance values (each taken over 1 second) to the pipette resistance observed before reducing the pipette pressure to 30 mBar, and finally resuming the pipette z steps if the resistance increase was less than 1.5% or if the standard deviation of current pulse resistances was less than 0.1 MΩ (see Figure S2, “closed-loop real-time image-guided pipette positioning,” for details).
After checking the pipette tip-cell membrane contact at 30 mBar, a ramp of suction (−20 mBar/s) was applied to reach a final value of −20 mBar, to achieve a gigaohm seal. If the rate of resistance increase was slow (i.e., the pipette resistance measured 10 seconds after the suction reached the final value of −20 mBar was less than 2 times the resistance measured right before applying the ramp of suction), stronger suction up to −100 mBar was applied in a ramp at −20 mBar/s; negative pressure stronger than −100 mBar was not implemented based on our observation during manual trials that suction levels more negative than −100 mBar did not help to form a gigaseal, but instead led to premature and leaky break-ins. The holding voltage was also set to −65 mV as the seal was being formed. If the pipette resistance did not reach 300 MΩ within 5 seconds after applying the hyperpolarizing voltage, the pressure level was modulated among 0, 25, and −20 mBar to “coax” the cell membrane into forming a gigaseal. This coaxing process was developed from our manual patching experiments in which alternating the pipette pressure among 0, 25, and −20 mBar led to a gigaseal formation for some of the trials that seemed to be failing (with the pipette resistance increasing very slowly or becoming stagnant following the suction and hyperpolarization), and it was required for 8 out of 42 gigaseals when targeting PV-positive neurons (in 16 PV-Cre x Ai14 mice) and for 4 out of 19 gigaseals when targeting CaMKIIα-positive neurons (in 10 CaMKIIα-Cre x Ai14 mice) during the imagepatching algorithm validation. If more than 5 minutes were needed for the pipette resistance to exceed a gigaohm, then the pipette was retracted from the brain to start a new trial. Otherwise, the pipette resistance was continuously recorded until it reached a stable value, not increasing by more than 15% over 10 seconds (see Figure S2, “gigaseal formation,” for details).
The break-in process was developed based on a combination of previous developed protocols (Häusser and Margrie, 2014; Komai et al., 2006; Margrie et al., 2003) and our experience with manual patching. Three consecutive pulses of suction were applied, with each pulse increasing in suction from 0 mBar to −25 mBar at −20 mBar/s and then returning to 0 mBar. These suction pulses were repeatedly applied every 5 seconds until the whole-cell configuration was verified with both visual (i.e., the target cell being filled with the pipette dye, indicated by an increase of at least 15% in the mean pixel intensity inside the patched cell boundary in the pipette dye channel) and electrical (i.e., resistance recorded < 300 MΩ) indications. If the whole-cell state could not be achieved after three consecutive pulses, the suction endpoint was lowered by 25 mBar, and suction pulses were again applied to the cell. If a suction endpoint lower than −350 mBar had to be applied, the pipette was retracted to start a new trial. With this algorithm, we were able to achieve the whole-cell configuration successfully from a gigaseal ~57% of the time when targeting PV-positive neurons (n = 24 out of 42 gigaseals in 16 PV-Cre x Ai14 mice) and ~68% of the time when targeting CaMKIIα-positive neurons (n = 13 out of 19 gigaseals in 10 CaMKIIα-Cre x Ai14 mice) during the imagepatching algorithm validation, where a successful whole-cell state is defined as that with less than 500 pA of leakage current when held at −65 mV in voltage-clamp mode.
Derivation of dye-ejection based pipette blockage test
We derived an algorithm for determining the amount of dye being ejected at the pipette tip, which indicates the pipette quality (Komai et al., 2006), from test images of clean and contaminated pipettes inside the somatosensory and motor cortices of anesthetized PV-Cre x Ai14 mice (3 mice). We used 12 pipettes, angled at ~30º below the horizontal, positioned ~150 – 200 μm deep inside the brain, applying ~300 mBar at the tip, to obtain test images (17× zoom), from which characteristic features were analyzed. We captured images of these pipettes both outside the brain (at ~6 – 8 mBar to make the pipette tip visible) and inside the brain (at ~300 mBar, right after entering the brain at ~600 μm/s). Out of 12 pipettes, 7 were “clean”, clearly showing the dye being ejected at the tip, while the rest were occluded. By comparing these two groups, we found that all of the images capturing a clean pipette possessed either or both of the following characteristics: (i) the maximum pixel intensity of the image captured inside the brain was at least 2 times higher than the maximum pixel intensity of the image captured outside the brain; (ii) the maximum pixel intensity of the image captured inside the brain was at least as high as the maximum pixel intensity of the image captured outside the brain, and the median of the pixel intensities of the image captured inside the brain was at least 40% of the maximum pixel intensity of the image captured inside the brain. In contrast, none of the blocked pipette images showed the above characteristics. We thus decided to use conditions (i) and (ii) for the pipette tip quality check during the imagepatching operation.
Quantification of PV-positive and CaMKIIα-positive cell densities
Z-stacks of two-photon images (each z-stack with five 223.5 × 223.5 μm2 images and 10 μm step between consecutive images) were acquired at a depth of ~100 – 250 μm in somatosensory or motor cortex of anesthetized PV-Cre x Ai14 mice or CaMKIIα-Cre x Ai14 mice. One or more z-stacks were acquired per mouse, with each stack at different depth and lateral location within the craniotomy. Cells expressing tdTomato were counted manually from each of the z-stacks and then scaled to give the number of cells in a volume of 200 × 200 × 100 μm3.
Input resistance and spontaneous firing rates of imagepatched cells
Input resistance (Figure S4D) of an imagepatched cell was determined by repeatedly injecting a hyperpolarizing current pulse (−100 pA, 1 second long) to the cell right after achieving the whole-cell configuration, then calculating the average of membrane voltage over two 100 ms periods (one right before current injection and another at the end of current injection), and finally dividing the absolute value of the difference between the average values of two 100 ms periods by 100 pA. Spontaneous firing rate (Figure S4E) of an imagepatched cell was determined by calculating the frequency of action potentials about 4 to 5 minutes after break-in over a period of one minute.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical details for comparing the recording quality metrics between the imagepatched and fully manually patched PV-positive neurons (Figures S4A–S4C) are provided in the Results section. The p-values associated with the Student’s t-Test were calculated using ttest() function in Excel 2013, with Tails parameter = 2 for two-tailed distribution and Type parameter = 3 for two-samples with unequal variance.
DATA AND SOFTWARE AVAILABILITY
The Imagepatcher software and the user guide are included as Methods S1.
Supplementary Material
Methods S1. MATLAB files and user manual for the Imagepatcher
Acknowledgments
We thank S. Komai (Nara Institute of Science and Technology, Japan), L. Gentet (Lyon Neuroscience Research Center, France), A. Pala (Georgia Tech., USA), I-W. Chen (Université René Descartes, France) for advice and information on manual two-photon image-guided patch clamp recordings in vivo. We would also like to acknowledge D. Park (MIT, USA) for assistance with manual image-guided patch clamp recordings in acute brain slices. We thank Li-Huei Tsai and Chinnakkaruppan Adaikkan (MIT, USA) for supplying CaMKIIα-Cre x Ai14 mice. We thank T. Diefenbach (Harvard University, USA) for advice and information on the Ultima IV two-photon microscope, and C. Deister (Brown University, USA) as well as C. Sun (MIT, USA) for assistance with operating the Ultima IV two-photon microscope with ScanImage 3.8. We also thank I. Wickersham (MIT, USA) for advice on the autopatcher control box assembly. H.-J.S. acknowledges the Samsung Scholarship. E.S.B. acknowledges support by Jeremy and Joyce Wertheimer, NIH 1R01NS102727, NIH 1R01EY023173, NIH 1R01MH103910, NIH Director’s Pioneer Award 1DP1NS087724, the MIT Synthetic Intelligence Project, the MIT Media Lab, the HHMI-Simons Faculty Scholars Program, and the New York Stem Cell Foundation–Robertson Award. All of the authors on the paper are inventors on a patent application describing this invention.
Footnotes
AUTHOR CONTRIBUTIONS
H.-J.S., I.v.W., S.B.K., C.R.F., and E.S.B conceived strategies for automating image-guided patch clamp recordings in vivo. H.-J.S. and I.v.W. conducted experiments, analyzed data, and conceived the imagepatching algorithm. H.-J.S. wrote the MATLAB program for the imagepatcher. B.A. assisted with experiments and optimization of portions of the imagepatching algorithm. E.S.B. supervised the project. H.-J.S., I.v.W., and E.S.B. wrote the paper, with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altamura C, Dell’Acqua ML, Moessner R, Murphy DL, Lesch KP, Persico AM. Altered neocortical cell density and layer thickness in serotonin transporter knockout mice: A quantitation study. Cereb Cortex. 2007;17:1394–1401. doi: 10.1093/cercor/bhl051. [DOI] [PubMed] [Google Scholar]
- Atallah BV, Bruns W, Carandini M, Scanziani M. Parvalbumin-Expressing Interneurons Linearly Transform Cortical Responses to Visual Stimuli. Neuron. 2012;73:159–170. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IW, Helmchen F, Lutcke H. Specific Early and Late Oddball-Evoked Responses in Excitatory and Inhibitory Neurons of Mouse Auditory Cortex. J Neurosci. 2015;35:12560–12573. doi: 10.1523/JNEUROSCI.2240-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SD. Regulation of fatty acid synthase gene expression: an approach for reducing fat accumulation. J Anim Sci. 1993;71:1957–1965. doi: 10.2527/1993.7171957x. [DOI] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentet LJ, Avermann M, Matyas F, Staiger JF, Petersen CCH. Membrane Potential Dynamics of GABAergic Neurons in the Barrel Cortex of Behaving Mice. Neuron. 2010;65:422–435. doi: 10.1016/j.neuron.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Gentet LJ, Kremer Y, Taniguchi H, Huang ZJ, Staiger JF, Petersen CC. Unique functional properties of somatostatin-expressing GABAergic neurons in mouse barrel cortex. Nat Neurosci. 2012;15:607–612. doi: 10.1038/nn.3051. [DOI] [PubMed] [Google Scholar]
- Häusser M, Margrie TW. Two-photon targeted patching and electroporation in vivo. Cold Spring Harb Protoc. 2014;2014:78–85. doi: 10.1101/pdb.prot080143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:0878–0890. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhanneau JS, Kremkow J, Dorrn AL, Poulet JFA. In Vivo Monosynaptic Excitatory Transmission between Layer 2 Cortical Pyramidal Neurons. Cell Rep. 2015;13:2098–2106. doi: 10.1016/j.celrep.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR. Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nat Methods. 2012;9:585–587. doi: 10.1038/nmeth.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandaramaiah SB, Holst GL, Wickersham IR, Singer AC, Franzesi GT, McKinnon ML, Forest CR, Boyden ES. Assembly and operation of the autopatcher for automated intracellular neural recording in vivo. Nat Protoc. 2016;11:634–654. doi: 10.1038/nprot.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb I, Stoy WA, Rousseau EB, Moody OA, Jenkins A, Forest CR. Cleaning patch-clamp pipettes for immediate reuse. Sci Rep. 2016;6:35001. doi: 10.1038/srep35001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai S, Denk W, Osten P, Brecht M, Margrie T. Two-photon targeted patching (TPTP) in vivo. Nat Protoc. 2006;1:647–652. doi: 10.1038/nprot.2006.100. [DOI] [PubMed] [Google Scholar]
- Lee AK, Epsztein J, Brecht M. Head-anchored whole-cell recordings in freely moving rats. Nat Protoc. 2009;4:385–392. doi: 10.1038/nprot.2009.5. [DOI] [PubMed] [Google Scholar]
- Li LY, Xiong XR, Ibrahim LA, Yuan W, Tao HW, Zhang LI. Differential Receptive Field Properties of Parvalbumin and Somatostatin Inhibitory Neurons in Mouse Auditory Cortex. Cereb Cortex. 2015;25:1782–1791. doi: 10.1093/cercor/bht417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B, Li L, Knoblich U, Zeng H, Peng H. 3D Image-Guided Automatic Pipette Positioning for Single Cell Experiments in vivo. Sci Rep. 2015;5:18426. doi: 10.1038/srep18426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margrie TW, Brecht M, Sakmann B. In vivo, low-resistance, whole-cell recordings from neurons in the anaesthetized and awake mammalian brain. Pflugers Arch Eur J Physiol. 2002;444:491–498. doi: 10.1007/s00424-002-0831-z. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Meyer AH, Caputi A, Monyer H, Hasan MT, Schaefer AT, Denk W, Brecht M. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Mateo C, Avermann M, Gentet LJ, Zhang F, Deisseroth K, Petersen CCH. In vivo optogenetic stimulation of neocortical excitatory neurons drives brain-state-dependent inhibition. Curr Biol. 2011;21:1593–1602. doi: 10.1016/j.cub.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Mountcastle VB. Introduction. Computation in cortical columns. Cereb Cortex. 2003;13:2–4. doi: 10.1093/cercor/13.1.2. [DOI] [PubMed] [Google Scholar]
- Pala A, Petersen CCHCH. In Vivo Measurement of Cell-Type-Specific Synaptic Connectivity and Synaptic Transmission in Layer 2/3 Mouse Barrel Cortex. Neuron. 2015;85:68–75. doi: 10.1016/j.neuron.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin R, Markram H. A Computer-assisted Multi-electrode Patch-clamp System. J Vis Exp. 2013:e50630. doi: 10.3791/50630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyan CA, Schummers J, Van Wart A, Kuhlman SJ, Wilson NR, Huang ZJ, Sur M. Response Features of Parvalbumin-Expressing Interneurons Suggest Precise Roles for Subtypes of Inhibition in Visual Cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmeyer JD, Yanik MF. High-throughput single-cell manipulation in brain tissue. PLoS One. 2012;7:e35603. doi: 10.1371/journal.pone.0035603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- van Welie I, Roth A, Ho SSN, Komai S, Häusser M. Conditional Spike Transmission Mediated by Electrical Coupling Ensures Millisecond Precision-Correlated Activity among Interneurons In Vivo. Neuron. 2016;90:810–823. doi: 10.1016/j.neuron.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NR, Schummers J, Runyan CA, Yan SX, Chen RE, Deng Y, Sur M. Two-way communication with neural networks in vivo using focused light. Nat Protoc. 2013;8:1184–1203. doi: 10.1038/nprot.2013.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Kolb I, Callahan BM, Su Z, Stoy W, Kodandaramaiah SB, Neve RL, Zeng H, Boyden ES, Forest CR, et al. Integration of autopatching with automated pipette and cell detection in vitro. J Neurophysiol. 2016;116:1564–1578. doi: 10.1152/jn.00386.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S1. MATLAB files and user manual for the Imagepatcher