Abstract
Human mPGES-1 is recognized as a promising target for next generation of anti-inflammatory drugs. Although various mPGES-1 inhibitors have been reported in literature, few have entered clinical trials and none has been proven clinically useful so far. It is highly desired for developing the next generation of therapeutics for inflammation-related diseases to design and discover novel inhibitors of mPGES-1 with new scaffolds. Here, we report the identification of a series of new, potent and selective inhibitors of human mPGES-1 with diverse scaffolds through combined computational and experimental studies. The computationally modeled binding structures of these new inhibitors with mPGES-1 provide some interesting clues for rational design of modified structures of the inhibitors to more favorably bind with mPGES-1.
Keywords: Prostaglandin, inflammation, selective inhibitor, inhibitor identification
Graphical Abstract
A combined use of virtual screening and wet experimental activity assays has led to identification of new, potent and selective inhibitors of human mPGES-1 with diverse scaffolds.
Prostaglandin E2 (PGE2) is known as the principal proinflammatory prostanoid and plays an important role in nociception.[1] The biosynthesis[2] of PGE2 starts from arachidonic acid (AA) which is converted by cyclooxygenase COX-1 or COX-2 to prostaglandin H2 (PGH2).[2] Then, PGH2 is converted to PGE2 by the prostaglandin E synthase (PGES) enzymes,[3] including microsomal PGES-1 (mPGES-1), an inducible enzyme.[4] It is known that mPGES-1 and COX-2 together[5, 6] play a key role in a number of inflammation-related diseases.[7–13] Hence, human mPGES-1 is recognized as a promising target for next generation of drugs to treat the inflammation-related diseases.[14]
There are a number of non-steroidal anti-inflammatory drugs (NSAIDs) available for current clinical practice. The available NSAIDs inhibit COX-1 and/or COX-2.[15] All of the available COX-1/2 inhibitors have significant adverse side effects.[16] The serious side effects led to withdrawal of rofecoxib (Vioxx), a selective COX-2 inhibitor. So, it is interesting to develop novel, improved anti-inflammatory drugs.[15] Through the action of the COX inhibitors, all prostaglandins downstream of PGH2 cannot be produced, resulting in a variety of problems. For example, blocking the production of prostaglandin-I2 (PGI2) will cause significant cardiovascular problems.[17] Inducible enzyme mPGES-1 is a more promising target for anti-inflammatory drugs, because the mPGES-1 inhibition will only block the PGE2 production without affecting PGI2 and other prostaglandins for their production, as confirmed by reported knock-out studies.[18, 19] Thus, mPGES-1 inhibitors are expected to retain the anti-inflammatory effect of COX inhibitors, but without the side effects caused by the COX inhibition.
Although various mPGES-1 inhibitors have been reported in literature,[20–40] few have entered clinical trials[41] and none has been proven clinically useful so far due to various problems with the compounds themselves. It is highly desired for developing the next generation of therapeutics for inflammation-related diseases to design and discover new inhibitors of mPGES-1 with different scaffolds. Here, we report the identification of a set of new, potent and selective inhibitors of human mPGES-1 with various scaffolds through combined computational and experimental studies.
Our virtual screening was based on the X-ray crystal structure (PDB ID: 4BPM)[42] of human mPGES-1 and performed on the Development Therapeutics Program (DTP) Release 4 compound library including ~265,000 compounds available at the National Cancer Institute (https://cactus.nci.nih.gov/download/nci/). The virtual screening procedure used to screen the compounds in the library is similar to that we previously used to identify small-molecule inhibitors of various protein targets.[43, 44] First, the ~265,000 compounds were screened by performing receptor-rigid docking using AutoDock Vina,[45] leading to identification of top-100,000 compounds. Then, each of the top-100,000 compounds was further optimized using a four-step procedure (including 2,000 steps of energy-minimization, 20 ps of molecular dynamic simulation, 4,000 steps of energy-minimization, and then Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) binding energy calculation using AMBER 12 software package)[46, 47] similar to the known binding estimation after refinement (BEAR) protocol.[48, 49] The top-40 compounds were selected according to the ascending order of the MM/PBSA binding energies.
The computationally selected 40 compounds were tested for their inhibitory activity against human mPGES-1. Our protocol for the protein preparation and in vitro activity assays were the same as what we described previously.[50–52] All of the 40 compounds were assayed first for their inhibitory activity at a concentration of 10 µM. Then, the most active compounds were tested further for the dose-dependent inhibition in order to determine their IC50 values (Table 1) against mPGES-1. Finally, the most promising compounds were also assayed for their inhibitory activities against COX-1/2 (mixed COX-1 and COX-2) in order to know their selectivity for mPGES-1 over COX-1/2. The COX-1/2 assays were performed by using the COX (ovine/human) Inhibitor Screening Assay Kit (Item No. 560131) ordered from Cayman Chemical Company (Ann Arbor, MI). According to the kit, the COX activity assay utilizes the competition between prostaglandins (PGs) and a PG tracer, i.e. a PG-acetylcholinesterase (PG-AChE) conjugate, for a fixed amount of PG antiserum.[53, 54] Following the assay using the kit, we used a mixture of COX-1 and COX-2 (denoted as COX-1/2) with equal amount of each enzyme. The efficacies of tested compounds were determined as % inhibition against the COX enzymes at the concentration of 100 µM. All of the enzyme activity assays were carried out in triplicate.
Table 1.
In vitro inhibitory activities of the newly identified mPGES-1 inhibitors.
| Com- pound |
%Inhibition of mPGES-1 at 10 µMa |
IC50 (nM) for mPGES-1b |
%Inhibition of COX-1/2 at 100 µMc |
|---|---|---|---|
| 1 | 99 | 276 ± 60 | 14 ±13 |
| 2 | 98 | 284 ± 81 | 8 ± 20 |
| 3 | 99 | 370 ± 79 | 1 ± 3 |
| 4 | 100 | 439 ± 84 | 9 ± 22 |
| 5 | 94 | 664 ± 106 | 0 ± 3 |
| 6 | 100 | 889 ± 186 | 37 ± 4 |
| 7 | 75 | 917 ± 99 | 15 ± 2 |
| 8 | 71 | N.D. | N.D. |
| 9 | 70 | N.D. | N.D. |
| 10 | 70 | N.D. | N.D. |
| 11 | 69 | N.D. | N.D. |
| 12 | 65 | N.D. | N.D. |
| 13 | 65 | N.D. | N.D. |
| 14 | 64 | N.D. | N.D. |
| 15 | 59 | N.D. | N.D. |
| 16 | 59 | N.D. | N.D. |
| 17 | 59 | N.D. | N.D. |
| 18 | 57 | N.D. | N.D. |
| 19 | 53 | N.D. | N.D. |
| 20 | 50 | N.D. | N.D. |
| 21 | 49 | N.D. | N.D. |
| 22 | 49 | N.D. | N.D. |
| 23 | 48 | N.D. | N.D. |
| 24 | 47 | N.D. | N.D. |
| 25 | 46 | N.D. | N.D. |
| 26 | 46 | N.D. | N.D. |
| 27 | 46 | N.D. | N.D. |
| 28 | 44 | N.D. | N.D. |
| 29 | 43 | N.D. | N.D. |
| 30 | 40 | N.D. | N.D. |
| 31 | 37 | N.D. | N.D. |
| 32 | 36 | N.D. | N.D. |
| 33 | 32 | N.D. | N.D. |
| 34 | 30 | N.D. | N.D. |
| 35 | 29 | N.D. | N.D. |
| 36 | 28 | N.D. | N.D. |
| 37 | 26 | N.D. | N.D. |
| 38 | 25 | N.D. | N.D. |
| 39 | 15 | N.D. | N.D. |
| 40 | 10 | N.D. | N.D. |
The % inhibition of the compound at a concentration of 10 µM against human mPGSE-1.
The determined IC50 against human mPGES-1 based on the data depicted in Figure 2.
The % inhibition of the compound at a concentration of 100 µM against the COX-1/2 (mixed COX-1 and COX-2). The enzyme mixture contained equal amounts of COX-1 and COX-2 in terms of their enzyme activities. In this way, when a compound can significantly inhibit either COX-1 or COX-2, it will show the significant inhibitory effects against the mixed COX-1 and COX-2.
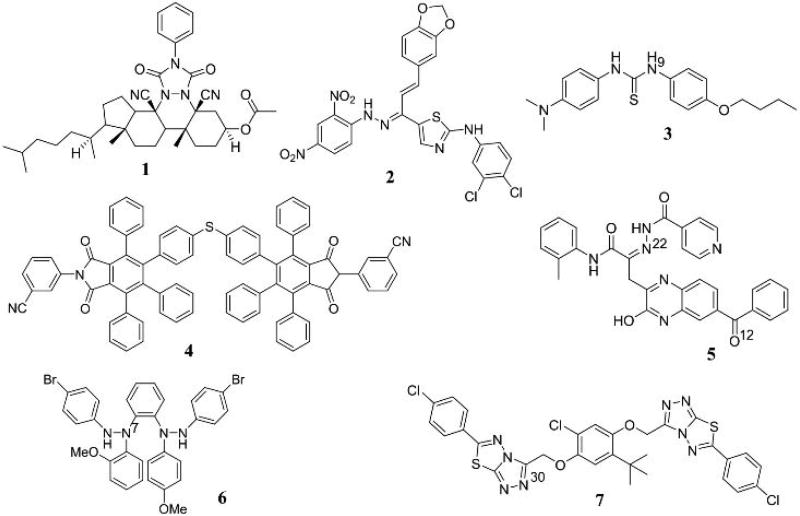
According to the activity assays, all of the computationally selected 40 compounds showed significant inhibitory activity against human mPGES-1, with 10% to 100% inhibition at a concentration of 10 µM (see Table 1). Molecular structures of the most active compounds (top-7) are depicted in Figure 1, and those of the remaining compounds are provided in Supporting Information.
Figure 1.
Molecular structures of the top-7 inhibitors of human mPGES-1 identified. Some atoms with the numbering as superscripts are mentioned in the text for convenience of the discussion.
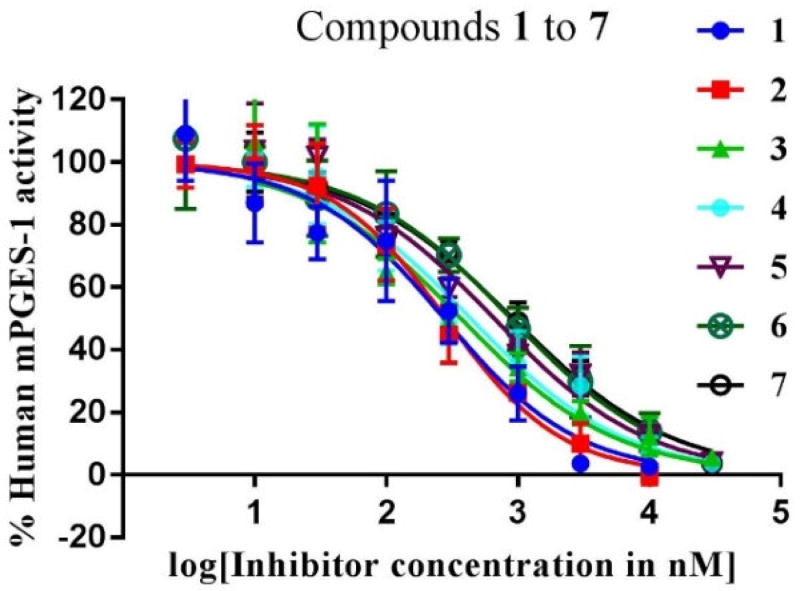
Based on the activity data summarized in Table 1, compounds 1 to 7 at a concentration of 10 µM inhibited the mPGES-1 activity by at least 75%. All of these compounds showed nanomolar IC50 values, 276 to 917 nM. Depicted in Figure 2 are their dose-response curves. The data in Table 1 also revealed that all of the top-7 compounds are highly selective for mPGES-1 over COX-1/2, as these compounds at a very high concentration (100 µM) showed no significant inhibition against COX-1 or COX-2, except for compound 6. Even for compound 6, the inhibition at 100 µM was only ~37%, suggesting that IC50 > 100 µM for compound 6 against COX-1/2.
Figure 2.
Dose-dependent inhibition of human mPGES-1 by compounds 1 to 7: plots of the remaining enzyme activity vs the inhibitor concentration.
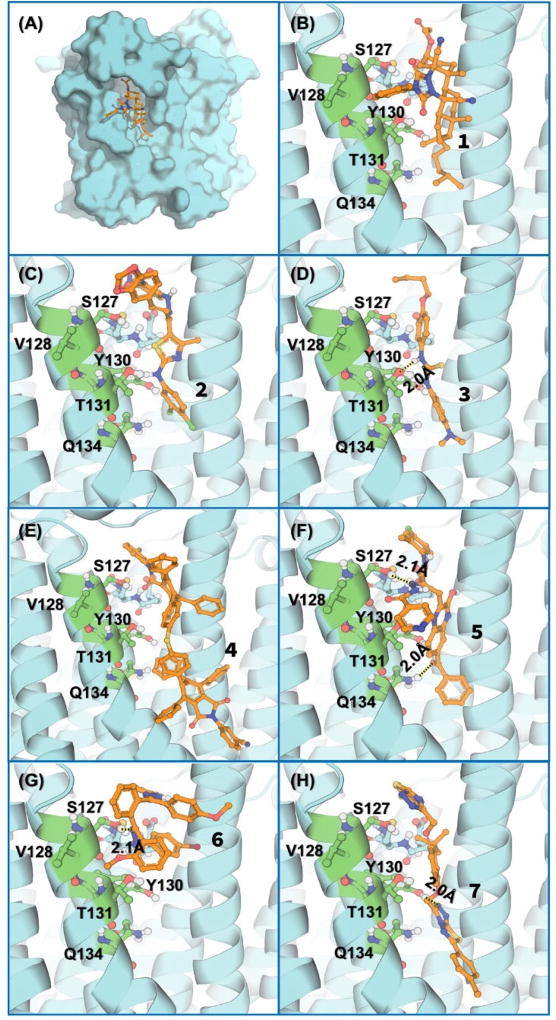
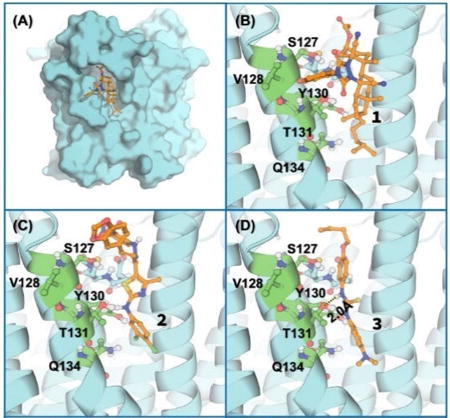
Depicted in Figure 3 are the energy-minimized structures of human mPGES-1 binding with the top-7 compounds. In general, each of these compounds binds with the enzyme at the substrate-binding site and fit the binding site well. Figure 3(A) depicts the overall complex of the enzyme with 1, and Figure 3(B) shows the structural detail of the binding site, showing that the main scaffold of 1 binds very well with the hydrophobic groove of the substrate-binding site of mPGES-1. The extended hydrocarbon side chain has hydrophobic interaction with the protein environment.
Figure 3.
Energy-minimized structures of human mPGES-1 binding with the identified inhibitors (1 to 7 depicted in Figure 1): (A) and (B) Compound 1; (C) 2; (D) 3; (E) 4; (F) 5; (G) 6; (H) 7. The protein is shown in cyan cartoon, and the key residues are shown in green ball-and-stick models. The ligand is shown in orange ball-and-stick models. Important polar interactions are shown in dashed lines.
As shown in Figure 3(C), 2,4-dinitrobenzyl group of compound 2 stays in the bottom of the substrate-binding pocket of mPGES-1. The thiazole and dichlorobenzyl groups have the hydrophobic interaction with the protein. Compound 3 fits very well into the substrate-binding site of mPGES-1, as seen in Figure 3(D) showing a hydrogen bond (HB) between the NH group (including N9) and the hydroxyl oxygen on the side chain of residue T131. Compound 4 is huge in size, but it fits well in the substrate-binding site as seen in Figure 3(E). It is interesting to know that the binding site of the enzyme can accommodate a ligand as large as compound 4.
As shown in Figure 3(F), there are two HBs between the protein and compound 5. One HB is between N22 of 5 and the hydroxyl group of S127 side chain, and the other forms between and O12 of 5 and the hydroxyl group of T131 side chain. In addition, the benzyl rings of 5 have the hydrophobic interaction with the protein.
Figure 3(G) shows that, unlike the other compounds discussed above, compound 6 binds with the protein on the upper part of the substrate-binding groove of mPGES-1, with a HB between N7 of 6 and the hydroxyl group of S127 side chain. As seen in Figure 3(H), compound 7 occupies the substrate-binding pocket with both of the phenyltriazolothiadiazole rings. N30 of compound 7 forms a HB with the hydroxyl group of Y130 side chain.
In summary, through structure-based virtual screening followed by in vitro activity assays, we have identified a series of new, potent and selective inhibitors of human mPGES-1 with diverse scaffolds. In addition, the diverse binding structures of these highly selective inhibitors with mPGES-1 depicted in Figure 3 provide some interesting clues concerning how to design modified structures of the inhibitors to more favorably bind with mPGES-1. Based on the structures in Figure 3, each inhibitor has some unique interaction with the protein. A more potent inhibitor/ligand could be designed to have more of these favorable protein-ligand interactions.
Supplementary Material
Acknowledgments
This work was supported in part by the funding of the Molecular Modeling and Biopharmaceutical Center at the University of Kentucky College of Pharmacy, the National Science Foundation (NSF grant CHE-1111761), and the National Institutes of Health via the National Center for Advancing Translational Sciences (UL1TR001998) grant. Z.Z. thanks the China Scholarship Council for a scholarship support for his graduate studies at the University of Kentucky. The authors also acknowledge the Computer Center at University of Kentucky for supercomputing time on a Dell Supercomputer Cluster consisting of 388 nodes or 4,816 processors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Molecular structures of compounds 8 to 40 listed in Table 1.
References
- 1.Serhan CN, Levy B. Success of prostaglandin E-2 in structure-function is a challenge for structure-based therapeutics. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8609–8611. doi: 10.1073/pnas.1733589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudo I, Murakami M. Prostaglandin E synthase, a terminal enzyme for prostaglandin E-2 biosynthesis. Journal of Biochemistry and Molecular Biology. 2005;38:633–638. doi: 10.5483/bmbrep.2005.38.6.633. [DOI] [PubMed] [Google Scholar]
- 3.Fahmi H. MPGES-1 as a novel target for arthritis. Current Opinion in Rheumatology. 2004;16:623–627. doi: 10.1097/01.bor.0000129664.81052.8e. [DOI] [PubMed] [Google Scholar]
- 4.Park JY, Pillinger MH, Abramson SB. Prostaglandin E-2 synthesis and secretion: The role of PGE(2) synthases. Clinical Immunology. 2006;119:229–240. doi: 10.1016/j.clim.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Murakami M, Nakatani Y, Tanioka T, Kudo I. Prostaglandin E synthase. Prostaglandins & Other Lipid Mediators. 2002;68-9:383–399. doi: 10.1016/s0090-6980(02)00043-6. [DOI] [PubMed] [Google Scholar]
- 6.Murakami M, Naraba H, Tanioka T, Semmyo N, Nakatani Y, Kojima F, Ikeda T, Fueki M, Ueno A, Oh-ishi S, Kudo I. Regulation of prostaglandin E-2 biosynthesis by inducible membrane-associated prostaglandin E-2 synthase that acts in concert with cyclooxygenase-2. Journal of Biological Chemistry. 2000;275:32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 7.Uematsu S, Matsumoto M, Takeda K, Akira S. Lipopolysaccharide-dependent prostaglandin E-2 production is regulated by the glutathione-dependent prostaglandin E-2 synthase gene induced by the toll-like receptor 4/MyD88/NF-IL6 pathway. Journal of Immunology. 2002;168:5811–5816. doi: 10.4049/jimmunol.168.11.5811. [DOI] [PubMed] [Google Scholar]
- 8.Kamei D, Murakami M, Nakatani Y, Ishikawa Y, Ishii T, Kudo I. Potential role of microsomal prostaglandin E synthase-1 in tumorigenesis. Journal of Biological Chemistry. 2003;278:19396–19405. doi: 10.1074/jbc.M213290200. [DOI] [PubMed] [Google Scholar]
- 9.Kamei D, Yamakawa K, Takegoshi Y, Mikami-Nakanishi M, Nakatani Y, Oh-ishi S, Yasui H, Azuma Y, Hirasawa N, Ohuchi K, Kawaguchi H, Ishikawa Y, Ishii T, Uematsu S, Akira S, Murakami M, Kudo I. Reduced pain hypersensitivity and inflammation in mice lacking microsomal prostaglandin E synthase-1. Journal of Biological Chemistry. 2004;279:33684–33695. doi: 10.1074/jbc.M400199200. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda-Matsuo Y, Ota A, Fukada T, Uematsu S, Akira S, Sasaki Y. Microsomal prostaglandin E synthase-1 is a critical factor of stroke-reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11790–11795. doi: 10.1073/pnas.0604400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami M, Kudo I. Recent advances in molecular biology and physiology of the prostaglandin E-2-biosynthetic pathway. Progress in Lipid Research. 2004;43:3–35. doi: 10.1016/s0163-7827(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 12.Claveau D, Sirinyan M, Guay J, Gordon R, Chan CC, Bureau Y, Riendeau D, Mancini JA. Microsomal prostaglandin E synthase-1 is a major terminal synthase that is selectively up-regulated during cyclooxygenase-2-dependent prostaglandin E-2 production in the rat adjuvant-induced arthritis model. Journal of Immunology. 2003;170:4738–4744. doi: 10.4049/jimmunol.170.9.4738. [DOI] [PubMed] [Google Scholar]
- 13.Oshima H, Oshima M, Inaba K, Taketo MM. Hyperplastic gastric tumors induced by activated macrophages in COX-2/mPGES-1 transgenic mice. EMBO Journal. 2004;23:1669–1678. doi: 10.1038/sj.emboj.7600170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friesen RW, Mancini JA. Microsomal prostaglandin E-2 synthase-1 (mPGES-1): A novel anti-inflammatory therapeutic target. Journal of medicinal chemistry. 2008;51:4059–4067. doi: 10.1021/jm800197b. [DOI] [PubMed] [Google Scholar]
- 15.Samuelsson B, Morgenstern R, Jakobsson PJ. Membrane prostaglandin E synthase-1: A novel therapeutic target. Pharmacological Reviews. 2007;59:207–224. doi: 10.1124/pr.59.3.1. [DOI] [PubMed] [Google Scholar]
- 16.Scholich K, Geisslinger G. Is mPGES-1 a promising target for pain therapy? Trends in Pharmacological Sciences. 2006;27:399–401. doi: 10.1016/j.tips.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Wang M, Yu Y, Lawson J, Funk CD, FitzGerald GA. Cyclooxygenases, microsomal prostaglandin E synthase-1, and cardiovascular function. Journal of Clinical Investigation. 2006;116:1391–1399. doi: 10.1172/JCI27540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engblom D, Saha S, Engstrom L, Westman M, Audoly LP, Jakobsson PJ, Blomqvist A. Microsomal prostaglandin E synthase-1 is the central switch during immune-induced pyresis. Nature Neuroscience. 2003;6:1137–1138. doi: 10.1038/nn1137. [DOI] [PubMed] [Google Scholar]
- 19.Trebino CE, Stock JL, Gibbons CP, Naiman BM, Wachtmann TS, Umland JP, Pandher K, Lapointe JM, Saha S, Roach ML, Carter D, Thomas NA, Durtschi BA, McNeish JD, Hambor JE, Jakobsson PJ, Carty TJ, Perez JR, Audoly LP. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9044–9049. doi: 10.1073/pnas.1332766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiffler MA, Antonysamy S, Bhattachar SN, Campanale KM, Chandrasekhar S, Condon B, Desai PV, Fisher MJ, Groshong C, Harvey A, Hickey MJ, Hughes NE, Jones SA, Kim EJ, Kuklish SL, Luz JG, Norman BH, Rathmell RE, Rizzo JR, Seng TW, Thibodeaux SJ, Woods TA, York JS, Yu XP. Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors. J. Med. Chem. 2016;59:194–205. doi: 10.1021/acs.jmedchem.5b01249. [DOI] [PubMed] [Google Scholar]
- 21.Hieke M, Greiner C, Dittrich M, Reisen F, Schneider G, Schubert-Zsilavecz M, Werz O. Discovery and biological evaluation of a novel class of dual microsomal prostaglandin E2 synthase-1/5-lipoxygenase inhibitors based on 2-[(4,6-diphenethoxypyrimidin-2-yl)thio]hexanoic acid. J. Med. Chem. 2011;54:4490–4507. doi: 10.1021/jm200092b. [DOI] [PubMed] [Google Scholar]
- 22.Hanke T, Dehm F, Liening S, Popella SD, Maczewsky J, Pillong M, Kunze J, Weinigel C, Barz D, Kaiser A, Wurglics M, Lammerhofer M, Schneider G, Sautebin L, Schubert-Zsilavecz M, Werz O. Aminothiazole-featured pirinixic acid derivatives as dual 5-lipoxygenase and microsomal prostaglandin E2 synthase-1 inhibitors with improved potency and efficiency in vivo. J. Med. Chem. 2013;56:9031–9044. doi: 10.1021/jm401557w. [DOI] [PubMed] [Google Scholar]
- 23.Terracciano S, Lauro G, Strocchia M, Fischer K, Werz O, Riccio R, Bruno I, Bifulco G. Structural Insights for the Optimization of Dihydropyrimidin-2(1H)-one Based mPGES-1 Inhibitors. ACS Med. Chem. Lett. 2015;6:187–191. doi: 10.1021/ml500433j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiro T, Kakiguchi K, Takahashi H, Nagata H, Tobe M. 7-Phenyl-imidazoquinolin-4(5H)-one derivatives as selective and orally available mPGES-1 inhibitors. Bioorg. Med. Chem. 2013;21:2868–2878. doi: 10.1016/j.bmc.2013.03.069. [DOI] [PubMed] [Google Scholar]
- 25.Shiro T, Kakiguchi K, Takahashi H, Nagata H, Tobe M. Synthesis and biological evaluation of substituted imidazoquinoline derivatives as mPGES-1 inhibitors. Bioorg. Med. Chem. 2013;21:2068–2078. doi: 10.1016/j.bmc.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Shiro T, Takahashi H, Kakiguchi K, Inoue Y, Masuda K, Nagata H, Tobe M. Synthesis and SAR study of imidazoquinolines as a novel structural class of microsomal prostaglandin E(2) synthase-1 inhibitors. Bioorg. Med. Chem. Lett. 2012;22:285–288. doi: 10.1016/j.bmcl.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Liedtke AJ, Keck PR, Lehmann F, Koeberle A, Werz O, Laufer SA. Arylpyrrolizines as inhibitors of microsomal prostaglandin E2 synthase-1 (mPGES-1) or as dual inhibitors of mPGES-1 and 5-lipoxygenase (5-LOX) J. Med. Chem. 2009;52:4968–4972. doi: 10.1021/jm900481c. [DOI] [PubMed] [Google Scholar]
- 28.Shang E, Wu Y, Liu P, Liu Y, Zhu W, Deng X, He C, He S, Li C, Lai L. Benzo[d]isothiazole 1,1-dioxide derivatives as dual functional inhibitors of 5-lipoxygenase and microsomal prostaglandin E(2) synthase-1. Bioorg. Med. Chem. Lett. 2014;24:2764–2767. doi: 10.1016/j.bmcl.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Wu TY, Juteau H, Ducharme Y, Friesen RW, Guiral S, Dufresne L, Poirier H, Salem M, Riendeau D, Mancini J, Brideau C. Biarylimidazoles as inhibitors of microsomal prostaglandin E2 synthase-1. Bioorg. Med. Chem. Lett. 2010;20:6978–6982. doi: 10.1016/j.bmcl.2010.09.129. [DOI] [PubMed] [Google Scholar]
- 30.Wiegard A, Hanekamp W, Griessbach K, Fabian J, Lehr M. Pyrrole alkanoic acid derivatives as nuisance inhibitors of microsomal prostaglandin E2 synthase-1. Eur. J. Med. Chem. 2012;48:153–163. doi: 10.1016/j.ejmech.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Chini MG, De Simone R, Bruno I, Riccio R, Dehm F, Weinigel C, Barz D, Werz O, Bifulco G. Design and synthesis of a second series of triazole-based compounds as potent dual mPGES-1 and 5-lipoxygenase inhibitors. Eur. J. Med. Chem. 2012;54:311–323. doi: 10.1016/j.ejmech.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Giroux A, Boulet L, Brideau C, Chau A, Claveau D, Cote B, Ethier D, Frenette R, Gagnon M, Guay J, Guiral S, Mancini J, Martins E, Masse F, Methot N, Riendeau D, Rubin J, Xu D, Yu H, Ducharme Y, Friesen RW. Discovery of disubstituted phenanthrene imidazoles as potent, selective and orally active mPGES-1 inhibitors. Bioorg. Med. Chem. Lett. 2009;19:5837–5841. doi: 10.1016/j.bmcl.2009.08.085. [DOI] [PubMed] [Google Scholar]
- 33.Xu D, Rowland SE, Clark P, Giroux A, Cote B, Guiral S, Salem M, Ducharme Y, Friesen RW, Methot N, Mancini J, Audoly L, Riendeau D. MF63 [2-(6-chloro-1H-phenanthro[9,10-d]imidazol-2-yl)-isophthalonitrile], a selective microsomal prostaglandin E synthase-1 inhibitor, relieves pyresis and pain in preclinical models of inflammation. J. Pharmacol. Exp. Ther. 2008;326:754–763. doi: 10.1124/jpet.108.138776. [DOI] [PubMed] [Google Scholar]
- 34.Lee K, Pham VC, Choi MJ, Kim KJ, Lee KT, Han SG, Yu YG, Lee JY. Fragment-based discovery of novel and selective mPGES-1 inhibitors Part 1: identification of sulfonamido-1,2,3-triazole-4,5-dicarboxylic acid. Bioorg. Med. Chem. Lett. 2013;23:75–80. doi: 10.1016/j.bmcl.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Cote B, Boulet L, Brideau C, Claveau D, Ethier D, Frenette R, Gagnon M, Giroux A, Guay J, Guiral S, Mancini J, Martins E, Masse F, Methot N, Riendeau D, Rubin J, Xu D, Yu H, Ducharme Y, Friesen RW. Substituted phenanthrene imidazoles as potent, selective, and orally active mPGES-1 inhibitors. Bioorg. Med. Chem. Lett. 2007;17:6816–6820. doi: 10.1016/j.bmcl.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 36.Riendeau D, Aspiotis R, Ethier D, Gareau Y, Grimm EL, Guay J, Guiral S, Juteau H, Mancini JA, Methot N, Rubin J, Friesen RW. Inhibitors of the inducible microsomal prostaglandin E2 synthase (mPGES-1) derived from MK-886. Bioorg. Med. Chem. Lett. 2005;15:3352–3355. doi: 10.1016/j.bmcl.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Bruno A, Di Francesco L, Coletta I, Mangano G, Alisi MA, Polenzani L, Milanese C, Anzellotti P, Ricciotti E, Dovizio M, Di Francesco A, Tacconelli S, Capone ML, Patrignani P. Effects of AF3442 [N-(9-ethyl-9H-carbazol-3-yl)-2-(trifluoromethyl)benzamide], a novel inhibitor of human microsomal prostaglandin E synthase-1, on prostanoid biosynthesis in human monocytes in vitro. Biochem. Pharmacol. 2010;79:974–981. doi: 10.1016/j.bcp.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Koeberle A, Haberl EM, Rossi A, Pergola C, Dehm F, Northoff H, Troschuetz R, Sautebin L, Werz O. Discovery of benzo[g]indol-3-carboxylates as potent inhibitors of microsomal prostaglandin E(2) synthase-1. Bioorg. Med. Chem. 2009;17:7924–7932. doi: 10.1016/j.bmc.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 39.Walker DP, Arhancet GB, Lu HF, Heasley SE, Metz S, Kablaoui NM, Franco FM, Hanau CE, Scholten JA, Springer JR, Fobian YM, Carter JS, Xing L, Yang S, Shaffer AF, Jerome GM, Baratta MT, Moore WM, Vazquez ML. Synthesis and biological evaluation of substituted benzoxazoles as inhibitors of mPGES-1: use of a conformation-based hypothesis to facilitate compound design. Bioorg. Med. Chem. Lett. 2013;23:1120–1126. doi: 10.1016/j.bmcl.2012.11.107. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Limburg D, Carter J, Mbalaviele G, Gierse J, Vazquez M. Selective inducible microsomal prostaglandin E(2) synthase-1 (mPGES-1) inhibitors derived from an oxicam template. Bioorg. Med. Chem. Lett. 2010;20:1604–1609. doi: 10.1016/j.bmcl.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y, Smith CL, Hu L, Campanale KM, Stoltz R, Huffman LG, Jr, McNearney TA, Yang XY, Ackermann BL, Dean R, Regev A, Landschulz W. Pharmacodynamic comparison of LY3023703, a novel microsomal prostaglandin e synthase 1 inhibitor, with celecoxib. Clin. Pharmacol. Ther. 2016;99:274–284. doi: 10.1002/cpt.260. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Howe N, Dukkipati A, Shah ST, Bax BD, Edge C, Bridges A, Hardwicke P, Singh OM, Giblin G, Pautsch A, Pfau R, Schnapp G, Wang M, Olieric V, Caffrey M. Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy. Crystal growth & design. 2014;14:2034–2047. doi: 10.1021/cg500157x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W, AbdulHameed MDM, Hamza A, Zhan C-G. New inhibitor of 3-phosphoinositide dependent protein kinase-1 identified from virtual screening. Bioorg. Med. Chem. Letters. 2012;22:1629–1632. doi: 10.1016/j.bmcl.2011.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamza A, Zhao X, Tong M, Tai HH, Zhan CG. Novel human mPGES-1 inhibitors identified through structure-based virtual screening. Bioorg. Med. Chem. 2011;19:6077–6086. doi: 10.1016/j.bmc.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Case DA, Darden TA, Cheatham TE, Iii, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 47.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degliesposti G, Portioli C, Parenti MD, Rastelli G. BEAR, a novel virtual screening methodology for drug discovery. J. Biomol. Screen. 2011;16:129–133. doi: 10.1177/1087057110388276. [DOI] [PubMed] [Google Scholar]
- 49.Rastelli G, Degliesposti G, Del Rio A, Sgobba M. Binding estimation after refinement, a new automated procedure for the refinement and rescoring of docked ligands in virtual screening. Chem. Biol. Drug Des. 2009;73:283–286. doi: 10.1111/j.1747-0285.2009.00780.x. [DOI] [PubMed] [Google Scholar]
- 50.Hamza A, Tong M, AbdulHameed MD, Liu J, Goren AC, Tai HH, Zhan CG. Understanding microscopic binding of human microsomal prostaglandin E synthase-1 (mPGES-1) trimer with substrate PGH2 and cofactor GSH: insights from computational alanine scanning and site-directed mutagenesis. J Phys Chem B. 2010;114:5605–5616. doi: 10.1021/jp100668y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang XQ, Yan WL, Gao DQ, Tong M, Tai H-H, Zhan C-G. Structural and functional characterization of human microsomal prostaglandin E synthase-1 by computational modeling and site-directed mutagenesis. Bioorg. Med. Chem. 2006;14:3553–3562. doi: 10.1016/j.bmc.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Hamza A, Zhao X, Tong M, Tai H-H, Zhan C-G. Novel human mPGES-1 inhibitors identified through structure-based virtual screening. Bioorg. Med. Chem. 2011;19:6077–6086. doi: 10.1016/j.bmc.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maclouf J, Grassi J, Pradelles P. Development of Enzyme-Immunoassay Techniques for Measurement of Eicosanoids. In: Walden TL, Hughes HN, editors. Prostaglandin and Lipid Metabolism in Radiation Injury. Springer US; Boston, MA: 1987. pp. 355–364. [Google Scholar]
- 54.Pradelles P, Grassi J, Maclouf J. Enzyme immunoassays of eicosanoids using acetylcholine esterase as label: an alternative to radioimmunoassay. Anal. Chem. 1985;57:1170–1173. doi: 10.1021/ac00284a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.