Abstract
Ligation-Mediated Polymerase Chain Reaction (LMPCR) is the most sensitive sequencing technique available to map single-stranded DNA breaks at the nucleotide level of resolution using genomic DNA. LMPCR has been adapted to map DNA damage and reveal DNA–protein interactions inside living cells. However, the sequence context (GC content), the global break frequency and the current combination of DNA polymerases used in LMPCR affect the quality of the results. In this study, we developed and optimized an LMPCR protocol adapted for Pyrococcus furiosus exo– DNA polymerase (Pfu exo–). The relative efficiency of Pfu exo– was compared to T7-modified DNA polymerase (Sequenase 2.0) at the primer extension step and to Thermus aquaticus DNA polymerase (Taq) at the PCR amplification step of LMPCR. At all break frequencies tested, Pfu exo– proved to be more efficient than Sequenase 2.0. During both primer extension and PCR amplification steps, the ratio of DNA molecules per unit of DNA polymerase was the main determinant of the efficiency of Pfu exo–, while the efficiency of Taq was less affected by this ratio. Substitution of NaCl for KCl in the PCR reaction buffer of Taq strikingly improved the efficiency of the DNA polymerase. Pfu exo– was clearly more efficient than Taq to specifically amplify extremely GC-rich genomic DNA sequences. Our results show that a combination of Pfu exo– at the primer extension step and Taq at the PCR amplification step is ideal for in vivo DNA analysis and DNA damage mapping using LMPCR.
INTRODUCTION
Unlike purified or cloned DNA, DNA inside living cells exists in a very dynamic environment where it interacts with many proteins, adopting particular structures, and forming more or less condensed chromatin. The principle of in vivo DNA analysis is to assess the local reactivity of DNA towards modifying agents, e.g., dimethylsulfate (DMS), ultraviolet light (UV) and DNase I, inside living cells, compared to that of purified DNA. In vivo DNA analysis is possible because the modifying agents produce different DNA damage distributions, depending upon whether they are applied to purified DNA (in vitro) or to living cells (in vivo) (1,2). Ligation-Mediated Polymerase Chain Reaction (LMPCR) is an extremely sensitive and specific genomic sequencing technique which has been successfully applied for over a decade by many groups to in vivo DNA–protein interaction analysis, DNA damage mapping, methylation analysis and nucleosome positioning (3–10). This frequent utilization is attributable to the fact that LMPCR is orders of magnitude more sensitive in mapping DNA single-strand breaks (SSBs) than original genomic sequencing methods. However, LMPCR is a complex technique to employ and, depending on the sequence context, reproducibility and consistency between experiments may not be achievable. In order to obtain more consistent and reliable results, facilitating interpretation, it is critical to develop improved LMPCR protocols that are widely applicable.
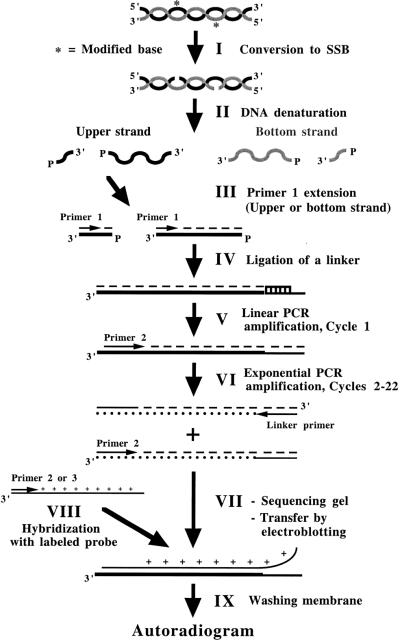
DNA polymerases are required at two steps in LMPCR procedure. The complete LMPCR procedure can be divided into nine steps (Fig. 1): (I) conversion of modified bases to SSBs; (II) heat denaturation of genomic DNA; (III) hybridization and extension of a gene-specific oligonucleotide (primer 1) for the bottom or upper DNA strand to produce DNA molecules with an unknown double-stranded blunt 3′-end; (IV) ligation of an asymmetrical double-stranded DNA linker to provide a common known sequence; (V–VI) linear and exponential PCR amplifications using a gene-specific nested oligonucleotide (primer 2) and the linker-specific oligonucleotide (linker primer); (VII) size-fractionation of the PCR products on a sequencing polyacrylamide gel and transfer of the DNA to a nylon membrane by electroblotting; (VIII) hybridization with a gene-specific labeled probe generated using primer 2 or a nested oligonucleotide (primer 3) with a PCR product corresponding to the sequence to be analyzed; and (IX) washing of the membrane and revealing of the sequence ladder by autoradiography.
Figure 1.
Overview of the different steps in the LMPCR protocol.
Primer extension and PCR amplification steps are particularly critical because both are key steps for the accurate autoradiographic representation of the initial frequency distribution of DNA SSBs along the DNA sequence to be analyzed. For example, if all the single-stranded DNA molecules having an SSB at the same nucleotide position are not fully elongated to form the blunt extremity at the primer extension step, the corresponding band will appear faint or missing on the autoradiogram. For an optimal outcome, the DNA polymerase used at the primer extension step should be: (i) thermostable; (ii) free of any terminal transferase activity; (iii) processive even on very GC-rich DNA template; and (iv) able to resolve particular secondary structures of the DNA. During the PCR amplification step, the initial relative representation/frequency distribution of ligated DNA molecules must be maintained through 20–22 cycles, regardless of the sequence context. During the PCR amplification step, the ideal polymerase should: (i) possess a high thermostability; (ii) quantitatively amplify a mixture of DNA fragments of different sizes regardless of their GC composition; and (iii) resolve particular secondary structures of the DNA. Sequenase 2.0 is thermolabile and possesses a terminal transferase activity. The low temperature permissiveness of Sequenase 2.0 might reduce its polymerization efficiency for sequences requiring high polymerization temperatures like GC-rich sequences. Despite its thermostability, poor amplification of very GC-rich sequences with Taq is relatively common. Thus, when using Sequenase 2.0 and Taq, unwanted gaps in the DNA sequence ladder are often observed on the autoradiogram leading to inconclusive results (11). In addition, this polymerase combination poorly amplifies very GC-rich DNA templates. Because of these deficiencies, DNA–protein interactions occurring in living cells might be missed. Since the first two articles describing LMPCR (3,4), other investigators have tried to improve the original protocol (12,13). However, the initial DNA polymerase combination using Sequenase 2.0 for the primer extension step and Taq for the PCR amplification step remained the most efficient combination (14,15).
Pfu exo– has successfully amplified DNA fragments that other DNA polymerases failed to amplify (16). Pfu exo– is highly thermostable, free of any terminal transferase activity and can amplify very GC-rich DNA templates of various lengths (16–18). Our first objective was to develop and optimize the conditions for using Pfu exo– during primer extension and PCR amplification steps in LMPCR. Because KCl stabilizes complex secondary DNA structures in vitro more efficiently than NaCl (19), our second objective was to evaluate whether or not the substitution of NaCl for KCl in the DNA polymerase buffers improved LMPCR amplification efficiency. Indeed, these complex DNA structures can induce premature polymerization arrest that substantially affects the yield of PCR products (20,21).
MATERIALS AND METHODS
Chemicals, enzymes and equipment
Taq DNA polymerase and T4 DNA ligase were purchased from Roche Molecular Biochemicals (Laval, Canada). Sequenase 2.0 and Pfu exo– DNA polymerases were respectively ordered from Amersham Pharmacia Biotech (Baie d’Urfé, Canada) and Stratagene (La Jolla, CA). T4 endonuclease V and photolyase were kindly provided by R. Stephen Lloyd (University of Texas Medical Branch, Galveston, TX) and Aziz Sancar (University of North Carolina, Chapel Hill, NC), respectively. [∝-32P]dCTP was supplied by NEN (Boston, MA). DMS, piperidine, K2PdCl4 and hydrazine were purchased from Sigma-Aldrich Canada (Guelph, Canada). The desalted oligonucleotide primers were synthesized by the Analysis and Synthesis Nucleic Acids Service of Laval University. All primer extensions and PCR amplifications were carried out on a Thermocycler PTC-100 or PTC-200 from MJ Research (Waltham, MA). Band intensities on autoradiograms were quantified using Fuji BAS 1000 phosphorimager from Fuji Medical Systems (Stamford, CT) and analyzed using software Image Gauge v3.0.
Oligonucleotides
The lower strand primers MH1 (5′-CTTTGCTGTCTGAGGGCG-3′) and MH2 (5′-GCGTCTGGCTGTGGAGCTGAAGGAGGCG-3′) with Tm values of 59.5 and 72.2°C, respectively, were selected in the promoter region of the autosomal human COX2 (cyclooxygenase 2) gene. Primers A1 and A2 were selected in the human PGK1 (phosphoglycerate kinase 1) gene promoter (6), and X1 and X2 were selected in the human FMR1 (fragile X mental retardation 1) gene promoter (8). Primers U1 (5′-AAGTACCTTGTAGAAAGCGC-3′), U2 (5′-CACTTCCACCACCAGCTCCTCCAT-3′) and U3 (5′-CCTCCATCTTCTCTTCAGCCCTGC-3′) hybridize to the upper strand on the 3′ side of the CGG repeats in the exon 1 of the FMR1 gene. Primers U1, U2 and U3 have Tm values of 55.7, 66.9 and 65.3°C, respectively. All Tm values were determined using Gene Jockey software.
Cell culture and DNA preparation
DNA was purified from peripheral blood lymphocytes of normal human males as described previously (8). DNA was exposed to 3 Jm–2s–1 for 10 s under a 254-nm UV germicidal lamp (9) or treated according to Maxam–Gilbert cleavage reactions (14) or with K2PdCl4 (22). Following UV exposure, cyclobutane pyrimidine dimers (CPD) were converted into ligatable SSBs using T4 endonuclase V and photolyase (23). The DNA concentration was measured using a spectrophotometer at 260 nm. The SSB frequency in the total genomic DNA was determined by alkaline agarose gel electrophoresis (24).
Ligation-Mediated Polymerase Chain Reaction
Primer extension and ligation steps. Purified genomic DNA and 1.25 pmol of gene-specific primer 1 in 15 µl of Sequenase 2.0 buffer (40 mM Tris–HCl pH 7.7, 50 mM KCl or 50 mM NaCl) were denatured using a thermocycler at 98°C for 3 min then cooled on ice. Samples were subsequently annealed at 48°C for 20 min then cooled on ice. Nine microliters of a freshly prepared primer extension mix [16.7 mM MgCl2, 16.7 mM dithiothreitol (DTT), 312.5 µM of each dNTP (dNTP, Na-salt, PCR Grade; Roche Molecular Biochemicals), 5.2 U Sequenase 2.0] were added to the samples, which were then incubated at: (i) 48°C for 5 min; (ii) 50°C for 1 min; (iii) 51°C for 1 min; (iv) 52°C for 1 min; (v) 54°C for 1 min; (vi) 56°C for 1 min; (vii) 58°C for 1 min; and (viii) 60°C for 1 min. The samples were cooled to 4°C, 6 µl of 310 mM Tris–HCl pH 7.7 was added and incubated at 67°C for 15 min, then cooled on ice. Forty-five microliters of a freshly prepared ligation mix [30 mM DTT, 1.1 mM ATP, 83.3 µg/ml bovine serum albumin (BSA), 100 pmol linker (100 pmol L11 5′-GAATTCAGATC-3′ and 100 pmol L25 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ in 250 mM Tris–HCl pH 7.7 containing 120 mM MgCl2), 3.25 U T4 DNA ligase] were added to the samples, which were incubated overnight at 18°C.
Purified genomic DNA and 1.25 pmol of gene-specific primer 1 in 30 µl of freshly prepared primer extension mix [1× cloned buffer (20 mM Tris–HCl pH 8.8, 2 mM MgSO4, 10 mM KCl or NaCl, 10 mM (NH4)2SO4, 0.1% (v/v) Triton X-100, 100 µg/ml BSA), 250 µM of each dNTP, 1.5 U Pfu exo–] were denatured on a thermocycler and under a hot bonnet at 98°C for 5 min and annealed for 2 min at the melting temperature of the primer minus 4°C (Tm – 4°C). Still under the hot bonnet, the temperature was gradually increased (1°C per 3 s) until it reached 75°C. Primer elongation was then carried out for 15 min at 75°C. The samples were cooled to 4°C, and 45 µl of a freshly prepared ligation mix (30 mM DTT, 1.1 mM ATP, 16.7 µg/ml BSA, 100 pmol linker, 50 mM Tris–HCl pH 7.4, 3.25 U T4 DNA ligase) were added to the samples and incubated overnight at 18°C.
PCR amplification step. Following ligation, 30 µl of a freshly prepared stop mix (7 M NH4Ac, 5 mM EDTA pH 8, 667 ng/µl glycogen) were added to the ligated DNA and precipitated with 2.5 vol of cold absolute ethanol, washed once with 80% ethanol. The DNA was dissolved in 50 µl H2O.
The dissolved ligated DNA was mixed with 50 µl of Taq amplification mix {2× Taq buffer [20 mM Tris–HCl pH 8.9, 80 mM KCl or NaCl, 0.02% (w/v) gelatin], 4 mM MgCl2, 500 µM of each dNTP, 10 pmol of linker primer (5′-GCGGTGACCCGGGAGATCTGAATTC-3′), 10 pmol of gene-specific primer 2, 3 U Taq}. The samples were processed as described in Table 1 (standard program).
Table 1. The standard and GC-rich PCR amplification programs.
| Steps | Denaturationa | Annealingb | Polymerizationc | |||||||
| GC-rich | Standard | GC-rich | Standard | GC-rich | Standard | |||||
| |
Temperature (°C) |
Duration (s) |
Temperature (°C) |
Duration (s) |
Tm (°C) |
Duration (s) |
Tm (°C) |
Duration (s) |
|
|
| 0 | – | – | 93 | 120 | – | – | – | – | – | – |
| 1 | 98 | 300 | 98 | 150 | –3°C | 60 | 180 | 120 | 180 | |
| 2 | 98 | 120 | 95 | 60 | –4°C | 60 | –1°C | 150 | 120 | 180 |
| 3 | 98 | 60 | 95 | 60 | –5°C | 60 | –2°C | 120 | 120 | 180 |
| 4 | 98 | 30 | 95 | 60 | –6°C | 60 | –3°C | 120 | 120 | 180 |
| 5d | 98 | 20 | 95 | 60 | –7°C | 60 | –4°C | 90 | 120 | 150 |
| 6 | 98 | 20 | 95 | 60 | –6°C | 60 | –3°C | 240 | 120 | 240 |
| 7 | 98 | 20 | 95 | 60 | –5°C | 60 | –2°C | 240 | 120 | 240 |
| 8 | 98 | 20 | 95 | 60 | –4°C | 60 | –1°C | 240 | 120 | 240 |
| 9 | 98 | 20 | 95 | 60 | –3°C | 60 | 240 | 600 | 600 |
aTemperature and duration of the step.
bAnnealing temperature of primer and duration of the step.
cDuration of the step. Temperatures were 74°C for Taq and 75°C for Pfu exo–.
dRepeat step 5, 13 more times (5 s per cycle are added in both annealing and polymerization steps of the PCR program standard).
The ligated DNA suspension was mixed with 50 µl of Pfu exo– amplification mix {2× cloned buffer [40 mM Tris–HCl pH 8.8, 4 mM MgSO4, 20 mM KCl or NaCl, 20 mM (NH4)2SO4, 0.2% (v/v) Triton X-100, 200 µg/ml BSA], 500 µM of each dNTP, 10 pmol of linker primer, 10 pmol of gene-specific primer 2, 3.5 U Pfu exo–}. When the GC content was <70% of the sequence being analyzed (primers for COX2 and PGK1), the samples were processed according to the standard program (Table 1). When this content was >70% of the sequence (primers for FMR1), the samples were cycled according to the GC-rich program (Table 1). In the region covered by the primer set U, modifications were made to the Pfu exo– amplification mix: 20 mM NaCl, 20% (v/v) dimethyl sulfoxide (DMSO), 125 µM dGTP, 375 µM 7-deaza-dGTP. Step 5 was repeated 21 times instead of 13 (Table 1).
Sequencing gel, electroblotting and hybridization. Following the PCR amplification step, DNA was ethanol precipitated, run on a sequencing gel, transferred to a nylon membrane and hybridized with a radio-labeled gene-specific probe as described previously (25).
Phosphorimager data treatment
Phosphorimager analyses were performed to measure precisely the intensity of individual bands as described previously (26). LMPCR protocols were repeated twice. For each protocol, a total of eight sets of bands from both autoradiograms were selected for each experimental condition and the intensity of each band was measured: (i) the area covered by each band was delimited and the counts per minute (c.p.m.) were quantified in this area and (ii) the measured value, representing density, was obtained by dividing the c.p.m. by the area covered by the band. Corresponding bands (or areas) for each experimental condition were measured likewise and were altogether called a set. In each set, an area corresponding to the experimental condition that could not generate a band (either experimental condition without DNA polymerase or without DNA) was delimited and defined as ‘background’. This background was subtracted from all the measured bands of a set. The band intensity value (minus the background) was then divided by the total value of band intensities for one set to obtain the relative intensity value of each band within a set. A total of four sets of bands were selected from a duplicate of LMPCR autoradiograms. A mean of the relative intensities was also calculated for all four bands measured for each experimental condition. Mean relative intensity values for each experimental condition were plotted against amounts of DNA or DNA polymerase.
RESULTS
Primer extension step
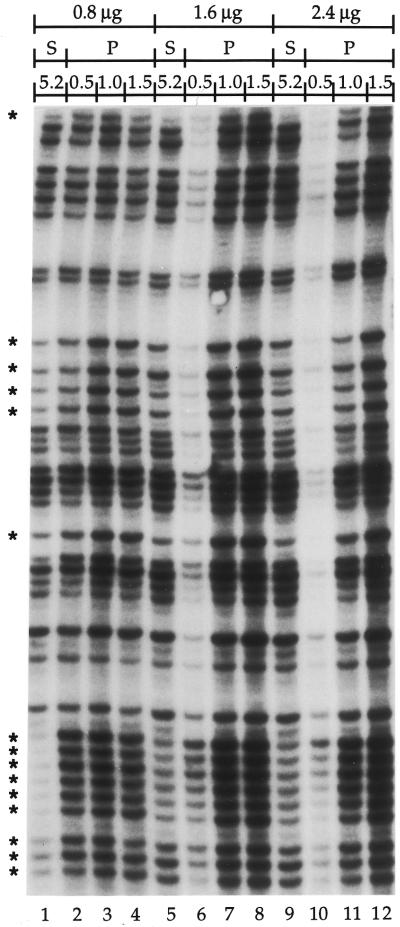
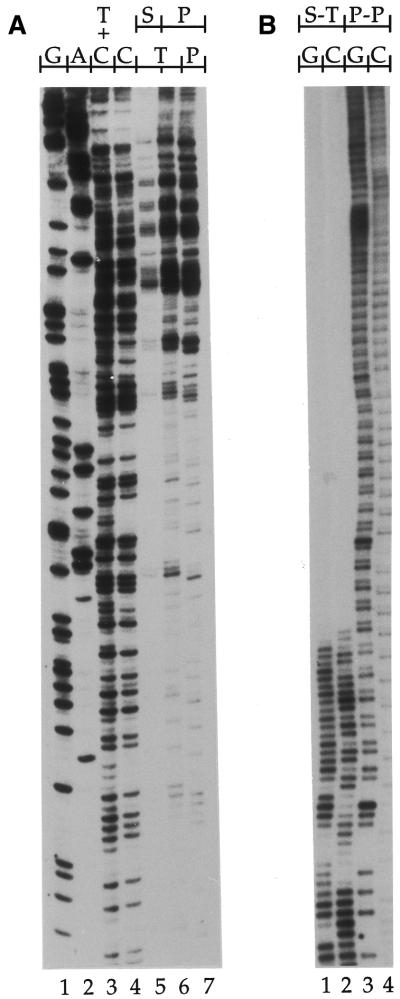
The efficiency of Pfu exo– for the primer extension step was compared with the efficiency of Sequenase 2.0 using Taq at the PCR amplification step. Three different amounts of purified genomic DNA treated with DMS (global SSB frequency: 1 break/400 bases, see Materials and Methods) were processed using Sequenase 2.0 or Pfu exo– at the primer extension step. A small promoter region in each of the FMR1, COX2 and PGK1 genes was mapped using LMPCR. Both Sequenase 2.0 and Pfu exo– extended primers to a minimum of 200 bp even along highly GC-rich DNA sequences like the FMR1 gene. The results shown in Figure 2 are from a region of the FMR1 promoter that has a GC content up to 75%. However, the portion of the 200 bp region shown reveals that the intensities of the bands observed with Sequenase 2.0 were far less homogeneous than with Pfu exo–, especially within guanine runs (Fig. 2). This lack of homogeneity in the band intensities was also observed in the other genes tested (data not shown). These results suggest that, in contrast to Sequenase 2.0, Pfu exo– efficiently produces ligatable ends with all DNA molecules independently of the sequence context; specific bands were faint, if not completely missing, in corresponding lanes using Sequenase 2.0. (Fig. 2, compare lanes 1 and 4).
Figure 2.
Comparison of the efficiency of Sequenase 2.0 and Pfu exo– with different amounts of DNA at the primer extension step of LMPCR. This autoradiogram shows a representative sequence that was produced using primer set X (primers X1, X2 and X3) from the FMR1 gene promoter. Every PCR amplification step was done using 3 U of Taq. LMPCR was performed on increasing quantities of purified genomic DNA treated with standard Maxam–Gilbert guanine cleavage reaction (global SSB frequency: 1 break/400 bases); 0.8, 1.6 and 2.4 µg of DNA was used (lanes 1–4, 5–8 and 9–12, respectively). Lanes 1, 5 and 9 show LMPCR protocols done using 5.2 U of Sequenase 2.0 (S) at the primer extension step; lanes 2–4, 6–8 and 10–12 show LMPCR protocols done using 0.5 (lanes 2, 6 and 10), 1.0 (lanes 3, 7 and 11) or 1.5 U (lanes 4, 8 and 12) of Pfu exo– (P) at the primer extension step. An asterisk indicates a band in the Sequenase 2.0 track that shows an intensity markedly different compared to the rest of the bands in the track.
In the original LMPCR protocol, 5.2 U of Sequenase 2.0 were optimally required for the primer extension step (3,4). With Pfu exo–, as little as 1 U was able to produce a sufficient number of ligatable DNA molecules from 0.8–2.4 µg of DMS-treated genomic DNA (Fig. 2, lanes 3, 7 and 11). When using 0.5 U of Pfu exo–, the maximum band intensity was obtained with 0.8 µg of DNA. Optimal band intensity using 1 U of Pfu exo– was obtained in combination with 0.8 µg of DNA, whereas optimal band intensity using 1.5 U of Pfu exo– was achieved when combined with both 1.6 and 2.4 µg of DNA (Fig. 2, lanes 3, 8 and 12). More Pfu exo– was necessary to produce similar band intensities starting from equal amounts of hydrazine treated genomic DNA (data not shown). Consequently, we set 1.5 U as the amount of Pfu exo– required when using <3 µg of treated genomic DNA to ensure optimal primer extension in any LMPCR protocols. These results show the importance of the ratio of DNA molecules per unit of Pfu exo– at primer extension.
PCR amplification step
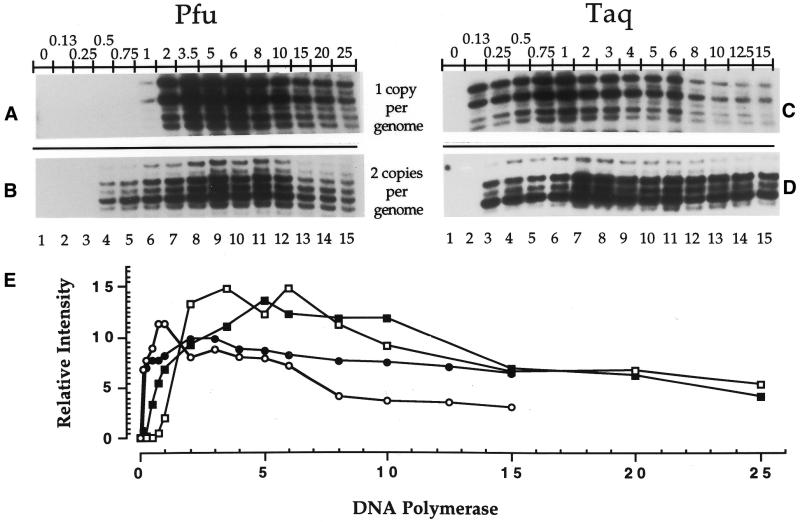
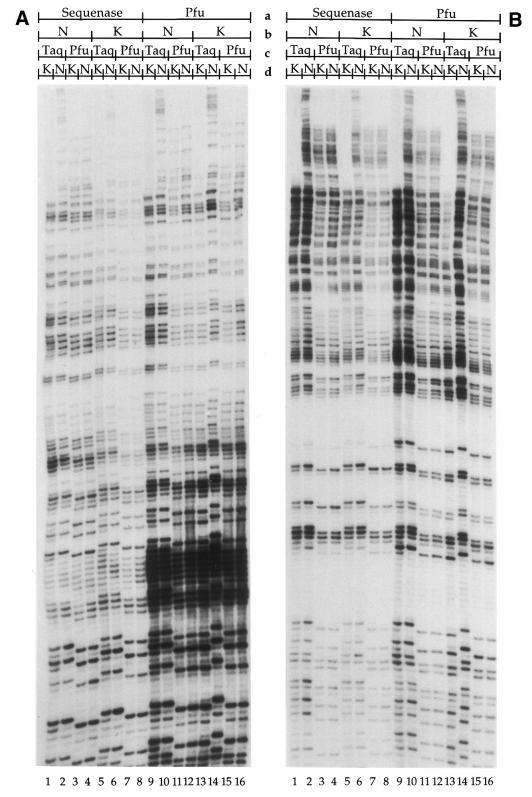
Using Sequenase 2.0 at the primer extension step, the efficiency of both Pfu exo– and Taq at the PCR amplification step was studied as a function of two parameters, i.e., varying (i) the number of units of DNA polymerase (Fig. 3) or (ii) the amount of DNA (Fig. 4). For each of the parameters studied, two DNA polymerases (Pfu exo– and Taq) and two gene promoters (COX2 and PGK1) were compared, totaling four protocols. Each of the four protocols was repeated twice using DNA with a global SSB frequency of 1 break/400 bases. Thus, eight autoradiograms were obtained. Eight sets of bands (one band for each of the 15 experimental conditions) were selected from each duplicated protocol and quantified using the phosphorimager. One representative strip of the 15 experimental conditions from each LMPCR protocol is shown (Fig. 3A–D) and a graph was produced from the analysis of these eight bands per experimental condition (Fig. 3E). We observed that 0.5 U of Pfu exo– and 0.13 U of Taq, for a one-copy gene per genome (PGK1), and 1 U of Pfu exo– and 0.13 U of Taq, for a two-copy gene per genome (COX2), were repeatedly the lowest amounts of DNA polymerase capable of producing bands on the autoradiogram (Fig. 3). We used both COX2, an autosomal gene, and PGK1, an X-linked gene, to study the effect of the number of DNA molecules on the efficiency of the DNA polymerases. Indeed, for a given amount of genomic DNA with identical break frequency, the number of DNA molecules corresponding to the COX2 gene will be two times higher than the number of DNA molecules corresponding to the PGK1 gene. Overall, band intensity patterns of both PGK1 and COX2 were similar and maximal amplification efficiency was reached using 5 U of Pfu exo– (Fig. 3A and B, lane 9, and E) and ∼2 U of Taq (Fig. 3C and D, lane 7, and E). More units of both DNA polymerases caused a decrease in band intensity. Unexpectedly, when using high amounts of Taq, the bands from PGK1 were weaker than those from COX2 (Fig. 3C, D and E). We have no explanation for this latter result.
Figure 3.
Comparison of PCR amplification efficiency using different amounts of either Taq or Pfu exo– at the PCR amplification step. Sequenase 2.0 was used for each primer extension step and 1 µg of purified genomic DNA treated with standard Maxam–Gilbert guanine cleavage reaction (global SSB frequency: 1 break/400 bases) was used. Each of the four protocols was repeated twice. All the stripes shown are representative samples from the analyzed autoradiograms. (A) The short representative sequence shown was analyzed using primer set A (primers A1, A2 and A3) selected from the PGK1 gene promoter (one copy per genome). For (A) and (B), the amount of Pfu exo– varied from 0–25 U (lanes 1–15). (B) The short representative sequence shown was analyzed using primer set MH (primers MH1 and MH2) selected from the COX2 gene promoter (two copies per genome). (C) The short representative sequence shown was analyzed using primer set A (primers A1, A2 and A3) selected from the PGK1 gene promoter. For (C) and (D), the amount of Taq varied from 0–15 U (lanes 1–15). (D) The short representative sequence shown was analyzed using primer set MH (primers MH1 and MH2) selected from the COX2 gene promoter. (E) Graph showing the effects of different amounts of Pfu exo– and Taq from (A) (open square), (B) (filled square), (C) (open circle) and (D) (filled circle) on genomic DNA. The calculated value at each point represents the relative intensity of the corresponding band against the total intensity value obtained by the addition of the intensity from every band of a lane from an autoradiogram.
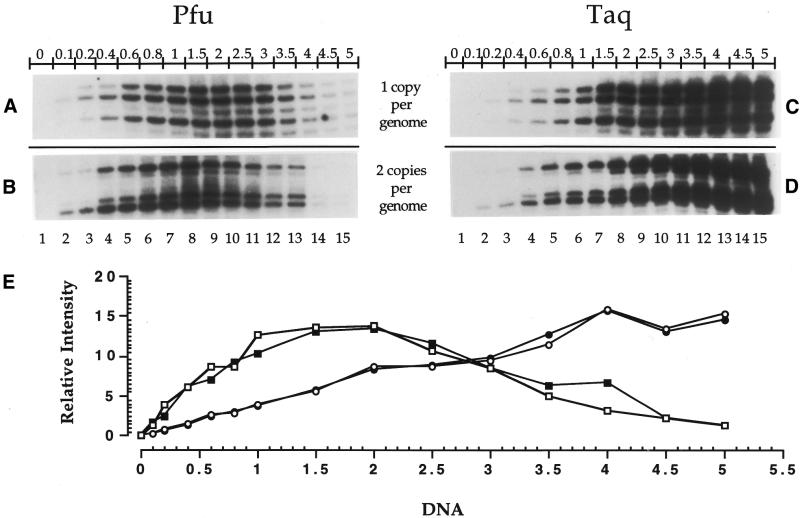
Figure 4.
Comparison of PCR amplification efficiency using different amounts of purified genomic DNA. Sequenase 2.0 was used at every primer extension step and purified genomic DNA treated with standard Maxam–Gilbert guanine cleavage reaction (global SSB frequency: 1 break/400 bases). Each of the four protocols was repeated twice. All the stripes shown are representative samples from the analyzed autoradiograms. The initial amount of DNA varied from 0–5 µg (lanes 1–15). (A) The short representative sequence shown was analyzed using primer set A (primers A1, A2 and A3) selected from the PGK1 gene promoter (one copy per genome). In (A) and (B), 3.5 U of Pfu exo– were used at the PCR amplification step. (B) The short representative sequence shown was analyzed using primer set MH (primers MH1 and MH2) selected from the COX2 gene promoter (two copies per genome). (C) The short representative sequence shown was analyzed using primer set A (primers A1, A2 and A3) selected from the PGK1 gene promoter. In (C) and (D), 3 U of Taq were used at the PCR amplification step. (D) The short representative sequence shown was analyzed using primer set MH (primers MH1 and MH2) selected from the COX2 gene promoter. (E) Graph showing the effects of different amounts of genomic DNA on the polymerization efficiency of Pfu exo– and Taq from (A) (open square), (B) (filled square), (C) (open circle) and (D) (filled circle). The calculated value at each point represents the relative intensity of the corresponding band against the total intensity value obtained by the addition of the intensity from every band of a lane from an autoradiogram.
Similarly, when various starting amounts of DNA were used, mixed with either 3.5 U of Pfu exo– or 3 U of Taq at the PCR amplification step, band intensity increased proportionally to reach a maximum at 2 and 5 µg of DNA using Pfu exo– and Taq, respectively (Fig. 4). We noticed that when starting with >2 µg of DNA, the band intensity decreased gradually using Pfu exo– (Fig. 4A, B and E). No decrease in band intensity was observed using Taq (Fig. 4C, D and E).
Stabilization of unusual secondary DNA structures by salts
In vitro, KCl stabilizes complex secondary DNA structures more efficiently than NaCl (19–21). This stabilization would negatively affect the yield of PCR products. In vitro DMS- and hydrazine-treated human genomic DNA (global SSB frequency: 1 break/400 bases) were processed by LMPCR with four combinations of DNA polymerases (Sequenase 2.0/Taq, Sequenase 2.0/Pfu exo–, Pfu exo–/Taq and Pfu exo–/Pfu exo–) using either NaCl- or KCl-based buffers on the FMR1 gene promoter (Fig. 5). Figure 5 shows a representative autoradiogram selected from repeated experiments. We observed that bands were always slightly more intense using NaCl-based buffers, independently of the DNA polymerase used in both primer extension and PCR amplification steps. NaCl markedly improved the quality of the results using Taq as shown with bands of higher molecular weight being observed in the uppermost portion of the autoradiograms (Fig. 5A and B, lanes 2, 6, 10 and 14). Interestingly, the addition of a non-templated extra nucleotide found in PCR products amplified with Taq did not occur using Pfu exo– and KCl at the primer extension step, in combination with Taq and KCl at the PCR amplification step (Fig. 5A and B, lanes 13). This was not observed using either Sequenase 2.0 or NaCl-based buffers. Moreover, the intensity of shadow bands (artifacts present beneath expected bands) was much lower using Taq and a NaCl-based buffer instead of a KCl one (Fig. 5A and B, lanes 1 and 2, 5 and 6, and 9 and 10). The combination of Pfu exo– at the primer extension step and Taq at the PCR amplification step, using a NaCl-based buffer, generated the best results, especially with DMS treated DNA (Fig. 5, lanes 10 and 14).
Figure 5.
Comparison of the effects of KCl and NaCl on band intensities at primer extension and PCR amplification steps of LMPCR. The region shown was analyzed using primer set X (primers X1, X2 and X3) selected from the FMR1 gene promoter. The starting amount of DNA was 1 µg with a SSB frequency of 1 break/400 bases. (A) Purified genomic DNA was treated with standard Maxam–Gilbert guanine cleavage reaction and processed by LMPCR. In lanes 1–4, Sequenase 2.0 was used with NaCl in the DNA polymerase buffer (Sequenase 2.0/NaCl) at the primer extension step with Taq/KCl (lane 1), Taq/NaCl (lane 2), Pfu exo–/KCl (lane 3) and Pfu exo–/NaCl (lane 4) at the PCR amplification step. In lanes 5–8, Sequenase 2.0/KCl was used at the primer extension step with Taq/KCl (lane 5), Taq/NaCl (lane 6), Pfu exo–/KCl (lane 7) and Pfu exo–/NaCl (lane 8) at the PCR amplification step. In lanes 9–12, Pfu exo–/NaCl was used at the primer extension step with Taq/KCl (lane 9), Taq/NaCl (lane 10), Pfu exo–/KCl (lane 11) and Pfu exo–/NaCl (lane 12) at the PCR amplification step. In lanes 13–16, Pfu exo–/KCl was used at the primer extension step with Taq/KCl (lane 13), Taq/NaCl (lane 14), Pfu exo–/KCl (lane 15) and Pfu exo–/NaCl (lane 16) at the PCR amplification step. (B) Purified genomic DNA was treated with standard Maxam–Gilbert pyrimidine (T+C) cleavage reaction. Lanes 1–16 are as described in (A). a, DNA polymerase at primer extension step; b, cation at primer extension step; c, DNA polymerase at PCR amplification step; d, cation at PCR amplification step.
Low break frequency without piperidine treatment
The LMPCR amplification procedure is often less efficient with low break frequency genomic DNA. To test the efficiency of Pfu exo– in a context of low break frequency, purified DNA was 254-nm UV-irradiated at a low dose to provide an SSB frequency of 1 break/5 kb following conversion of CPDs. The DNA was processed by LMPCR using either Sequenase 2.0 or Pfu exo– at the primer extension step and either Taq or Pfu exo– at the PCR amplification step on the PGK1 gene promoter (Fig. 6A). Figure 6A shows a representative autoradiogram selected from repeated experiments. Bands were more intense using Pfu exo– at the primer extension step than Sequenase 2.0. We observed that Pfu exo– produced a band pattern similar to that of Taq with an identical overall intensity. Furthermore, we observed that the band pattern using Taq shifted one base up from the band pattern of Pfu exo–, clearly confirming that Taq possesses a terminal transferase activity (Fig. 6A, lanes 6 and 7). The efficiencies of the DNA polymerases used in this study employing DNA with different break frequencies is summarized in Table 2.
Figure 6.
Efficiency of Pfu exo– on low break frequency DNA and extremely GC-rich DNA in LMPCR. (A) LMPCR of purified genomic DNA with a low global SSB frequency (1 break/5 kb) produced by irradiation with 30 Jm–2 UVC (254 nm). The sequence shown was analyzed using primer set A (primers A1, A2 and A3) selected from the PGK1 gene promoter. The starting amount of DNA was 1 µg. Lanes 1–4, LMPCR of purified genomic DNA treated with standard Maxam–Gilbert cleavage reactions with Pfu exo– used for both primer extension and PCR amplification steps; lane 5, LMPCR of purified DNA treated with 254-nm UV using Sequenase 2.0 at the primer extension step and Taq at the PCR amplification step; lane 6, LMPCR of purified DNA treated with UVC using Sequenase 2.0 at the primer extension step and Pfu exo– at the PCR amplification step; lane 7, LMPCR of purified DNA treated with UVC using Pfu exo– at both primer extension and PCR amplification steps. (B) LMPCR of purified genomic DNA with a high global SSB frequency (1 break/400 bp) on the CGG triplet repeat of the FMR1 gene. The sequence shown was analyzed using primer set U (primers U1, U2 and U3) selected from the FMR1 gene promoter. The starting amount of DNA was 1 µg. Purified genomic DNA from a normal male who has a FMR1 gene with a haplotype of 33 CGG triplet repeats (∼100% GC-rich) was treated with standard Maxam–Gilbert guanine or cytosine cleavage reaction. Lanes 1–2, LMPCR using Sequenase 2.0 at the primer extension step and Taq at the PCR amplification step (lane 1, guanine; lane 2 cytosine). Lanes 3–4, LMPCR using Pfu exo– at both primer extension and PCR amplification steps (lane 3, guanine; lane 4 cytosine).
Table 2. Qualitative evaluation of the efficiency of DNA polymerases at the primer extension and amplification steps on 1 µg of genomic DNA of variable global SSB frequencies.
| Choice of DNA polymerase |
High-break frequency (>1/kb) + piperidine
treatment |
Low-break frequency (<1/kb) + piperidine
treatment |
High-break frequency (>1/kb)
w/o piperidine treatment |
Low-break frequency (<1/kb) w/o
piperidine treatment |
| Sequenase 2.0 in primer extension | +++ | ++ | ++ | + |
| Pfu exo– in primer extension | +++ | +++ | +++ | +++ |
| Pfu exo– in PCR amplification | ++ | ++ | ++ | +++ |
| Taq in PCR amplification | +++ | +++ | +++ | +++ |
+, good; ++, very good; +++, excellent.
Highly GC-rich sequences
The LMPCR amplification procedure is often much less efficient when the DNA is very GC-rich. Therefore, we developed an LMPCR protocol using Pfu exo– with a NaCl-based buffer, DMSO and the 7-deaza-dGTP base analog to sequence extremely GC-rich DNA. Exon 1 of the FMR1 gene contains a polymorphism for the CGG repeat that spans from a few to several hundred repeats (8). We compared the LMPCR of a 33 CGG repeat haplotype of the FMR1 gene using Pfu exo– only versus the Sequenase 2.0/Taq combination (Fig. 6B). While the CGG repeat and the sequence next to the CGG polymorphism was repeatedly decoded using Pfu exo–, only 6–10 CGG triplets were decoded using Sequenase 2.0 and Taq (Fig. 6B).
DISCUSSION
Primer extension step
Since each guanine residue of a DNA sequence has the same probability of being methylated in vitro by DMS, all band intensities from a sample should appear similar on the autoradiogram. The lack of similarity in the Sequenase 2.0 lanes could not result from premature polymerization arrest or annealing problems, since bands corresponding to longer DNA molecules were much more intense than bands corresponding to smaller DNA molecules. Inefficient ligation and PCR amplification were not determining factors because these steps were identical in both Sequenase 2.0 and Pfu exo– LMPCR protocols. Our results suggest that Sequenase 2.0 was unable to produce ligatable blunt extremities with the same efficiency for every cleaved DNA molecule. Suppression of ligatable ends by DNA polymerases depends on either their inability to extend to the end of the template or their terminal transferase activity. The suppression of ligatable ends by Sequenase 2.0 is most probably caused by its terminal transferase activity (17). Hu (17) showed that the addition of extra non-templated nucleotides by Sequenase 2.0 happened more frequently when DNA molecules were terminated by a cytosine, indicating a sequence-dependent terminal transferase activity. We observed a similar phenomenon while bands representing guanines (especially in guanine runs) were more frequently missing than those representing pyrimidines (data not shown). Because a G is matched during the extension of the primer and the non-extended DNA sequence is decoded on LMPCR autoradiograms, it appears that the guanines are missing and additional nucleotides are added following cytosines on the extended strand. In LMPCR, the addition of an extra non-templated nucleotide at the extremity of blunt double-stranded DNA molecules prevents ligation of the asymmetric linker, and subsequently the amplification of these molecules, thereby modifying the quantitative representation of the corresponding band on the autoradiogram. The probable absence of sequence-dependent terminal transferase activity using Pfu exo– and its apparent ability to extend DNA molecules to the end of the template resulted in a more similar signal intensity among the bands of the same sequence ladder and rendered the interpretation of in vivo genomic mapping, using LMPCR, more reliable. Unexpectedly, more Pfu exo– was required to observe equal band intensities with hydrazine-treated DNA than required for DMS-treated DNA using an equivalent amount of DNA with a similar break frequency. It has been documented that native Pfu possesses no terminal transferase activity but rather a strong 3′→5′ exonuclease activity (17). However, no publication has confirmed or rejected the fact that Pfu exo– has any terminal transferase activity. It has been postulated that an equilibrium between 3′→5′ exonuclease and polymerization activities existed in DNA polymerases (27). T7 DNA polymerase produces blunt extremities after primer elongation. Sequenase 2.0, a modified version of the T7 DNA polymerase, has lost the 3′→5′ exonuclease activity and consequently unbalanced polymerization activity resulted in the addition of a terminal transferase activity (27). Our results suggest that Pfu exo– might possess a similar terminal transferase activity affecting predominantly DNA molecules ending with a purine (viewed as a pyrimidine on the autoradiogram due to probe hybridization revealing the non-extended strand). Alternatively, Pfu exo– could have difficulty extending DNA molecules ending with a purine until a break is reached.
PCR amplification step
Our results showed that more Pfu exo– than Taq was required to reach similar band intensity using 1 µg of DNA. Unlike Taq, the polymerization efficiency of Pfu exo– was modulated by the amount of DNA. Furthermore, we observed that Taq has a higher DNA molecules/unit of DNA polymerase ratio than Pfu exo–, Taq being much less affected by the number of DNA molecules available than Pfu exo–. Like Taq, Pfu exo– faithfully maintained the starting SSB distribution throughout the PCR amplification step cycles whatever the number of units of DNA polymerase assayed as displayed on autoradiograms (Fig. 3). The distinct DNA molecules/unit of DNA polymerase ratio for Taq and Pfu exo– could be explained by the different processivity of both DNA polymerases. With an average of 1 kb of DNA polymerized/min, Taq takes half the time of Pfu exo– to polymerize the same length of DNA. However, two different situations seem to affect the polymerization efficiency of Pfu exo–. First, the availability of a large number of DNA molecules could bring about a competition between DNA molecules and prevent the Pfu exo– from settling on one DNA molecule to begin polymerization (Fig. 4E). Secondly, small amounts of DNA molecules in the first PCR cycles could lead to a competition between molecules of Pfu exo– and prevent the Pfu exo– from settling on one DNA molecule and beginning polymerization. Under identical conditions, time exposure of the hybridized membranes were consistently longer using Pfu exo– at the PCR amplification step compared to the use of Taq, meaning that less PCR products were generated (Fig. 5). Nevertheless, using Pfu exo–, additional cycles can be incorporated in the PCR amplification step program in order to get similar time exposure without significantly affecting the signal-to-noise ratio (Fig. 6B). We limit the number of PCR cycles to 22 with Taq at the PCR amplification step because additional cycles drastically decrease the signal-to-noise ratio (data not shown). Using amounts of DNA within optimal range of the DNA molecules/unit of DNA polymerase ratio, Pfu exo– is an alternative to Taq for the PCR amplification step in LMPCR.
Salt conditions and GC-rich sequences
Exon 1 of the FMR1 gene contains a DNA sequence of between 6 and over 250 CGG triplet repeats (28). This particular DNA sequence forms complex secondary DNA structures in vitro. Sequencing through these structures often produces shorter extension products than anticipated because DNA polymerases often have difficulty in resolving them. Modifications of PCR conditions and DNA polymerase buffers can improve the efficiency of PCR reactions on highly GC-rich sequences (29–31). It has been shown that the K+ cation promotes the stabilization of unusual secondary structures of the DNA (ex: tetrahelix) better than the Na+ cation in vitro (19). Figures 5 and 6B show that the use NaCl instead of KCl in DNA polymerase buffers improved the mapping of GC-rich sequences using LMPCR with any combinations of DNA polymerases. Interestingly, Taq lost its terminal transferase activity in one particular experimental condition (Fig. 5A and B, lanes 13). It seems that somehow, the combination of Pfu exo– and KCl buffers affects either the terminal transferase activity of Taq or the equilibrium between its polymerization and 3′→5′ exonuclease activities. This is consistent with the fact that using any DNA polymerases during the primer extension step, more shadow bands were produced using Taq with KCl (Fig. 5). Since this phenomenon does not occur with Sequenase 2.0, it must be in some ways associated with the processing activities of Pfu exo–. Following the primer extension step, Pfu exo– remains active during the PCR amplification step whereas Sequenase 2.0 is heat inactivated (data not shown). KCl, found in the majority of DNA polymerase buffers, might preserve more efficiently the activities of Pfu exo– than NaCl. Still active during the PCR amplification step, Pfu exo– could conjointly work with Taq, in a KCl buffer that already seems to reduce the terminal transferase activity of Taq (shadow bands). There are many examples of two DNA polymerases working conjointly in a single PCR reaction. Many biotechnological companies now offer a wide range of new DNA polymerases like Roche’s Expand DNA polymerases. Expand systems are a combination of Taq and Pyrococcus woesei DNA polymerases. We assayed the Expand DNA polymerases of the PCR amplification step in LMPCR according to the manufacturer’s specifications, but further testing was abandoned since they did not bring about any improvements when comparing the PCR amplification step using Taq (data not shown). Pfu exo– can withstand denaturing temperatures of 98°C, without losing much of its activity, and can incorporate modified nucleotides (16). Since Sequenase 2.0 is thermolabile and Taq could not resist denaturing temperatures of 98°C over few cycles, this combination of DNA polymerases was not able to sequence through highly GC-rich DNA sequences despite the use of DMSO, NaCl and modified nucleotides.
Effects of break frequency in LMPCR
Depending on the method used to produce SSB, identical DNA sequences can be more or less difficult to process by LMPCR. Treated DNA can be classified into four categories, in increasing order of processing difficulty: (i) high-break frequency (>1 break/kb) using hot piperidine treatment; (ii) low-break frequency (<1 break/kb) using hot piperidine treatment; (iii) high-break frequency (>1 break/kb) using enzymatic treatment; and (iv) low-break frequency (<1 break/kb) using enzymatic treatment (Table 2). In DNA repair studies, the conversion of rare DNA damage into SSB by highly specific repair enzymes produces DNA that is difficult to process by LMPCR. Prior to our study, Sequenase 2.0 and Taq was the only combination of DNA polymerases that efficiently mapped these rare DNA damage with LMPCR (14). A DNA polymerase has to be as efficient on a low-break frequency genomic DNA as it is on a high-break frequency DNA to be widely applicable in LMPCR. Even though Pfu exo– might possess a terminal transferase activity on ending purines (pyrimidines on autoradiograms), it was still more efficient than Sequenase 2.0 at the primer extension step. Pfu exo– can also be used as an alternative to Taq on low-break frequency DNA that was not converted into SSB by hot piperidine.
CONCLUSION
In this paper, we have demonstrated that Pfu exo– can be an excellent DNA polymerase for LMPCR. In all conditions assayed, Pfu exo– was more efficient than Sequenase 2.0 at the primer extension step (Table 2). Moreover, extremely GC-rich DNA sequences could only be sequenced through using Pfu exo–. However, the efficiency of Pfu exo– was more affected by the number of DNA molecules available than Taq was, and hybridized membranes needed a longer exposure time. We also showed that NaCl-based buffers improved the quality of the results. The presence of NaCl in the Taq amplification buffer significantly lowered the occurring shadow bands. NaCl was also necessary to sequence 100 bp of nearly 100% GC-rich DNA using Pfu exo–.
For most LMPCR protocols, we suggest using Pfu exo– at the primer extension step and Taq at the PCR amplification step, using NaCl-based buffers during both steps. Alternatively, Pfu exo– should be used instead of Taq when the DNA sequences are highly GC-rich.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Dr Jean-Philippe Therrien for his assistance with the phosphorimager and to Mrs Geneviève Chevalier and Dr Elliot A. Drobetsky for carefully editing the manuscript. This study was supported by grant CM063375 from the Medical Research Council of Canada (MRC) and by a grant from the Canadian Genetic diseases Network (MRC/NSERC NCE Program). Jean-François Cloutier was a recipient of a studentship from Park-Davis Canada and Régen Drouin is a research scholar (Junior II level) of the ‘Fonds de la Recherche en Santé du Québec’.
References
- 1.Pfeifer G.P., Singer-Sam,J. and Riggs,A.D. (1993) Analysis of methylation and chromatin structure. Methods Enzymol., 225, 567–583. [DOI] [PubMed] [Google Scholar]
- 2.Cartwright I.L. and Kelly,S.E. (1991) Probing the nature of chromosomal DNA–protein contacts by in vivo footprinting. Biotechniques, 11, 188–196. [PubMed] [Google Scholar]
- 3.Mueller P.R. and Wold,B. (1989) In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science, 246, 780–786. [Published erratum appears in Science (1990) 248, 802] [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer G.P., Steigerwald,S.D., Mueller,P.R., Wold,B. and Riggs,A.D. (1989) Genomic sequencing and methylation analysis by ligation mediated PCR. Science, 246, 810–813. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer G.P., Tanguay,R.L., Steigerwald,S.D. and Riggs,A.D. (1990) In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev., 4, 1277–1287. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer G.P. and Riggs,A.D. (1991) Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev., 5, 1102–1113. [DOI] [PubMed] [Google Scholar]
- 7.Hornstra I.K., Nelson,D.L., Warren,S.T. and Yang,T.P. (1993) High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum. Mol. Genet., 2, 1659–1665. [DOI] [PubMed] [Google Scholar]
- 8.Drouin R., Angers,M., Dallaire,N., Rose,T.M., Khandjian,E.W. and Rousseau,F. (1997) Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum. Mol. Genet., 6, 2051–2060. [DOI] [PubMed] [Google Scholar]
- 9.Drouin R. and Therrien,J.-P. (1997) UVB-induced cyclobutane pyrimidine dimer frequency correlates with skin cancer mutational hotspots in p53. Photochem. Photobiol., 66, 719–726. [DOI] [PubMed] [Google Scholar]
- 10.Grange T., Bertrand,E., Espinas,M.L., Fromont-Racine,M., Rigaud,G., Roux,J. and Pictet,R. (1997) In vivo footprinting of the interaction of proteins with DNA and RNA. Methods, 11, 151–163. [DOI] [PubMed] [Google Scholar]
- 11.Quivy J.P. and Becker,P.B. (1993) An improved protocol for genomic sequencing and footprinting by ligation-mediated PCR. Nucleic Acids Res., 21, 2779–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity P.A. and Wold,B.J. (1992) Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl Acad. Sci. USA, 89, 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Cam L., Polanowska,J., Fajas,L., Fabbrizio,E. and Sardet,C. (1999) Improved LM-PCR procedure for in vivo footprinting analysis of GC-rich promoters. Biotechniques, 26, 840–843. [DOI] [PubMed] [Google Scholar]
- 14.Pfeifer G.P., Chen,H.H., Komura,J. and Riggs,A.D. (1999) Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol., 304, 548–571. [DOI] [PubMed] [Google Scholar]
- 15.Dai S.M., Chen,H.H., Chang,C., Riggs,A.D. and Flanagan,S.D. (2000) Ligation-mediated PCR for quantitative in vivo footprinting. Nat. Biotechnol., 18, 1108–1111. [DOI] [PubMed] [Google Scholar]
- 16.Chong S.S., Eichler,E.E., Nelson,D.L. and Hughes,M.R. (1994) Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am. J. Med. Genet., 51, 522–526. [DOI] [PubMed] [Google Scholar]
- 17.Hu G. (1993) DNA polymerase-catalyzed addition of nontemplated extra nucleotides to the 3‘ end of a DNA fragment. DNA Cell Biol., 12, 763–770. [DOI] [PubMed] [Google Scholar]
- 18.Cline J., Braman,J.C. and Hogrefe,H.H. (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res., 24, 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen D.B. and Eckstein,F. (1989) Incomplete primer extension during in vitro DNA amplification catalyzed by Taq polymerase: exploitation for DNA sequencing. Nucleic Acids Res., 17, 9613–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tombline G., Bellizzi,D. and Sgaramella,V. (1996) Heterogeneity of primer extension products in asymmetric PCR is due both to cleavage by a structure-specific exo/endonuclease activity of DNA polymerases and to premature stops. Proc. Natl Acad. Sci. USA, 93, 2724–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iverson B.L. and Dervan,P.B. (1987) Adenine specific DNA chemical sequencing reaction. Nucleic Acids Res., 15, 7823–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeifer G.P. and Tornaletti,S. (1997) Footprinting with UV irradiation and LMPCR. Methods, 11, 189–196. [DOI] [PubMed] [Google Scholar]
- 24.Drouin R., Gao,S. and Holmquist,G.P. (1996) Agarose gel electrophoresis for DNA damage analysis. In Pfeifer,G.P. (ed.), Technologies for Detection of DNA Damage and Mutations. Plenum Press, New York, NY, pp. 37–43.
- 25.Drouin R., Rodriguez,H., Holmquist,G.P. and Akman,S.A. (1996) Ligation-mediated PCR for analysis of oxidative DNA damage. In Pfeifer,G.P. (ed.), Technologies for Detection of DNA Damage and Mutations. Plenum Press, New York, NY, pp. 211–225.
- 26.Therrien J.-P., Drouin,R., Baril,C. and Drobetsky,E.A. (1999) Human cells compromised for p53 function exhibit defective global and transcription-coupled nucleotide excision repair, whereas cells compromised for pRb function are defective only in global repair. Proc. Natl Acad. Sci. USA, 96, 15038–15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabor S. and Richardson,C.C. (1989) Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J. Biol. Chem., 264, 6447–6458. [PubMed] [Google Scholar]
- 28.Fu Y.H., Kuhl,D.P., Pizzuti,A., Pieretti,M., Sutcliffe,J.S., Richards,S., Verkerk,A.J., Holden,J.J., Fenwick,R.G.,Jr, Warren,S.T. et al. (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell, 67, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 29.Hung T., Mak,K. and Fong,K. (1990) A specificity enhancer for polymerase chain reaction. Nucleic Acids Res., 18, 4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner S.L. and Jenkins,F.J. (1995) Use of deoxyinosine in PCR to improve amplification of GC-rich DNA. Biotechniques, 19, 48–52. [PubMed] [Google Scholar]
- 31.Condorelli D.F., Milana,G., Dell’Albani,P., Roccazzello,A.M., Insirello,E., Pavone,L. and Mollica,F. (1996) Routine clinical application of the FRAXA Pfu PCR assay: limits and utility. Clin. Genet., 50, 366–371. [DOI] [PubMed] [Google Scholar]