Abstract
The ~70 protocadherins comprise the largest group within the cadherin superfamily. Their diversity, the complexity of the mechanisms through which their genes are regulated, and their many critical functions in nervous system development have engendered a growing interest in elucidating the intracellular signaling pathways through which they act. Recently, multiple protocadherins across several subfamilies have been implicated as modulators of Wnt signaling pathways, and through this as potential tumor suppressors. Here, we review the extant data on the regulation by protocadherins of Wnt signaling pathways and components, and highlight some key unanswered questions that could shape future research.
Keywords: planar cell polarity, cell adhesion, cancer, tumor suppressor, epigenetics extracellular cadherin (EC)
1. Introduction
The ~70 protocadherins (Pcdhs) have, since the turn of the 21st century, emerged as some of the most interesting regulators of neural development. Pcdhs make up the largest group within the broader cadherin superfamily of cell adhesion molecules, which also includes the canonical classical cadherins, the seven-transmembrane domain cadherins, and atypical cadherins such as Fat and Dachsous (for more information, see the other reviews in this special issue). Functional studies have implicated numerous Pcdhs in the regulation of neuronal survival, axon outgrowth and targeting, dendrite arbor complexity, the self-avoidance of sister axon and dendrite branches, and synaptogenesis. Several Pcdh genes also have been implicated, either by mutation or epigenetic dysregulation, in a wide variety of neurological and neurodevelopmental disorders, including epilepsy, mood disorders, autism, and schizophrenia (see reviews by Peek, et al. [1] and Keeler et al. [2], as well as the other reviews in this special issue).
Despite this progress in identifying functional roles for many Pcdhs, less is understood about the intracellular signaling pathways with which they engage in order to play these roles. Recently, we and others have uncovered interactions between Pcdh molecules and Wnt signaling pathways, which are known to be critical for embryonic development in general, and neural development in particular, as well as in the etiology and progression of multiple types of cancer. In this brief review, we will summarize the studies that have begun to elucidate links between Pcdh adhesion molecules and Wnt signaling components.
2. Wnt Signaling Pathways
We will begin by providing a brief overview of Wnt signaling pathways; multiple aspects of these complicated pathways are discussed more fully in a number of recent reviews [3–10].
2.1 Wnt proteins and their receptors
Wnts are cysteine-rich proteins that are evolutionarily conserved; humans and mice have 19 Wnt ligands [11]. Originally named Int-1, the mouse Wnt1 gene was identified in the early 1980s by Nusse and Varmus as a preferential integration site of mouse mammary tumor virus (MMTV), an oncogenic retrovirus [12]. Subsequently, it was determined that Drosophila wingless (wg), which plays a role in segment polarity during larval development, is a homolog of Wnt1 [13]. Wnts possess an N-terminal signal peptide sequence for secretion and are subject to a number of protein modifications, most prominently glycosylation and acylation, both of which are crucial for their function [14–18]. While Wnts have been studied extensively, to date crystal structures of only 2 Wnts, XWnt8 (Xenopus) and WntD (Drosophila), have been elucidated [19, 20].
The main Wnt receptors are Frizzleds (Fzd), a family of seven-transmembrane G-protein-coupled receptors that possess a large extracellular cysteine-rich domain that mediates Wnt binding [20, 21]. Mammals have 10 Fzd receptors [22]. The intracellular domain (ICD) of Fzd binds Dishevelled proteins (Dvls) through a conserved KTXXXW motif; Dvl interacts with a large number of Wnt co-receptors to activate the different Wnt pathways [23]. Fzds have been found to be phosphorylated at the ICD, an event that downregulates their functions [24]. Furthermore, ubiquitination and de-ubiquitination of Fzds are important for the regulation of events downstream of Wnt-Fzd binding [25].
Two highly homologous proteins, Lrp5 and Lrp6 in vertebrates, as well as their Drosophila homolog Arrow, act as Wnt co-receptors with Fzds to initiate canonical Wnt signaling (see below, 2.2). Lrp6 remains the best studied Lrp (Low-density lipoprotein-related rececptor) and is a large, single transmembrane domain protein with greater affinity to Wnts complexed with Fzd than to Wnts on its own. The extracellular domain of Lrp6 contains many independent Wnt-binding sites, allowing simultaneous interaction with many Wnt-Fzd complexes [26]. A key event in the regulation of Lrp6 function is the phosphorylation of several sites within its ICD, with the first event being phosphorylation of the 5 PPPSP repeats by several proline targeted kinases; this then primes Lrp6 for a second phosphorylation event by the casein kinase I (CKI) family at neighboring Ser residues (PPPSPXS) [27–29].
Additionally, ROR1 and ROR2 (Receptor tyrosine kinase-like Orphan Receptor) have been shown to be co-receptors for Wnt5a to facilitate planar-cell polarity (PCP) signaling in verterbrates [30, 31] (see below, 2.2). The binding of Wnt5a leads to the homodimerization of ROR2, which together forms a ternary complex with Fzd [32, 33]. This leads to the recruitment of the actin-binding protein filamin A and activation of c-Jun N-terminal kinase (JNK) [34]. ROR receptors are also phosphorylated, in a Wnt5a-dependent manner, to activate PCP signaling [32, 35]. A final Wnt-binding co-receptor is the receptor tyrosine kinase Ryk, which has been implicated in multiple Wnt signaling pathways [36–38].
2.2 Canonical, PCP, and Wnt/Ca2+ pathways
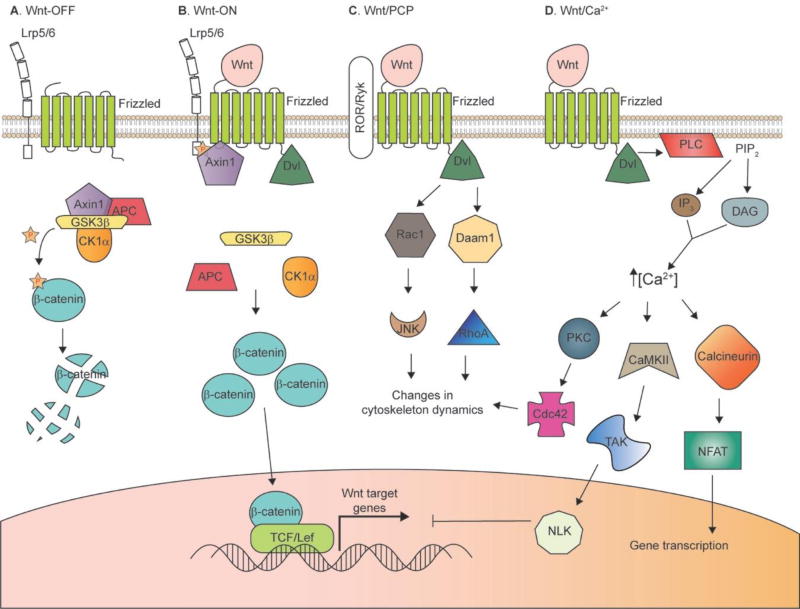
The binding of secreted Wnts to Fzd, Lrp5/6, Ryk, and ROR co-receptors has been shown to activate at least three distinct pathways: the “canonical” (β-catenin-dependent) pathway, the Wnt/PCP pathway, and the Wnt/Ca2+ pathway. The canonical Wnt/β-catenin pathway, the one that is best characterized molecularly, is dependent on the proteolytic state of cytoplasmic β-catenin (Figure 1). In the Wnt-OFF state, a large proportion of β-catenin is found at the plasma membrane, complexed with classical cadherin cytoplasmic domains, where it regulates cell adhesion at adherens junctions and synapses via its partner α-catenin’s ability to bind actin filaments [39]. In the Wnt-OFF state, cytoplasmic β-catenin levels are kept low by the constitutive activity of a “β-catenin destruction complex” consisting of Axin1, adenomatous polyposis coli (APC), CK1α, and glycogen synthase kinase 3β (GSK3β; reviewed by Clevers and Nusse [5]; Nusse and Clevers [8]). Axin1 serves as a scaffold for the destruction complex, and associated CK1α and GSK3β sequentially phosphorylate β-catenin, targeting it for ubiquitin-dependent degradation by the proteasome [40, 41].
Figure 1. Wnt signaling pathways.
A and B: Canonical (β-catenin-dependent) pathway. In the “Wnt-OFF” state (A), β-catenin levels are kept low in the cytoplasm by the action of the “destruction complex”. GSK3β phosphorylates β-catenin, which targets it for desctruction by the proteasome. Wnt binding to Frizzled and Lrp5/6 co-receptors results in disruption of the destruction complex, allowing β-catenin to accumulate in the cytoplasm. β-catenin can then translocate to the nucleus and promote the activation of Wnt target genes by displacing co-repressors, and recruiting co-activators, of TCF/Lef. C: Wnt/PCP pathway. Binding of Wnt to Frizzled and ROR or Ryk co-receptors leads to recruitment of Dvl, which can act through Rac1 or Daam1 to initate changes in cytoskeletal dynamics important for cell orientation and movement. D: Wnt/Ca2+ pathway. Wnt binding and recruitment of Dvl leads to activation of PLC, which cleaves PIP2 to generate IP3 and DAG. This leads to release of Ca2+ from intracellular stores, and downstream signaling through a number of Ca2+-dependent kinases and phosphatases.
When Wnt ligands bind to Fzd and Lrp5/6, the Wnt-ON state is triggered. This results in Dvl binding to the C-terminus of Fzd [42], which recruits Axin1 to the cytoplasmic tail of Lrp5/6, facilitating the phosphorylation of Lrp5/6 by GSK3β and CK1α [27–29, 43, 44]. The fully phosphorylated Lrp6 can then bind Axin1, which recruits the β-catenin destruction complex. The activity of GSK3β in the destruction complex is inhibited by phosphorylated Lrp5/6, resulting in decreased phosphorylation of both β-catenin and Axin1 [45–47]. Dephosphorylated Axin1 is dissociated from the receptor complex and also from β-catenin, which inactivates the destruction complex until Axin1 is again phosphorylated [48, 49]. This cascade of events causes β-catenin to accumulate in the cytoplasm and to translocate to the nucleus, where it interacts with members of the T-cell factor (TCF)/lymphoid enhancer factor (Lef) family of transcription factors to activate a wide variety of Wnt target genes [50] (Figure 1).
A separate Wnt-receptor interaction outcome that is β-catenin independent is the Wnt/PCP pathway. In this pathway, Wnt-Fzd binding, along with its coreceptors ROR or Ryk, recruits Dvl, which can: 1) form a complex with Dishevelled-associated activator of morphogenesis 1 (Daam1) to activate RhoA; or 2) activate JNK through Rac1 to affect cytoskeletal dynamics and cell polarity [7]. The PCP pathway is critical in establishing cell polarity in morphogenetic processes such as the regulation of cell movements during gastrulation, neural tube closure and the orientation of stereocilia in the inner ear in vertebrates [23, 51]. Finally, a distinct Wnt/Ca2+ pathway has been implicated in cancer, inflammation and neurodegeneration, as well as a variety of critical events in embryonic development (reviewed by Slusarski and Peligri [9]; De [10]). In this pathway, Wnt-Fzd binding activates phospholipase C (PLC) in a Dvl-dependent manner. This leads to a release of Ca2+ from intracellular stores as PLC hydrolyzes PIP2 to form IP3/DAG, which then activates calmodulin-dependent kinase II (CaMKII), protein kinase C (PKC) and calcineurin [10]. CaMKII activates TAK, which promotes Nemo-like kinase (NLK) activity to inhibit Wnt/β-catenin transcriptional activity [52]; thus, the Wnt/Ca2+ pathway can antagonize the canonical Wnt pathway. PKC phosphorylates the small GTPase Cdc42, a major mediator of actin cytoskeleton reorganization [53], while calcineurin dephosphorylates nuclear factor of activated T cell (NFAT), promoting its translocation to the nucleus to upregulate genes controlling cell fate and cell migration [54–57].
Wnt signaling pathways have long been associated with cancer and carcinogenesis. The link between Wnt and cancer was established with their initial discovery, as enhanced expression of the int1 (Wnt1) gene due to MMTV insertion caused mammary hyperplasia and tumors in mice [12, 58, 59]. Subsequently, two studies identified mutations in the APC gene (whose gene product, as mentioned above, interacts with β-catenin as part of the destruction complex) as the underlying cause of hereditary colon cancer syndrome [60, 61]. Others have identified mutations of other components of Wnt signaling pathways in various cancers: for example, β-catenin mutation in gastric cancer [62], Axin1 mutations in hepatocellular carcinomas and medulloblastomas [63, 64], and β-catenin, Axin1, Axin2, and TCF4 mutations in colon cancer [65]. While the canonical Wnt pathway remains best defined in its role in cancer, the relatively more diverse and less understood non-canonical Wnt pathways have also been implicated in metastasis formation and cell migration of cancer cells [66, 67]. Other reviews can be consulted for insights on the misregulation of canonical and non-canonical Wnt signaling in cancer cells [68–70].
Since the initial discovery of Wnt signaling’s role in tumorigenesis, we have learned that Wnt signaling plays a vast array of roles in embryonic development, including cell fate determination, early patterning events, and organ morphogenesis, as well as in the adult, where Wnt signaling regulates stem cell renewal and tissue homeostasis [71–77]. The variety of intracellular pathways, some of which are mutually antagonistic, downstream of Wnt-Fzd binding allows for a wide range of cellular outcomes mediated by common signals. Understanding the signaling partners, such as Pcdhs, that can influence signaling downstream of Wnts will be important for understanding the many roles they play.
3. Protocadherins
3.1 Pcdh gene and protein structure
In this review, we will focus only on the “true” protocadherins (Pcdhs): the ~60 members of the clustered α-, β-, and γ-Pcdh families and the 10 non-clustered δ-Pcdhs. Other “atypical” cadherin molecules (sometimes referred to colloquially as Pcdhs), including the seven-transmembrane Flamingo/Celsr family and the large Fat and Dachsous cadherins, have been implicated extensively in the control of PCP pathways. Flamingo, in particular, is known to interact physically and functionally with Frizzled receptors in order to regulate PCP [78]. Thorough discussions of the seven-transmembrane cadherins and the Fat cadherins can be found, respectively, in the articles by Goffinet and Tissir (reference to be added later in revision) and by Avilés and Goodrich [79] in this special issue. Here, we will confine our discussion to the emerging roles for clustered and non-clustered Pcdhs in the regulation of the canonical (primarily) and non-canonical Wnt pathways.
Pcdhs represent the largest group within the cadherin superfamily of cell adhesion molecules known to play critical roles in several biological processes, including embryonic morphogenesis, neural circuit formation, angiogenesis, and cancer [80–82]. Members of the cadherin superfamily are characterized by extracellular cadherin (EC) motifs that are approximately 100 amino acids long and that mediate trans-interactions between cells. Shintaro Suzuki and colleagues used degenerate PCR to search for additional cadherin-related molecules, and discovered and named the first Pcdhs (including one of the clustered Pcdhs and one of the δ1-Pcdhs) in the early 1990s [83]. Like “classical” cadherins, Pcdhs are type I transmembrane proteins; however, unlike cadherins which have five EC domains, Pcdhs have six (clustered Pcdhs, δ2-Pcdhs) or seven (δ1-Pcdhs) EC domains, and distinct cytoplasmic domains that lack catenin-binding sites, allowing for integration into distinct signaling pathways [83–94].
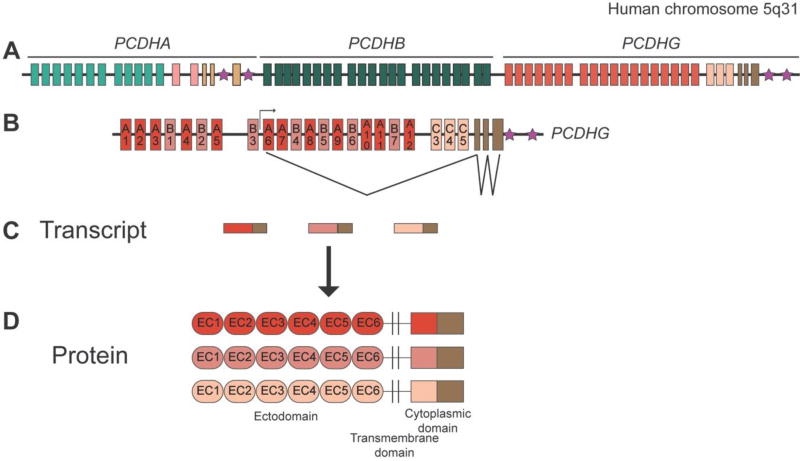
The clustered Pcdhs consist of ~60 proteins, termed α-, β-, and γ-Pcdhs, that are encoded by three tandem gene clusters (Pcdha, Pcdhb, and Pcdhg) encompassing about 1 MB at human chromosome 5q31 and mouse chromosome 18 [93, 95, 96]. Eight of these clustered Pcdh genes had been identified previously as “Cadherin-related Neuronal Receptor” (CNR) proteins by Kohmura et al. [97]. Each large “variable” exon (expressed from its own promoter) encodes six EC domains, a transmembrane domain and a variable cytoplasmic domain of approximately 90 amino acids, and is spliced to three constant exons that encode a shared ~125 amino acid C-terminal domain for the Pcdha and Pcdhg clusters. The Pcdhb cluster does not contain such constant exons and thus are expressed as a single-exon transmembrane molecule [98, 99] (Figure 2). The Pcdhg cluster encodes 22 γ-Pcdh proteins that can be grouped into three subfamilies on the basis of sequence similarity, termed γ-Pcdh-A, -B, and –C; γ-Pcdh-C3, -C4 and –C5 are more similar to α-Pcdh-C1 and –C2, found within the Pcdha cluster, than they are to any other γ-Pcdhs [93].
Figure 2. The protocadherin gene clusters.
A: Schematic of the human PCDHA, PCDHB, and PCDHG gene clusters on chromosome 5q31. A very similar structure is observed for the mouse clusters at chromosome 18. B: The exon structure of the PCDHG cluster is expanded below, with an example of the transcription initiation and splicing pattern (for A6, in this instance). C: Schematic of the PCDHG spliced transcripts generated by the cluster; each mature transcript consists of one large variable exon and the three small constant exons. D: Protein structure of the γ-Pcdhs (α-Pcdhs are identical in structure; β-Pcdhs lack any constant domain). Six extracellular cadherin (EC) repeats, a transmembrane domain, and a variable cytoplasmic domain are encoded by each variable exon; the constant exons encode a 125 amino acid C-terminal domain. Stars indicate the sites of “cluster control regions”, enhancers required for normal expression patterns of the Pcdh clusters.
Many nonclustered Pcdh genes have an exon structure similar to the clustered Pcdhs: In most δ-Pcdh genes, a single large first exon encodes the N-terminal signal peptide, multiple EC domains, a single transmembrane domain, and a small part of the intracellular domain, while the remainder of the intracellular domain is encoded by several remaining exons. Exceptions are Pcdh1 and Pcdh11 (a.k.a. PcdhX/Y; a Pcdh11 gene is found on the homologous region of both the X and Y chromosomes in humans, but only on the X chromosome in other mammals; [100]), the EC domains of which are encoded by 2 exons [91]. While alternative splicing of small exons is observed in all δ1-Pcdh transcripts, this is particularly prominent within Pcdh11 and Pcdh19, resulting in numerous splice variants for these genes [91, 101–103].
3.2 Regulation of clustered Pcdh gene expression
Pcdh genes are predominantly expressed in the developing and adult nervous system [83, 91, 97, 99, 104–111], though they are also expressed at low levels in other organs such as lung and kidney [107, 108, 112, 113]. A number of studies have uncovered a complex mechanism of transcription for the Pcdha, Pcdhb, and Pcdhg clusters that results in each neuron expressing a semi-stochastic repertoire of isoforms. Each variable exon has its own promoter region that includes a ~20 base pair conserved sequence element (CSE) required for expression. A variable exon promoter is “chosen” for activation by a DNA looping mechanism involving enhancers outside of the clusters, in a manner dependent on the transcription factor CTCF, cohesin, and the methyltransferase SETDB1 ([114–119], reviewed by Hirayama and Yagi [120] in this special issue). A long transcript through the remainder of the cluster, including all downstream variable exons as well as (for the Pcdha and Pcdhg clusters) the three constant exons, is generated. The 5’ variable exon is cis-spliced to the three downstream constant exons to generate a mature Pcdha or Pcdhg transcript (note that Pcdhb transcripts include only a single variable exon; [98, 99]. Based on single-cell RT-PCR studies, it is believed that the majority of Pcdha, Pcdhb, and Pcdhg variable exons are monoallelically expressed, while PcdhaC1, PcdhC2, PcdhgC3, PcdhgC4, and PcdhgC5, the C isoforms of α-Pcdh and γ-Pcdh, are biallelically expressed. Both clusters are transcriptionally active in any given cell, but a particular variable exon promoter is “chosen” from only one of the two alleles. This produces a stochastic expression pattern for most of the α-, β-, and γ-Pcdh family members, while the C isoforms are ubiquitously expressed [121–123]. Single-cell RT-PCR analysis of cerebellar Purkinje neurons found that each cell expressed approximately 4 α-Pcdh isoforms, 2 β-Pcdh isoforms, and 7 γ-Pcdh isoforms [124, 125].
It has been found that this differential expression of clustered Pcdh genes is controlled by epigenetic modifications, specifically silencing by methylation ([98, 126–128]; reviewed by Hirayama and Yagi [120], in this special issue). Work from Takeshi Yagi and colleagues, in particular, has demonstrated that the degree of methylation of Pcdha promoters, as well as the 5’ regions of each variable exon, was negatively correlated with that exon’s expression level; PcdhaC1 and PcdhaC2, both of which are ubiquitously expressed, possess hypomethylated promoters [127, 129]. Demethylation through the application of 5-azacytidine was sufficient to increase the transcription of Pcdha genes, and experimental hypermethylation of a promoter repressed its transcriptional activity [127]. In lines of transgenic mice harboring deletions or duplications of exons within the Pcdha cluster, a decrease in methylation of any variable exon situated at the 3’ end of the cluster was observed. These hypomethylated exons subsequently were found to be ubiquitously expressed, while duplicated copies of the normally ubiquitous PcdhaC1 and PcdhaC2 became hypermethylated and stochastically expressed when situated farther 5’ in the cluster [129, 130]. Work from the same laboratory identified Dnmt3b as the DNA methyltransferase responsible for regulating methylation patterns of stochastically expressed Pcdh isoforms in neural cells at early embryonic stages [131]. As will be seen below (section 4), such mechanisms are relevant to the epigenetic dysregulation of Pcdh genes in cancer and other disorders (reviewed by El Hajj et al., [132], in this special issue).
3.3 Pcdh functions and signaling
Most studies on Pcdhs have focused on their roles in neurodevelopment. The clustered Pcdh proteins have been detected on axons, in dendrites, and in some, but far from all, synapses [97, 109, 113, 133–139]. These observations are consistent with functions revealed by analyses of Pcdha or Pcdhg mutant mice, which indicated that α- and γ-Pcdhs play critical roles in neuronal survival, axon and dendrite arborization, self-avoidance, and tiling, and synaptogenesis, depending on the neuronal subtype examined [109, 137, 140–152]. Comparison of mice lacking all three Pcdh gene clusters with those lacking only the Pcdha, Pcdhb, or Pcdhg clusters indicates overlapping and synergistic functions for the clustered Pcdh families, though the most severe phenotypes are generally attributable to loss of the γ-Pcdhs [153, 154]. Individual non-clustered δ-Pcdhs are expressed by discrete neuronal subsets throughout the brain, and functional studies collectively have revealed roles in axon outgrowth and pathfinding, synaptic plasticity, and synapse elimination [84, 86, 110, 155–163]. Though not the focus of this review, the many roles of Pcdhs in the nervous system have been discussed extensively in several recent reviews ([1, 2, 164–166]; from this special issue, see Light and Jontes, 2017; El Hajj et al., [132]; Aviles and Lefebvre, [79], Rubinstein et al., 2017; Phillips et al., [167]; Hirayama and Yagi, [120]).
Pcdhs have been found to engage with a number of intracellular signaling partners of potential relevance to multiple Wnt pathways. The α- and γ-Pcdhs interact with focal adhesion kinase (FAK) and Pyk2 (also known as FAK2) via their respective constant regions [94]. Interaction with these Pcdhs inhibits activity of these kinases by suppressing their autophosphorylation, which is the first step for kinase activation [94]. Consistent with this, analyses of Pcdhg mutant cerebral cortex revealed hyperactivation of a pathway including PLC, FAK and protein kinase C (PKC) [85]. This report is bolstered by concurrent work showing hyperactivation of Pyk2 and FAK, and reduced activity of both Rac1 and RhoA in animals with a deletion of the Pcdha gene cluster [168]. Recent work identified a serine residue within the γ-Pcdh constant domain that is phosphorylated by PKC in vitro and in vivo; this phosphorylation event reduces inhibition of FAK by γ-Pcdhs [87]. Together, these studies provide the strongest evidence thus far for a signaling pathway downstream of the clustered Pcdhs; notably, PLC, PKC, Rac1 and RhoA are all of relevance to multiple Wnt signaling pathways (Figure 1).
Yeast two-hybrid experiments revealed an interaction of the intracellular domain of Pcdh7 with protein phosphatase 1 alpha (PP1α), a protein implicated in synaptic plasticity [169, 170]. It was later determined that this interaction is conserved for all δ1-Pcdhs, facilitated through the CM3 motif, which is absent in δ2-Pcdhs [91]. Several δ2-Pcdhs (Pcdh10, Pcdh17, Pcdh18b, Pcdh19), on the other hand, have been found to interact with Nap1, a component of the WAVE complex, through the WRC interacting receptor sequence (WIRS) [84, 86, 88, 90, 171]. The WAVE protein complex comprises WAVE1, Cyfip1, Abi2, Nap1 and HSPC300, and is activated by Rac1 and Arf GTPases. The WAVE complex interacts with the Arp2/3 complex to promote actin assembly [172]. The WIRS, a conserved motif, is not exclusive to δ2-Pcdhs, but is also present in α-Pcdhs and Pcdh9, as well as many other adhesion molecules [171]. Note that these pathways leading to actin rearrangements may have several points of contact with components of Wnt/PCP signaling as well.
4. Regulation of Wnt signaling by Pcdhs
4.1 Epigenetic dysregulation of Pcdhs in cancer
Considering that Pcdhs are thought of primarily as neuronal cell adhesion molecules, and neurons are intrinsically postmitotic and terminally differentiated, it is perhaps surprising that several studies have also implicated Pcdhs in many types of cancer. Several groups have reported silencing of δ-Pcdh expression, due to promoter hypermethylation, in many primary tumors or cell lines: Pcdh1 in breast cancer [173]; Pcdh7 in bladder cancer [174]; Pcdh8 in renal cell carcinoma [175], nonmuscle invasive bladder [176], hematologic [177] and breast [178] cancers; Pcdh9 in glioblastoma [179] and hepatocellular carcinoma [180]; Pcdh10 in breast [181], lung, nasopharyngeal, esophageal [182], hepatocellular carcinoma [182, 183], hematologic [184], colorectal, pancreatic [185], gastric [185, 186], cervical [187, 188], prostate [189], and testicular [190] cancers; Pcdh17 in laryngeal and esophageal squamous cell carcinoma [191, 192], urological [193, 194], gastric, and colorectal cancers [195]; and Pcdh20 in non-small-cell lung [196] and hepatocellular carcinoma [197]. A smaller number of studies has implicated dysregulation of the clustered Pcdh genes in cancer as well. A microarray-based methylation study of astrocytomas revealed that Pcdhga11 is hypermethylated in these cells, which resulted in decreased transcription. Transcript levels of Pcdhga11 were restored when the authors treated these astrocytomas with a demethylating agent [198]. The Pcdhb gene cluster has been associated with “CpG island methylator phenotype”, a term used to describe concordant methylation of multiple loci in various cancers [199], as the pattern of methylation within the cluster was able to distinguish two groups of neuroblastoma patients at opposite ends of the International Neuroblastoma Risk Group classification system [200]. Other studies have found clustered Pcdhs to be differentially methylated in prostate cancers, and hypermethylated in breast cancers [201, 202]. There is now increasing evidence that Pcdhs are potential tumor suppressor genes, as reexpression of Pcdh8, Pcdh10, Pcdh17 and Pcdh20 suppresses tumor cell proliferation, inhibits cell migration, and induces apoptosis and autophagy in cancer cell lines [178, 192, 195, 196].
4.2 Clustered Pcdhs
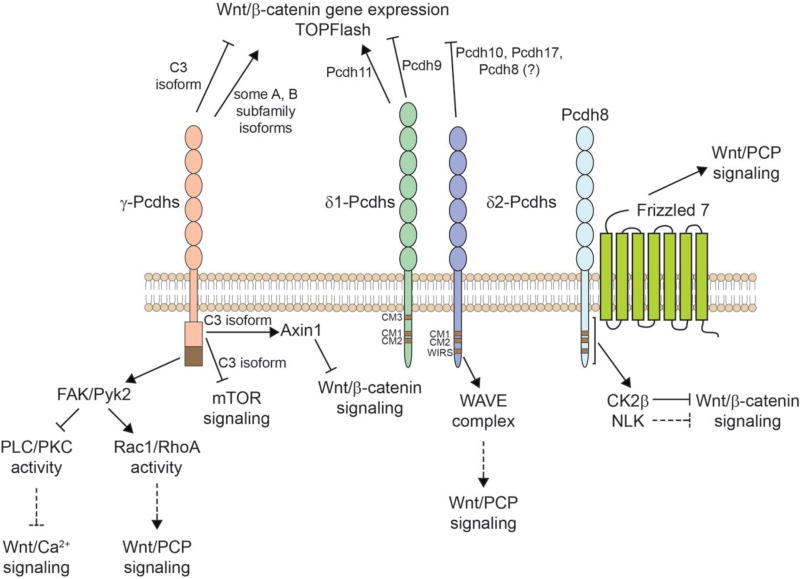
The well-established role of Wnt signaling in tumorigenecity thus suggests that Pcdhs could act as tumor suppressor genes via Wnt pathway regulation. A growing body of literature has, in fact, begun to identify mechanisms by which Pcdhs can regulate Wnt signaling (Figure 3). A genome-wide analysis of promoter methylation in Wilm’s tumor (WiT), a pediatric kidney cancer, identified a region spanning 800 kilobases at chromosome 5q31 that was hypermethylated; this is the region containing the ~60 genes of the Pcdh gene clusters [112]. Consistent with this hypermethylation, the authors demonstrated silencing of Pcdhg gene expression in WiT. While there is extensive hypermethylation across the Pcdhg gene cluster in WiT, some individual Pcdh genes (such as Pcdhga6 and Pcdhgc3) are not hypermethylated and remain expressed; therefore, the authors performed a knockdown of all Pcdhg genes using an siRNA targeting the constant exons in a WiT cell line. This led to an increase in β-catenin/TCF/Lef reporter gene activity and a corresponding increase in expression of target genes in the Wnt signaling pathway [112]. In addition, overexpression of individual γ-Pcdh isoforms in HEK293 and WiT cell lines led to a decrease in Wnt signaling activity and inhibition of colony formation and tumor cell growth in vitro [112]. This group subsequently expanded on this study to show that long range epigenetic silencing of the Pcdhg cluster is also observed in colorectal cancer, and is associated with the early stages of colorectal tumorigenesis [203]. Focusing on the most abundantly expressed γ-Pcdh isoform in the colon, Dallosso et al. [203] also observed that Pcdhgc3 expression is silenced in colorectal cancer cells. Overexpression of γ-Pcdh-C3 in human colon cancer cell lines increased apoptosis, inhibited growth, and led to a reduction of β-catenin/TCF/Lef reporter activity as well as a decrease in the levels of endogenous “active” β-catenin. The authors go on to implicate the mTOR pathway, as siRNA knockdown of Pcdhgc3 led to an increase in phosphophorylation of mTOR at serine residue 2448 (S2448) as well as an increase in total mTOR levels as assayed by Western blot; the opposite effects on mTOR were observed when Pcdhgc3 was overexpressed [203]. Together, this work provided the first firm evidence that clustered Pcdhs could regulate tumor cell behavior by inhibiting canonical Wnt signaling.
Figure 3. Regulation of Wnt pathways by Pcdhs.
Summary of results implicating γ-Pcdhs (left) and δ-Pcdhs (right) in the regulation of Wnt signaling, as discussed in the main text. The γ-Pcdh-C3 isoform, and the δ-Pcdhs Pcdh8, 9, 10, and 17 have been reported to suppress Wnt-induced expression of target genes, while some other γ-Pcdh isoforms and the δ1 protein Pcdh11 have been reported to have the opposite effect (top). Some Pcdhs have known cytoplasmic interactors that have been shown to, or that potentially could, impinge upon Wnt signaling pathways (bottom). Pcdh8 has been shown to interact with Frizzeld7 to promote Wnt/PCP signaling (far right). Long lines with a short perpendicular line indicate inhibition, while arrows indicate activation. Dashed lines indicate possible signaling connections, based on the literature, that remain to be demonstrated directly.
We subsequently sought to elucidate the specificity of Wnt pathway suppression by γ-Pcdh isoforms, and to identify the molecular mechanisms through which it is achieved [204]. Using the TOPFLASH assay, in which HEK293 cells transfected with reporter constructs that yield a quantifiable luciferase signal upon exposure to Wnt3a, as well as quantitative PCR (qPCR) for Wnt target genes, we confirmed that the γ-Pcdh-C3 isoform, specifically, inhibits the canonical transcriptional pathway. Surprisingly, however, we found that 13 other γ-Pcdh isoforms can actually potentiate β-catenin/TCF/Lef luciferase reporter activity in response to Wnt3a. We determined that the variable cytoplasmic domain (VCD), unique to each γ-Pcdh isoform, is important in this regulation of Wnt signaling: expression of constructs encoding only the VCD of C3 or A1 isoforms was sufficient to, respectively, suppress or potentiate Wnt signaling [204]. We identified Axin1, a key component of the destruction complex, as an evolutionarily-conserved physical interactor of the γ-Pcdh-C3 VCD, and showed that the C3 VCD competes with Dvl for binding to the DIX domain of Axin1. This interaction stabilized Axin1 at the membrane, and reduced phosphorylation of Lrp6 [204].
We were also able to confirm that β-catenin/TCF/Lef reporter activity can be modulated up (by overexpression of γ-Pcdh-A1) or down (by overexpression of γ-Pcdh-C3) in the mouse cerebral cortex in vivo, using conditional transgenic alleles [204]. Our data suggest a novel mechanism in which the interaction of γ-Pcdh-C3 with Axin1 can potentially sequester it away from other Wnt signaling components, which leads to Lrp6 hypophosphorylation and reduced Wnt target gene expression through an as-yet undetermined pathway. It is likely that γ-Pcdhs act as modulators or buffers for Wnt signaling activity, rather than as primary co-factors, as a grossly normal cerebral cortex still forms in mice overexpressing γ-Pcdh-C3 or –A1 [204]; major disruption of Wnt signaling in embryonic telencephalon (e.g., through ablation of the Wnt-producing cortical hem; [205]) leads to a severe disruption of cortical development. As it stands, it will be important to determine if any of the many roles that γ-Pcdhs play in the brain are dependent on its modulation of Wnt pathways. In this respect, the interaction of the C3 VCD with Axin1 is particularly interesting: both Axin1 [206] and the γ-Pcdhs [85, 147, 168] are required for complex dendrite arborization in cortical or hippocampal neurons.
4.3 Non-clustered Pcdhs
Like some γ-Pcdhs, a number of non-clustered δ-Pcdhs have been implicated as potential tumor suppressors through regulation of Wnt signaling pathways (Figure 3). Reduction of Pcdh10 expression due to promoter hypermethylation has been reported in several tumors [207], while its experimental reexpression inhibits cell growth, decreases colony formation, prevents cell invasion and promotes cell apoptosis, all of which indicate a possible role for Pcdh10 as a tumor suppressor. It appears that Pcdh10 can act through the canonical Wnt/β-catenin pathway, as Zhao et al. [208] reported that the expression of Pcdh10 negatively regulated Wnt transcriptional response, as shown using TOPFLASH and qPCR. One Wnt target gene that was shown to be suppressed is a long noncoding RNA called MALAT1; overexpression of Pcdh10 decreased β-catenin binding at MALAT1’s promoter through an unknown mechanism [208]. Another study implicated Pcdh10 in the negative regulation of Wnt/β-catenin signaling [209]. Pcdh10 overexpression was found to inhibit myeloma cell proliferation, even when cells were treated with LiCl to activate Wnt signaling [209]. Pcdh10 negatively regulated a canonical Wnt pathway, as assayed by TOPFLASH and qPCR for several Wnt target genes (Xu et al., 2015); additionally, the expression of several Wnt pathway proteins, including β-catenin, was reduced while GSK3β was upregulated [209]. Lastly, the authors observed a decrease in the expression of B-cell CLL/lymphoma 9 (BCL-9), a coactivator of β-catenin, in myeloma cell lines that overexpress Pcdh10, confirming the negative regulation of the Wnt/β-catenin signaling by Pcdh10 [209].
Pcdh8 (for simplicity, we will use this gene name here regardless of organism; original papers reported this gene as paraxial protocadherin (PAPC) in amphibians [210] and arcadlin in rats [161]) has been intensively studied for its roles in development mediated through the Wnt/PCP pathway. Pcdh8 was first discovered as a gene expressed in the Spemann organizer of Xenopus, and later in the paraxial mesoderm during embryo gastrulation [210]. Pcdh8 expression was found to be upregulated as a result of signaling through a non-canonical Wnt5a/ROR2-JNK (PCP) pathway in early Xenopus development [211]. The overexpression of Pcdh8 RNA was sufficient to trigger gastrulation movements in Xenopus animal cap explants [210], and Pcdh8 has also been shown to coordinate tissue separation and convergent extension in Xenopus development by acting through the Wnt/PCP pathway to simultaneously activate JNK via Rho A while inactivating Rac1 [212, 213]. It has recently been determined that glycosylation of Wnt5a is required in vivo for Pcdh8 synthesis, further lending support to Pcdh8’s links to the Wnt/PCP pathway [214].
Much progress has been made in identifying key interactors of Pcdh8 in its regulation of the PCP pathway. Pcdh8 physically interacts with Fzd7 through its extracellular region, as determined by co-immunoprecipitation and bimolecular fluorescence complementation. This interaction regulates tissue separation in the mesoderm of Xenopus embryos [212, 215]. It has also been found that the intracellular domain of Pcdh8 recruits Sprouty, an inhibitor of convergence-extension movements, to the membrane, and antagonizes Sprouty’s ability to inhibit the PCP pathway [216]. Concurrent work revealed that Xenopus ankyrin repeats domain protein 5 (xANR5) physically interacts with Pcdh8 to promote the PCP pathway indirectly, by regulating JNK and Rho activity [217]. Jung et al. [218] discovered that the activation of RhoA signaling through the binding of Pcdh8 and Fzd7, in conjunction with the Wnt/PCP pathway, regulates invagination of the ear placode in Xenopus. Finally, Pcdh8 stability appears to be a regulation point for Wnt signaling pathways. The regulation of Pcdh8 localization and stability is determined by its phosphorylation by GSK3, and subsequent polyubiquitination by the E3 ubiquitin ligase, β-TrCP [219]. A recent study also demonstrated a physical interaction between the Pcdh8 intracellular domain and NLK, which feeds back onto the canonical β-catenin transcription pathway; this interaction stabilizes both proteins by inhibiting their ubiquitination and is required for Pcdh8’s promotion of Wnt/PCP signaling [220]. Finally, in a study by Kietzmann et al. [221], it was found that the intracellular domain of Pcdh8 interacts with casein kinase 2β (CKIIβ) and recruits it to the membrane. This prevents CKIIβ from forming a tetrameric complex consisting of 2 CKIIα and 2 CKIIβ subunits that is required for CK2 phosphorylation of β-catenin, which potentiates Wnt signaling by stabilizing β-catenin [221, 222]. This shift in CKIIβ localization mediated by Pcdh8 decreases Wnt/β-catenin signaling activity and target gene expression [221].
This general down-regulation of canonical Wnt/β-catenin-dependent signaling pathways by non-clustered Pcdhs is supported by several other studies. As noted above, Pcdh9 has been found to be downregulated in hepatocellular carcinoma (HCC) [180]. Over-expression of Pcdh9 in HCC-derived cell lines decreases migration and results in reduced phosphorylation of GSK3β at serine residue 9 (and thus presumably higher GSK3β activity); however, this did not seem to suppress the transcription of Wnt target genes [180], so a distinct pathway may be involved. Overexpression of Pcdh17 in tumor cells led to a reduction of active (that is, non-GSK3β phosphorylated) β-catenin, decreased levels of β-catenin mRNA and of proteins encoded by Wnt target genes, and a suppression of tumor growth [223]. Pcdh20 (a δ-Pcdh-related protein sometimes classified as its own δ0 subgroup) was also found to negatively regulate Wnt signaling activity in some HCC-derived cells as well as in HEK293T [197]. Pcdh20 overexpression suppresses HCC cell migration in vitro and cell growth both in vitro and in vivo; the authors posit that Pcdh20 activates GSK3β by modulating the Erk and Akt pathway [197]. This is consistent with a concurrent study demonstrating lower levels of active β-catenin, translocation of β-catenin from the nucleus to the cytoplasm and membrane, and reduced expression of Wnt target genes in cells transfected with Pcdh20 [224]. Interestingly, there is one Pcdh, Pcdh11Y, that may upregulate canonical Wnt signaling. Unusually for Pcdhs, Pcdh11Y has a β-catenin binding site localized within its COOH terminus and was found to interact with β-catenin by immunoprecipitation assay [225]. Overexpression of Pcdh11Y in human prostate or colon cancer cell lines was found to activate Wnt signaling, as measured by a TOPFlash assay and semi-quantitative PCR for Wnt target genes [226].
5. Conclusions
From the forgoing discussion, we hope it is clear that the varied groups of both clustered and non-clustered Pcdh adhesion molecules are increasingly becoming implicated in regulation of Wnt signaling pathways, particularly as this relates to cancer and morphological development of embryos. Nevertheless, this is still an emerging interaction and much remains unclear about how Pcdhs might impinge upon the multiple pathways activated downstream of Wnt binding to its receptors. In closing, we highlight a few of the questions that should be explored in future studies.
First, why do some Pcdhs suppress canonical (Wnt/β-catenin-dependent gene transcription) signaling, while others appear to potentiate it? Most reports thus far demonstrate the former rather than the latter, but this may simply reflect the small number of Pcdhs and cellular contexts that have been examined. Presumably, as we have shown for the γ-Pcdhs, a major component of this is differential interaction of Pcdh cytoplasmic domains with particular signaling partners. For example, we showed that the variable cytoplasmic domain of γ-Pcdh-C3, but not several other γ-Pcdh isoforms, competes with Dvl for binding to Axin1, leading to suppression of canonical Wnt signaling [203]. The challenge now is to identify other signaling proteins that might mediate the opposite effect on Wnt target genes that we found when overexpressing other γ-Pcdh isoforms. In addition to the variable cytoplasmic domains, which are unique to each γ-Pcdh isoform, all γ-Pcdhs share a constant cytoplasmic domain; any role for this domain in Wnt signaling remains to be determined, though its binding to, and inhibition of, FAK and Pyk2 affects signaling pathways of potential relevance (Figure 3). In the case of δ-Pcdhs, all of those thus far examined inhibit the canonical pathway except Pcdh11Y. In this respect, it is interesting to note that among the δ-Pcdhs, only Pcdh11 harbors a cytoplasmic β-catenin binding site resembling that found in classical cadherins [225]. It remains to be shown, however, whether this somehow underlies Pcdh11’s ability to upregulate the canonical pathway. Clearly, a more complete catalog of Pcdh cytoplasmic interactors is needed in order to generate hypotheses about mechanisms of Wnt pathway modulation. Additionally, it is totally unknown whether homophilic engagement of Pcdh extracellular domains causes conformational changes that may activate intracellular signaling. One interesting thing to note is that γ-Pcdh-C3, the only isoform that we found to inhibit canonical Wnt signaling [204], appears to be ubiquitously expressed, at least among neurons [123]. Thus, it may be that a basal level of homophilic interaction, mediated by C3, obtains between any two cells. If homophilic interaction potentiates signaling via Axin1, then C3 may mediate a basal suppression of the canonical pathway. Introduction of matching between other γ-Pcdh isoforms, many of which seem to potentiate Wnt signaling (e.g., A1 or B1), could strengthen cell-cell adhesion while counterbalancing C3’s inhibition of the canonical pathway. Though it is too early to speculate further on such a mechanism, it is clear that a major question is whether Pcdhs regulate Wnt signaling pathways constitutively, or whether this is dependent on cell-cell interaction; if so, the isoform composition of adhesion complexes may determine ultimate signaling outcomes.
Second, it remains unclear the extent to which changes in TOPFlash output or altered expression of Wnt target genes induced by Pcdh experimental over-expression or down-regulation due to altered methylation in vivo represents true canonical Wnt signaling. In our own unpublished experiments, for example, we have found that γ-Pcdh-C3 inhibits the expression of Wnt target genes or TCF/Lef reporter constructs without significantly altering the levels or localization of active β-catenin. Similarly, overexpression of Pcdh9 could reduce phosphorylation of GSK3β at Ser9, which should increase its activity, without a concomitant increase in assayed Wnt target genes [179]. Like many intracellular signaling proteins, Wnt components can interact with many partners and link into many distinct pathways. It is quite possible that several Pcdhs can modulate the downstream activity of β-catenin/TCF/Lef-dependent gene transcription through novel “non-canonical” pathways, including but not limited to regulation of Wnt/PCP and/or Wnt/Ca2+ branches of signaling. An example may be Pcdh8’s interaction with NLK [219], which can negatively feedback on β-catenin/TCF/Lef-dependent gene transcription (Figure 3).
Finally, most of the evidence linking Pcdhs in to Wnt signaling pathways comes from the study of cancer cells, and yet most major functional roles for Pcdhs identified thus far in vivo concern aspects of neuronal development, including axon outgrowth, dendrite arborization, and synaptogenesis (reviewed by [1]). Wnt signaling is known to play many important roles in the developing nervous system (reviewed by [227]); thus it will be important to determine whether altered Wnt signaling plays any role in the many neural functions identified for Pcdh proteins. Certainly, major disruption of Wnt signaling is incompatible with the formation of a structurally normal nervous system, so the discrete phenotypes observed when clustered or non-clustered Pcdhs are deleted in mice could only reflect fairly subtle modulation of Wnt pathways. Additionally, some Wnt pathway proteins can affect neural circuit formation on their own (e.g., Axin1 is important for the formation of dendritic arbors in hippocampal neurons [206]), reinforcing the point above that the Pcdhs may affect Wnt pathway components without acting through the accepted “canonical” signaling pathways.
Acknowledgments
Work in the Weiner Laboratory described herein has been supported by the following grants to J.A.W.: R01 NS055272 and R21 NS090030
Abbreviations
- Pcdhs
protocadherins
- PP1a
protein phosphatase 1a
- WIRS
WRC interacting receptor sequence
- FAK
focal adhesion kinase
- PKC
protein kinase C
- MMTV
mouse mammary tumor virus
- Fzd
Frizzleds
- ICD
Intracellular domain
- Dvls
Dishevelled proteins
- Lrp
Low-density lipoprotein-related rececptor
- CKI
Casein kinase I
- ROR
Receptor tyrosine kinase-like Orphan Receptor
- PCP
planar-cell polarity
- JNK
c-Jun N-terminal kinase
- APC
adenomatous polyposis coli
- GSK3β
glycogen synthase kinase 3β
- TCF
T-cell factor
- Lef
lymphoid enhancer factor
- Daam1
Dishevelled-associated activator of morphogenesis 1
- PLC
phospholipase C
- CaMKII
calmodulin-dependent kinase II
- PKC
protein kinase C
- NLK
Nemo-like kinase
- NFAT
nuclear factor of activated T cell
- CSE
conserved sequence element
- qPCR
quantitative PCR
- VCD
variable cytoplasmic domain
- BCL-9
B-cell CLL/lymphoma 9
- PAPC
Paraxial protocadherin
- xANR5
Xenopus ankyrin repeats domain protein 5
- HCC
hepatocellular carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peek SL, Mah KM, Weiner JA. Regulation of neural circuit formation by protocadherins. Cellular and Molecular Life Sciences. 2017:1–25. doi: 10.1007/s00018-017-2572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeler AB, Molumby MJ, Weiner JA. Protocadherins branch out: Multiple roles in dendrite development. Cell Adh Migr. 2015;9(3):214–26. doi: 10.1080/19336918.2014.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nature reviews Molecular cell biology. 2009;10(7):468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harbor perspectives in biology. 2009;1(2):a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niehrs C. The complex world of WNT receptor signalling. Nature reviews Molecular cell biology. 2012;13(12):767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 8.Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Slusarski DC, Pelegri F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Developmental biology. 2007;307(1):1–13. doi: 10.1016/j.ydbio.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De A. Wnt/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin (Shanghai) 2011;43(10):745–756. doi: 10.1093/abbs/gmr079. [DOI] [PubMed] [Google Scholar]
- 11.Willert K, Nusse R. Wnt proteins. Cold Spring Harbor perspectives in biology. 2012;4(9):a007864. doi: 10.1101/cshperspect.a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 13.Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, Nusse R. The Drosophila homology of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987;50(4):649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 14.Ching W, Hang HC, Nusse R. Lipid-independent secretion of a Drosophila Wnt protein. Journal of Biological Chemistry. 2008;283(25):17092–17098. doi: 10.1074/jbc.M802059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komekado H, Yamamoto H, Chiba T, Kikuchi A. Glycosylation and palmitoylation of Wnt-3a are coupled to produce an active form of Wnt-3a. Genes to Cells. 2007;12(4):521–534. doi: 10.1111/j.1365-2443.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 16.Kurayoshi M, Yamamoto H, Izumi S, Kikuchi A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochemical Journal. 2007;402(3):515–523. doi: 10.1042/BJ20061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Developmental cell. 2006;11(6):791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423(6938):448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 19.Chu ML-H, Ahn VE, Choi H-J, Daniels DL, Nusse R, Weis WI. Structural studies of Wnts and identification of an LRP6 binding site. Structure. 2013;21(7):1235–1242. doi: 10.1016/j.str.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh J-C, Rattner A, Smallwood PM, Nathans J. Biochemical characterization of Wnt-frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proceedings of the National Academy of Sciences. 1999;96(7):3546–3551. doi: 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Chang H, Rattner A, Nathans J. Chapter Seven-Frizzled Receptors in Development and Disease. Current topics in developmental biology. 2016;117:113–139. doi: 10.1016/bs.ctdb.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. New insights into the mechanism of Wnt signaling pathway activation. Int Rev Cell Mol Biol. 2011;291(21):e71. doi: 10.1016/B978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 24.Yanfeng WA, Tan C, Fagan RJ, Klein PS. Phosphorylation of frizzled-3. Journal of Biological Chemistry. 2006;281(17):11603–11609. doi: 10.1074/jbc.M600713200. [DOI] [PubMed] [Google Scholar]
- 25.Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. The EMBO Journal. 2010;29(13):2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a frizzled8· Wnt3a· LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. Journal of Biological Chemistry. 2010;285(12):9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 γ couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438(7069):867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- 28.Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Molecular cell. 2004;13(1):149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 29.Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438(7069):873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hikasa H, Shibata M, Hiratani I, Taira M. The Xenopus receptor tyrosine kinase Xror2 modulates morphogenetic movements of the axial mesoderm and neuroectoderm via Wnt signaling. Development. 2002;129(22):5227–5239. doi: 10.1242/dev.129.22.5227. [DOI] [PubMed] [Google Scholar]
- 31.Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes to Cells. 2003;8(7):645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 32.Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R, Vijayakumar S, Economides AN, Aaronson SA. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes & development. 2010;24(22):2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato A, Yamamoto H, Sakane H, Koyama H, Kikuchi A. Wnt5a regulates distinct signalling pathways by binding to Frizzled2. The EMBO journal. 2010;29(1):41–54. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witte F, Bernatik O, Kirchner K, Masek J, Mahl A, Krejci P, Mundlos S, Schambony A, Bryja V, Stricker S. Negative regulation of Wnt signaling mediated by CK1-phosphorylated Dishevelled via Ror2. The FASEB Journal. 2010;24(7):2417–2426. doi: 10.1096/fj.09-150615. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto H, Yoo SK, Nishita M, Kikuchi A, Minami Y. Wnt5a modulates glycogen synthase kinase 3 to induce phosphorylation of receptor tyrosine kinase Ror2. Genes to Cells. 2007;12(11):1215–1223. doi: 10.1111/j.1365-2443.2007.01128.x. [DOI] [PubMed] [Google Scholar]
- 36.Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/beta-catenin signaling. J Cell Biol. 2011;194(5):737–50. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu W, Yamamoto V, Ortega B, Baltimore D. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119(1):97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Macheda ML, Sun WW, Kugathasan K, Hogan BM, Bower NI, Halford MM, Zhang YF, Jacques BE, Lieschke GJ, Dabdoub A. The Wnt receptor Ryk plays a role in mammalian planar cell polarity signaling. Journal of Biological Chemistry. 2012;287(35):29312–29323. doi: 10.1074/jbc.M112.362681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes & development. 2006;20(23):3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 40.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. The EMBO journal. 1998;17(5):1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272(40):24735–8. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 42.Wong H-C, Bourdelas A, Krauss A, Lee H-J, Shao Y, Wu D, Mlodzik M, Shi D-L, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Molecular cell. 2003;12(5):1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilić J, Huang Y-L, Davidson G, Zimmermann T, Cruciat C-M, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316(5831):1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 44.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development. 2004;131(8):1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 45.Piao S, Lee S-H, Kim H, Yum S, Stamos JL, Xu Y, Lee S-J, Lee J, Oh S, Han J-K. Direct inhibition of GSK3β by the phosphorylated cytoplasmic domain of LRP6 in Wnt/β-catenin signaling. PloS one. 2008;3(12):e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu G, Huang H, Abreu JG, He X. Inhibition of GSK3 phosphorylation of β-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PloS one. 2009;4(3):e4926. doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto H, Kishida S, Kishida M, Ikeda S, Takada S, Kikuchi A. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3β regulates its stability. Journal of Biological Chemistry. 1999;274(16):10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
- 48.Kim S-E, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV, MacDonald BT, Zhang X, Abreu JG, Peng L, He X. Wnt Stabilization of β-Catenin Reveals Principles for Morphogen Receptor-Scaffold Assemblies. Science. 2013;340(6134):867–870. doi: 10.1126/science.1232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Wang S, Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein & cell. 2014;5(3):186–193. doi: 10.1007/s13238-014-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor perspectives in biology. 2012;4(11):a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annual review of genetics. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishitani T, Ninomiya-Tsuji J, Nagai S-i, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H. The TAK1–NLK–MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature. 1999;399(6738):798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 53.Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes & development. 2009;23(3):265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- 54.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes & development. 2003;17(18):2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 55.Okamura H, Aramburu J, García-Rodríguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Molecular cell. 2000;6(3):539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 56.Porter CM, Havens MA, Clipstone NA. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. Journal of Biological Chemistry. 2000;275(5):3543–3551. doi: 10.1074/jbc.275.5.3543. [DOI] [PubMed] [Google Scholar]
- 57.Shaw K, Ho AM, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan PG. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proceedings of the National Academy of Sciences. 1995;92(24):11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nusse R, van Ooyen A, Cox D, Fung YKT, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 59.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55(4):619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 60.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, Mckechnie D, Finniear R, Markham A, Groffen J, Boguski MS, Altschul SF, Horii A, Ando H, Miyoshi Y, Miki Y, Nishisho I, Nakamura Y. Identification of Fap Locus Genes from Chromosome-5q21. Science. 1991;253(5020):661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 61.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253(5020):665–9. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 62.Clements WM, Wang J, Sarnaik A, Kim OJ, MacDonald J, Fenoglio-Preiser C, Groden J, Lowy AM. β-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Research. 2002;62(12):3503–3506. [PubMed] [Google Scholar]
- 63.Dahmen R, Koch A, Denkhaus D, Tonn J, Sörensen N, Berthold F, Behrens J, Birchmeier W, Wiestler O, Pietsch T. Deletions of AXIN1, a component of the WNT/wingless pathway, in sporadic medulloblastomas. Cancer research. 2001;61(19):7039–7043. [PubMed] [Google Scholar]
- 64.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nature genetics. 2000;24(3):245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 65.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25(57):7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 66.Endo M, Nishita M, Fujii M, Minami Y. Chapter Three-Insight into the Role of Wnt5a-Induced Signaling in Normal and Cancer Cells. International review of cell and molecular biology. 2015;314:117–148. doi: 10.1016/bs.ircmb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 67.Nomachi A, Nishita M, Inaba D, Enomoto M, Hamasaki M, Minami Y. Receptor tyrosine kinase Ror2 mediates Wnt5a-induced polarized cell migration by activating c-Jun N-terminal kinase via actin-binding protein filamin A. Journal of Biological Chemistry. 2008;283(41):27973–27981. doi: 10.1074/jbc.M802325200. [DOI] [PubMed] [Google Scholar]
- 68.Asem MS, Buechler S, Wates RB, Miller DL, Stack MS. Wnt5a Signaling in Cancer. Cancers. 2016;8(9) doi: 10.3390/cancers8090079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sedgwick AE, D'Souza-Schorey C. Wnt Signaling in Cell Motility and Invasion: Drawing Parallels between Development and Cancer. Cancers. 2016;8(9) doi: 10.3390/cancers8090080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2016 doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 72.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Current opinion in cell biology. 2013;25(2):254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(14):4042–5. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 74.Kretzschmar K, Clevers H. Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Developmental Biology. 2017 doi: 10.1016/j.ydbio.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 75.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 76.Miki T, Yasuda S-y, Kahn M. Wnt/β-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Reviews and Reports. 2011;7(4):836–846. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 77.Mohammed MK, Shao C, Wang J, Wei Q, Wang X, Collier Z, Tang S, Liu H, Zhang F, Huang J, Guo D, Lu M, Liu F, Liu J, Ma C, Shi LL, Athiviraham A, He TC, Lee MJ. Wnt/beta-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes & diseases. 2016;3(1):11–40. doi: 10.1016/j.gendis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98(5):585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 79.Avilés EC, Goodrich LV. Seminars in Cell & Developmental Biology. Elsevier; 2017. Configuring a robust nervous system with Fat cadherins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nature reviews. Molecular cell biology. 2005;6(8):622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 81.Sotomayor M, Gaudet R, Corey DP. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol. 2014;24(9):524–36. doi: 10.1016/j.tcb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nature reviews. Neuroscience. 2007;8(1):11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 83.Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, Taketani S, Suzuki S. Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12(6):2249–56. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biswas S, Emond MR, Duy PQ, Hao lT, Beattie CE, Jontes JD. Protocadherin-18b interacts with Nap1 to control motor axon growth and arborization in zebrafish. Mol Biol Cell. 2014;25(5):633–42. doi: 10.1091/mbc.E13-08-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garrett AM, Schreiner D, Lobas MA, Weiner JA. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74(2):269–76. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H, Tanaka Y, Takeichi M. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell. 2014;30(6):673–87. doi: 10.1016/j.devcel.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 87.Keeler AB, Schreiner D, Weiner JA. Protein Kinase C Phosphorylation of a γ-protocadherin C-terminal lipid binding domain regulates focal adhesion kinase inhibition and dendrite arborization. Journal of Biological Chemistry. 2015;290(34):20674–20686. doi: 10.1074/jbc.M115.642306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol. 2008;182(2):395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299(3):551–72. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 90.Tai K, Kubota M, Shiono K, Tokutsu H, Suzuki ST. Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 2010;1344:13–24. doi: 10.1016/j.brainres.2010.04.065. [DOI] [PubMed] [Google Scholar]
- 91.Vanhalst K, Kools P, Staes K, van Roy F, Redies C. delta-Protocadherins: a gene family expressed differentially in the mouse brain. Cell Mol Life Sci. 2005;62(11):1247–59. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vanhalst K, Kools P, Vanden Eynde E, Van Roy F. The human and murine protocadherin-β one-exon gene families show high evolutionary conservation, despite the difference in gene number. FEBS letters. 2001;495(1–2):120–125. doi: 10.1016/s0014-5793(01)02372-9. [DOI] [PubMed] [Google Scholar]
- 93.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–90. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 94.Chen J, Lu Y, Meng S, Han M-H, Lin C, Wang X. α-and γ-Protocadherins negatively regulate PYK2. Journal of biological chemistry. 2009;284(5):2880–2890. doi: 10.1074/jbc.M807417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, Yagi T. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics. 2000;63(1):75–87. doi: 10.1006/geno.1999.6066. [DOI] [PubMed] [Google Scholar]
- 96.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, Maniatis T. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11(3):389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20(6):1137–51. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 98.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10(1):21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 99.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16(15):1890–905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kahr I, Vandepoele K, van Roy F. Delta-protocadherins in health and disease. Prog Mol Biol Transl Sci. 2013;116:169–92. doi: 10.1016/B978-0-12-394311-8.00008-X. [DOI] [PubMed] [Google Scholar]
- 101.Ahn K, Huh JW, Kim DS, Ha HS, Kim YJ, Lee JR, Kim HS. Quantitative analysis of alternative transcripts of human PCDH11X/Y genes. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2010;153b(3):736–44. doi: 10.1002/ajmg.b.31041. [DOI] [PubMed] [Google Scholar]
- 102.Blanco-Arias P, Sargent CA, Affara NA. Protocadherin X (PCDHX) and Y (PCDHY) genes; multiple mRNA isoforms encoding variant signal peptides and cytoplasmic domains. Mammalian Genome. 2004;15(1):41–52. doi: 10.1007/s00335-003-3028-7. [DOI] [PubMed] [Google Scholar]
- 103.Blevins CJ, Emond MR, Biswas S, Jontes JD. Differential expression, alternative splicing, and adhesive properties of the zebrafish δ1-protocadherins. Neuroscience. 2011;199:523–534. doi: 10.1016/j.neuroscience.2011.09.061. [DOI] [PubMed] [Google Scholar]
- 104.Aoki E, Kimura R, Suzuki ST, Hirano S. Distribution of OL-protocadherin protein in correlation with specific neural compartments and local circuits in the postnatal mouse brain. Neuroscience. 2003;117(3):593–614. doi: 10.1016/s0306-4522(02)00944-2. [DOI] [PubMed] [Google Scholar]
- 105.Kim SY, Chung HS, Sun W, Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147(4):996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 106.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144(2):579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 107.Gaitan Y, Bouchard M. Expression of the delta-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr Patterns. 2006;6(8):893–9. doi: 10.1016/j.modgep.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Redies C, Heyder J, Kohoutek T, Staes K, Van Roy F. Expression of protocadherin-1 (Pcdh1) during mouse development. Dev Dyn. 2008;237(9):2496–505. doi: 10.1002/dvdy.21650. [DOI] [PubMed] [Google Scholar]
- 109.Garrett AM, Weiner JA. Control of CNS synapse development by {gamma}-protocadherin-mediated astrocyte-neuron contact. J Neurosci. 2009;29(38):11723–31. doi: 10.1523/JNEUROSCI.2818-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T, Yokoyama K, Tezuka T, Sagara H, Hirano S, Kiyonari H, Takada M, Kobayashi K, Watanabe M, Kano M, Nakazawa T, Yamamoto T. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron. 2013;78(5):839–54. doi: 10.1016/j.neuron.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 111.Hirano S, Yan Q, Suzuki ST. Expression of a Novel Protocadherin, OL-Protocadherin, in a Subset of Functional Systems of the Developing Mouse Brain. The Journal of Neuroscience. 1999;19(3):995–1005. doi: 10.1523/JNEUROSCI.19-03-00995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C, Pritchard-Jones K, Royer-Pokora B, Lee SB, Owen C, Malik S, Feng Y, Frank M, Ward A, Brown KW, Malik K. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 2009;5(11):e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, Kemler R. Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci. 2005;29(4):603–16. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 114.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(51):21081–6. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2(2):345–57. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 116.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):17195–200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proceedings of the National Academy of Sciences. 2012;109(23):9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286(36):31885–95. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, Labonte B, Ho SM, Chandrasekaran S, Do C, Ramirez BR, Peter CJ, C WJ, Safaie BM, Morishita H, Roussos P, Nestler EJ, Schaefer A, Tycko B, Brennand KJ, Yagi T, Shen L, Akbarian S. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet. 2017 doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirayama T, Yagi T. Regulation of clustered protocadherin genes in individual neurons. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 121.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat Genet. 2005;37(2):171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 122.Hirano S, Takeichi M. Cadherins in Brain Morphogenesis and Wiring. Physiological Reviews. 2012;92(2):597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- 123.Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic Gene Regulation of Pcdh- and Pcdh- Clusters Involving Both Monoallelic and Biallelic Expression in Single Purkinje Cells. Journal of Biological Chemistry. 2006;281(41):30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 124.Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, Yagi T. Single-neuron diversity generated by Protocadherin-beta cluster in mouse central and peripheral nervous systems. Frontiers in molecular neuroscience. 2012;5:90. doi: 10.3389/fnmol.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Frontiers in molecular neuroscience. 2012;5:45. doi: 10.3389/fnmol.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaneko R, Kawaguchi M, Toyama T, Taguchi Y, Yagi T. Expression levels of Protocadherin-alpha transcripts are decreased by nonsense-mediated mRNA decay with frameshift mutations and by high DNA methylation in their promoter regions. Gene. 2009;430(1–2):86–94. doi: 10.1016/j.gene.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 127.Kawaguchi M, Toyama T, Kaneko R, Hirayama T, Kawamura Y, Yagi T. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. 2008;283(18):12064–75. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- 128.Toyoda S, Kawaguchi M, Kobayashi T, Tarusawa E, Toyama T, Okano M, Oda M, Nakauchi H, Yoshimura Y, Sanbo M, Hirabayashi M, Hirayama T, Hirabayashi T, Yagi T. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82(1):94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 129.Kaneko R, Abe M, Hirabayashi T, Uchimura A, Sakimura K, Yanagawa Y, Yagi T. Expansion of stochastic expression repertoire by tandem duplication in mouse Protocadherin-alpha cluster. Sci Rep. 2014;4:6263. doi: 10.1038/srep06263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Noguchi Y, Hirabayashi T, Katori S, Kawamura Y, Sanbo M, Hirabayashi M, Kiyonari H, Nakao K, Uchimura A, Yagi T. Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J Biol Chem. 2009;284(46):32002–14. doi: 10.1074/jbc.M109.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell. 2014;158(5):1045–59. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.El Hajj N, Dittrich M, Haaf T. Epigenetic Dysregulation of Protocadherins in Human Disease. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 133.Junghans D, Heidenreich M, Hack I, Taylor V, Frotscher M, Kemler R. Postsynaptic and differential localization to neuronal subtypes of protocadherin beta16 in the mammalian central nervous system. The European journal of neuroscience. 2008;27(3):559–71. doi: 10.1111/j.1460-9568.2008.06052.x. [DOI] [PubMed] [Google Scholar]
- 134.Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan WS, Arndt K, Frank M, Gordon RE, Gawinowicz MA, Zhao Y, Colman DR. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32(1):63–77. doi: 10.1016/s0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 135.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci. 2003;23(12):5096–104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Puller C, Haverkamp S. Cell-type-specific localization of protocadherin beta16 at AMPA and AMPA/Kainate receptor-containing synapses in the primate retina. The Journal of comparative neurology. 2011;519(3):467–79. doi: 10.1002/cne.22528. [DOI] [PubMed] [Google Scholar]
- 137.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36(5):843–54. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 138.Nuhn JS, Fuerst PG. Developmental localization of adhesion and scaffolding proteins at the cone synapse. Gene Expr Patterns. 2014;16(1):36–50. doi: 10.1016/j.gep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Andrade GB, Kunzelman L, Merrill MM, Fuerst PG. Developmentally dynamic colocalization patterns of DSCAM with adhesion and synaptic proteins in the mouse retina. Molecular vision. 2014;20:1422. [PMC free article] [PubMed] [Google Scholar]
- 140.Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, Smith C, Thu CA, Tapia JC, Tasic B, Sanes JR, Maniatis T. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75(3):402–9. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, Yamamoto T, Yamamoto H, Hirabayashi T, Yagi T. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. The European journal of neuroscience. 2008;28(7):1362–76. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 142.Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, Uchiyama Y, Hirabayashi T, Mombaerts P, Yagi T. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Molecular and cellular neurosciences. 2008 doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 143.Hasegawa S, Hirabayashi T, Kondo T, Inoue K, Esumi S, Okayama A, Hamada S, Yagi T. Constitutively expressed Protocadherin-alpha regulates the coalescence and elimination of homotypic olfactory axons through its cytoplasmic region. Frontiers in molecular neuroscience. 2012;5:97. doi: 10.3389/fnmol.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li Y, Xiao H, Chiou TT, Jin H, Bonhomme B, Miralles CP, Pinal N, Ali R, Chen WV, Maniatis T, De Blas AL. Molecular and functional interaction between protocadherin-gammaC5 and GABAA receptors. J Neurosci. 2012;32(34):11780–97. doi: 10.1523/JNEUROSCI.0969-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Meguro R, Hishida R, Tsukano H, Yoshitake K, Imamura R, Tohmi M, Kitsukawa T, Hirabayashi T, Yagi T, Takebayashi H, Shibuki K. Impaired clustered protocadherin-alpha leads to aggregated retinogeniculate terminals and impaired visual acuity in mice. Journal of neurochemistry. 2015;133(1):66–72. doi: 10.1111/jnc.13053. [DOI] [PubMed] [Google Scholar]
- 146.Molumby MJ, Anderson RM, Newbold DJ, Koblesky NK, Garrett AM, Shreiner D, Radley JJ, Weiner JA. α-Protocadherins interact with neuroligin-1 and negatively regulate dendritic spine morphogenesis. Cell Rep. 2017;(18):1–13. doi: 10.1016/j.celrep.2017.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Molumby MJ, Keeler AB, Weiner JA. Homophilic protocadherin cell-cell interactions promote dendrite complexity. Cell reports. 2016;15(5):1037–1050. doi: 10.1016/j.celrep.2016.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Prasad T, Wang X, Gray PA, Weiner JA. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development. 2008;135(24):4153–64. doi: 10.1242/dev.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Prasad T, Weiner JA. Direct and Indirect Regulation of Spinal Cord Ia Afferent Terminal Formation by the gamma-Protocadherins. Frontiers in molecular neuroscience. 2011;4:54. doi: 10.3389/fnmol.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(1):8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen WV, Nwakeze CL, Denny CA, O'Keeffe S, Rieger MA, Mountoufaris G, Kirner A, Dougherty JD, Hen R, Wu Q, Maniatis T. Pcdhalphac2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science. 2017;356(6336):406–411. doi: 10.1126/science.aal3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mountoufaris G, Chen WV, Hirabayashi Y, O'Keeffe S, Chevee M, Nwakeze CL, Polleux F, Maniatis T. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science. 2017;356(6336):411–414. doi: 10.1126/science.aai8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hasegawa S, Kobayashi H, Kumagai M, Nishimaru H, Tarusawa E, Kanda H, Sanbo M, Yoshimura Y, Hirabayashi M, Hirabayashi T, Yagi T. Clustered Protocadherins Are Required for Building Functional Neural Circuits. Frontiers in molecular neuroscience. 2017;10:114. doi: 10.3389/fnmol.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hasegawa S, Kumagai M, Hagihara M, Nishimaru H, Hirano K, Kaneko R, Okayama A, Hirayama T, Sanbo M, Hirabayashi M, Watanabe M, Hirabayashi T, Yagi T. Distinct and Cooperative Functions for the Protocadherin-alpha, -beta and -gamma Clusters in Neuronal Survival and Axon Targeting. Frontiers in molecular neuroscience. 2016;9:155. doi: 10.3389/fnmol.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci. 2008;28(1):100–5. doi: 10.1523/JNEUROSCI.4490-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Leung LC, Harris WA, Holt CE, Piper M. NF-Protocadherin Regulates Retinal Ganglion Cell Axon Behaviour in the Developing Visual System. PLoS One. 2015;10(10):e0141290. doi: 10.1371/journal.pone.0141290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Leung LC, Urbančič V, Baudet ML, Dwivedy A, Bayley TG, Lee AC, Harris WA, Holt CE. Coupling of NF-protocadherin signaling to axon guidance by cue-induced translation. Nat Neurosci. 2013;16(2):166–73. doi: 10.1038/nn.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 2007;10(9):1151–9. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- 159.Williams EO, Sickles HM, Dooley AL, Palumbos S, Bisogni AJ, Lin DM. Delta Protocadherin 10 is Regulated by Activity in the Mouse Main Olfactory System. Front Neural Circuits. 2011;5:9. doi: 10.3389/fncir.2011.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu C, Niu L, Yan Z, Wang C, Liu N, Dai Y, Zhang P, Xu R. Pcdh11x Negatively Regulates Dendritic Branching. Journal of molecular neuroscience : MN. 2015;56(4):822–8. doi: 10.1007/s12031-015-0515-8. [DOI] [PubMed] [Google Scholar]
- 161.Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, Miki N, Hayashi Y, Yoshioka M, Kaneko K, Kato H, Worley PF. Arcadlin Is a Neural Activity-regulated Cadherin Involved in Long Term Potentiation. Journal of Biological Chemistry. 1999;274(27):19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- 162.Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, De Robertis EM, Yamagata K. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56(3):456–71. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151(7):1581–94. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mah KM, Weiner JA. The Cadherin Superfamily. Springer; 2016. Clustered Protocadherins; pp. 195–221. [Google Scholar]
- 165.Hayashi S, Takeichi M. Emerging roles of protocadherins: from self-avoidance to enhancement of motility. J Cell Sci. 2015;128(8):1455–64. doi: 10.1242/jcs.166306. [DOI] [PubMed] [Google Scholar]
- 166.Coughlin G, Kurrasch D. Protocadherins and hypothalamic development: do they play an unappreciated role? Journal of neuroendocrinology. 2015;27(6):544–555. doi: 10.1111/jne.12280. [DOI] [PubMed] [Google Scholar]
- 167.Phillips GR, LaMassa N, Nie YM. Seminars in Cell & Developmental Biology. Elsevier; 2017. Clustered protocadherin trafficking. [DOI] [PubMed] [Google Scholar]
- 168.Suo L, Lu H, Ying G, Capecchi MR, Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. Journal of molecular cell biology. 2012;4(6):362–76. doi: 10.1093/jmcb/mjs034. [DOI] [PubMed] [Google Scholar]
- 169.Hoffman A, Taleski G, Sontag E. The protein serine/threonine phosphatases PP2A, PP1 and calcineurin: A triple threat in the regulation of the neuronal cytoskeleton. Mol Cell Neurosci. 2017 doi: 10.1016/j.mcn.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 170.Yoshida K, Watanabe M, Kato H, Dutta A, Sugano S. BH-protocadherin-c, a member of the cadherin superfamily, interacts with protein phosphatase 1 alpha through its intracellular domain. FEBS Letters. 1999;460(1):93–98. doi: 10.1016/s0014-5793(99)01309-5. [DOI] [PubMed] [Google Scholar]
- 171.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156(1):195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468(7323):533–8. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis. 2013;34(6):1196–207. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beukers W, Hercegovac A, Vermeij M, Kandimalla R, Blok AC, van der Aa MM, Zwarthoff EC, Zuiverloon TC. Hypermethylation of the polycomb group target gene PCDH7 in bladder tumors from patients of all ages. J Urol. 2013;190(1):311–6. doi: 10.1016/j.juro.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 175.Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, Clarke N, Latif F, Maher ER. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30(12):1390–401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- 176.Lin Y-L, Wang Y-L, Ma J-G, Li W-P. Clinical significance of protocadherin 8 (PCDH8) promoter methylation in non-muscle invasive bladder cancer. Journal of Experimental & Clinical Cancer Research. 2014;33(1):68. doi: 10.1186/s13046-014-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, Suh KS, Goy A, Wilson W, Verma A, Braunschweig I, Muthusamy N, Kahl BS, Byrd JC, Wiestner A, Melnick A, Parekh S. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–34. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, Keniry M, Memeo L, Rojtman A, Mansukhani M, Hibshoosh H, Tycko B, Parsons R. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27(34):4657–65. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.de Tayrac M, Etcheverry A, Aubry M, Saikali S, Hamlat A, Quillien V, Le Treut A, Galibert MD, Mosser J. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer. 2009;48(1):55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 180.Zhu P, Lv J, Yang Z, Guo L, Zhang L, Li M, Han W, Chen X, Zhuang H, Lu F. Protocadherin 9 inhibits epithelial-mesenchymal transition and cell migration through activating GSK-3beta in hepatocellular carcinoma. Biochem Biophys Res Commun. 2014;452(3):567–74. doi: 10.1016/j.bbrc.2014.08.101. [DOI] [PubMed] [Google Scholar]
- 181.Miyamoto K, Fukutomi T, Akashi-Tanaka S, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116(3):407–14. doi: 10.1002/ijc.21054. [DOI] [PubMed] [Google Scholar]
- 182.Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25(7):1070–80. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 183.Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med. 2013;13(2):127–34. doi: 10.1007/s10238-012-0182-9. [DOI] [PubMed] [Google Scholar]
- 184.Ying J, Gao Z, Li H, Srivastava G, Murray PG, Goh HK, Lim CY, Wang Y, Marafioti T, Mason DY, Ambinder RF, Chan AT, Tao Q. Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br J Haematol. 2007;136(6):829–32. doi: 10.1111/j.1365-2141.2007.06512.x. [DOI] [PubMed] [Google Scholar]
- 185.Yu B, Yang H, Zhang C, Wu Q, Shao Y, Zhang J, Guan M, Wan J, Zhang W. High-resolution melting analysis of PCDH10 methylation levels in gastric, colorectal and pancreatic cancers. Neoplasma. 2010;57(3):247–252. doi: 10.4149/neo_2010_03_247. [DOI] [PubMed] [Google Scholar]
- 186.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, Jin H, To KF, Chan FK, Sung JJ. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136(2):640–51. e1. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 187.Narayan G, Scotto L, Neelakantan V, Kottoor SH, Wong AH, Loke SL, Mansukhani M, Pothuri B, Wright JD, Kaufmann AM, Schneider A, Arias-Pulido H, Tao Q, Murty VV. Protocadherin PCDH10, involved in tumor progression, is a frequent and early target of promoter hypermethylation in cervical cancer. Genes Chromosomes Cancer. 2009;48(11):983–92. doi: 10.1002/gcc.20703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang KH, Liu HW, Lin SR, Ding DC, Chu TY. Field methylation silencing of the protocadherin 10 gene in cervical carcinogenesis as a potential specific diagnostic test from cervical scrapings. Cancer Sci. 2009;100(11):2175–80. doi: 10.1111/j.1349-7006.2009.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li Z, Li W, Xie J, Wang Y, Tang A, Li X, Ye J, Gui Y, Cai Z. Epigenetic inactivation of PCDH10 in human prostate cancer cell lines. Cell biology international. 2011;35(7):671–676. doi: 10.1042/CBI20100568. [DOI] [PubMed] [Google Scholar]
- 190.Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102(2):419–27. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Giefing M, Zemke N, Brauze D, Kostrzewska-Poczekaj M, Luczak M, Szaumkessel M, Pelinska K, Kiwerska K, Tonnies H, Grenman R, Figlerowicz M, Siebert R, Szyfter K, Jarmuz M. High resolution ArrayCGH and expression profiling identifies PTPRD and PCDH17/PCH68 as tumor suppressor gene candidates in laryngeal squamous cell carcinoma. Genes Chromosomes Cancer. 2011;50(3):154–66. doi: 10.1002/gcc.20840. [DOI] [PubMed] [Google Scholar]
- 192.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T, Inazawa J. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31(6):1027–36. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 193.Costa VL, Henrique R, Danielsen SA, Eknaes M, Patricio P, Morais A, Oliveira J, Lothe RA, Teixeira MR, Lind GE, Jeronimo C. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6(9):1120–30. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 194.Wang XB, Lin YL, Li ZG, Ma JH, Li J, Ma JG. Protocadherin 17 promoter methylation in tumour tissue from patients with bladder transitional cell carcinoma. J Int Med Res. 2014;42(2):292–9. doi: 10.1177/0300060513504364. [DOI] [PubMed] [Google Scholar]
- 195.Hu X, Sui X, Li L, Huang X, Rong R, Su X, Shi Q, Mo L, Shu X, Kuang Y, Tao Q, He C. Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229(1):62–73. doi: 10.1002/path.4093. [DOI] [PubMed] [Google Scholar]
- 196.Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, Ohki M, Hirohashi S, Inazawa J. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66(9):4617–26. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 197.Lv J, Zhu P, Yang Z, Li M, Zhang X, Cheng J, Chen X, Lu F. PCDH20 functions as a tumour-suppressor gene through antagonizing the Wnt/beta-catenin signalling pathway in hepatocellular carcinoma. J Viral Hepat. 2015;22(2):201–11. doi: 10.1111/jvh.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Waha A, Guntner S, Huang TH, Yan PS, Arslan B, Pietsch T, Wiestler OD, Waha A. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7(3):193–9. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hughes LA, Melotte V, De Schrijver J, De Maat M, Smit VT, Bovée JV, French PJ, Van Den Brandt PA, Schouten LJ, De Meyer T. The CpG island methylator phenotype: what's in a name? Cancer research. 2013 doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 200.Banelli B, Brigati C, Di Vinci A, Casciano I, Forlani A, Borzì L, Allemanni G, Romani M. A pyrosequencing assay for the quantitative methylation analysis of the PCDHB gene cluster, the major factor in neuroblastoma methylator phenotype. Laboratory Investigation. 2011;92(3):458–465. doi: 10.1038/labinvest.2011.169. [DOI] [PubMed] [Google Scholar]
- 201.Kobayashi Y, Absher DM, Gulzar ZG, Young SR, McKenney JK, Peehl DM, Brooks JD, Myers RM, Sherlock G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21(7):1017–27. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009;69(12):5251–8. doi: 10.1158/0008-5472.CAN-08-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dallosso AR, Oster B, Greenhough A, Thorsen K, Curry TJ, Owen C, Hancock AL, Szemes M, Paraskeva C, Frank M, Andersen CL, Malik K. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31(40):4409–19. doi: 10.1038/onc.2011.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mah KM, Houston DW, Weiner JA. The gamma-Protocadherin-C3 isoform inhibits canonical Wnt signalling by binding to and stabilizing Axin1 at the membrane. Sci Rep. 2016;6:31665. doi: 10.1038/srep31665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Caronia-Brown G, Yoshida M, Gulden F, Assimacopoulos S, Grove EA. The cortical hem regulates the size and patterning of neocortex. Development. 2014;141(14):2855–65. doi: 10.1242/dev.106914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen Y, Liang Z, Fei E, Chen Y, Zhou X, Fang W, Fu WY, Fu AK, Ip NY. Axin Regulates Dendritic Spine Morphogenesis through Cdc42-Dependent Signaling. PLoS One. 2015;10(7):e0133115. doi: 10.1371/journal.pone.0133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tang X, Yin X, Xiang T, Li H, Li F, Chen L, Ren G. Protocadherin 10 is frequently downregulated by promoter methylation and functions as a tumor suppressor gene in non-small cell lung cancer. Cancer Biomarkers. 2013;12(1):11–19. doi: 10.3233/CBM-2012-00280. [DOI] [PubMed] [Google Scholar]
- 208.Zhao Y, Yang Y, Trovik J, Sun K, Zhou L, Jiang P, Lau TS, Hoivik EA, Salvesen HB, Sun H, Wang H. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014;74(18):5103–17. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 209.Xu Y, Yang Z, Yuan H, Li Z, Li Y, Liu Q, Chen J. PCDH10 inhibits cell proliferation of multiple myeloma via the negative regulation of the Wnt/beta-catenin/BCL-9 signaling pathway. Oncol Rep. 2015;34(2):747–54. doi: 10.3892/or.2015.4056. [DOI] [PubMed] [Google Scholar]
- 210.Kim SH, Yamamoto A, Bouwmeester T, Agius E, Robertis EM. The role of paraxial protocadherin in selective adhesion and cell movements of the mesoderm during Xenopus gastrulation. Development. 1998;125(23):4681–90. doi: 10.1242/dev.125.23.4681. [DOI] [PubMed] [Google Scholar]
- 211.Schambony A, Wedlich D. Wnt-5A/Ror2 regulate expression of XPAPC through an alternative noncanonical signaling pathway. Dev Cell. 2007;12(5):779–92. doi: 10.1016/j.devcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 212.Medina A, Swain RK, Kuerner KM, Steinbeisser H. Xenopus paraxial protocadherin has signaling functions and is involved in tissue separation. The EMBO journal. 2004;23(16):3249–3258. doi: 10.1038/sj.emboj.7600329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Unterseher F, Hefele JA, Giehl K, De Robertis EM, Wedlich D, Schambony A. Paraxial protocadherin coordinates cell polarity during convergent extension via Rho A and JNK. The EMBO journal. 2004;23(16):3259–3269. doi: 10.1038/sj.emboj.7600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Himmelreich N, Kaufmann LT, Steinbeisser H, Korner C, Thiel C. Lack of phosphomannomutase 2 affects Xenopus laevis morphogenesis and the non-canonical Wnt5a/Ror2 signalling. J Inherit Metab Dis. 2015;38(6):1137–46. doi: 10.1007/s10545-015-9874-0. [DOI] [PubMed] [Google Scholar]
- 215.Kraft B, Berger CD, Wallkamm V, Steinbeisser H, Wedlich D. Wnt-11 and Fz7 reduce cell adhesion in convergent extension by sequestration of PAPC and C-cadherin. J Cell Biol. 2012;198(4):695–709. doi: 10.1083/jcb.201110076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang Y, Janicki P, Koster I, Berger CD, Wenzl C, Grosshans J, Steinbeisser H. Xenopus Paraxial Protocadherin regulates morphogenesis by antagonizing Sprouty. Genes Dev. 2008;22(7):878–83. doi: 10.1101/gad.452908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chung HA, Yamamoto TS, Ueno N. ANR5, an FGF target gene product, regulates gastrulation in Xenopus. Curr Biol. 2007;17(11):932–9. doi: 10.1016/j.cub.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 218.Jung B, Köhler A, Schambony A, Wedlich D. PAPC and the Wnt5a/Ror2 pathway control the invagination of the otic placode in Xenopus. BMC developmental biology. 2011;11(1):36. doi: 10.1186/1471-213X-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kai M, Ueno N, Kinoshita N. Phosphorylation-dependent ubiquitination of paraxial protocadherin (PAPC) controls gastrulation cell movements. PLoS One. 2015;10(1):e0115111. doi: 10.1371/journal.pone.0115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kumar R, Ciprianidis A, Theiss S, Steinbeisser H, Kaufmann LT. Nemo-like kinase 1 (Nlk1) and paraxial protocadherin (PAPC) cooperatively control Xenopus gastrulation through regulation of Wnt/planar cell polarity (PCP) signaling. Differentiation. 2017;93:27–38. doi: 10.1016/j.diff.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 221.Kietzmann A, Wang Y, Weber D, Steinbeisser H. Xenopus paraxial protocadherin inhibits Wnt/beta-catenin signalling via casein kinase 2beta. EMBO Rep. 2012;13(2):129–34. doi: 10.1038/embor.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Song DH, Dominguez I, Mizuno J, Kaut M, Mohr SC, Seldin DC. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J Biol Chem. 2003;278(26):24018–25. doi: 10.1074/jbc.M212260200. [DOI] [PubMed] [Google Scholar]
- 223.Yin X, Xiang T, Mu J, Mao H, Li L, Huang X, Li C, Feng Y, Luo X, Wei Y. Protocadherin 17 functions as a tumor suppressor suppressing Wnt/β-catenin signaling and cell metastasis and is frequently methylated in breast cancer. Oncotarget. 2016;7(32):51720. doi: 10.18632/oncotarget.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chen T, Long B, Ren G, Xiang T, Li L, Wang Z, He Y, Zeng Q, Hong S, Hu G. Protocadherin20 Acts as a Tumor Suppressor Gene: Epigenetic Inactivation in Nasopharyngeal Carcinoma. J Cell Biochem. 2015;116(8):1766–75. doi: 10.1002/jcb.25135. [DOI] [PubMed] [Google Scholar]
- 225.Chen MW, Vacherot F, De La Taille A, Gil-Diez-De-Medina S, Shen R, Friedman RA, Burchardt M, Chopin DK, Buttyan R. The emergence of protocadherin-PC expression during the acquisition of apoptosis-resistance by prostate cancer cells. Oncogene. 2002;21(51):7861–71. doi: 10.1038/sj.onc.1205991. [DOI] [PubMed] [Google Scholar]
- 226.Yang X, Chen M-W, Terry S, Vacherot F, Chopin DK, Bemis DL, Kitajewski J, Benson MC, Guo Y, Buttyan R. A human-and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Research. 2005;65(12):5263–5271. doi: 10.1158/0008-5472.CAN-05-0162. [DOI] [PubMed] [Google Scholar]
- 227.Inestrosa NC, Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015;359(1):215–23. doi: 10.1007/s00441-014-1996-4. [DOI] [PubMed] [Google Scholar]